Abstract
Expression of replication-dependent histone genes at the posttranscriptional level is controlled by stem-loop binding protein (SLBP). One function of SLBP is to bind the stem-loop structure in the 3′ untranslated region of histone pre-mRNAs and facilitate 3′ end processing. Interaction of SLBP with the stem-loop is mediated by the centrally located RNA binding domain (RBD). Here we identify several highly conserved amino acids in the RBD mutation of which results in complete or substantial loss of SLBP binding activity. We also identify residues in the RBD which do not contribute to binding to the stem-loop RNA but instead are required for efficient recruitment of U7 snRNP to histone pre-mRNA. Recruitment of the U7 snRNP to the pre-mRNA also depends on the 20-amino-acid region located immediately downstream of the RBD. A critical region of the RBD contains the sequence YDRY. The tyrosines are required for RNA binding, and the DR dipeptide is essential for processing but not for RNA binding. It is likely that the RBD of SLBP interacts directly with both the stem-loop RNA and other processing factor(s), most likely the U7 snRNP, to facilitate histone pre-mRNA processing.
Replication-dependent histone genes comprise a unique group of genes whose expression is coordinately regulated with DNA synthesis (15). Expression of these genes peaks during S phase of the cell cycle and rapidly declines upon completion of DNA replication at the end of S phase (10, 30). In contrast to all other mRNAs, replication-dependent histone mRNAs are not polyadenylated and instead terminate with a highly conserved stem-loop structure (15). The stem-loop structure, consisting of six base pairs and a four nucleotide loop, associates with a protein termed the stem-loop binding protein (SLBP) or the hairpin binding protein (14, 29). Mammalian SLBP is a 30-kDa protein containing 270 amino acids and can be divided into three domains: the centrally located RNA binding domain (RBD) and the flanking N-terminal and C-terminal domains. The RBD of SLBP does not resemble any previously identified motifs involved in RNA recognition (29).
Replication-dependent histone mRNAs are formed from longer pre-mRNA transcripts by an endonucleolytic cleavage (3, 8). The processing reaction depends on two sequence elements in the histone pre-mRNA: the stem-loop structure (27) and a purine-rich element, termed the histone downstream element (HDE), located 10 to 15 nucleotides further downstream (2). The stem-loop is recognized by SLBP, whereas the HDE associates with the U7 snRNP, containing the 60-nucleotide U7 snRNA and associated proteins (20, 22). Binding of U7 snRNP to the pre-mRNA occurs via base pairing between the HDE and the 5′ end of U7 snRNA (2, 18) and is likely also strengthened by interactions between a U7-specific protein(s) and the SLBP–stem-loop (SLBP/SL) complex (5). This additional interaction is especially important in processing of histone pre-mRNAs containing HDEs which can form only a weak duplex with the U7 snRNA and thus are unable to efficiently recruit U7 snRNP to the pre-mRNA (5, 17, 23). Stable binding of SLBP and U7 snRNP to their respective targets in the pre-mRNA leads to a subsequent association of additional trans-acting factors, including a poorly characterized heat-labile factor (9), followed by cleavage of the pre-mRNA four to five nucleotides downstream from the stem-loop. After 3′ end processing, SLBP remains associated with the terminal stem-loop and assists the mature histone mRNA to the cytoplasm, where it likely plays an important role in histone mRNA translation and stability (7, 25, 31). SLBP is cell cycle regulated and therefore may be a key factor responsible for cell cycle regulation of histone mRNA levels (30).
Two proteins that bind the stem-loop structure at the 3′ end of histone mRNA have been isolated from Xenopus laevis oocytes (28). One protein, referred to as xSLBP1, is homologous to mammalian SLBPs and is also involved in 3′ end processing of histone pre-mRNAs. A second SLBP found in Xenopus oocytes, designated xSLBP2, does not participate in 3′ end processing and probably functions in storage of histone mRNA during oogenesis (28). xSLBP2 is similar to xSLBP1 only in the central RBD. Replacement of the RBD of xSLBP1 with the corresponding domain of xSLBP2 resulted in a chimeric protein that retained high affinity for the stem-loop. Surprisingly, this chimeric protein was inactive in processing although it contains the C-terminal region from xSLBP1 required for processing (11). Thus, binding to the stem-loop of histone pre-mRNA is not the only function of the RBD in 3′ end processing. Here we identify residues in the RBD of human SLBP that are not required for binding to the RNA target and instead play another role in processing. Our experiments indicate that these residues together with the 20-amino-acid region in the C terminus are each important for efficient recruitment of the U7 snRNP to the histone pre-mRNA. In addition, by aligning RBD sequences of various SLBPs, we identified absolutely conserved amino acids and demonstrated that these residues are required for binding to the stem-loop RNA. Changes in some of these residues results in a reduction in affinity for the stem-loop, accompanied by a decrease in processing activity, demonstrating the importance of high-affinity binding of SLBP to the stem-loop for efficient 3′ end processing.
MATERIALS AND METHODS
Construction of the mutant and chimeric SLBP clones.
Substitutions of the amino acid residues within the RBD of SLBP were carried out in the pGEM3 vector containing the human SLBP cDNA spanning from the initiation codon to the MscI site in the 3′ untranslated region, 57 nucleotides downstream from the stop codon. Substitution of these residues was facilitated by the presence of the following unique restriction sites located within the cDNA region encoding the RBD: NgoMIV, MfeI, BsmI, SalI, and BamHI. The appropriate restriction fragments in the wild-type SLBP cDNA were replaced by double-stranded oligonucleotides terminating with compatible sticky ends and containing the desired mutations. The following combinations of restriction enzymes were used during mutagenesis: NgoMIV and MfeI (to insert mutations located between amino acids 1 and 21), MfeI and BsmI (to insert mutations located between amino acids 22 and 39), BsmI and SalI (to insert mutations located between amino acids 40 and 52), and SalI and BamHI (to insert mutations located between amino acids 53 and 72). To facilitate identification of the desired clones, where possible, oligonucleotides were designed so that one of the two restriction sites was disrupted upon ligation of the insert into the pGEM3 vector, without changing the amino acid sequence of the RBD. The double mutants QPF, RHLF, QPQVA, and RHLQVA were constructed by recombining appropriate fragments containing the individual mutations. Details of each construct and sequences of the oligonucleotides are available on request. For in vitro protein synthesis, the SLBP cDNAs containing appropriate mutations were subcloned into the pSP64T vector. This vector was modified from the pSP64 Poly(A) vector (Promega) by introducing the 5′ and 3′ untranslated regions from the rabbit β-globin gene and a fragment encoding the poly(A) tail (12). The mutant SLBP cDNAs were inserted into the pSP64T vector downstream from the 5′ untranslated region using NcoI and XbaI restriction sites. For expression of the mutant forms of SLBP in Sf9 insect cells using the Bac-to-Bac expression system, mutant cDNAs were cloned into one of the pFastBac vectors (Gibco-BRL).
