Inflammasomes are cytosolic sensing machineries that play a critical role in antimicrobial defense. In recent years, great progress has been made in characterizing the molecular mode of action and functional role of the human inflammasome sensor NLRP1.
Abstract
In response to infection or cell damage, inflammasomes form intracellular multimeric protein complexes that play an essential role in host defense. Activation results in the maturation and subsequent secretion of pro-inflammatory cytokines of the IL-1 family and a specific cell death coined pyroptosis. Human NLRP1 was the first inflammasome-forming sensor identified at the beginning of the millennium. However, its functional relevance and its mechanism of activation have remained obscure for many years. Recent discoveries in the NLRP1 field have propelled our understanding of the functional relevance and molecular mode of action of this unique inflammasome sensor, which we will discuss in this perspective.
Introduction
Our bodies are under constant threat from exogenous and endogenous dangers, including infection with pathogenic bacteria, viruses, parasites, or developing cancer cells. To protect against these perpetual dangers, vertebrates rely on both an innate and adaptive immune system. The innate immune system builds on germline encoded pattern recognition receptors (PRRs) as its nonself sensing interface. Upon engagement, PRRs of a certain family assemble into a multimeric protein complex, coined the inflammasome (Broz and Dixit, 2016).
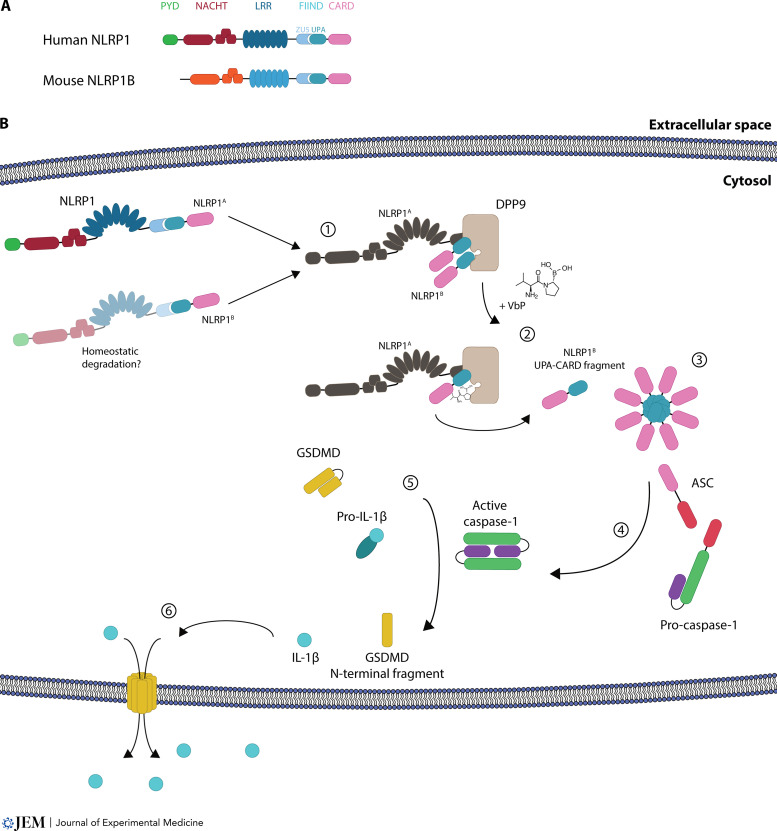
The inflammasome consists of a sensor protein, which can either directly, or through an adapter protein called apoptosis-associated speck-like protein containing a CARD (ASC), activate pro–caspase-1. Signaling in the inflammasome complex takes place via homotypic protein–protein domain interactions. For instance, the pyrin domain (PYD) of a sensor can interact with the N-terminal PYD of ASC, which then recruits pro–caspase-1 via its C-terminal caspase activation and recruitment domain (CARD). Thus-activated caspase-1 then matures cytokines of the IL-1 family, most prominently the pro-inflammatory cytokine pro–IL-1β (Broz and Dixit, 2016). Furthermore, caspase-1 cleaves the protein gasdermin D (GSDMD). This resolves an inherent autoinhibitory function within GSDMD, resulting in the assembly of the N-terminal moieties of GSDMD molecules into a pore at the plasma membrane that leads to the subsequent rupture of the cell, a process known as pyroptosis (Fig. 1; Kayagaki et al., 2015; Shi et al., 2015). Activation of this signaling complex can be regarded as a last-resort mechanism due to the demise of the cell and the highly inflammatory nature of its outcome. Most characterized inflammasome sensors are part of the nucleotide-binding domain and leucine-rich repeat (LRR) containing (NLR) protein family, and the most studied member of this family is NLR family pyrin domain containing 3 (NLRP3). Inflammasome sensors can directly be activated by encountering a ligand, for instance AIM2 inflammasome activation by cytosolic double-stranded DNA stemming from viral or bacterial infection, or indirectly through the perturbation of cellular processes, such as membrane rupture and subsequent potassium efflux, which activates the NLRP3 inflammasome (Bürckstümmer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Muñoz-Planillo et al., 2013; Pétrilli et al., 2007). Here we will discuss recent discoveries surrounding the inflammasome sensor NLRP1 and its modes of action.
Figure 1.
NLRP1 inflammasome activation and its downstream signaling pathway. (A) Overview of the domain structure of human NLRP1 and mouse NLRP1B. (B) Overview of human NLRP1 activation by the DPP8/9 inhibitor VbP. (1) Generation of NLRP1A and NLRP1B by homeostatic degradation and assembly of the ternary DPP9-NLRP1A-NLRP1B complex. (2) Direct displacement of NLRP1B from the DPP9 substrate tunnel leads to release of the C-terminal UPA-CARD fragments and (3) generation of an inflammasome seed. (4) Via the adaptor ASC, caspase-1 is activated. (5) Caspase-1 cleaves its substrates pro–IL-1β, which matures to pro-inflammatory IL-1β, and GSDMD, which (6) generates pores into the membrane and leads to a pyroptotic cell death.
