Abstract
Angiotensin-converting enzyme 2 (ACE2) is the first target of SARS-CoV-2 and a key functional host receptor through which this virus hooks into and infects human cells. The necessity to block this receptor is one of the essential means to prevent the outbreak of COVID-19. This study was conducted to determine the most eligible natural compound to suppress ACE2 to counterfeit its interaction with the viral infection. To do this, the most known compounds of sixty-six Iraqi medicinal plants were generated and retrieved from PubChem database. After preparing a library for Iraqi medicinal plants, 3663 unique ligands’ conformers were docked to ACE2 using the GLIDE tool. Results found that twenty-three compounds exhibited the highest binding affinity with ACE2. The druglikeness and toxicity potentials of these compounds were evaluated using SwissADME and Protox servers respectively. Out of these virtually screened twenty-three compounds, epicatechin and kempferol were predicted to exert the highest druglikeness and lowest toxicity potentials. Extended Molecular dynamics (MD) simulations showed that ACE2-epicatechin complex exhibited a slightly higher binding stability than ACE2-kempferol complex. In addition to the well-known ACE2 inhibitors that were identified in previous studies, this study revealed for the first time that epicatechin from Hypericum perforatum provided a better static and dynamic inhibition for ACE2 with highly favourable pharmacokinetic properties than the other known ACE2 inhibiting compounds. This study entailed the ability of epicatechin to be used as a potent natural inhibitor that can be used to block or at least weaken the SARS-CoV-2 entry and its subsequent invasion. In vitro experiments are required to validate epicatechin effectiveness against the activity of the human ACE2 receptor.
Keywords: ACE2, COVID-19, Drug design, Epicatechin, Medicinal plants, SARS-CoV-2
1. Introduction
The Angiotensin-converting enzyme 2 (ACE2) protein, is one of the angiotensin-converting enzyme family of dipeptidyl carboxydipeptidases. It plays a crucial role in the pathogenesis of SARS-CoV-2, as it provides a route of entry of viral particles, establishing it as a functional receptor for this newly emerged outbreak [1]. This protein is encoded by the ACE2 gene, which has recently attracted high scientific attention since emerging of the viral pandemic. The ACE2 gene is positioned on chromosome X, within the Xp22.2 arm. It consists of 19 exons, with an open reading frame encoding up to 805 amino acids. The mature product of the ACE2 protein has a molecular weight of 110–120 KD [2]. It is well-established that ACE2 protein contains an extracellular domain (starting from the first to 740 amino acid residues), a transmembrane region (741–768 amino acid residues), and an intracellular tail (769–805 amino acid residues) [3].
SARS-CoV-2 particles attach and enter the host cells through the binding of the receptor-binding domain (RBD) of the spike (S) proteins with the ectodomain portion of ACE2 [4]. This sort of direct interaction between SARS-CoV-2 and ACE2 is thought to be the key essence of the efficient spread of this viral infection among humans [[5], [6], [7]]. Based on the interaction of the viral RBD with the host ACE2 receptor, small compounds can be utilized to target the rapid proliferation of the virus through inhibiting its cognate receptor. Once the host ACE2 is being occupied by these compounds, the viral RBD loses its attachment to this receptor before entering inside the host cell. Once the ACE2 receptor is blocked, it would be more difficult for SARS-CoV-2 to gain access to the host cells, which could at least slow the onset of the epidemic until the invading virus disappears [8]. Since the binging affinity of the newly emerged SARS-CoV-2 is 10–20 folds stronger than that SARS-CoV to the host ACE2 [9], the inhibition of ACE2 is one of the best regimens to prevent the outbreak of these newly emerged viral particles. Thus, blocking this receptor may be effective in preventing this highly emerged viral particle from entering the cell and performing its scheduled role of infection.
COVID-19 is currently being treated with some anti-infective drugs, such as antiviral drugs [10], antimalarial drugs [11], and immunosuppressive drugs [12]. However, the effectiveness of these drugs in treating patients with SARS-CoV-2 has not been approved yet [13]. Several synthetic compounds have recently been suggested to inhibit ACE2 by binding with the amino acid residues that are involved in direct interaction with the viral receptor-binding domain (RBD). However, it is well-known that each suggested synthetic may take several years of validation before its become commercially available [14]. Alternatively, several natural compounds have recently been suggested to inhibit ACE2, such as flavonoids [15], proanthocyanidins [16], secoiridoids [17], xanthones [18]. These natural compounds of have high biological availability and low cytotoxicity are the most recommended for the possible treatment of SARS-CoV-2 patients [19]. Given the importance of natural compounds for inhibiting ACE2, several pieces of research have been conducted to prevent the binding of ACE2 with RBD using many natural compounds derived from a variety of plants around the world, such as the flavonoid isothymol from Ammoides verticillata [20], the steroid glycyrrhizin from Glycyrrhiza glabra [21], the alkaloid nicotianamine from soybean [22], the phytocompound withanone from Indian ginseng (Withania somnifera) [23], resveratrol from grape skins (Vitis vinifera) [24], and the quinolone rilapladib from Diplocyclos palmatus [25]. However, further investigations of other medicinal plants resources in other portions of the world are rather important to confirm these findings and to find out the most eligible compound to inhibit this receptor. Therefore, this study has been conducted to find out natural alternative agents for preventing ACE2 from direct binding with SARS-CoV-2 with more eligible physiochemical properties and better oral bioavailability.
2. Materials and methods
2.1. Study design
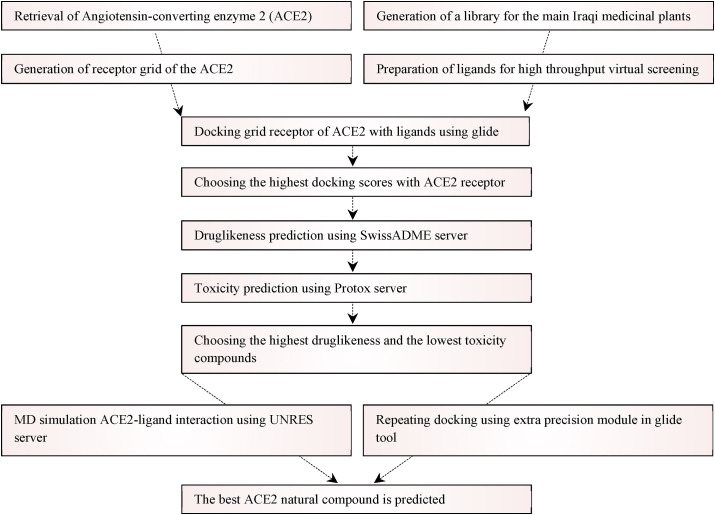
Both protein and ligands were retrieved, prepared, and docked. The investigated compounds with the highest docking scores with the ACE2 receptor were further screened via several druglikeness and toxicity predictions. The best-suited compounds in terms of high docking scores, favourable druglikeness properties, and absence of toxicity were analyzed by further docking and MD simulation to find out the best natural compound for the ACE2 receptor. A schematic diagram detailing the main tools employed in the study is shown in Fig. 1 .
Fig. 1.
A schematic diagram for the main steps conducted in this study.
2.2. Generation of a library for Iraqi medicinal plants
Sixty-six well-known Iraqi medicinal plants were obtained from the literature [[26], [27], [28]] (Suppl. Table 1). The chemical compounds of each involved medicinal plant were obtained from Dr. Duke's phytochemical and ethnobotanical database (http://phytochem.nal.usda.gov/). In each involved medicinal plant, the ubiquitous chemicals were excluded and only unique compounds were retrieved and documented in an excel sheet. Subsequently, the SDF structures of 3392 compounds were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). All the repeated compounds were deleted and all retrieved SDF structures were stored in the same file before being prepared for docking.
2.3. Retrieval of protein
The UniProtKB accession number for the ACE2 is Q9BYF1, and its NCBI reference sequence is NP_068576.1. The 3D structure of the targeted native human ACE2 receptor was retrieved from Protein Data Bank (PDB) server (https://www.rcsb.org/). The PDB ID of the retrieved ACE2 crystal was 1R42, which was deposited in a bound state with 2-acetamido-2-deoxy-β-d-glucopyranose (formula: C8H15O6) [29]. The resolution of this native structure was 2.20 Å, and the number of the deposited residues of this structure was 597 which covered up to 72% of ACE2, starting from 19-Ser amino acid residue and ending with the Ala-614 amino acid residue. Within this region, all the amino acid residues involved in the direct interaction with the viral RBD moiety of SARS-CoV-2 were involved.
2.4. Preparation of ligands for docking
Based on ionic states, stereochemistry, and ring conformations, different conformers of the ligand structures were prepared using ligand preparation script of Grid-Based Ligand Docking with Energetics (GLIDE) software [30]. Partial atomic charges were calculated by using OPLS4 force field. At most, thirty-two low-energy conformers were generated per ligand at pH 7.0 ± 2.0. The maximum limit of ligand size was set at 500 atoms. The generated ligand structures were utilized in downstream virtual screening.