Synthesis of SLBP by in vitro translation.
The in vitro coupled transcription and translation reaction was carried out in the rabbit reticulocyte extract using a Promega TnT kit according to the manufacturer's protocol. Each reaction (total volume of 25 μl) contained 12.5 μl of the extract, 5 U of SP6 RNA polymerase (Promega), and 1.0 μg of the DNA template encoding either the wild-type or mutant version of the human SLBP subcloned into the pSP64T vector. To determine the amount of SLBP made during the TnT reactions, the samples were supplemented with [35S]methionine (NEN) and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
Expression and purification of SLBP using the baculovirus system.
The wild-type and mutant forms of SLBP were expressed in Sf9 insect cells using the Bac-to-Bac baculovirus expression system (Gibco-BRL) as recommended by the manufacturer. The His-tagged proteins were purified by affinity chromatography on Ni-nitrilotriacetic acid agarose (Qiagen).
Mobility shift assay.
The ability to bind the histone stem-loop RNA by the mutant SLBP proteins synthesized either in vitro using the TnT kit or in vivo using the baculovirus system was determined by band shift assay as previously described (5, 11). Routinely, 2.5 μl of the TnT reaction or 0.1 μg of the protein expressed in the baculovirus-infected cells was mixed on ice in a total volume of 10 μl with 100 fmol of the 5′-labeled stem-loop RNA, 20 mM EDTA, and 2.5 μl of buffer D used for dialysis of the nuclear extract (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 0.5 mM dithiothreitol, 0.2 mM EDTA [pH 8.0], 20% glycerol). The samples were immediately resolved on a 7.5% native polyacrylamide gel (37.5 parts of acrylamide to 1 part of bisacrylamide) containing Tris-borate-EDTA buffer. The gel was dried, and radioactive bands were detected by autoradiography and/or by PhosphorImager.
Preparation of RNA.
In vitro 3′ end processing was carried out using 86-nucleotide pre-mRNAs containing the stem-loop structure and the U7 binding site from either the H2a-614 or H1t pre-mRNA. A 30-nucleotide RNA containing the wild-type stem-loop structure and the 5-nucleotide flanks was used in the band shift assay. Sequences of all three RNA species and methods for their synthesis and labeling were described previously (5). The U7 snRNA was detected by hybridization with the anti-U7 RNA probe, synthesized, and labeled as described elsewhere (5).
Preparation of nuclear extract and 3′ end processing.
Preparation of the nuclear extract from mouse myeloma cells, immunodepletion of the SLBP, and complementation of the depleted extract with the recombinant SLBP were performed according to protocols described previously (4, 5, 16). Each processing reaction contained 5 μl of the nuclear extract corresponding to approximately 50 μg of total protein, 180 fmol of the pre-mRNA substrate labeled at the 5′ end with [γ-32P]ATP, and 20 mM EDTA. Samples were incubated at 32°C for 1 h and processed as previously described (5).
Western blots.
Nuclear protein (50 μg) was resolved on an SDS–12% polyacrylamide gel and transferred to a nitrocellulose membrane. SLBP was detected with an antibody raised against the C-terminal 13 amino acids of the protein using an enhanced chemiluminescence system.
Immunoprecipitation of processing complexes.
Processing complexes containing the U7 snRNP were formed and subsequently precipitated by anti-SLBP essentially as described elsewhere (5). Each reaction contained 50 ng of unlabeled pre-mRNA substrate, 50 μl of the nuclear extract, and 20 mM EDTA (pH 8) in a total volume of 100 μl and was supplemented with 0.01 to 0.1 ng of the substrate labeled at the 5′ end with [γ-32PO4]ATP (500 to 5,000 cpm respectively). The radiolabeled pre-mRNA was used to monitor the overall efficiency of immunoprecipitation of the pre-mRNA substrate by the SLBP antibody. In some experiments, the SLBP-depleted nuclear extract supplemented with different recombinant SLBPs was used instead of the undepleted nuclear extract. In all cases, the reaction samples were prepared on ice followed by a short (3- to 5-min) incubation at 22°C to allow formation of the processing complexes. The affinity-purified SLBP antibody (10 μl at 1.0 μg/μl) was subsequently added to each reaction, and the samples were rotated at 4°C for 1.5 h and then transferred to a new tube containing 15 μl of protein A-agarose beads (Gibco-BRL). Prior to use, the protein A-agarose beads were incubated for 2 h in an extract from sea urchin blastula nuclei (containing a U7 snRNA which does not cross-react with the anti-mouse U7 snRNA probe), to reduce nonspecific binding of components of the mouse nuclear extract. Subsequent steps in the immunoprecipitation procedure and Northern blot analysis of U7 snRNA were performed as previously described (5).
RESULTS
Amino acids conserved in the RBDs of all SLBPs are involved in RNA recognition.
In Xenopus oocytes there are two different SLBPs, xSLBP1 and xSLBP2, which bind the stem-loop structure at the 3′ end of replication-dependent histone mRNAs with the same affinity (28). xSLBP1 is a homologue of human and mouse SLBPs and is involved in processing of histone pre-mRNAs in the nucleus. Similar to mammalian SLBPs, xSLBP1 is also found in the cytoplasm, where it may function in regulating translation of histone mRNA. xSLBP2 is found only in the cytoplasm and is inactive in 3′ end processing of histone pre-mRNA both in vivo and in vitro. Expression of xSLBP2 is restricted to oogenesis and the protein is degraded during oocyte maturation, indicating that xSLBP2 may function in translational repression of histone mRNA (28). This protein shares amino acid similarity with processing-specific SLBPs only within the central RBD.