The NLRP1 inflammasome: Early days
Human NLRP1 was the first inflammasome-forming sensor discovered and described in the landmark paper of Tschopp and coworkers in 2002 (Martinon et al., 2002). In an in vitro system, they observed that NLRP1 is found in a high-molecular-weight complex serving as a platform to activate caspase-1, which in turn subsequently cleaves and matures IL-1β. Although the discovery of human NLRP1 as an inflammasome activator founded inflammasome research per se, the activation of NLRP1 itself remained enigmatic for a long time. A first report that used recombinant NLRP1 protein and reconstituted the inflammasome in vitro suggested in 2007 that NLRP1 is activated by the bacterial cell wall component muramyl dipeptide (MDP; Faustin et al., 2007). It was shown that recombinant NLRP1 oligomerizes after the encounter with MDP, leading to the activation of caspase-1. However, no loss-of-function data were provided showing NLRP1-dependent inflammasome activation following MDP stimulation in cells. As such, the MDP-driven inflammasome response seen in THP-1 cells, a monocytic cell line, could also stem from NLRP3 inflammasome activation, since MDP is known to prime this inflammasome (Martinon et al., 2004). Indeed, no follow-up studies substantiated MDP-dependent activation of human NLRP1. More indicative studies were published the same year, reporting that UVB irradiation of keratinocytes led to IL-1β secretion, which was diminished in cells that were silenced for NLRP1 (Feldmeyer et al., 2007; Watanabe et al., 2007). Only recently, those observations were revisited using keratinocytes, in which NLRP1 was deleted using CRISPR/Cas9 (Fenini et al., 2018). These aforementioned and following reports also established that the core inflammasome signaling components and pro–IL-1β are expressed in the human epithelium (Sand et al., 2018). This clearly contrasts with the expression pattern of NLRP3 and murine NLRP1, which are mainly found in myeloid cells. Moreover, genetic association studies have implied a role for certain NLRP1 haplotypes in vitiligo and various associated autoimmune diseases (Jin et al., 2007; Levandowski et al., 2013).
Of mice and men
In the following years, more decisive insight on NLRP1 biology came from the murine system. However, a crucial aspect of NLRP1 biology is the difference between mice and men. For other inflammasome sensors such as NLRP3, the activation trigger and activation mechanisms (e.g., efflux of potassium) are generally concordant across different species. However, this is not necessarily the case for human and mouse NLRP1, which are highly divergent. An obvious difference between mice and men is already visible when investigating the gene loci coding for NLRP1: in the human system, there is only a single gene for NLRP1 on chromosome 17, while seven transcript variants have been described. The mouse Nlrp1 gene locus on the syntenic region on chromosome 11, however, displays several Nlrp1 paralogs that are located in tandem to one another (Lilue et al., 2018). Analyzing different inbred mouse strains, up to seven different paralogs for mouse Nlrp1 have been identified while, depending on the mouse strain, the number of paralogs encoded can range from three to five Nlrp1 family members. For instance, C57BL/6 mice harbor three Nlrp1 paralogs (Nlrp1a, Nlrp1b, and Nlrp1c; Lilue et al., 2018). In addition to this multi-gene configuration, the individual Nlrp1 mouse alleles, most prominently Nlrp1b alleles, are highly polymorphic. As such, five different Nlrp1b alleles have been characterized in various inbred mouse strains (designated Nlrp1b1–Nlrp1b5) that differ in their functionality. Nlrp1a and Nlrp1c alleles, on the other hand, show a lesser degree of polymorphism, while little is known about the recently described novel Nlpr1d-f alleles (Lilue et al., 2018; Sastalla et al., 2013). Since most of our knowledge on murine NLRP1 stems from work on the Nlrp1b allele, which encodes the NLRP1B protein, we will mostly draw interspecies comparisons to this Nlrp1 family member.
Regarding their domain architecture, human NLRP1 and mouse NLRP1B share similar features: they both have a central domain present in NAIP, CIITA, HET-E, and TP-1 (NACHT) followed by LRRs, a function-to-find domain (FIIND) that can be subdivided into ZU5 (found in ZO-1 and UNC5 domain) and UPA (conserved in UNC5, PIDD, and Ankyrin domain) subdomains, and a CARD domain. The NACHT domain, sometimes also called nucleotide-binding oligomerization domain, contains Walker A and Walker B motifs, which are reported for ATP binding and ATP hydrolysis, respectively (Danot et al., 2009). The unusual feature of the NLRP1 protein is its FIIND domain, which undergoes constitutive autocleavage and thus generates an N-terminal and a C-terminal fragment in cells that remain noncovalently associated (D’Osualdo et al., 2011). After activation of the sensor and subsequent release of the C-terminal fragment, both the mouse NLRP1B and human NLRP1 inflammasome employ their C-terminal CARD domain for downstream signaling and activation. While human NLRP1 critically requires the adapter protein ASC for downstream signaling, mouse NLRP1B can both directly engage caspase-1 and also employ ASC for this purpose (Broz et al., 2010; de Vasconcelos et al., 2019; Zhong et al., 2016). However, regarding differences in domain architecture, only human NLRP1 harbors an N-terminal PYD, which is considered to act in an autoinhibitory fashion (Chavarría-Smith et al., 2016; Zhong et al., 2016). This contrasts with other NLRP proteins, for instance NLRP3, which use their N-terminal PYD to interact with the adaptor ASC. Further, human NLRP1 contains a disordered region between its PYD and the NACHT domain (Chui et al., 2020). It is also worth mentioning that in humans, but not in mice, another FIIND-containing and inflammasome-forming protein is present: CARD8 (Johnson et al., 2018). In contrast to NLRP1, CARD8 only consists of a disordered N terminus, followed by a FIIND and a C-terminal CARD domain. CARD8 employs its CARD domain to directly interact with and activate caspase-1 (Gong et al., 2021; Hollingsworth et al., 2021a; Linder et al., 2020).