2.5. Preparation of protein for docking
Protein preparation wizard script was used for the optimization and refinement of ACE2 receptors for virtual screening. Bond orders were assigned and hydrogens were added to the imported ACE2 structure, and zero-order bonds to metals were created. The water molecules beyond 5 Å were removed. At pH 7.0, the energy minimization was conducted at the default cut-off Root Mean Square Deviation (RMSD) value of 0.30 Å using Optimized Potentials for Liquid Simulation 4 (OPLS4) force field.
2.6. Docking protocol
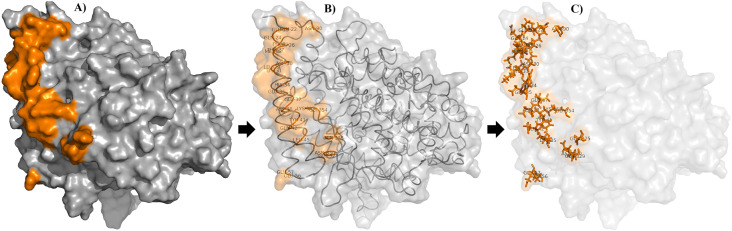
Based on the literature [31], twenty-five amino acid residues were found to be involved in the binding with the viral RBD, namely Glu22, Glu23, Gln24, Lys26, Thr27, Asp30, Lys31, His34, Glu35, Glu37, Asp38, Tyr41, Gln42, Leu45, Glu56, Glu57, Leu79, Met82, Tyr83, Asp90, Gln325, Glu329, Asn330, Lys353, and Gly354 (Fig. 2 ). Using GLIDE receptor grid generation, these residues were selected as possible receptors for the screened ligands. The van der Waal radii of the atoms of the receptor were scaled by 1.00 Å with a partial atomic charge of 0.25 Å. A grid box with coordinates X, Y, Z = 30 Å was generated at the centroid of the ACE2 active site. Using docking ligands script, 3663 ligands’ conformers were docked to the generated receptor grid of the ACE2 receptor. The screened ligands were docked flexibly by High Throughput Virtual Screening (HTVS) using force field OPLS4. The best conformer of each screened complex was selected on the basis of the observed docking score.
Fig. 2.
Critical amino acid residues involved in the direct RBD-ACE2 interface. All these residues were included in the grid receptor generation of the ACE2. The critical amino acid residues are represented as solid surfaces, ribbons, and sticks in A), B), and C) respectively.
2.7. Druglikeness prediction
Many possible medicinal compounds could not reach the clinical trials due to their unfavorable absorption, distribution, metabolism, and elimination (ADME) parameters. SwissADME webserver was utilized to assess the ADME parameters in the high docking scores drug-like compounds [32]. SMILES (Simplified molecular-input line-entry system) structures of these ligands were used as input data in SwissADME to predicts several drug-related features, such as pharmacokinetics, druglikeness, and toxicity. GI absorption and BBB permeant were investigated in pharmacokinetics. Druglikeness of the ligands with high docking scores were assessed following Lipinski's rule of five, Ghose's rule, Veber's rule, Egan's rule, and Muegge's rule. The potential toxicity of these ligands was also predicted using the Protox web server [33]. It was employed to predict the possible hepatoxicity, carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity.
2.8. MD simulation
Downloaded PDB files of the targeted ACE2, being static and devoid of H-atom, thus, they were optimized and modified to understand dynamic and functional behavior after being conjugated with ligands. Molecular dynamic (MD) simulation of the best protein-ligand interactions was conducted to analyze the dynamic stability of the ACE2 upon being docked with the best ligands using UNRES online server [34]. This server is recently developed to predict the dynamic and thermodynamic of proteins based on physics-based coarse-grain simulations. It can be used to predict protein structure, dynamics, and interactions with high accuracy at larger times [[35], [36], [37]]. The coarse-grain-based MD method was conducted at the temperatures of 300 K with the Langevin thermostat that was implemented to perform the conformational search. The total number of steps was 200,000 that were applied per simulation. Each MD trajectory was run with an integration time of 48.9 fs and other parameters were set at defaults to ensure better simulation of the system. MD runs were performed to predict root mean square deviation (RMSD), potential energy, the radius of gyration, and root mean square fluctuation (RMSF). The trajectories of both simulations were saved in 250 ns intervals. Output files were retrieved from the UNRES server and Qtgrace software, ver. 2.6., was utilized to visualize and annotate simulations.
2.9. Protein-protein interaction
To give a further indication of the extent of epicatechin inhibitory effect on ACE2/RBD interaction, protein-protein docking was conducted ACE2 and RBD in the presence and absence of epicatechin. Accordingly, two independent molecular docking experiments between ACE2 and RBD were conducted using PatchDock online server [38]. The outputs of PatchDock files were refined using Fast Interaction REfinement in molecular DOCKing (FireDock) online server [39]. The interaction energies, the participation of van Der Waals interactions, and hydrogen bonds in the observed global energy in the topmost poses in both docking cases were recorded and compared. Molecular visualization of the docking output was done with PyMol, ver. 7.0.1 (The PyMOL Molecular Graphics System, Schrödinger, LLC.).
3. Results
The main approach of this study was taken place by conducting many computations to set out more eligible anti-ACE2 natural inhibitors having better pharmacokinetics and medicinal chemistry properties. Thus, the utilization of these compounds offers superiority over the recently suggested medicines with no or minimal side effects. The vast importance of variable computational tools was employed to screen a wide range of compounds to reveal their potential to be used as inhibitors against the ACE2. Graphical representations were visualized using Maestro suit Schrödinger, LLC).
3.1. Virtual screening by high throughput molecular docking
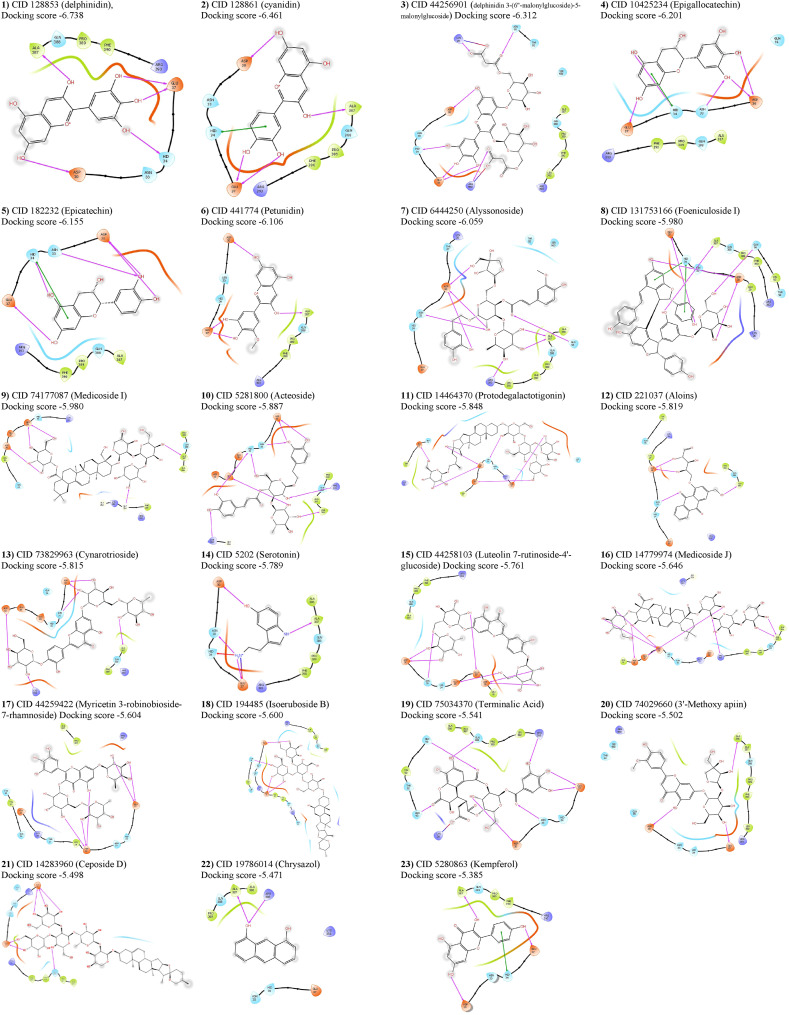
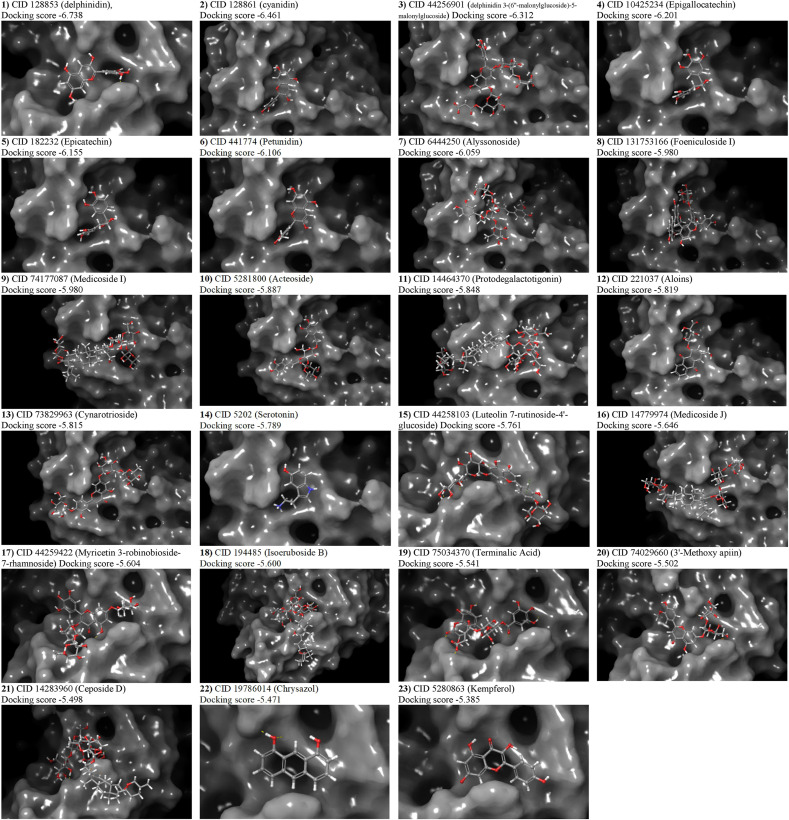