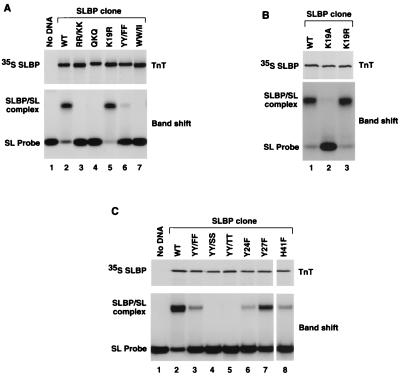
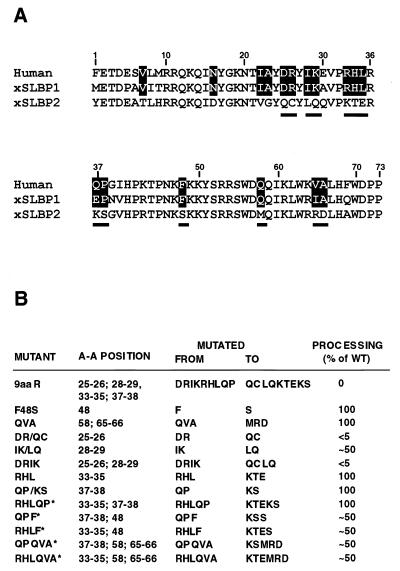
Figure 1A presents a sequence alignment of the 73-amino-acid RBD from human SLBP, both Xenopus SLBPs, and Drosophila SLBP (24). In the four SLBPs, 36 of the 73 amino acids of the RBD are identical and 13 other amino acids are replaced by similar residues. The strong evolutionary conservation of these amino acids suggested that they are all involved in recognition of the common target for each protein; the stem-loop structure at the 3′ end of histone mRNAs. We tested this hypothesis by substituting some of the conserved amino acids with either similar, neutral, or biochemically different residues (Fig. 1B) and by determining the relative binding affinity of mutant proteins in the band shift assay. All mutant proteins were expressed in the presence of [35S]methionine using the TnT system (Promega) and resolved by SDS-polyacrylamide gel electrophoresis (Fig. 2, top row of each panel). The gels were used for autoradiography and PhosphorImager analysis, allowing precise monitoring of both the quality of SLBP synthesized in vitro (e.g., presence of possible prematurely terminated or improperly initiated products) and the quantity of the full-length protein subsequently used for the band shift assay. All mutant SLBPs were generated in vitro at relatively low concentrations and are likely to be properly folded in the reticulocyte extract. This approach ensured that any changes in the ability to shift the RNA probe likely resulted from differences in the binding affinity between the tested mutant proteins. All amino acids subjected to mutagenesis, with the exception of the tryptophan at position 56, which is replaced by phenylalanine in Drosophila melanogaster SLBP, are absolutely conserved in the four SLBPs shown in Fig. 1A.
FIG. 1.
Conserved amino acids within the RBD of the SLBP. (A) Schematic of the vertebrate SLBPs, with the position of the RBD indicated. The RBDs of the human, two X. laevis, and Drosophila (Fly) SLBPs were aligned. The amino acids conserved in all four proteins are highlighted and the residues subjected to mutagenesis are underlined. N-ter and C-ter, N and C termini. (B) Amino acid substitutions made within the RBD of the human SLBP and their relative effect on binding to the RNA stem-loop structure. The binding affinity of each mutant SLBP is reported as a percentage of wild-type (WT) binding affinity (100%). The values represent the range of affinities obtained from at least two independent experiments.
FIG. 2.
The conserved amino acids in the RBD are required for efficient binding of the protein to the stem-loop structure. Wild-type (WT) and mutant forms of the human SLBP containing various amino acid substitutions of the conserved residues, as indicated above each lane and listed in Fig. 1, were synthesized in vitro using the rabbit reticulocyte TnT system (Promega) and analyzed by electrophoresis in SDS–10% polyacrylamide gels (top). Each mutant protein was subsequently tested for binding efficiency in the band shift assay using the 30-nucleotide stem-loop RNA labeled at the 5′ end as a probe (bottom). Lane 1 of panels A and C represents a negative control in which the TnT reaction was carried out in the absence of exogenous DNA.
The wild-type human SLBP expressed in vitro using the TnT system efficiently binds to the stem-loop RNA probe. As shown in Fig. 2, 10% of the entire TnT reaction consistently shifted between 75 and 90% of the RNA probe. The complex formed by the SLBP expressed in the reticulocyte lysate has a mobility identical to that of the complex formed in nuclear extracts (29) or with baculovirus protein (not shown). As determined by the same assay, replacement of the two conserved arginines at positions 10 and 11 of the RBD with lysines (RR/KK mutant) completely abolished binding to the stem-loop RNA probe (SL probe) (Fig. 2A, lane 3). No band corresponding to the SLBP/SL complex was detected even after a long exposure of the film (not shown). Arginines 10 and 11 are adjacent to a conserved QKQ tripeptide at positions 12 to 14 of the RBD. Substitution of this tripeptide with alanines resulted in a protein (QKQ mutant) that was unable to bind the stem-loop RNA (Fig. 2A, lane 4). Lysine 19 is absolutely conserved in all known SLBPs, including those of sea urchin, Caenorhabditis elegans (13), and Chlamydomonas (unpublished results). Interestingly, changing this amino acid to arginine did not reduce binding to the stem-loop RNA (K19R mutant) (Fig. 2A, lane 5; Fig. 2B, lane 3). However, replacing lysine 19 with a neutral residue, alanine, resulted in a protein severely impaired in stem-loop RNA binding (Fig. 2B, lane 2).
In addition to changing the basic residues and the QKQ tripeptide, we mutated two sets of conserved aromatic residues, the two tryptophans at positions 56 and 63 and the two tyrosines at positions 24 and 27. As determined by the band shift assay, replacement of the two tryptophans with isoleucines produced an SLBP mutant unable to bind the stem-loop structure (WW/II mutant) (Fig. 2A, lane 7). A very conservative mutation replacing the two tyrosines at position 24 and 27 with phenylalanines (YY/FF mutant) resulted in a protein that retains only 5 to 10% of the wild-type binding affinity (Fig. 2A, lane 6; Fig. 2C, lane 3). Replacement of the same tyrosines with either serines (YY/SS mutant) or threonines (YY/TT mutant), other amino acids containing an OH group, abolished binding (Fig. 2C, lane 4 or 5, respectively). We next made two single mutations by individually substituting each tyrosine with phenylalanine. The tyrosine at position 24 is more important for RNA recognition, since replacement of this residue with phenylalanine (Y24F mutant) resulted in a more severe decrease in binding than the same replacement of the tyrosine at position 27 (Y27F mutant). The Y24F and Y27F SLBPs retained less than 10 and approximately 50%, respectively, of the wild-type binding efficiency (Fig. 2C, lanes 6 and 7). Binding of SLBP to the stem-loop RNA was also dramatically reduced by replacing histidine at position 41 with phenylalanine (H41F mutant) (Fig. 2C, lane 8). The effect of this mutation on binding was similar to that of the Y24F mutation.
Complementation of the SLBP-depleted nuclear extract with mutant SLBPs.