A comparison of NLRP1 from diverse primates revealed signs of positive selection for NLRP1 based on a high dN/dS ratio, which accounts for a higher rate of nonsynonymous over synonymous substitutions (George et al., 2011). This directional selection of the NLRP1 gene within primates has been attributed to result from a strong evolutionary pressure, hinting to NLRP1 being involved in an evolutionary arms race (Chavarría-Smith et al., 2016). Furthermore, comparing rodents with humans, NLRP1 shows the lowest level of identity among all known inflammasome-forming NLRs, which explains their different modes of action, as outlined below. Another important difference between the human and mouse homologues is their functionality in different tissues: functional data on mouse NLRP1 activation mainly stems from experiments with cells of the myeloid lineage, e.g., classical immune cells such as macrophages. Moreover, a gain-of-function (GOF) mutation in murine NLRP1A results in leukopenia due to pyroptosis of hematopoietic progenitor cells (Masters et al., 2012). In humans, however, functional NLRP1 is largely confined to epithelial barrier tissues such as keratinocytes and bronchial epithelial cells (Drutman et al., 2019; Linder et al., 2020). This is also reflected in the occurrence of tissue-specific phenotypes of GOF mutations for human NLRP1 that manifest in the tissue of the epithelial barrier (Zhong et al., 2016). In contrast, a functionality for the human CARD8 inflammasome has been shown for bone marrow–derived cells (Johnson et al., 2020; Johnson et al., 2018; Linder et al., 2020). Due to these tissue-specific differences, there appears to be no redundancy between CARD8 activation and NLRP1 activation. All in all, because of these differences between mice and men, the results of mouse studies could only partially contribute to decipher the role of human NLRP1.
Mouse NLRP1B
Activation of mouse NLRP1B by lethal factor (LF)
Unbeknownst at that time, research on mouse NLRP1B was driven by studying host–pathogen interaction with the Gram-positive bacterium Bacillus anthracis in the 1950s (Smith and Keppie, 1954). It was noted that the proteinaceous bacterial virulence factors protective antigen and the zinc-dependent metalloproteinase LF, which together make up the anthrax lethal toxin (LT), are among the key determinants in the pathogenicity of B. anthracis. As such, direct LT administration can induce a rapid inflammatory response in mice that culminates in systemic shock and death, which is dependent on macrophages and IL-1 signaling (Hanna et al., 1993). Using inbred mouse strains, the Nlrp1 gene region was identified in 2006 as the susceptibility factor for LT-induced macrophage cell death (Boyden and Dietrich, 2006). Of the five highly polymorphic alleles for Nlrp1b, only Nlrp1b1 and Nlrp1b5 respond to LT stimulation (Boyden and Dietrich, 2006; Yu et al., 2018). However, although Nlrp1 is responsible for cytotoxicity and death after high doses of LT delivery, the Nlrp1 locus confers resistance in the context of an intravenous or subcutaneous spore infection of mice with B. anthracis (Moayeri et al., 2010). While it was shown that mice carrying a LT-sensitive allele were able to restrict B. anthracis infection and that this was dependent on expression of caspase-1 and IL-1β, the exact activation mechanism of NLRP1B stayed enigmatic for some time (Moayeri et al., 2010). In 2012, it could be shown that LF proteolytically processes NLRP1B, leading to inflammasome activation (Hellmich et al., 2012; Levinsohn et al., 2012). However, it could not be proven if the proteolytic event on NLRP1B itself is necessary or sufficient for NLRP1B activation, or if perhaps other substrates, which could inhibit NLRP1B, required cleavage by LF for inflammasome activation.
Activation of mouse NLRP1B by “functional degradation”
Ultimately it was shown that cleavage of NLRP1B itself can activate the inflammasome through testing an engineered system, in which NLRP1B was equipped with an N-terminal GFP followed by a synthetically introduced TEV (tobacco etch virus) cleavage site. After expression of TEV, resulting in NLRP1B cleavage, such a construct could be activated to trigger caspase-1 activation (Chavarría-Smith and Vance, 2013). However, it remained enigmatic why activation of NLRP1B by LF required the proteasome for activation (Squires et al., 2007). In 2018, several studies came to a unifying concept of NLRP1B activation, the “functional degradation” model: cleavage of NLRP1B on its N terminus exposes a destabilizing neo–N terminus, which is recognized and ubiquitinated by the ubiquitin ligase Ubr2 (Chui et al., 2019; Sandstrom et al., 2019; Xu et al., 2019). Subsequently, the N-terminal fragment of NLRP1B is degraded by the proteasome, but the C-terminal UPA-CARD fragment, generated by autoproteolysis within the FIIND, is not degraded and can thus trigger inflammasome activation. Based on these considerations, it can be speculated that the N terminus of NLRP1B has evolved to sense pathogenic protease activity and acts as a kind of tripwire. Further supporting the “functional degradation,” model it was shown that the enzymatic activity of the Shigella flexneri E3 ubiquitin ligase IpaH7.8 is also sensed by NLRP1B (Sandstrom et al., 2019). In this case, the N terminus of NLRP1B appears to function as a decoy and is decorated by the ligase with ubiquitin and therefore subjected to degradation, which activates the inflammasome. Thus, by sensing a pathogen's proteolytic activity or ubiquitination activity, mouse NLRP1B activation follows the concept of effector-triggered immunity (Dangl and Jones, 2001).