A total of 3663 unique ligands’ conformers of sixty-six Iraqi medicinal plants were docked to the SARS-CoV-2 attachment regions in the ACE2 receptor. The highest docking score of each protein-ligand complex that emerged from molecular docking was compared with the best docking scores of the most known anti-ACE2 compounds. In addition to the highest score that delphinidin (PubChem CID 128853) showed in terms of its docking with ACE2, twelve previously known anti-ACE2 compounds were also considered as positive controls in the conducted docking experiments. These compounds were also found in the Iraqi medicinal plants and therefore they are considered as positive controls alongside the other compounds that are not known to possess any anti-ACE2 activity. These compounds are cyanidin (PubChem CID 128861), kempferol (PubChem CID 5280863), quercetin (PubChem CID 5280343), luteolin (PubChem CID 5280445), emodin (PubChem CID 3220), rhein (PubChem CID 10168), rhoifolin (PubChem CID 5282150), rutoside (PubChem CID 5280805), chrysin (PubChem CID 5281607), nicotianamine (CID 9882882), apigenin (PubChem CID 5280443), and acetylglucosamine (PubChem CID 24139). Our generated library showed that these known anti-ACE2 compounds were all available in the Iraqi medicinal plants, and their presence provided a high throughput validation for our conducted docking experiments. Within these highlighted controls, delphinidin, cyanidin, and kempferol were respectively represented the best three well-known anti-ACE2 controls in terms of their high docking scores inferred from the GLIDE tool (Suppl. Table 2). Accordingly, twenty-three compounds starting from delphinidin (docking score −6.738) until reaching kempferol (docking score −5.385) were submitted to druglikeness predictions. Whereas the other controls alongside other docked compounds were omitted from downstream screening due to their lower docking scores. The presence of a variable of amino acid residues with hydrogen bonding, polar, van der Waals, and Pi-Alkyl interactions was proved variable intensities of connections between the analyzed ligands and ACE2. The binding conformations of all ligands with docking scores starting from delphinidin and ending with kempferol are shown in Table 1 . Despite its high docking score, serotonin showed unfavourable bindings with the His34 (Fig. 3 ). Due to the importance of His34 in the direct interaction with the viral RBD, this sort of unfavourable binding of serotonin – His34 interactions had lowered the effectiveness of this compound and the consequent stability of its interaction with ACE2. Thus, serotonin should not be suggested as an optimum natural inhibitor against ACE2. The binding modes of all candidate compounds showed variable positioning and intensities within the targeted pocket of the ACE2 receptor (Fig. 4 ). Hence, it is mandatory to combine these observed variable ligands – ACE2 interactions with the druglikeness possibility of each compound to find out the most eligible one to act as a natural anti-ACE2 compound.
Table 1.
Chemical structures and docking scores of the best-suited ligands to the active site of angiotensin converting enzyme type-2 receptor.
| No. | Name | PubChem code | MW | Isomeric SMILES | Docking scores (Kcal/mol) |
|---|---|---|---|---|---|
| 1 | Delphinidin | CID 128853 | 303.24 | C1=C(C C(C(=C1O)O)O)C2 = [O+]C3=CC(=CC(=C3C C2O)O)O | −6.738 |
| 2 | Cyanidin | CID 128861 | 287.24 | C1=CC(=C(C C1C2 = [O+]C3=CC(=CC(=C3C C2O)O)O)O)O | −6.461 |
| 3 | delphinidin 3-(6″-malonylglucoside)-5-malonylglucoside) | CID 44256901 | 799.600 | C1=C(C C(C(=C1O)O)O)C2=C(C C3C(=CC(=CC3 = [O+]2)O)O[C@H]4C([C@H]([C@@H](C(O4)COC(=O)CC(=O)O)O)O)O)O[C@H]5C(C([C@@H](C(O5)COC(=O)CC(=O)O)O)O)O | −6.312 |
| 4 | Epigallocatechin | CID 10425234 | 306.27 | C1[C@@H]([C@@H](OC2=CC(=CC(=C21)O)O)C3=CC(=C(C(=C3)O)O)O)O | −6.201 |
| 5 | Epicatechin | CID 182232 | 290.27 | C1[C@@H]([C@@H](OC2=CC(=CC(=C21)O)O)C3=CC(=C(C C3)O)O)O | −6.155 |
| 6 | Petunidin | CID 441774 | 317.27 | COC1=CC(=CC(=C1O)O)C2 = [O+]C3=CC(=CC(=C3C C2O)O)O | −6.106 |
| 7 | Alyssonoside | CID 6444250 | 770.700 | CC1C(C(C(C(O1)OC2C(C(OC(C2OC(=O)/C C/C3=CC(=C(C C3)O)OC)COC4C(C(CO4)(CO)O)O)OCCC5=CC(=C(C C5)O)O)O)O)O)O | −6.059 |
| 8 | Foeniculoside I | CID 131753166 | 842.800 | C1=CC(=CC C1/C C/C2=C3C(C(OC3=CC(=C2)O)C4=CC C(C C4)O)C5=C6C(C(OC6=CC(=C5)O)C7=CC C(C C7)O)C8=CC(=CC(=C8)OC9C(C(C(C(O9)CO)O)O)O)O)O | −5.980 |
| 9 | Medicoside I | CID 74177087 | 1061.20 | CC1(CCC2(CCC3(C(=CCC4C3(CCC5C4(CCC(C5(C)CO)OC6C(C(C(CO6)O)O)OC7C(C(C(C(O7)CO)O)O)OC8C(C(C(CO8)O)O)O)C)C)C2C1)C)C(=O)OC9C(C(C(C(O9)CO)O)O)O)C | −5.930 |
| 10 | Acteoside | CID 5281800 | 624.6 | C[C@H]1[C@@H]([C@H]([C@H]([C@@H](O1)O[C@@H]2[C@H]([C@@H](O[C@@H]([C@H]2OC(=O)/C C/C3=CC(=C(C C3)O)O)CO)OCCC4=CC(=C(C C4)O)O)O)O)O)O | −5.887 |
| 11 | Protodegalactotigonin | CID 14464370 | 1215.30 | CC1C2C(CC3C2(CCC4C3CCC5C4(CCC(C5)OC6C(C(C(C(O6)CO)OC7C(C(C(C(O7)CO)O)OC8C(C(C(CO8)O)O)O)OC9C(C(C(C(O9)CO)O)O)O)O)O)C)C)OC1(CCC(C)COC1C(C(C(C(O1)CO)O)O)O)O | −5.848 |
| 12 | Aloins | CID 221037 | 386.4 | C1=CC C2C(=C1)C(=O)C3=C(C2=O)C(=CC(=C3)CO)OCC(C(C(C O)O)O)O | −5.819 |
| 13 | Cynarotrioside | CID 73829963 | 756.700 | CC1C(C(C(C(O1)OCC2C(C(C(C(O2)OC3=CC(=C4C(=C3)OC(=CC4=O)C5=CC(=C(C C5)OC6C(C(C(C(O6)CO)O)O)O)O)O)O)O)O)O)O)O | −5.815 |
| 14 | Serotonin | CID 5202 | 176.21 | C1=CC2=C(C C1O)C(=CN2)CCN | −5.789 |
| 15 | Luteolin 7-rutinoside-4′-glucoside | CID 44258103 | 756.700 | CC1[C@@H]([C@@H](C([C@@H](O1)OCC2[C@H]([C@@H](C([C@@H](O2)OC3=CC(=C4C(=C3)OC(=CC4=O)C5=CC(=C(C C5)O[C@H]6C(C([C@@H]([C@@H](O6)CO)O)O)O)O)O)O)O)O)O)O)O | −5.761 |
| 16 | Medicoside J | CID 14779974 | 1075.20 | CC1C(C(C(C(O1)OC2C(C(COC2OC(=O)C34CCC(CC3C5 = CCC6C(C5(CC4)C)(CCC7C6(CC(C(C7(C)C(=O)O)OC8C(C(C(C(O8)CO)O)O)O)O)C)C)(C)C)O)O)O)O)OC9C(C(C(CO9)O)O)O | −5.646 |
| 17 | Myricetin 3-robinobioside-7-rhamnoside | CID 44259422 | 772.700 | CC1[C@@H]([C@@H](C([C@@H](O1)OCC2[C@@H](C(C([C@@H](O2)OC3=C(OC4=CC(=CC(=C4C3 = O)O)O[C@H]5C([C@H]([C@H](C(O5)C)O)O)O)C6=CC(=C(C(=C6)O)O)O)O)O)O)O)O)O | −5.604 |
| 18 | Isoeruboside B | CID 194485 | 1081.20 | CC1CCC2(C(C3C(O2)CC4C3(CCC5C4CC(C6C5(CCC(C6)OC7C(C(C(C(O7)CO)OC8C(C(C(C(O8)CO)O)OC9C(C(C(C(O9)CO)O)O)O)OC2C(C(C(C(O2)CO)O)O)O)O)O)C)O)C)C)OC1 | −5.600 |
| 19 | Terminalic Acid | CID 75034370 | 652.500 | C1=C(C C(C(=C1O)O)O)C(=O)OC2C3C(C(C(O2)CO)OC(=O)C(C4C(C(=O)OC5=C4C(=CC(=C5O)O)C(=O)O3)O)CC(=O)O)O | −5.541 |
| 20 | 3′-Methoxy apiin | CID 74029660 | 594.500 | COC1=C(C CC(=C1)C2=CC(=O)C3=C(C C(C C3O2)OC4C(C(C(C(O4)CO)O)O)OC5C(C(CO5)(CO)O)O)O)O | −5.502 |
| 21 | Ceposide D | CID 14283960 | 1179.30 | CC1CCC2(C(C3C(O2)CC4C3(CCC5C4CC = C6C5(CCC(C6)OC7C(C(C(CO7)O)O)OC8C(C(C(C(O8)C)OC9C(C(C(C(O9)CO)O)OC2C(C(C(C(O2)CO)O)O)O)OC2C(C(C(C(O2)CO)O)O)O)O)O)C)C)C)OC1 | −5.498 |
| 22 | Chrysazol | CID 19786014 | 210.23 | C1=CC2=CC3=C(C C2C(=C1)O)C(=CC C3)O | −5.471 |
| 23 | Kempferol | CID 5280863 | 286.24 | C1=CC(=CC]C1C2 = C(C(=O)C3=C(C]C(C]C3O2)O)O)O)O | −5.385 |
Fig. 3.
Two-dimensional representations of the best pose interactions between the ligands and ACE2 receptor. Twenty-three compounds are shown attached within the ACE2 pocket.
Fig. 4.
Three-dimensional representations of the best pose interactions between the ligands and ACE2. Twenty-three compounds are shown attached within the ACE2 pocket.
3.2. Druglikeness prediction