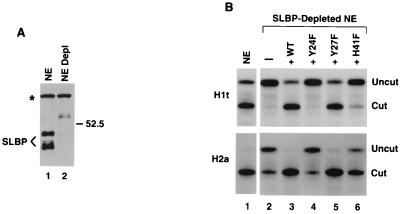
We and others have previously shown that high-affinity binding of SLBP to the stem-loop structure in histone pre-mRNA is critically important for efficient 3′ end processing of histone pre-mRNA (5, 17, 19, 27). Mutations of the stem-loop structure that reduced SLBP binding more than 10-fold resulted in a complete inhibition of processing. Other changes in the stem-loop reducing affinity to SLBP 5- to 10-fold had a less drastic effect on the efficiency of 3′ end processing (5, 19). Here we address whether processing efficiency is affected in the same fashion by changes in SLBP that decrease its affinity for histone pre-mRNA. Using the baculovirus expression system, we expressed three mutant SLBPs that had reduced binding activity and evaluated their ability to rescue processing activity of the SLBP-depleted nuclear extract. As determined by Western blotting, immunodepletion of the nuclear extract with the SLBP antibody resulted in complete removal of the protein (Fig. 3A, lane 2). We used a histone pre-mRNA containing the HDE from the H1t gene, since processing of this pre-mRNA is completely dependent on SLBP (5). Incubation of H1t pre-mRNA in the nuclear extract resulted in processing of 75% of the input pre-mRNA, and removal of SLBP from the extract abolished processing (Fig. 3B, top, lanes 1 and 2, respectively). Addition of 100 ng of recombinant wild-type SLBP restored processing of the H1t substrate to the initial level (Fig. 3B, top, lane 3). A similar increase in processing efficiency was achieved upon addition of the Y27F mutant SLBP, which binds the stem-loop about 50% as efficiently as the wild-type protein (Fig. 3B, top, lane 5). Addition of the same amount of the weakly binding Y24F mutant protein did not stimulate processing, and addition of the H41F mutant protein, which has similar stem-loop binding activity, resulted in accumulation of only a trace amount of the cleavage product (Fig. 3B, top, lanes 4 and 6, respectively). These results are consistent with our earlier studies in which weakening the interaction between SLBP and the RNA by mutations in the stem-loop structure resulted in a decrease in processing efficiency both in vivo (19) and in vitro (5). The fact that the Y27F mutant SLBP does not significantly affect the processing efficiency of SLBP in spite of a twofold reduction in RNA binding suggests that there is a threshold level of affinity above which processing occurs with maximal efficiency.
FIG. 3.
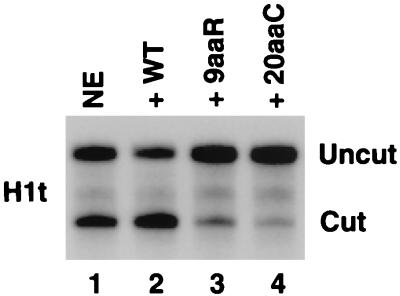
High-affinity stem-loop binding is required for efficient processing of histone H1t pre-mRNA but not H2a pre-mRNA. (A) The nuclear extract (NE) was immunodepleted of SLBP using the SLBP antibody, and the efficiency of depletion was determined by Western blotting carried out with the same antibody (lane 2). Lane 1 represents an undepleted control nuclear extract. The asterisk indicates a protein that cross-reacts with the SLBP antibody when denatured but not in the nuclear extract and serves as a control for specificity of depletion. The position of a 52.5-kDa marker is indicated. (B) Ability of wild-type (WT) and mutant forms of SLBP with reduced binding activity to support processing of the H1t (top) and H2a (bottom) pre-mRNAs. The indicated mutant proteins were expressed in the baculovirus system and used to complement the SLBP-deleted nuclear extract. The processing activity of the nuclear extract before and after depletion is shown in lanes 1 and 2, respectively. The uncut band corresponds to the input histone pre-mRNA, and the cut band corresponds to the mature product of 3′ end processing.
We tested the same proteins for their ability to restore processing of the H2a-614 pre-mRNA, which contains an HDE capable of forming a relatively strong duplex (12 base pairs interrupted by only one mismatch) with the 5′ end of U7 snRNA. Depending on the batch of nuclear extract used, this processing substrate is cleaved in vitro with 10 to 20% efficiency even in the absence of SLBP (Fig. 3B, bottom, lane 2; references 4 and 29). The basal level of processing was elevated to virtually 100% by addition of either the wild-type SLBP or the Y27F mutant protein (Fig. 3B, bottom, lanes 3 and 5, respectively). The Y24F mutant protein did not stimulate processing of the H2a pre-mRNA substrate (Fig. 3B, lane 4). Surprisingly, the H41F SLBP increased processing of the H2a-614 pre-mRNA as much as fivefold (Fig. 3B, bottom, lane 6), although it has similar affinity for the stem-loop as Y24F SLBP. It is likely that the region of SLBP around tyrosine 24 may be involved in both RNA binding and other interactions essential for processing (see below).
Residues in the RBD essential for processing.
We have previously shown that replacement of the RBD from xSLBP2 in xSLBP1 resulted in a protein, 1-2-1, that binds the stem-loop with the same affinity as xSLBP1 but is inactive in processing, both in frog oocytes and in vitro (11). Thus, the RBD must have other functions in histone pre-mRNA processing in addition to binding the pre-mRNA. Here we define residues in the RBD that are important for efficient histone pre-mRNA processing in vitro. The RBDs of SLBP from more distantly related metazoans, C. elegans, and D. melanogaster, also do not substitute for the mammalian RBD in histone pre-mRNA processing (unpublished results). This is likely due to incompatibility of the vertebrate and invertebrate processing machineries as has been shown for Xenopus and sea urchins (6). Thus, the SLBP sequences from C. elegans and D. melanogaster were not informative in identification of amino acids required for processing.