A common denominator: DPP8/9 inhibition activates NLRP1
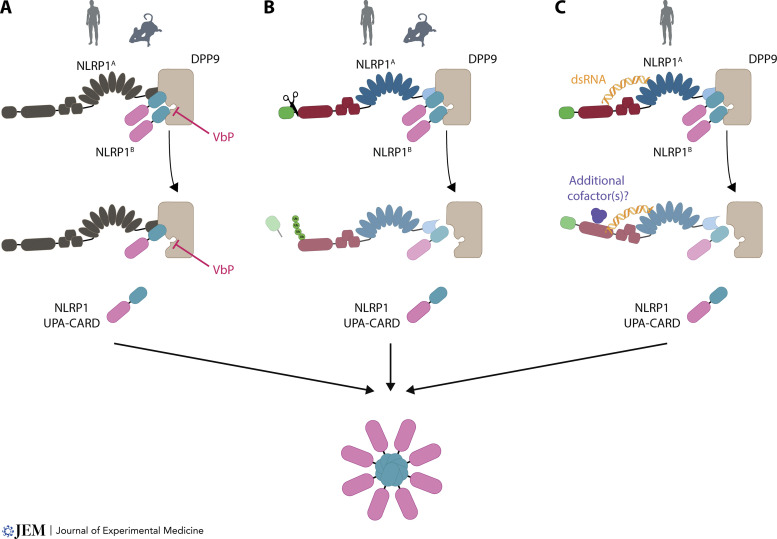
It came as a surprise in 2018 that dipeptidylpeptidase (DPP) inhibition by the small molecule Val-boroPro (VbP), a broad DPP4/7/8/9 inhibitor, activates the NLRP1B inflammasome (Okondo et al., 2018). The year before, the same authors had already proposed the induction of a “pro–caspase-1–dependent” pyroptosis in mouse macrophages by inhibition of DPP8/9, with a yet unclear activation mechanism (Okondo et al., 2017). As such, it was shown that VbP, also named talabostat or PT-100, as well as the DPP8/9-selective inhibitors 1G244 and 8j, activate the NLRP1 inflammasome (Okondo et al., 2017; Okondo et al., 2018; Zhong et al., 2018). DPP8/9 are intracellular proteases that exhibit dipeptidylaminopeptidase activity and cleave after proline residues at the penultimate position P1. How the inhibition of these DPPs would activate the NLRP1 inflammasome was at first quite puzzling. However, around the same time, it was independently found that human NLRP1 interacts with DPP9 through immunoprecipitation studies followed by mass spectrometry (Zhong et al., 2018). Subsequently it could be shown that binding of DPP9 as well as its catalytic activity are required for inhibiting the activation of the NLRP1 inflammasome (Zhong et al., 2018). Thus, the inhibition of DPP8/9 by small molecules restrains this inhibitory effect on NLRP1 and leads to downstream inflammasome activation. The binding of DPP9 is common to both human and murine NLRP1, since this interaction occurs via their FIIND domain, which is their most homologous region. Moreover, the FIIND domain of the inflammasome sensor CARD8 is likewise bound by DPP8/9 and kept in check by a mechanism analogous to NLRP1 (Johnson et al., 2018; Sharif et al., 2021). Because of this, the protein can also be activated and form an inflammasome following DPP8/9 inhibition (Johnson et al., 2018).
Subsequent studies investigated the interaction of NLRP1 with DPP9 from a structural point of view by cryo-electron microscopy (cryo-EM; Hollingsworth et al., 2021b; Huang et al., 2021). Here, a ternary DPP9-NLRP1A-NLRP1B complex was identified that consists of a full-length NLRP1A molecule and a second NLRP1B molecule, of which only the C-terminal fragment, consisting of UPA-CARD, was visible in the cryo-EM map density. For the NLRP1A molecule, the first β-strand of the UPA subdomain folds into the ZU5 fold, and this keeps the N-terminal and C-terminal fragments associated and in an autoinhibited state. For the NLRP1B molecule, the UPA-CARD fragment binds, via its N terminus, into DPP9’s substrate tunnel and therefore is sequestered and kept in an inhibited state by DPP9. Interestingly, patients with the autoinflammatory syndrome autoinflammation with arthritis and dyskeratosis carry a P1214R mutation of NLRP1 (Zhong et al., 2018). This mutation abrogates the interaction of NLRP1 with DPP9 and this subsequently leads to inflammasome activation. Furthermore, mutations in human DPP9 were also found in patients, which partially resembled symptoms of arthritis and dyskeratosis (Harapas et al., 2021 Preprint). Those mutations either generated a catalytically inactive protein due to stop-gain mutations or affected DPP9’s substrate binding via missense mutations and thus hampered inhibition of NLRP1 by DPP9. Although the interaction of DPP9 with NLRP1B resembles a substrate-bound structure and NLRP1 also having a proline at P1, which would thus be recognized as a classical substrate for DPP9 cleavage, it is not processed by DPP9 on its N terminus (Griswold et al., 2019; Hollingsworth et al., 2021b; Zhong et al., 2018). Interestingly, however, it has also been claimed that binding alone is not sufficient for efficient inhibition of the NLRP1 inflammasome but also requires the catalytic activity of DPP9 (Huang et al., 2021).
In conclusion, the activation of NLRP1 through inhibition of DPP8/9 is thought to act through direct competition and displacement of NLRP1B from the DPP9 substrate tunnel by VbP, thereby unleashing the inflammatory C-terminal part of NLRP1 (see Fig. 1 for an overview of inflammasome activation by VbP). Following the displacement, the C-terminal fragment assembles via its UPA subdomain into an inflammasome seed and thus leads to activation of downstream signaling, as shown through structural studies (Gong et al., 2021; Hollingsworth et al., 2021a). Intriguingly, in cryo-EM structures of a ternary DPP9-CARD8A-CARD8B complex, the C-terminal CARD8B is not directly bound by DPP9 via its N terminus interacting with the substrate tunnel of DPP9 but via another interaction site (Sharif et al., 2021 ). In in vitro structural studies, VbP did not displace CARD8B from DPP9, yet it could do so in cells.
How the NLRP1B molecule, which is missing its N-terminal fragment, is generated in the first place and how this process is regulated are not clear and require further studies. Regulating the generation of the NLRP1B molecule could be another way to control inflammasome activation for NLRP1 and could act like a licensing step for activation.
Of note, no physiological counterpart has been found for NLRP1 or CARD8 activation in response to DPP8/9 inhibition so far. However, it is conceivable that NLRP1 and CARD8 act as sensors for cellular perturbation in a guard-like fashion, as known from plant defense systems (van Wersch et al., 2020). As such, they could survey the inhibition of DPP8/9 activity, which could be a result of pathogenic activity. In addition to being activated by VbP, there are reports on human and rodent NLRP1 activation by Toxoplasma gondii (Cirelli et al., 2014; Ewald et al., 2014; Witola et al., 2011). T. gondii–dependent activation of NLRP1 has been best studied in the rat, where certain inbred strains exhibit T. gondii infection–dependent inflammasome activation, whereas others do not. Interestingly, in these different inbred rat strains, the sensitivity to T. gondii is reciprocal to the sensitivity to anthrax LF. Moreover, comparison of different rat alleles suggests that this functionality maps to a small N-terminal region of NLRP1 (Cirelli et al., 2014). Hence, it is conceivable that this particular region is directly or indirectly impacted by T. gondii infection. However, it has also been noted that the sensitivity of different Nlrp1 alleles toward T. gondii infection parallels the sensitivity toward DPP9 inhibition (Gai et al., 2019).