The optimum pharmacokinetics properties of any suggested drug-like compounds, such as high GI absorption, intracellular metabolism, and excretion, alongside low toxicity, are essential to validate their adequacy as a lead compound during the drug design. SwissADME tool showed that seven compounds (delphinidin, cyaniding, epigallocatechin, epicatechin, petunidin, and kempferol) were found with high GI absorption properties out of the other investigated candidates (Table 2 ). Furthermore, Lipinski, Ghose, Veber, Egan, and Muegge rules were utilized as filters to provide a sufficient rationale for the grouping of each compound as a drug or nondrug. Any virtually screened compound should pass these main filters before being suggested for oral administration and any possible reduction in druglikeness of a lead compound may occur due to more violation of this compound by any one of these filters [40]. It was found that cyanidin, epicatechin, petunidin, chrysazol, and kempferol were the most submissive for druglikeness filtrations, while the other applied compounds exerted variable violation(s) for these rules. Further filtration was applied to figure out the potential toxicity of these compounds on the body. In addition to druglikeness properties, further filtrations were conducted for these compounds using Protox server to find out the possible biological risk of these compounds upon administration. Results showed that epicatechin (PubChem CID 182232) and kempferol (PubChem CID 5280863) were passed these filters since both exhibited no hepatoxicity, carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity. Whereas, variable degrees of violations were witnessed for other compounds (including cyanidin, petunidin, and chrysazol), and therefore they were eliminated from further in-depth analyses. Since epicatechin and kempferol showed all the desirable properties in terms of pharmacokinetics, druglikeness, and the absence of any possible toxicity, further in-depth molecular docking should be conducted for each compound to unravel the ligand-receptor interactions with further details to decide which one represent the lead molecule.
Table 2.
Druglikeness of the most suited ligands to the active site of angiotensin converting enzyme type-2 receptor. The compound no. 5 (epicatechin) and compound no. 22 (kempferol) proved the best pharmacokinetics, druglikeness, and toxicity features over all other tested compounds.
| No. | Compound | GI absorption |
Druglikeness |
Toxicity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipinski rules violations | Ghose rules | Veber rules | Egan rules | Muegge rules | Hepatoxicity | Carcinogenicity | Immunotoxicity | Mutagenicity | Cytotoxicity | ||||
| 1 | Delphinidin | High | 1 violation | Yes | Yes | 1 violation | 1 violation | Inactive | Active | Inactive | Inactive | Inactive | |
| 2 | Cyanidin | High | Yes | Yes | Yes | Yes | Yes | Inactive | Active | Inactive | Inactive | Inactive | |
| 3 | delphinidin 3-(6″-malonylglucoside)-5-malonylglucoside) | Low | 3 violations | 4 violations | 2 violations | 1 violation | 5 violations | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 4 | Epigallocatechin | High | 1 violation | Yes | Yes | Yes | 1 violation | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 5 | Epicatechin | High | Yes | Yes | Yes | Yes | Yes | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 6 | Petunidin | High | Yes | Yes | Yes | Yes | Yes | Inactive | Active | Active | Inactive | Inactive | |
| 7 | Alyssonoside | Low | 3 violations | 4 violations | 2 violations | 1 violation | 4 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 8 | Foeniculoside I | Low | 3 violations | 3 violations | 1 violation | 1 violation | 6 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 9 | Medicoside I | Low | 3 violations | 4 violations | 2 violations | 1 violation | 5 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 10 | Acteoside | Low | 3 violations | 4 violations | 2 violations | 1 violation | 4 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 11 | Protodegalactotigonin | Low | 3 violations | 4 violations | 2 violations | 1 violation | 7 violations | Active | Inactive | Active | Inactive | Inactive | |
| 12 | Aloins | Low | Yes | 1 violation | 1 violation | 1 violation | Yes | Active | Inactive | Active | Inactive | Inactive | |
| 13 | Cynarotrioside | Low | 3 violations | 4 violations | 1 violation | 1 violation | 4 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 14 | Serotonin | High | Yes | Yes | Yes | Yes | 1 violation | Inactive | Inactive | Inactive | Inactive | Inactive | |
| 15 | Luteolin 7-rutinoside-4′-glucoside | Low | 3 violations | 4 violations | 1 violation | 1 violation | 4 violations | Active | Inactive | Active | Inactive | Inactive | |
| 16 | Medicoside J | Low | 3 violations | 4 violations | 2 violations | 1 violation | 5 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 17 | Myricetin 3-robinobioside-7-rhamnoside | Low | 3 violations | 4 violations | 1 violation | 1 violation | 4 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 18 | Isoeruboside B | Low | 3 violations | 4 violations | 2 violations | 1 violation | 5 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 19 | Terminalic Acid | Low | 3 violations | 3 violations | 1 violation | 1 violation | 4 violations | Inactive | Inactive | Active | Active | Inactive | |
| 20 | 3′-Methoxy apiin | Low | 3 violations | 4 violations | 1 violation | 1 violation | 3 violations | Inactive | Inactive | Active | Inactive | Inactive | |
| 21 | Ceposide D | Low | 3 violations | 4 violations | 2 violations | 1 violation | 5 violations | Active | Inactive | Active | Inactive | Inactive | |
| 22 | Chrysazol | Low | Yes | Yes | Yes | Yes | Yes | Inactive | Inactive | Inactive | Active | Inactive | |
| 23 | Kempferol (control) | High | Yes | Yes | Yes | Yes | Yes | Inactive | Inactive | Inactive | Inactive | Inactive | |
3.3. Extra precision molecular docking
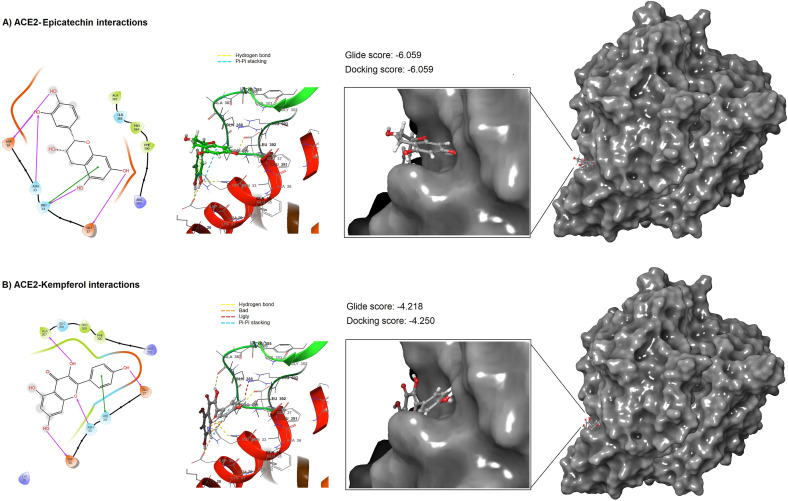