There are 15 identical and 2 similar amino acids (highlighted in Fig. 4A) in the human SLBPs and xSLBP1 which are different in xSLBP2. Of the 15 identical residues, 10 are clustered between amino acids 22 and 38 of the RBD, while 3 of the remaining 5 are in the C-terminal portion of the RBD. The absence of all or some of these amino acids in the chimeric protein 1-2-1 must be responsible for its inability to function in 3′ end processing in spite of efficient binding to the histone pre-mRNA. To determine which of these 17 amino acids are critical for 3′ end processing, we systematically mutated most of them either individually or in small groups in the human SLBP and tested the ability of the mutant proteins to bind RNA and to restore processing to an SLBP-depleted extract. We replaced the amino acids in human SLBP with the amino acids found in the same position in xSLBP2, since these residues are likely to be neutral for RNA binding (Fig. 4). In the first round of this scanning mutagenesis, we altered nine amino acids near the center of the RBD (9aaR mutation). In addition, we replaced the phenylalanine at position 48 with serine (F48S mutation) and the QVA in the C-terminal part of the RBD with MRD (QVA mutation). As determined by band shift assay, all of the mutant proteins expressed in baculovirus-infected insect cells efficiently bound to the stem-loop RNA (Fig. 5A). Subsequently, the same preparation of each mutant SLBP was tested for the ability to restore in vitro processing activity to the SLBP-depleted nuclear extract using the H1t histone pre-mRNA substrate. In each experiment, approximately 100 ng of the baculovirus-expressed SLBP was added to the depleted extract. This amount of protein is sufficient to bind the vast majority of the input substrate (not shown). The 9aaR mutant protein was completely inactive in processing of the H1t histone pre-mRNA (Fig. 5B, lane 4), demonstrating that the 14-residue region from amino acids 25 to 38 of the human RBD is critical for processing. The F48S and QVA mutant proteins complemented the depleted extract as efficiently as the wild-type SLBP (Fig. 5B, lanes 5 and 6, respectively). We next prepared a second set of mutations by altering a small number of the amino acids within the 14-amino-acid cluster. The DR dipeptide at positions 25 and 26 that is flanked by the two tyrosines critical for RNA recognition was replaced with QC found in xSLBP2 (DR/QC mutation). This substitution reduced the processing activity on the H1t substrate by more than 95% (Fig. 5C, lane 3), although it did not affect the ability of the SLBP to form a stable complex with the stem-loop (Fig. 5A, lane7). There was approximately a 50% reduction in processing activity as a result of the IK/LQ mutation, replacing the IK dipeptide (amino acids 28 and 29) with LQ found in xSLBP2 (Fig. 5C, lane 4). The DRIK mutant protein in which all four amino acids were mutated had even less processing activity than the DR mutant, consistent with an additive effect of the DR and IK mutations (Fig. 5C, lane 5). Both IK/LQ (not shown) and DRIK (Fig. 5A, lane 8) efficiently bound the stem-loop RNA in the band shift assay. Partial or complete alteration of the RHLQP region revealed that these five remaining amino acids of the original 9aaR mutation do not play a major role in the processing activity of human SLBP. The RHL and QP/KS mutant proteins (Fig. 5C, lanes 8 and 9, respectively) as well as the RHLQP mutant protein (data not shown and Fig. 4B) were as active in processing as the wild-type SLBP.
FIG. 4.
Amino acids conserved in the RBD of human SLBP and xSLBP1 but not in xSLBP2. (A) Alignment of the RBDs from the three different SLBPs. Amino acids conserved or similar between the human SLBP and the xSLBP1 that differ from the amino acids in xSLBP2 are highlighted. The residues subjected to mutagenesis are underlined. (B) Amino acid substitutions made within the RBD of human SLBP and their effect on the activity of the protein in the 3′ end processing of histone H1t pre-mRNA. The processing efficiency of each mutant SLBP is reported as a percentage of the processing efficiency of wild-type (WT) SLBP (100%). The values represent the approximate efficiencies obtained from at least two independent experiments. ∗, data not shown.
FIG. 5.
Substitution of nine amino acids within the RBD of human SLBP abolishes processing of H1t pre-mRNA but not binding activity of the protein. The RBD of human SLBP was mutated in three different regions by replacing the amino acids (underlined in Fig. 4) with the corresponding amino acids found in xSLBP2. (A) Ability of wild-type (WT) and mutant (indicated above each lane) SLBPs to bind the RNA stem-loop probe, determined by band shift assay. The probe is shown in lanes 1 and 6. (B and C) Activity of the nuclear extract (NE) before (panel B, lane 1) and after (panel B, lane 2; panel C, lanes 1 and 6) depletion of SLBP in processing of the H1t pre-mRNA. The effect of addition of 100 ng of the different SLBPs (indicated above each lane) to the SLBP-depleted extract on 3′ end processing is shown in panel B, lanes 3 to 6, and panel C, lanes 2 to 5 and 7 to 9.
We combined mutations that alone resulted in no reduction in processing activity (RHL, QP/KS, QVA, and F48S). The F48S mutation in conjunction with either the QP/KS or RHL mutation resulted in about a 50% reduction in processing activity (Fig. 4B). A similar reduction of SLBP processing activity was also observed when the QP/KS or RHL mutation was combined with the other neutral mutation, QVA (Fig. 4B).
Activity of mutant SLBPs in processing is substrate dependent.
We have previously reported that there is a region of 20 amino acids in human SLBP immediately downstream of the RBD that contains residues required for efficient processing of the histone H1t pre-mRNA (5). Replacing this region with an unrelated sequence of 20 amino acids found in the same place in xSLBP2 abolished the ability of human SLBP to support in vitro processing (5). This mutant was originally designated 20aa but here is referred to as 20aaC. We have thus identified two relatively short regions of SLBP which are absolutely required for its activity in processing of H1t pre-mRNA and not for RNA binding: a cluster of amino acids in the amino-terminal half of the RBD including the DR dipeptide, and the first 20 amino acids of the C-terminal domain.
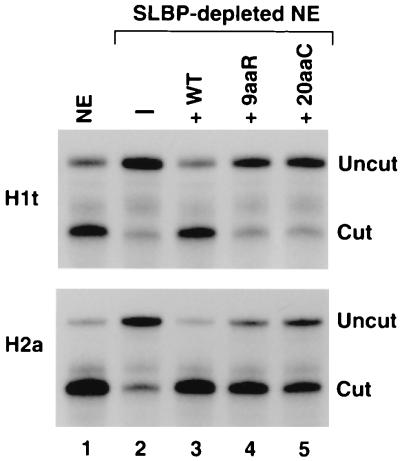
We tested the ability of the 9aaR and the 20aaC mutant proteins, which were inactive in processing of the H1t pre-mRNA (Fig. 6, top), to process the H2a-614 pre-mRNA. As discussed above, the SLBP-depleted extract itself can cleave between 10 and 20% of the H2a-614 pre-mRNA (Fig. 6, bottom, lane 2), consistent with only partial dependence of processing of this substrate on SLBP (4, 5, 23, 29). Addition of the wild-type SLBP to the depleted extract elevated this basal level of processing to more than 95% (Fig. 6, bottom, lane 3), restoring the initial processing efficiency of the nuclear extract (Fig. 6, bottom, lane 1). In the same assay, the 9aaR (Fig. 6, bottom, lane 4) and 20aaC (Fig. 6, bottom, lane 5) mutant proteins increased processing efficiency to approximately 90% and 80%, respectively. The 20aaC mutant protein was reproducibly less active than the 9aaR protein in complementing the SLBP-depleted nuclear extract. Under the conditions of these assays, virtually all of the substrate added to the nuclear extract is stably associated with the mutant proteins, as determined by using either the SLBP antibody or Ni-nitrilotriacetic acid agarose beads to isolate the SLBP–pre-mRNA complex (not shown).