Human NLRP1
Physiological activation of human NLRP1 by viral infection
Human NLRP1 stepped into the spotlight in 2016, when its role as the primary inflammasome sensor in human skin was substantiated, and GOF mutations of NLRP1 were described: multiple self-healing palmoplantar carcinoma and familial keratosis lichenoides chronica, which result in increased susceptibility to skin cancer (Zhong et al., 2016). Interestingly, multiple self-healing palmoplantar carcinoma patients carry mutations in the PYD, and familial keratosis lichenoides chronica patients display mutations in the LRR domain of NLRP1; both domains are supposed to act in an autoinhibitory fashion on NLRP1, and those mutations are thought to perturb this autoinhibitory function. Further, in this study, the primary-like but immortalized N/TERT-1 keratinocyte cell line was introduced as a suitable cell model to study endogenously expressed NLRP1.
In 2020, two physiological activators of human NLRP1 were described. First, it was reported that NLRP1 is cleaved on its N terminus by viral 3C proteases (Robinson et al., 2020; Tsu et al., 2021). This was found through testing different pathogens or by bioinformatically predicting possible 3C protease cleavage sites. The mode of activation is well in line with the model of “functional degradation.” After cleavage by a protease, the N-terminal fragment is degraded, and the C-terminal UPA-CARD can form an inflammasome. As mentioned above, an activation of human NLRP1 by a cleavage event had already been described before, as a human NLRP1, similarly engineered as described for the mouse NLRP1B molecule with an N-terminal GFP and TEV cleavage site, could be activated by a TEV cleavage event (Chavarría-Smith et al., 2016). Noteworthy, human CARD8 can likewise be activated by a proteolytic event. Here, it has been shown that an HIV protease, which is usually only activated after budding of the virus from the cell, can be activated intracellularly by nonnucleoside reverse-transcriptase inhibitors and thus cleave and activate the CARD8 inflammasome (Wang et al., 2021). Second, we reported on the activation of NLRP1 by double-stranded RNA (dsRNA; Bauernfried et al., 2021). This was found by studying Semliki Forest Virus as an activator of human NLRP1 in a screen for pathogens to activate NLRP1. In this setting, activation of NLRP1 was pinpointed to the generation of dsRNA in the lifecycle of Semliki Forest Virus. By using recombinant NLRP1, a direct interaction of human NLRP1, but not mouse NLRP1B, with dsRNA was observed. Moreover, enhanced ATPase activity was detected after NLRP1 encountered dsRNA but not double-stranded DNA, indicating that NLRP1 responds to dsRNA with a conformational switch. However, further studies are needed to determine the exact mechanism by which dsRNA activates NLRP1 and if other factors are involved in this pathway (see Fig. 2 for an overview of activation modes of human and mouse NLRP1).
Figure 2.
Modes of NLRP1 inflammasome activation. (A) Activation of NLRP1 by DPP8/9 inhibition. Direct displacement of the UPA-CARD fragments by DPP9 inhibition leads to inflammasome activation. (B) Activation of NLRP1 by species-specific protease cleavage events followed by the “functional degradation” of the N-terminal fragment. Human NLRP1 is activated by 3C protease (depicted here) and mouse NLRP1B by LF cleavage. (C) Activation of NLRP1 by dsRNA and conformational rearrangement. See main text for more details. Icons are depicting activatability of human or mouse NLRP1.
Concluding remarks
Having reviewed the current knowledge about NLRP1 inflammasome biology, let’s delve into some speculations about the origin and primordial role of this molecule. As outlined in this review, NLRP1 has evolved to sense various types of perturbations or molecular entities. However, unlike a promiscuous receptor that serves different ligands through the same ligand-binding domain and the same molecular mode of action, the mechanisms by which NLRP1 senses these different modalities are more complex. Destabilization of the DPP9-NLRP1 complex, as it is achieved by DPP8/9 inhibition, is without doubt the most proximal mode of NLRP1 activation. Since this core functionality is already represented by CARD8 in humans, it appears that this activity does not suffice to cover the nonself-sensing capacity of this receptor family. As such, NLRP1 entertains a prominent “NLR module” that is N-terminal to its FIIND. While this component could be seen as the remnant or an adaption of an NLR that just serves to function as a decoy with its “tripwire” mode of activation, several findings argue against this notion. If these domains had no functional role, why would they have survived evolution? Indeed, NLRP1 is under strong positive selection, yet this is mainly attributable to the disordered region between the PYD and NACHT, as well as the LRRs. Motifs and residues relevant for nucleotide binding and/or hydrolysis in the NACHT of NLRP1 are preserved, and in line with this, the NACHT can exert ATP hydrolysis activity (Bauernfried et al., 2021; MacDonald et al., 2013). Moreover, mutations in ATP coordinating amino acid residues of the NACHT appear to result in spontaneous activity (Bauernfried et al., 2021). Further, if NLRP1’s sole function was to act as a protease sensor or DPP8/9 activity sensor, evolution could have trimmed NLRP1 into a protein similar to the size of CARD8, which can still serve these functions. From a reductionist perspective, this would greatly minimize the cost of expression of such a lengthy protein. In fact, as already outlined above, dsRNA-mediated NLRP1 activation is in line with a more complex role of the NLR portion of NLRP1 in nonself recognition.