Due to the high docking score, highly accepted druglikeness properties, and no toxicity, molecular docking was repeated for both epicatechin and kempferol using the most accurate Extra Precision (XP) module provided in the GLIDE tool. Results of high precision docking confirmed the virtual screening results and showed that epicatechin exerted a higher docking score (docking score −6.059 kcal/mol) with ACE2 than that found in the kempferol-ACE2 case (docking score −4.250 kcal/mol). Both ligands interacted with the pocket of ACE2 but in different modes. 2D analyses of ligand-protein interactions showed that epicatechin interacted with ACE2 by five hydrogen bonding with four amino acid residues (Asp30, Asn33, His34, and Glu37). In addition to hydrogen bonding with His34, epicatechin was also interacted with this residue by Pi-Pi stacking (Fig. 5 A). Interestingly, epicatechin only interacted with the amino acid residues that are involved in direct binding with the spike protein of SARS-CoV-2. Kempferol was also showed another interaction with ACE2 by forming four hydrogen bonding with four amino acid residues, three of them were found to be desirable since they were formed with three amino acid residues Asp30, Asn33, and Glu37 that involved in the direct binding with the SARS-CoV-2spike protein. Whereas the fourth hydrogen bond was formed with Ala387, which was not concerned with the direct ACE2-spike interactions. Moreover, the binding of kempferol with His34 was restricted with only one hydrophobic interaction (Fig. 5B). Thus, detailed 2D interactions of epicatechin and kempferol showed that epicatechin enjoyed more favourable binding with ACE2 due to the presence of more effective ligand-protein binding interactions. To support this observation, 3D interaction analysis has suggested the presence of remarkable interactions of epicatechin with four amino acid residues that are directly involved with the binding with the viral RBD. Whereas the binding of kempferol was only interacted with a less extent to these amino acid residues and involved with other interactions with the amino acid residue Ala387 and other amino acids that are not involved with the direct interaction with the viral RBD. Due to this observation and due to the lower docking score observed from kempferol, it can be stated that epicatechin is the lead molecule in inhibiting ACE2. However, further comparison using dynamic simulation may aid this observation.
Fig. 5.
Two- and three-dimensional representations for ACE2-epicatechin complex in A), and ACE2-kempferol complex in B) as they docked using the extra-precision module.
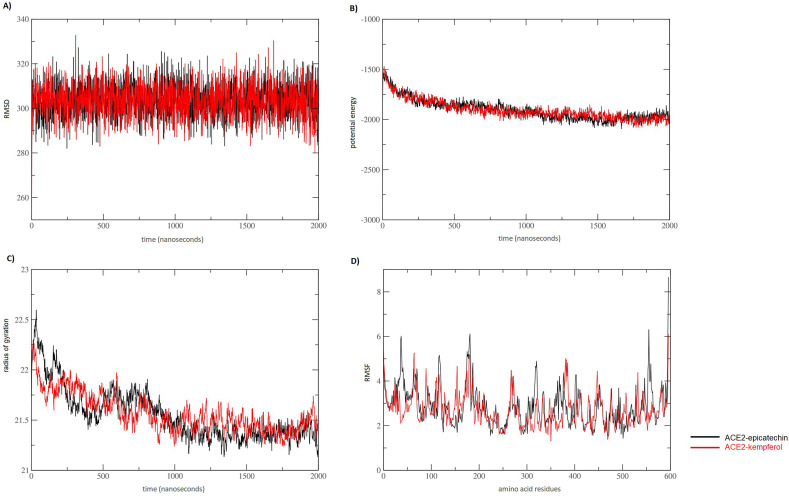
3.4. MD simulation
An extended time of MD was provided using the recently developed high-speed coarse-grain simulation [41]. Two independent simulations were conducted to evaluate structural fluctuation and functional stability of ACE2 upon being interacted with epicatechin and kempferol. The results of the RMSD plot indicated that both ACE2 complexed with epicatechin and kempferol showed comparable results in MD simulations as both complexes were quite consistent, suggesting the stability of both complexes in the entire time of simulation (Fig. 6 A). The values of RMSD suggested that the binding of both complexes has been stabilized without any conformational shift. Likewise, the general behavior of ACE2 was observed to be rather stable in the entire MD simulation period of 2000 ns in both simulated complexes. This observation entailed a comparable validity of epicatechin and kempferol in the binding with the targeted ACE2 in considerable stability. Potential energy calculations showed that all angles, bonds, and torsions are at their natural values according to the input force-field parameters. However, the ACE2-kempferol complex showed more values at the end of the simulation, which suggested exhibiting a slightly less stable complex than that observed in ACE2-epicatechin (Fig. 6B). The distribution of protein atoms around its axis, or radius gyration values, is proportional to the energy of the simulated complexes. Thus, radius gyration values were analyzed to compare the conformational changes in both complexes in the simulation time. Data obtained from potential energy were supported by observing higher values of radius gyration of ACE2-kempferol complex in comparison with ACE2-epicatechin complex. These higher values of ACE2-kempferol complex were particularly observed in the second half of the MD simulation (from 1000 ns to 2000 ns) (Fig. 6C). These data suggested that ACE2-epicatechin complex showed slightly more dynamic stability than that found in ACE2-kempferol complex. Another layer of confirmation for this observed stability was provided by calculating the RMSF of both investigated complexes. In the RMSF plot, more arbitrary fluctuations were observed, but no conformational shifting was seen in the positions occupied by residues involved in direct interactions with both compounds since the plotting of amino acid residues that involved in making direct interactions with epicatechin showed that all involved direct amino acid residues interactions were positioned in non-fluctuated areas (Fig. 6D). Thus, both RMSD and RMSF values were similar for both complexes, which suggested the validity of the epicatechin alongside the previously known kempferol compound. Once again, MD simulation results discovered that the epicatechin was the lead compound as it showed better binding stability with the soluble ACE2 receptor.
Fig. 6.
Comparative molecular dynamic (MD) simulation of ACE2 complexed with kempferol (black) and epicatechin (red). A) Root-mean-square deviation (RMSD), B) Potential energy, and C) radius of gyration in A° plotted against 2000 ns simulation time. D) Root-mean-square fluctuation (RMSF) in A° plotted against amino acid residues.
3.5. Protein-protein interaction
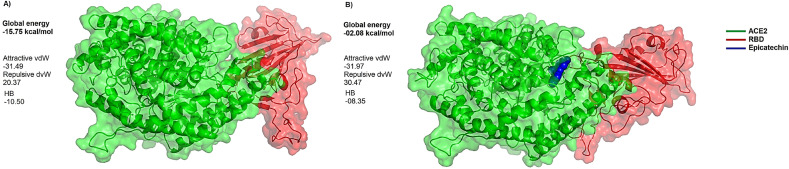
Further confirmation for the string inhibitory role predicted for epicatechin was provided from performing direct protein-protein interactions between ACE2 and the viral RBD in the presence and absence of the docked epicatechin. The predicted global binding energies for the topmost poses of ACE2-RBD docked complexes were −15.75 kcal/mol and −02.08 kcal/mol in the presence and absence of the docked epicatechin respectively. This significant reduction in binding energies refers to the possible ability of this compound to induce dramatic conformational changes in the interface area between ACE2 and RBD and a consequent drastic reduction in the binding energy between the interacted proteins (Fig. 7 ).
Fig. 7.
Comparative docking views for the binding of ACE2-RBD in the presence and absence of epicatechin. In presence of epicatechin (branch A), the global binding energy is −15.75 kcal/mol. In the absence of epicatechin (branch B), a drastic reduction in the affinity of binding between ACE2 and RBD is found as the global binding energy is only 02.09 kcal/mol. Letters vdW and HB refer to the respective participation of van Der Waals interactions and hydrogen bonds in the observed global energy in both docking cases.