FIG. 6.
Activity of 9aaR and 20aaC mutant SLBPs in 3′ end processing is substrate dependent. Processing activity of the nuclear extract (NE) using the H1t (top) and H2a-614 (bottom) histone pre-mRNAs before and after depletion of SLBP (lanes 1 and 2, respectively) and effect of addition of 100 ng of the different SLBPs (indicated above each lane) to the SLBP-depleted extract on 3′ end processing (lanes 3 to 5). WT, wild type.
We also tested the effect of addition of excess 9aaR and 20aaC mutant proteins to a standard nuclear extract on processing of the histone H1t pre-mRNA. The nuclear extract that had not been depleted of SLBP processed about 40% of the H1t pre-mRNA (Fig. 7, lane 1). Supplementing this extract with the exogenous wild-type SLBP (100 ng) increased the efficiency of processing to over 60% (Fig. 7, lane 2), while addition of the same amount of the 9aaR protein resulted in reduction of processing efficiency to less than 10% (Fig. 7, lane 3). A consistently stronger inhibition was caused by addition of the 20aaC mutant protein (Fig. 7, lane 4), which reduced processing efficiency to approximately 5%. This dominant negative effect is similar to that seen when xSLBP2 or 1-2-1 is added to the processing extract (11) and almost certainly results from interaction of the substrate with the excess mutant SLBP rather than with the endogenous SLBP in the extract.
FIG. 7.
Mutant proteins 9aaR and 20aaC inhibit H1t pre-mRNA processing when added to a nuclear extract. Lane 1 shows the processing activity of the nuclear extract (NE) using the H1t pre-mRNA as a substrate; in lanes 2 to 4, 250 ng of each protein was added to the nuclear extract, and processing of the H1t pre-mRNA was analyzed. WT, wild type.
The processing-deficient mutant SLBPs inefficiently recruit U7 snRNP to the processing complexes.
Binding of U7 snRNP to the substrate pre-mRNA occurs primarily by base pairing between the 5′ end of U7 snRNA and the HDE, located approximately 10 nucleotides downstream from the cleavage site. A major, if not the only, function of SLBP in 3′ end processing is stabilization of the interaction between U7 snRNP and the pre-mRNA substrate (5, 17). SLBP bound to the stem-loop structure may interact with one of the protein components of the U7 snRNP providing an additional support in anchoring the particle to the pre-mRNA. The stabilizing role of SLBP is critically important in processing of pre-mRNA substrates, including the H1t pre-mRNA, which contain an HDE that can form relatively few base pairs with U7 snRNA (5).
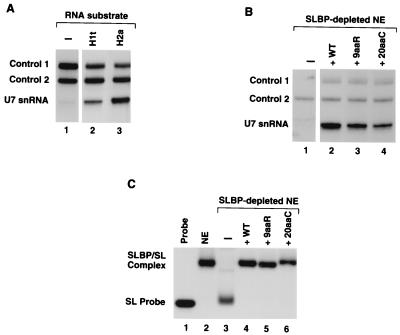
We have previously shown using anti-SLBP that less U7 snRNP coimmunoprecipitates with H1t pre-mRNA than with H2a-614 pre-mRNA, which contains a nearly perfect HDE (5). To confirm that this difference did not result from variability in the efficiency of immunoprecipitation of the pre-mRNA–SLBP complexes between samples, we carried out the following experiment. The same amounts (50 ng) of either the H1t or the H2a-614 histone pre-mRNA were mixed with an undepleted nuclear extract and incubated for 5 min at 22°C, allowing formation of the processing complexes, but not cleavage of the substrate to mature mRNA. A small amount of labeled RNA substrate (100 pg) was added simultaneously with the unlabeled substrate to monitor the efficiency of immunoprecipitation (control 1 bands in Fig. 8A and B). The reaction mixtures were subsequently incubated with the SLBP antibody at 4°C, and complexes assembled on the pre-mRNA were isolated on protein A-agarose beads. RNA was prepared from the immunoprecipitates and then analyzed for the presence of U7 snRNA by Northern blotting (5). During recovery of the RNA from agarose beads, a small amount of synthetic unlabeled U7 snRNA (containing additional nucleotides at the 5′ and 3′ ends and therefore migrating slower than the endogenous U7 snRNA) was added to each sample (control 2 bands in Fig. 8A and B) to monitor the efficiency of ethanol precipitation and hybridization with the anti-U7 probe. In the presence of the small amount of radioactive pre-mRNA, only a trace amount of U7 snRNA was recovered from the agarose beads (Fig. 8A, lane 1). In this reaction, nearly 100% of the radioactive pre-mRNA was immunoprecipitated, since SLBP is in molar excess of the pre-mRNA (control 1 band, Fig. 8A, lane 1). After addition of 50 ng of unlabeled H1t or H2a-614 pre-RNA, the efficiency of immunoprecipitation of the radioactive pre-mRNA was reduced to approximately 40% (Fig. 8A, lane 2) or 30% (Fig. 8A, lane 3), respectively, due to limiting amounts of the endogenous SLBP. Significantly, in spite of similar efficiency of pre-mRNA immunoprecipitation, the H2a-614 pre-mRNA bound four- to fivefold more U7 snRNP than the H1t pre mRNA (Fig. 8A, lanes 2 and 3). We conclude that in agreement with our previous results (5), these differences are due to the decreased ability of the U7 snRNP to form a stable complex with the H1t pre-mRNA.
FIG. 8.