It is tempting to speculate that the “NLR portion” of NLRP1 originates from a primordial NLR protein that has engaged in a pathogens arms race. The FIIND domain might have been added to such a primordial NLR later in evolution, so that this molecule would be protected from pathogen antagonism by its C-terminal “booby trap.” In fact, the fusion of a FIIND–death fold domain extension to the C terminus of a protein is seen for several proteins in the human system and it is even more widespread in other organisms (Mitchell et al., 2019). For example, lower vertebrates harbor a variety of proteins associated with immune functions that have a C-terminal FIIND–death fold domain extension, analogous to CARD8 or NLRP1 (Jin et al., 2013; Tyrkalska et al., 2016). One could speculate that the N-terminal part constitutes or constituted the actual antimicrobial function of these proteins. However, counteracting actions of the pathogen and degradation of this N-terminal “business part” of these proteins would inevitably unleash the C-terminal domain and thereby induce cell-autonomous defense mechanisms, effectuated by the death fold domain. As such, adding the C-terminal FIIND–death fold domain would be a means to preserve an important defense protein from pathogen attack. It will be interesting to address this hypothesis experimentally for other FIIND–death fold domain proteins and also to identify the evolutionary traces of the emergence of NLRP1 and CARD8. In this regard, it will be interesting to find out at which point in evolution the primordial NLRP1/CARD8 FIIND coopted DPP8/9 as its “partner in crime.” Connected to this question, it should be interesting to explore why the catalytic activity of DPP9 is so tightly guarded by NLRP1 and CARD8. Again, it is tempting to speculate that DPP9 serves a yet to be characterized antimicrobial function that is inhibited or overwhelmed by pathogens. Such a functionality could only be uncovered in NLRP1 or CARD8-deficient cells, in which the absence of DPP9 could be studied without triggering spontaneous inflammasome activation.
Overall, it is clear that NLRP1 has not yet revealed all its secrets and that there is still plenty of research ahead of us in this emerging field.
Acknowledgments
Icons for the human and the mouse are from SciDraw.
This work is funded by European Research Council 2020-ADG ENGINES (101018672).
Author contributions: S. Bauernfried and V. Hornung conceived and wrote the manuscript.
References
- Bauernfried, S., Scherr M.J., Pichlmair A., Duderstadt K.E., and Hornung V.. 2021. Human NLRP1 is a sensor for double-stranded RNA. Science. 371:eabd0811. 10.1126/science.abd0811 [DOI] [PubMed] [Google Scholar]
- Boyden, E.D., and Dietrich W.F.. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240–244. 10.1038/ng1724 [DOI] [PubMed] [Google Scholar]
- Broz, P., and Dixit V.M.. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16:407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Broz, P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer, T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K.L., and Superti-Furga G.. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266–272. 10.1038/ni.1702 [DOI] [PubMed] [Google Scholar]
- Chavarría-Smith, J., and Vance R.E.. 2013. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 9:e1003452. 10.1371/journal.ppat.1003452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarría-Smith, J., Mitchell P.S., Ho A.M., Daugherty M.D., and Vance R.E.. 2016. Functional and Evolutionary Analyses Identify Proteolysis as a General Mechanism for NLRP1 Inflammasome Activation. PLoS Pathog. 12:e1006052. 10.1371/journal.ppat.1006052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui, A.J., Okondo M.C., Rao S.D., Gai K., Griswold A.R., Johnson D.C., Ball D.P., Taabazuing C.Y., Orth E.L., Vittimberga B.A., and Bachovchin D.A.. 2019. N-terminal degradation activates the NLRP1B inflammasome. Science. 364:82–85. 10.1126/science.aau1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui, A.J., Griswold A.R., Taabazuing C.Y., Orth E.L., Gai K., Rao S.D., Ball D.P., Hsiao J.C., and Bachovchin D.A.. 2020. Activation of the CARD8 Inflammasome Requires a Disordered Region. Cell Rep. 33:108264. 10.1016/j.celrep.2020.108264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli, K.M., Gorfu G., Hassan M.A., Printz M., Crown D., Leppla S.H., Grigg M.E., Saeij J.P., and Moayeri M.. 2014. Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog. 10:e1003927. 10.1371/journal.ppat.1003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Osualdo, A., Weichenberger C.X., Wagner R.N., Godzik A., Wooley J., and Reed J.C.. 2011. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One. 6:e27396. 10.1371/journal.pone.0027396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones J.D.. 2001. Plant pathogens and integrated defence responses to infection. Nature. 411:826–833. 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- Danot, O., Marquenet E., Vidal-Ingigliardi D., and Richet E.. 2009. Wheel of Life, Wheel of Death: A Mechanistic Insight into Signaling by STAND Proteins. Structure. 17:172–182. 10.1016/j.str.2009.01.001 [DOI] [PubMed] [Google Scholar]
- de Vasconcelos, N.M., Vliegen G., Gonçalves A., De Hert E., Martín-Pérez R., Van Opdenbosch N., Jallapally A., Geiss-Friedlander R., Lambeir A.M., Augustyns K., et al. 2019. DPP8/DPP9 inhibition elicits canonical Nlrp1b inflammasome hallmarks in murine macrophages. Life Sci. Alliance. 2:e201900313. 10.26508/lsa.201900313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drutman, S.B., Haerynck F., Zhong F.L., Hum D., Hernandez N.J., Belkaya S., Rapaport F., de Jong S.J., Creytens D., Tavernier S.J., et al. 2019. Homozygous NLRP1 gain-of-function mutation in siblings with a syndromic form of recurrent respiratory papillomatosis. Proc. Natl. Acad. Sci. USA. 116:19055–19063. 10.1073/pnas.1906184116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald, S.E., Chavarria-Smith J., and Boothroyd J.C.. 2014. NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun. 82:460–468. 10.1128/IAI.01170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustin, B., Lartigue L., Bruey J.M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., and Reed J.C.. 2007. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell. 25:713–724. 10.1016/j.molcel.2007.01.032 [DOI] [PubMed] [Google Scholar]