4. Discussion
ACE2 receptor is one of the most interesting therapeutic options to neutralize COVID 19 infection [42]. The strong binding between SARS-CoV-2 spike glycoprotein with ACE2 receptor is considered to be the first critical step for the viral entry to the host cell. ACE2 receptors have also been involved in modulating blood pressure and maintaining blood pressure homeostasis. Several animal studies have shown a link between increased blood pressure and reduced levels of ACE2 [43]. However, it is unlikely that the in-hospital use of ACE inhibitors has any association with the increased mortality risk in patients with COVID 19 [44]. Thus, there is an urgent need for anti-ACE2 drugs to decrease the danger of COVID-19 infection. Two antimalarial agents (chloroquine and hydroxychloroquine) have been administered to patients with COVID-19 in order to bind with the ACE2 receptor. However, these agents have serious adverse impacts on patients with acute renal failure, cardiovascular disorder, hypertension, and diabetes and thus were rejected by the Food and Drug Administration (FDA) as anti-SARS-CoV-2 therapeutics [45]. Alternatively, computer-assisted drug designing would be an eminent approach toward COVID-19 treatment through implementing plant-derived natural agents to reduce the undesirable side effects of these synthetic compounds [46]. Natural products have for long been considered attractive sources of therapeutic agents and gained remarkable prominence. These natural products are noticeably available in low molecular weight formulas and the majority of them have proven to be safe [47]. Available evidence supports the inhibitory properties of natural products toward the biological activity of ACE2. Since many natural compounds have been recently reported to module RBD – ACE2 interface, sixty-six Iraqi medicinal plants were screened in this study to assess their anti-ACE2 activity through their ability to prevent RBD – ACE2 interaction, and its consequent SRAR-CoV-2 attachment, fusion, and entry.
Accordingly, strict in silico screening filters were conducted on 3663 ligands’ conformers to find out the most eligible one to inhibit RBD – ACE2 interface with high docking efficacy, high GI adsorption, high druglikeness, and low toxicity. Out of these conformers, only epicatechin and kempferol were passed these filters and proven to have all the desirable pharmacokinetics that should exist in the optimum drugs. The analyses of the ligand – ACE2 interactions showed that epicatechin was more specific in the binding with the targeted amino acid residues than kempferol. Furthermore, subsequent MD simulations proved slightly more stability of epicatechin-ACE2 complex over kempferol-ACE complex. For these reasons, this study found that epicatechin was the lead molecule in inhibiting the RBD-ACE2 interface. The importance of this compound may be originated from the strong interaction with several essential amino acid residues within ACE2. As it was shown in the literature [48], the amino acid residues of Asp30, Asn33, His34, and Glu37 that share hydrogen bonds with epicatechin were positioned in extremely important regions that participated in the binding interface with the spike protein of SARS-CoV-2. So, inhibiting these residues may act a crucial role in preventing ACE2 from direct binding with the spike of SARS-CoV-2.
In addition to epicatechin, the therapeutic potentials of the other top-nine investigated natural compounds were also explored to find out their activities against COVID 19 infection. These compounds were found to be available from variable sources of medicinal plants in Iraq. Delphinidin, cyanidin, delphinidin 3-(6″-malonylglucoside)-5-malonylglucoside) (PubChem CID 44256901), petunidin (PubChem CID 441774), foeniculoside I (PubChem CID 131753166), epigallocatechin (PubChem CID 10425234), and medicoside I (PubChem CID 74177087) were respectively obtained from Abelmoschus esculentus, Adiantum capillus-veneris, Cichorium intybus, Crocus sativus, Foeniculum vulgare, Juniperus communis, and Medicago sativa. Whereas both alyssonoside (PubChem CID 6444250), acteoside (PubChem CID 5281800) were obtained from Ballota nigra. Several reports have suggested some of these natural compounds to exhibit various degrees of inhibitions against the infection with COVID19 [49]. The aqueous extracts of delphinidin derivatives have been shown to exert obvious in vitro inhibitory effects against ACE by competing with its active site (IC50 value 84.5) [50]. A molecular docking study has recently shown that delphinidin 3-(6″-malonylglucoside)-5-malonylglucoside) has exerted encouraging binding affinity with the human ACE2 receptor [51]. The purified epigallocatechin has been found to generate an inhibition for the ACE2 with IC50 in the micromolar range [52]. However, the utilization of acteoside has only been restricted with the symptomatic treatment of the dry cough-associated oral and pharyngeal mucosa irritation [53]. Irrespective of these compounds, picatechin has been found to possess noticeable antiviral effects, which may be of remarkable importance in the onset of the pandemic situation [54]. The high effectiveness of this flavonoid is not surprising as it was found that epicatechin inhibits the onset of Mayaro and hepatitis C viral diseases by inhibiting their replication [55,56]. Interestingly, the majority of epicatechin interactions were restricted to the formation of hydrogen bonds with positively charged amino acid residues within the targeted ACE2 receptor. Such mode of interaction with the hydrophilic amino acid residues is one of the suggested mechanisms through which flavonoids bind with a strong affinity to their targeted proteins [57].
In this study, the lead molecule is obtained from one of the medicinal plants that have been widely used in Iraqi traditional medicine is Hypericum perforatum, also known as perforate St John's-wort. This plant is a flowering perennial herb that is used for versatile therapeutic attributes, such as anxiety [58], gastric lesions [59], depression [60], wounds [61], and burns [62]. In addition to its obvious antioxidant role [63], it has been suggested to include this herb in treating many other ailments, such as cancer [64], inflammation-related disorders [65], and viral diseases [4]. Very recently, it has been reported that one of the epicatechin derivatives (PubChem CID 72276) that also derived from the same plant has been involved in a strong binding with several amino acid residues of the SARS-CoV-2 protease [66]. Whereas other studies have found that catechin exhibited a high affinity to the ACE2 receptor [67]. However, in our study, we have also found the same results but epicatechin showed a remarkably stronger docking score with ACE2 than found in catechin. Noteworthy, both compounds have similar molecular weight but they only differ in their configurations. Epicatechin takes the cis configuration, while catechin takes the trans configuration. Thus, these cis-trans differences have made epicatechin superior in its binding with ACE2. This observation has indirectly supported another study that reported a strong impact of the stereochemical configuration on the activity of both compounds because the uptake and metabolism of epicatechin were found to be higher than catechin [68]. However, such tiny stereochemical differences between these natural compounds were not highlighted at least in terms of the binding with ACE2. Thus, it is rational to say that cis-trans alteration has an effective role in altering the ACE2 inhibitory activity of both natural compounds.
These data have cumulatively suggested that epicatechin has a noticeable binding efficiency with several critical amino acid residues and may be used as putative inhibitors to the ACE2. Thus, it can be stated that in addition to the biological importance of epicatechin interactions with ACE2, sufficient biological stability was ensured in this complex. This observation entailed the ability of this compound to ensure solid binding with amino acid residues of ACE2, and sufficient dynamic stability of the consequent complex upon formation.
Computational approaches have been achieving popularity in the medication industry by demonstrating they are essential in the discovery of drugs. Based on computational/simulation approaches, several medicines have been developed that are presently available for the treatment of different diseases [69]. In contrast to these encouraging data, it has recently been proven that the docking score is not good enough to fully support as a cutoff value for selecting possible inhibitors for SARS-CoV-2 infection because no good correlations have been found between docking scores and pIC50 values for these inhibitors [70]. Furthermore, it has been found that even though all tested docking protocols have a good pose prediction, their screening accuracy is quite limited as they fail to correctly rank a test set of compounds [71]. Though increasing docking search algorithms have recently been improved, it is still questioned if there are any reliable approach in which molecular docking have aided to bring a drug to the market. Accordingly, biomedical laboratories or pharmaceutical companies are still hesitant to implement this technology in drug screening. This is may be due to some concerns correlated with interacting energy and binding conformations, resolution of available structures, and the algorithms used to identify the possible binding poses between ligands and their corresponding receptors [72]. Despite these contradicting reports, the results of this work highlighted the importance of epicatechin in terms of its high affinity to bind with ACE2 at the site of SARS-CoV-2 interaction. However, the results from the high binding affinity between epicatechin and ACE2 should be supported by in vivo testing and reaffirmed by the results of in vivo studies. In a prospective view, comparative studies with experimental high-throughput screening should be conducted to enrich docking accuracy and to correspondingly illuminate any possible hypothetical results. Therefore, conducting an adequate wet lab-based validation of the epicatechin antiviral possibility is extremely necessary before drawing bold conclusions on this virtual screening approach.
5. Conclusion
In summary, we investigate the inhibitory activity of 3663 ligands’ conformers from sixty-six sources of Iraqi medicinal plants. The screening of all ligands to the targeted ACE2 receptor showed that the epicatechin from Hypericum perforatum can bind ACE2 excellently. Its desirable pharmacokinetic and druglikeness properties and the absence of any possible toxicity have prevailed over the other screened compounds. Add to that, the stable binding capacity to ACE2 and a high-quality MD profile have confirmed its superiority over other compounds. All these features collectively indicated that epicatechin could act as a potential inhibitory agent to prevent the binding of ACE2 with the SARS-CoV2 spike. Based on the data obtained from this research, it can be stated that this compound can be employed as a potent natural inhibitor for ACE2 that can prevent its interaction with SARS-CoV2 at the onset of infection. The therapeutic potentials of this new chemical compound against the highly emerged COVID19 can be exploited for in vitro validation in order to combat the deadly infection of these highly emerged viral particles in the nearest future.