9aaR and 20aaC mutant SLBPs have significantly reduced ability to recruit the U7 snRNP to the histone pre-mRNA. (A) A nuclear extract was incubated in the absence of unlabeled substrate (lane 1) or in the presence of 50 ng of either the H1t (lane 2) or H2a-614 (lane 3) histone pre-mRNA. Anti-SLBP was added, and RNA prepared from the immunoprecipitate was resolved on an 8% polyacrylamide–7M urea gel. U7 snRNA was detected by Northern blotting using an antisense U7 snRNA probe as described in Materials and Methods. Control 1 is the H2a-614 histone pre-mRNA radiolabeled at the 5′ end, which was added to each reaction (100 pg) to monitor the efficiency of immunoprecipitation of the unlabeled substrate RNA. Control 2 (1 ng) represents a synthetic unlabeled U7 snRNA which contains additional nucleotides at both 5′ and 3′ flanks and therefore migrates slower than the endogenous U7 snRNA. This control RNA is added to each sample prior to ethanol precipitation and allows monitoring of the efficiency of the total RNA recovery and hybridization. (B) A nuclear extract (NE) was depleted of SLBP (lane 1) and then supplemented with the indicated baculovirus-expressed proteins (lanes 2 to 4); 50 ng of unlabeled H1t histone pre-mRNA was added to each extract together with 10 pg of the labeled H1t substrate (control 1). Anti-SLBP was added, and the immunoprecipitates were processed for detection of U7 snRNA as in panel A; 0.1 ng of control 2 RNA was used as a control for precipitation of the RNA. WT, wild type. (C) Mobility shift assay to determine the binding activity in the control extract (lane 2), the SLBP-depleted nuclear extract (lane 3), and the SLBP-depleted nuclear extract supplemented with the baculovirus-expressed SLBPs (lanes 4 to 6), indicated above each lane. Lane 1 contains the probe alone.
The most likely explanation for the inability of both the 9aaR and 20aaC mutant proteins to function in H1t pre-mRNA processing was that each mutation disrupts the interaction of SLBP with U7 snRNP and thus causes inefficient recruitment of the U7 snRNP to the pre-mRNA substrate. We tested this possibility by directly measuring the ability of the 9aaR and 20aaC mutant proteins to recruit U7 snRNP to processing complexes assembled on the H1t pre-mRNA (Fig. 8B). H2a pre-mRNA was not used in this assay due to the high level of SLBP-independent recruitment of U7 snRNA to this substrate. Approximately 50 ng of unlabeled H1t pre-mRNA was mixed with the SLBP-depleted nuclear extract alone or supplemented with either the wild-type or mutant SLBPs. The amounts of both control RNAs used in this experiment, i.e., control 1 RNA (10 pg of the H1t substrate labeled at the 5′ end) and control 2 RNA (0.1 ng of cold synthetic U7 snRNA), were reduced 10 times compared to the previous experiment (Fig. 8A) to avoid possible interference of a strong radioactive signal generated by the control bands with the ability to detect the small amounts of U7 snRNA recruited by the H1t pre-mRNA. Incubation of the SLBP-depleted nuclear extract containing both labeled and unlabeled H1t pre-mRNA with the anti-SLBP did not result in immunoprecipitation of detectable amounts of the pre-mRNA (control 1) or the U7 snRNA (Fig. 8B, lane 1), demonstrating that the sample was free of SLBP. The essentially complete removal of SLBP from the nuclear extract was confirmed by band shift assay (Fig. 8C, lane 3) and Western blotting with the SLBP antibody (Fig. 3A). The synthetic U7 snRNA (control 2) was detected in this sample at the expected level, indicating that any immunoprecipitated RNA was quantitatively recovered during ethanol precipitation (Fig. 8B, lane 1). Addition of 1 μg of the baculovirus-expressed wild-type SLBP to the SLBP-depleted extract restored binding activity to the extract (Fig. 8C, lane 4) and allowed immunoprecipitation of a significant amount of processing complexes containing U7 snRNP (Fig. 8B, lane 2). The same amount of the 9aaR mutant protein, although as efficient as the wild-type SLBP in restoring binding activity to the depleted extract (Fig. 8C, lane 5) and in immunoprecipitating the substrate RNA, recruited only about 40% of the amount of U7 snRNA recruited by the wild-type protein (Fig. 8B, lane 3). The 20aaC mutant SLBP was even more impaired in recruiting U7 snRNP to the H1t pre-mRNA and allowed immunoprecipitation of 25% of the control amount of U7 snRNP (Fig. 8B, lane 4). Again, addition of the 20aaC protein to the SLBP-depleted extract restored binding activity to the regular level (Fig. 8C, lane 6) and resulted in efficient precipitation of the H1t pre-mRNA (Fig. 8B, lane 4). Thus, reduction in the amount of U7 snRNP immunoprecipitated in the presence of the 20aaC protein did not result from lower affinity of the mutant SLBP for the substrate RNA.
DISCUSSION
Human SLBP and one of the two SLBPs isolated from Xenopus, xSLBP1, are closely related to each other and are involved in 3′ end processing of replication-dependent histone pre-mRNA in the nucleus. The second form of SLBP in Xenopus, xSLBP2, does not function in processing and is similar to the other SLBPs only within the RBD (28). Although it binds to the same RNA target, the RBD of xSLBP2 cannot substitute for the RBD in the processing-specific SLBPs (11).
Residues of the RBD required for RNA recognition.
A minimal RBD required for efficient binding to the stem-loop structure was mapped in human SLBP to a 73-amino-acid region in the center of the protein, between amino acids 126 and 198 (29). There are two regions in the RBD highly conserved among different SLBPs, one located between amino acids 10 and 21 and the other located between amino acids 41 and 54. In addition, the RBD contains a number of conserved aromatic residues scattered throughout the domain, reminiscent of the conserved aromatic residues present in the RNA recognition motif (RRM) (26). Many of these residues in the RRM make contacts with the single-stranded regions of the RNA and may be involved in stacking interactions with the bases of the RNA target. A number of the highly conserved residues in the SLBP RBD are basic and are always either lysines or arginines. In several RNA binding proteins, arginine-rich regions in particular are important in binding and the arginines cannot be substituted with lysines (1). Basic residues in RNA binding proteins are also frequently involved in electrostatic interactions with the phosphate groups of the recognized RNA sequences. Some of these residues can be either lysine or arginine, but they cannot be changed to alanine without a major reduction in binding affinity.
Since SLBP has a novel, previously unknown RBD, we tested whether the same types of rules apply for binding of the stem-loop RNA by SLBP. Martin et al. showed that changing the two highly conserved arginines at position 10 and 11 of the RBD to alanines resulted in complete loss of binding activity (13), suggesting that the positive charge at both positions was important. We demonstrated here that changing the same arginines to lysines (RR/KK mutant) also abolished binding activity of SLBP. The guanidinium group of the arginines is likely to be involved in specific interactions with the phosphodiester backbone and cannot be replaced by the amino group of the lysine (1). In addition to the RR/KK mutation, we made a K19R mutation by replacing the absolutely conserved lysine at position 19 (also present in SLBPs of the sea urchin, Drosophila and Chlamydomonas [unpublished results]) with arginine. In contrast to the RR/KK mutation, replacement of lysine 19 with arginine did not reduce binding affinity of the protein. However, replacing lysine 19 with alanine resulted in virtually complete disruption of SLBP binding, suggesting that the presence of a positively charged residue in this position is essential for RNA binding. Changing the tyrosines at positions 24 and 27 of the RBD to phenylalanines greatly reduced binding affinity, suggesting that there is a major role for the hydroxyl group as well as for the aromatic ring in RNA binding. Perhaps the ring of tyrosine stacks against bases of the RNA target and the hydroxyl group is involved in formation of specific hydrogen bonds. In addition, in most SLBPs there are two tryptophans (residues 56 and 63) near the carboxyl end of the RBD. Replacement of these tryptophans with isoleucines completely abolished binding of SLBP to the stem-loop structure, confirming the importance of the conserved aromatic residues in sequence specific recognition of the RNA target.