- Feldmeyer, L., Keller M., Niklaus G., Hohl D., Werner S., and Beer H.D.. 2007. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr. Biol. 17:1140–1145. 10.1016/j.cub.2007.05.074 [DOI] [PubMed] [Google Scholar]
- Fenini, G., Grossi S., Contassot E., Biedermann T., Reichmann E., French L.E., and Beer H.D.. 2018. Genome Editing of Human Primary Keratinocytes by CRISPR/Cas9 Reveals an Essential Role of the NLRP1 Inflammasome in UVB Sensing. J. Invest. Dermatol. 138:2644–2652. 10.1016/j.jid.2018.07.016 [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri, T., Yu J.W., Datta P., Wu J., and Alnemri E.S.. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 458:509–513. 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai, K., Okondo M.C., Rao S.D., Chui A.J., Ball D.P., Johnson D.C., and Bachovchin D.A.. 2019. DPP8/9 inhibitors are universal activators of functional NLRP1 alleles. Cell Death Dis. 10:587. 10.1038/s41419-019-1817-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, R.D., McVicker G., Diederich R., Ng S.B., MacKenzie A.P., Swanson W.J., Shendure J., and Thomas J.H.. 2011. Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Res. 21:1686–1694. 10.1101/gr.121327.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Q., Robinson K., Xu C., Huynh P.T., Chong K.H.C., Tan E.Y.J., Zhang J., Boo Z.Z., Teo D.E.T., Lay K., et al. 2021. Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8. Nat. Commun. 12:188. 10.1038/s41467-020-20319-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold, A.R., Cifani P., Rao S.D., Axelrod A.J., Miele M.M., Hendrickson R.C., Kentsis A., and Bachovchin D.A.. 2019. A Chemical Strategy for Protease Substrate Profiling. Cell Chem. Biol. 26:901–907.e6. 10.1016/j.chembiol.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, P.C., Acosta D., and Collier R.J.. 1993. On the role of macrophages in anthrax. Proc. Natl. Acad. Sci. USA. 90:10198–10201. 10.1073/pnas.90.21.10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapas, C.R., Robinson K.S., Lay K., Wong J., Raas-Rothschild A., Boisson B., Drutman S.B., Laohamonthonkul P., Bonner D., Gorrell M., et al. 2021. DPP9 deficiency: an Inflammasomopathy which can be rescued by lowering NLRP1/IL-1 signaling. (Preprint posted June 9, 2021) 10.1101/2021.01.31.21250067 [DOI]
- Hellmich, K.A., Levinsohn J.L., Fattah R., Newman Z.L., Maier N., Sastalla I., Liu S., Leppla S.H., and Moayeri M.. 2012. Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One. 7:e49741. 10.1371/journal.pone.0049741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, L.R., David L., Li Y., Griswold A.R., Ruan J., Sharif H., Fontana P., Orth-He E.L., Fu T.M., Bachovchin D.A., and Wu H.. 2021a. Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. Nat. Commun. 12:189. 10.1038/s41467-020-20320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, L.R., Sharif H., Griswold A.R., Fontana P., Mintseris J., Dagbay K.B., Paulo J.A., Gygi S.P., Bachovchin D.A., and Wu H.. 2021b. DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature. 592:778–783. 10.1038/s41586-021-03350-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., and Fitzgerald K.A.. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 458:514–518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M., Zhang X., Toh G.A., Gong Q., Wang J., Han Z., Wu B., Zhong F., and Chai J.. 2021. Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature. 592:773–777. 10.1038/s41586-021-03320-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y., Mailloux C.M., Gowan K., Riccardi S.L., LaBerge G., Bennett D.C., Fain P.R., and Spritz R.A.. 2007. NALP1 in vitiligo-associated multiple autoimmune disease. N. Engl. J. Med. 356:1216–1225. 10.1056/NEJMoa061592 [DOI] [PubMed] [Google Scholar]
- Jin, T., Huang M., Smith P., Jiang J., and Xiao T.S.. 2013. Structure of the caspase-recruitment domain from a zebrafish guanylate-binding protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69:855–860. 10.1107/S1744309113015558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.C., Taabazuing C.Y., Okondo M.C., Chui A.J., Rao S.D., Brown F.C., Reed C., Peguero E., de Stanchina E., Kentsis A., and Bachovchin D.A.. 2018. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med. 24:1151–1156. 10.1038/s41591-018-0082-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.C., Okondo M.C., Orth E.L., Rao S.D., Huang H.C., Ball D.P., and Bachovchin D.A.. 2020. DPP8/9 inhibitors activate the CARD8 inflammasome in resting lymphocytes. Cell Death Dis. 11:628. 10.1038/s41419-020-02865-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki, N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Levandowski, C.B., Mailloux C.M., Ferrara T.M., Gowan K., Ben S., Jin Y., McFann K.K., Holland P.J., Fain P.R., Dinarello C.A., and Spritz R.A.. 2013. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1β processing via the NLRP1 inflammasome. Proc. Natl. Acad. Sci. USA. 110:2952–2956. 10.1073/pnas.1222808110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinsohn, J.L., Newman Z.L., Hellmich K.A., Fattah R., Getz M.A., Liu S., Sastalla I., Leppla S.H., and Moayeri M.. 2012. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 8:e1002638. 10.1371/journal.ppat.1002638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilue, J., Doran A.G., Fiddes I.T., Abrudan M., Armstrong J., Bennett R., Chow W., Collins J., Collins S., Czechanski A., et al. 2018. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat. Genet. 50:1574–1583. 10.1038/s41588-018-0223-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, A., Bauernfried S., Cheng Y., Albanese M., Jung C., Keppler O.T., and Hornung V.. 2020. CARD8 inflammasome activation triggers pyroptosis in human T cells. EMBO J. 39:e105071. 10.15252/embj.2020105071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, J.A., Wijekoon C.P., Liao K.C., and Muruve D.A.. 2013. Biochemical and structural aspects of the ATP-binding domain in inflammasome-forming human NLRP proteins. IUBMB Life. 65:851–862. 10.1002/iub.1210 [DOI] [PubMed] [Google Scholar]
- Martinon, F., Burns K., and Tschopp J.. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Martinon, F., Agostini L., Meylan E., and Tschopp J.. 2004. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14:1929–1934. 10.1016/j.cub.2004.10.027 [DOI] [PubMed] [Google Scholar]