Authors' contributions
MBSA conceived the study and performed data analyses and writing of the manuscript. HOH contributed to data analysis and the revision of the manuscript. JMBA prepared the Iraqi library of medicinal plants. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors declared that this research received no external funding from any agency or institution.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.105155.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D.M.E. Angiotensin‐converting enzyme‐2 (ACE2), SARS‐CoV‐2 and pathophysiology of coronavirus disease 2019 (COVID‐19) The Journal of Pathology. 2020;51:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wysocki J., Schulze A., Batlle D. Novel variants of angiotensin converting enzyme-2 of shorter molecular size to target the kidney renin angiotensin system. Biomolecules. 2019;9:886. doi: 10.3390/biom9120886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang F., Yang J., Zhang Y., Dong M., Wang S., Zhang Q., Liu F.F., Zhang K., Zhang C. Angiotensin-converting enzyme 2 and angiotensin 1–7: novel therapeutic targets. Nature Reviews Cardiology. 2014;11:413. doi: 10.1038/nrcardio.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Muhammad I., Zhang Y., Ren Y., Zhang R., Huang X., Diao L., Liu H., Li X., Sun X. Antiviral activity against infectious bronchitis virus and bioactive components of Hypericum perforatum L. Frontiers in Pharmacology. 2019;10:1272. doi: 10.3389/fphar.2019.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Scientific Reports. 2018;8:1–11. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathogens. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashim H.O., Mohammed M.K., Mousa M.J., Abdulameer H.H., Alhassnawi A.T.S., Hassan S.A., Al-Shuhaib M.B.S. Infection with different strains of SARS-CoV-2 in patients with COVID-19. Archives of Biological Sciences. 2020;72:575–585. [Google Scholar]
- 8.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 2021;39:3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacological Research. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe Covid-19. New England Journal of Medicine. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends. 2020;16:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 12.Contentti E.C., Correa J. Immunosuppression during the COVID-19 pandemic in neuromyelitis optica spectrum disorders patients: a new challenge. Multiple Sclerosis and Related Disorders. 2020;41:102097. doi: 10.1016/j.msard.2020.102097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H., Wang C., Han W., Geng C., Chen D., Wu B., Zhang J., Wang C., Jiang P. Association of leptin and leptin receptor polymorphisms with coronary artery disease in a North Chinese Han population. Revista Da Sociedade Brasileira de Medicina Tropical. 2020;53 doi: 10.1590/0037-8682-0388-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farooq S., Ngaini Z. Natural and synthetic drugs as potential treatment for coronavirus disease 2019 (COVID-2019) Chemistry Africa. 2021;4:1–13. [Google Scholar]
- 15.Liskova A., Samec M., Koklesova L., Samuel S.M., Zhai K., Al-Ishaq R.K., Abotaleb M., Nosal V., Kajo K., Ashrafizadeh M. Biomedicine & Pharmacotherapy; 2021. Flavonoids against the SARS-CoV-2 Induced Inflammatory Storm; p. 111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Fang S., Wu Y., Cheng X., Zhang L., Shen X., Li S., Xu J., Shang W., Gao Z. Discovery of SARS-CoV-2-E channel inhibitors as antiviral candidates. Acta Pharmacologica Sinica. 2021:1–7. doi: 10.1038/s41401-021-00732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thangavel N., Al Bratty M., Al Hazmi H.A., Najmi A., Alaqi R.O.A. Molecular docking and molecular dynamics aided virtual search of OliveNetTM Directory for Secoiridoids to combat SARS-CoV-2 infection and associated hyperinflammatory responses. Frontiers in Molecular Biosciences. 2020;7 doi: 10.3389/fmolb.2020.627767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasirzadeh A., Bazeli J., Hajavi J., Yavarmanesh N., Zahedi M., Abounoori M., Razavi A., Maddah M.M., Mortazavi P., Moradi M. 2021. Inhibiting IL-6 during Cytokine Storm in COVID-19: Potential Role of Natural Products. [Google Scholar]
- 19.Atanasov A.G., Waltenberger B., Pferschy-Wenzig E.-M., Linder T., Wawrosch C., Uhrin P., Temml V., Wang L., Schwaiger S., Heiss E.H. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnology Advances. 2015;33:1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdelli I., Hassani F., Bekkel Brikci S., Ghalem S. In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. Journal of Biomolecular Structure and Dynamics. 2021;39:3263–3276. doi: 10.1080/07391102.2020.1763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H., Du Q. 2020. Potential Natural Compounds for Preventing SARS-CoV-2 (2019-nCoV) Infection. [Google Scholar]
- 22.Takahashi S., Yoshiya T., Yoshizawa-Kumagaye K., Sugiyama T. Nicotianamine is a novel angiotensin-converting enzyme 2 inhibitor in soybean. Biomedical Research. 2015;36:219–224. doi: 10.2220/biomedres.36.219. [DOI] [PubMed] [Google Scholar]
- 23.Balkrishna A., Pokhrel S., Singh J., Varshney A. 2020. Withanone from Withania Somnifera May Inhibit Novel Coronavirus (COVID-19) Entry by Disrupting Interactions between Viral S-Protein Receptor Binding Domain and Host ACE2 Receptor. [Google Scholar]
- 24.Wahedi H.M., Ahmad S., Abbasi S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics. 2021;39:3225–3234. doi: 10.1080/07391102.2020.1762743. [DOI] [PubMed] [Google Scholar]
- 25.Alexpandi R., De Mesquita J.F., Pandian S.K., Ravi A.V. Quinolines-based SARS-CoV-2 3CLpro and RdRp inhibitors and Spike-RBD-ACE2 inhibitor for drug-repurposing against COVID-19: an in silico analysis. Frontiers in Microbiology. 2020;11:1796. doi: 10.3389/fmicb.2020.01796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-douri N.A. A survey of medicinal plants and their traditional uses in Iraq. Pharmaceutical Biology. 2000;38:74–79. doi: 10.1076/1388-0209(200001)3811-BFT074. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed H.M. Ethnopharmacobotanical study on the medicinal plants used by herbalists in Sulaymaniyah Province, Kurdistan, Iraq. Journal of Ethnobiology and Ethnomedicine. 2016;12:1–17. doi: 10.1186/s13002-016-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawarty A.M.A.M.A., Behçet L., Çakilcioğlu U. An ethnobotanical survey of medicinal plants in Ballakayati (Erbil, North Iraq) Turkish Journal of Botany. 2020;44:345–357. [Google Scholar]
- 29.Warner F.J., Smith A.I., Hooper N.M., Turner A.J. Angiotensin-converting enzyme-2: a molecular and cellular perspective. Cellular and Molecular Life Sciences: CMLS. 2004;61:2704–2713. doi: 10.1007/s00018-004-4240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 31.Muchtaridi M., Fauzi M., Khairul Ikram N.K., Mohd Gazzali A., Wahab H.A. Natural flavonoids as potential angiotensin-converting enzyme 2 inhibitors for anti-SARS-CoV-2. Molecules. 2020;25:3980. doi: 10.3390/molecules25173980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Research. 2018;46:W257–W263. doi: 10.1093/nar/gky318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czaplewski C., Karczyńska A., Sieradzan A.K., Liwo A. UNRES server for physics-based coarse-grained simulations and prediction of protein structure, dynamics and thermodynamics. Nucleic Acids Research. 2018;46:W304–W309. doi: 10.1093/nar/gky328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuncewicz K., Spodzieja M., Sieradzan A., Karczyńska A., Dąbrowska K., Dadlez M., Speiser D.E., Derre L., Rodziewicz-Motowidło S. A structural model of the immune checkpoint CD160–HVEM complex derived from HDX-mass spectrometry and molecular modeling. Oncotarget. 2019;10:536. doi: 10.18632/oncotarget.26570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krupa P., Karczyńska A.S., Mozolewska M.A., Liwo A., Czaplewski C. UNRES-Dock—protein–protein and peptide–protein docking by coarse-grained replica-exchange MD simulations. Bioinformatics. 2021;37:1613–1615. doi: 10.1093/bioinformatics/btaa897. [DOI] [PubMed] [Google Scholar]
- 37.Liwo A., Czaplewski C., Sieradzan A.K., Lipska A.G., Samsonov S.A., Murarka R.K. Theory and practice of coarse-grained molecular dynamics of biologically important systems. Biomolecules. 2021;11:1347. doi: 10.3390/biom11091347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Research. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Research. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pathania S., Singh P.K. 2021. Analyzing FDA-Approved Drugs for Compliance of Pharmacokinetic Principles: Should There Be a Critical Screening Parameter in Drug Designing Protocols? [DOI] [PubMed] [Google Scholar]