- Masters, S.L., Gerlic M., Metcalf D., Preston S., Pellegrini M., O’Donnell J.A., McArthur K., Baldwin T.M., Chevrier S., Nowell C.J., et al. 2012. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity. 37:1009–1023. 10.1016/j.immuni.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P.S., Sandstrom A., and Vance R.E.. 2019. The NLRP1 inflammasome: new mechanistic insights and unresolved mysteries. Curr. Opin. Immunol. 60:37–45. 10.1016/j.coi.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri, M., Crown D., Newman Z.L., Okugawa S., Eckhaus M., Cataisson C., Liu S., Sastalla I., and Leppla S.H.. 2010. Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS Pathog. 6:e1001222. 10.1371/journal.ppat.1001222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Planillo, R., Kuffa P., Martínez-Colón G., Smith B.L., Rajendiran T.M., and Núñez G.. 2013. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 38:1142–1153. 10.1016/j.immuni.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo, M.C., Johnson D.C., Sridharan R., Go E.B., Chui A.J., Wang M.S., Poplawski S.E., Wu W., Liu Y., Lai J.H., et al. 2017. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat. Chem. Biol. 13:46–53. 10.1038/nchembio.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo, M.C., Rao S.D., Taabazuing C.Y., Chui A.J., Poplawski S.E., Johnson D.C., and Bachovchin D.A.. 2018. Inhibition of Dpp8/9 Activates the Nlrp1b Inflammasome. Cell Chem. Biol. 25:262–267.e5. 10.1016/j.chembiol.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétrilli, V., Papin S., Dostert C., Mayor A., Martinon F., and Tschopp J.. 2007. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 14:1583–1589. 10.1038/sj.cdd.4402195 [DOI] [PubMed] [Google Scholar]
- Robinson, K.S., Teo D.E.T., Tan K.S., Toh G.A., Ong H.H., Lim C.K., Lay K., Au B.V., Lew T.S., Chu J.J.H., et al. 2020. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science. 370:eaay2002. 10.1126/science.aay2002 [DOI] [PubMed] [Google Scholar]
- Sand, J., Haertel E., Biedermann T., Contassot E., Reichmann E., French L.E., Werner S., and Beer H.D.. 2018. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 9:24. 10.1038/s41419-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom, A., Mitchell P.S., Goers L., Mu E.W., Lesser C.F., and Vance R.E.. 2019. Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science. 364:eaau1330. 10.1126/science.aau1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastalla, I., Crown D., Masters S.L., McKenzie A., Leppla S.H., and Moayeri M.. 2013. Transcriptional analysis of the three Nlrp1 paralogs in mice. BMC Genomics. 14:188. 10.1186/1471-2164-14-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, H., Hollingsworth L.R., Griswold A.R., Hsiao J.C., Wang Q., Bachovchin D.A., and Wu H.. 2021. Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment. Immunity. 54:1392–1404. 10.1016/j.immuni.2021.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F.. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526:660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Smith, H., and Keppie J.. 1954. Observations on experimental anthrax; demonstration of a specific lethal factor produced in vivo by Bacillus anthracis. Nature. 173:869–870. 10.1038/173869a0 [DOI] [PubMed] [Google Scholar]
- Squires, R.C., Muehlbauer S.M., and Brojatsch J.. 2007. Proteasomes control caspase-1 activation in anthrax lethal toxin-mediated cell killing. J. Biol. Chem. 282:34260–34267. 10.1074/jbc.M705687200 [DOI] [PubMed] [Google Scholar]
- Tsu, B.V., Beierschmitt C., Ryan A.P., Agarwal R., Mitchell P.S., and Daugherty M.D.. 2021. Diverse viral proteases activate the NLRP1 inflammasome. eLife. 10:e60609. 10.7554/eLife.60609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrkalska, S.D., Candel S., Angosto D., Gómez-Abellán V., Martín-Sánchez F., García-Moreno D., Zapata-Pérez R., Sánchez-Ferrer Á., Sepulcre M.P., Pelegrín P., and Mulero V.. 2016. Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat. Commun. 7:12077. 10.1038/ncomms12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch, S., Tian L., Hoy R., and Li X.. 2020. Plant NLRs: The Whistleblowers of Plant Immunity. Plant Commun. 1:100016. 10.1016/j.xplc.2019.100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Gao H., Clark K.M., Mugisha C.S., Davis K., Tang J.P., Harlan G.H., DeSelm C.J., Presti R.M., Kutluay S.B., and Shan L.. 2021. CARD8 is an inflammasome sensor for HIV-1 protease activity. Science. 371:eabe1707. 10.1126/science.abe1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, H., Gaide O., Pétrilli V., Martinon F., Contassot E., Roques S., Kummer J.A., Tschopp J., and French L.E.. 2007. Activation of the IL-1beta-processing inflammasome is involved in contact hypersensitivity. J. Invest. Dermatol. 127:1956–1963. 10.1038/sj.jid.5700819 [DOI] [PubMed] [Google Scholar]
- Witola, W.H., Mui E., Hargrave A., Liu S., Hypolite M., Montpetit A., Cavailles P., Bisanz C., Cesbron-Delauw M.F., Fournié G.J., and McLeod R.. 2011. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect. Immun. 79:756–766. 10.1128/IAI.00898-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Shi J., Gao H., Liu Y., Yang Z., Shao F., and Dong N.. 2019. The N-end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. EMBO J. 38:e101996. 10.15252/embj.2019101996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.H., Moecking J., Geyer M., and Masters S.L.. 2018. Mechanisms of NLRP1-Mediated Autoinflammatory Disease in Humans and Mice. J. Mol. Biol. 430:142–152. 10.1016/j.jmb.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Zhong, F.L., Mamaï O., Sborgi L., Boussofara L., Hopkins R., Robinson K., Szeverényi I., Takeichi T., Balaji R., Lau A., et al. 2016. Germline NLRP1 Mutations Cause Skin Inflammatory and Cancer Susceptibility Syndromes via Inflammasome Activation. Cell. 167:187–202.e17. 10.1016/j.cell.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Zhong, F.L., Robinson K., Teo D.E.T., Tan K.Y., Lim C., Harapas C.R., Yu C.H., Xie W.H., Sobota R.M., Au V.B., et al. 2018. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem. 293:18864–18878. 10.1074/jbc.RA118.004350 [DOI] [PMC free article] [PubMed] [Google Scholar]