- 41.Krupa P., Spodzieja M., Sieradzan A.K. Prediction of CD28-CD86 protein complex structure using different level of resolution approach. Journal of Molecular Graphics and Modelling. 2021;103:107802. doi: 10.1016/j.jmgm.2020.107802. [DOI] [PubMed] [Google Scholar]
- 42.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., Hou C., Wang H., Liu J., Yang D. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Critical Care. 2020;24:1–10. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosso M., Thanaraj T.A., Abu-Farha M., Alanbaei M., Abubaker J., Al-Mulla F. Molecular Therapy-Methods & Clinical Development; 2020. The Two Faces of ACE2: the Role of ACE2 Receptor and its Polymorphisms in Hypertension and COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P., Zhu L., Cai J., Lei F., Qin J.-J., Xie J., Liu Y.-M., Zhao Y.-C., Huang X., Lin L. Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circulation Research. 2020;126:1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. Journal of Biomolecular Structure and Dynamics. 2021;39:3092–3098. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shawan M.M.A.K., Halder S.K., Hasan M.A. Luteolin and abyssinone II as potential inhibitors of SARS-CoV-2: an in silico molecular modeling approach in battling the COVID-19 outbreak. Bulletin of the National Research Centre. 2021;45:1–21. doi: 10.1186/s42269-020-00479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abubakar M.B., Usman D., Batiha G.E.-S., Cruz-Martins N., Malami I., Ibrahim K.G., Abubakar B., Bello M.B., Muhammad A., Gan S.H. Natural products modulating Angiotensin Converting Enzyme 2 (ACE2) as potential COVID-19 therapies. Frontiers in Pharmacology. 2021;12 doi: 10.3389/fphar.2021.629935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghoran S.H., El-Shazly M., Sekeroglu N., Kijjoa A. Natural products from medicinal plants with anti-human coronavirus activities. Molecules. 2021;26:1754. doi: 10.3390/molecules26061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ojeda D., Jiménez-Ferrer E., Zamilpa A., Herrera-Arellano A., Tortoriello J., Alvarez L. Inhibition of angiotensin convertin enzyme (ACE) activity by the anthocyanins delphinidin-and cyanidin-3-O-sambubiosides from Hibiscus sabdariffa. Journal of Ethnopharmacology. 2010;127:7–10. doi: 10.1016/j.jep.2009.09.059. [DOI] [PubMed] [Google Scholar]
- 51.Sharma P., Shanavas A. Natural derivatives with dual binding potential against SARS-CoV-2 main protease and human ACE2 possess low oral bioavailability: a brief computational analysis. Journal of Biomolecular Structure and Dynamics. 2021;39:5819–5830. doi: 10.1080/07391102.2020.1794970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Actis-Goretta L., Ottaviani J.I., Fraga C.G. Inhibition of angiotensin converting enzyme activity by flavanol-rich foods. Journal of Agricultural and Food Chemistry. 2006;54:229–234. doi: 10.1021/jf052263o. [DOI] [PubMed] [Google Scholar]
- 53.Hausmann M., Obermeier F., Paper D.H., Balan K., Dunger N., Menzel K., Falk W., Schoelmerich J., Herfarth H., Rogler G. In vivo treatment with the herbal phenylethanoid acteoside ameliorates intestinal inflammation in dextran sulphate sodium‐induced colitis. Clinical & Experimental Immunology. 2007;148:373–381. doi: 10.1111/j.1365-2249.2007.03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernatova I., Liskova S. Mechanisms modified by (−)-Epicatechin and taxifolin relevant for the treatment of hypertension and viral infection: knowledge from preclinical studies. Antioxidants. 2021;10:467. doi: 10.3390/antiox10030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y.-T., Wu Y.-H., Tseng C.-K., Lin C.-K., Chen W.-C., Hsu Y.-C., Lee J.-C. Green tea phenolic epicatechins inhibit hepatitis C virus replication via cycloxygenase-2 and attenuate virus-induced inflammation. PloS One. 2013;8 doi: 10.1371/journal.pone.0054466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferreira P.G., Ferraz A.C., Figueiredo J.E., Lima C.F., Rodrigues V.G., Taranto A.G., Ferreira J.M.S., Brandão G.C., Vieira-Filho S.A., Duarte L.P. Detection of the antiviral activity of epicatechin isolated from Salacia crassifolia (Celastraceae) against Mayaro virus based on protein C homology modelling and virtual screening. Archives of Virology. 2018;163:1567–1576. doi: 10.1007/s00705-018-3774-1. [DOI] [PubMed] [Google Scholar]
- 57.Codorniu-Hernández E., Rolo-Naranjo A., Montero-Cabrera L.A. Theoretical affinity order among flavonoids and amino acid residues: an approach to understand flavonoid–protein interactions. Journal of Molecular Structure: THEOCHEM. 2007;819:121–129. [Google Scholar]
- 58.Zirak N., Shafiee M., Soltani G., Mirzaei M., Sahebkar A. Hypericum perforatum in the treatment of psychiatric and neurodegenerative disorders: current evidence and potential mechanisms of action. Journal of Cellular Physiology. 2019;234:8496–8508. doi: 10.1002/jcp.27781. [DOI] [PubMed] [Google Scholar]
- 59.Cayci M.K., Dayioglu H. Hypericum perforatum extracts healed gastric lesions induced by hypothermic restraint stress in Wistar rats. Saudi Med J. 2009;30:750–754. [PubMed] [Google Scholar]
- 60.Brattström A. Long-term effects of St. John's wort (Hypericum perforatum) treatment: a 1-year safety study in mild to moderate depression. Phytomedicine. 2009;16:277–283. doi: 10.1016/j.phymed.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 61.Altan A., Aras M.H., Damlar İ., Gökçe H., Özcan O., Alpaslan C. The effect of Hypericum Perforatum on wound healing of oral mucosa in diabetic rats. European Oral Research. 2018;52:143–149. doi: 10.26650/eor.2018.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seyhan N. Evaluation of the healing effects of Hypericum perforatum and curcumin on burn wounds in rats. Evidence-Based Complementary and Alternative Medicine. 2020:2020. doi: 10.1155/2020/6462956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozkan E.E., Ozden T.Y., Ozsoy N., Mat A. Evaluation of chemical composition, antioxidant and anti-acetylcholinesterase activities of Hypericum neurocalycinum and Hypericum malatyanum. South African Journal of Botany. 2018;114:104–110. [Google Scholar]
- 64.Kacerovská D., Pizinger K., Majer F., Šmíd F. Photodynamic therapy of nonmelanoma skin cancer with topical hypericum perforatum extract—a pilot study. Photochemistry and Photobiology. 2008;84:779–785. doi: 10.1111/j.1751-1097.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 65.Hammer K.D.P., Hillwig M.L., Solco A.K.S., Dixon P.M., Delate K., Murphy P.A., Wurtele E.S., Birt D.F. Inhibition of prostaglandin E2 production by anti-inflammatory Hypericum perforatum extracts and constituents in RAW264. 7 mouse macrophage cells. Journal of Agricultural and Food Chemistry. 2007;55:7323–7331. doi: 10.1021/jf0710074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yalçın S., Yalçınkaya S., Ercan F. Determination of potential drug candidate molecules of the Hypericum perforatum for COVID-19 treatment. Current Pharmacology Reports. 2021:1–7. doi: 10.1007/s40495-021-00254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jena A.B., Kanungo N., Nayak V., Chainy G.B.N., Dandapat J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: insights from computational studies. Scientific Reports. 2021;11:1–14. doi: 10.1038/s41598-021-81462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ottaviani J.I., Momma T.Y., Heiss C., Kwik-Uribe C., Schroeter H., Keen C.L. The stereochemical configuration of flavanols influences the level and metabolism of flavanols in humans and their biological activity in vivo. Free Radical Biology and Medicine. 2011;50:237–244. doi: 10.1016/j.freeradbiomed.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Mallipeddi P.L., Kumar G., White S.W., Webb T.R. Recent advances in computer-aided drug design as applied to anti-influenza drug discovery. Current Topics in Medicinal Chemistry. 2014;14:1875–1889. doi: 10.2174/1568026614666140929153812. [DOI] [PubMed] [Google Scholar]
- 70.Macip G., Garcia‐Segura P., Mestres‐Truyol J., Saldivar‐Espinoza B., Ojeda‐Montes M.J., Gimeno A., Cereto‐Massagué A., Garcia‐Vallvé S., Pujadas G. Haste makes waste: a critical review of docking‐based virtual screening in drug repurposing for SARS‐CoV‐2 main protease (M‐pro) inhibition. Medicinal Research Reviews. 2021 doi: 10.1002/med.21862. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Llanos M.A., Gantner M.E., Rodriguez S., Alberca L.N., Bellera C.L., Talevi A., Gavernet L. Strengths and weaknesses of docking simulations in the SARS-CoV-2 era: the main protease (Mpro) case study. Journal of Chemical Information and Modeling. 2021;61:3758–3770. doi: 10.1021/acs.jcim.1c00404. [DOI] [PubMed] [Google Scholar]
- 72.Phillips M.A., Stewart M.A., Woodling D.L., Xie Z.-R. Has molecular docking ever brought us a medicine. Molecular Docking. 2018:141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.