Based on these and other studies (13), it seems likely that binding of SLBP to the stem-loop structure involves a set of interactions similar to those identified in other RNA-protein complexes. However, details of these interactions will be known only after determining the structure of the SLBP-RNA complex by X-ray crystallography or nuclear magnetic resonance spectroscopy.
Role of the RBD and the C-terminal domain in histone pre-mRNA processing.
The RBDs of vertebrate processing-specific SLBPs, including mouse and human SLBPs and xSLBP1 from Xenopus, contain a number of residues that are not conserved in the processing-deficient xSLBP2. Systematic substitution of these amino acids in the RBD of human SLBP with the amino acids found in the same place in xSLBP2 allowed identification of a region of the RBD that plays an important role in processing. This region includes two key amino acids, an aspartic acid at position 25 (D25) and arginine at position 26 (R26), mutation of which makes SLBP almost completely inactive in H1t pre-mRNA processing. Also important is the nearby IK dipeptide, mutation of which results in a twofold reduction in processing efficiency. The remaining residues conserved in mammalian SLBPs and xSLBP1 but not in xSLBP2 make a much smaller contribution to the overall processing activity of SLBP.
The DR dipeptide is flanked by two tyrosines critical for RNA binding, Y24 and Y27. The proximity of these residues may be functionally important. It is likely that SLBP interacts with another component of the processing machinery only when it is bound to the histone pre-mRNA. Binding of SLBP to its RNA target, partially mediated by the two tyrosines, could for example trigger some conformational changes in the RBD, exposing the DR dipeptide thus facilitating its direct involvement in processing. The Y24F mutant is completely processing deficient, although it retains the same residual binding activity as the H41F protein that is capable of supporting substantial processing of the H2a pre-mRNA. This observation suggests that the Y24F mutation, adjacent to the DR dipeptide, affects both RNA binding and processing.
Immunoprecipitation experiments with the 9aaR mutant protein support the notion that the DR dipeptide and the other amino acids of the RBD conserved only in processing-specific SLBPs are involved in stabilization of the U7 snRNP on the pre-mRNA. The same role seems to be played by residues in the 20-amino-acid region immediately C terminal to the RBD. The 20aaC and 9aaR mutants recruit 4- and 2.5-fold, respectively, less U7 snRNP to the H1t pre-mRNA than the wild-type SLBP. Consistent with this result, the 20aaC mutant protein is also less efficient in rescuing processing of the H2a pre-mRNA in the SLBP-depleted nuclear extract and has a stronger dominant negative effect on histone H1t pre-mRNA processing in the presence of the endogenous SLBP. Combining both the 9aaR and the 20aaC mutations did not result in further reduction of the amount of immunoprecipitated U7 snRNP nor in further reduction of SLBP activity in H2a pre-mRNA processing (not shown). We conclude that both regions of SLBP participate in the same interaction that results in stabilizing U7 snRNP on the pre-mRNA. The 20aaC mutation results in more severe disruption of this interaction. It is likely that the residual processing activity of the 20aaC mutant SLBP on H2a-614 pre-mRNA is due to the high affinity of U7 snRNP for the H2a-614 HDE and remaining weak interactions between the mutant SLBP and U7 snRNP.
Surprisingly, although both 9aaR and 20aaC mutant SLBPs are inactive in processing of the H1t pre-mRNA, they can still recruit a significant amount of the U7 snRNP to this substrate at 4°C. A likely explanation for this observation is the lower temperature of the immunoprecipitation assay (22° followed by 4°C) than of the processing assay (32°C). It is possible that under these conditions, the HDE of the H1t pre-mRNA can form a relatively strong duplex with the U7 snRNA, which is not stable at 32°C. This interpretation is supported by previous studies which demonstrated that lowering the temperature of incubation favors stronger interaction between the pre-mRNA and U7 snRNA and decreases the SLBP dependence of in vitro processing (21).
Requirements for formation of a stable processing complex.
Although the mutant SLBP protein 9aaR containing nine amino acids of the RBD replaced with the xSLBP2 sequence was completely inactive in processing of H1t pre-mRNA, it showed significant activity when the H2a-614 pre-mRNA substrate was used. A similar result was obtained with the 20aaC mutant protein. These mutants bind the pre-mRNA with the same affinity as the wild-type SLBP but are impaired in recruitment of U7 snRNP. The same variable effect on processing of the two substrates was observed with the H41F mutant SLBP which binds the pre-mRNA weakly. Thus, strong interaction (either direct or indirect) between the SLBP and U7 snRNP as well as high-affinity binding of the SLBP to the histone pre-mRNA are critical for processing of histone pre-mRNAs that form relatively few base pairs with the U7 snRNA (e.g., the H1t pre-mRNA) and are less important for processing of pre-mRNAs that contain a strong HDE (e.g., the H2a pre-mRNA). Taken together, our results indicate that assembly of a stable processing complex on the histone pre-mRNA depends on the additive strength of at least three interactions: RNA-RNA interactions between U7 snRNA and the HDE, binding of SLBP to the stem-loop, and interaction of the SLBP-stem-loop complex with U7 snRNP. A similar requirement for multiple interactions to stabilize an RNA processing complex is seen in recognition of the 3′ splice site by U2aF65 and U2aF35 which interact with the polypyrimidine tract and the AG dinucleotide, respectively. The presence of a long and uninterrupted polypyrimidine tract tightly bound by U2aF65 diminishes the requirement for binding of U2aF35, which is indispensable for cleavage of the 3′ splice sites containing weak polypyrimidine tracts (32, 33).
SLBP does not interact with the U7 snRNP in the absence of the pre-mRNA substrate and, as judged by gel filtration chromatography, is not found in a complex with other nuclear proteins (unpublished results). It is likely that the first step in histone pre-mRNA processing is binding of SLBP to the stem-loop structure followed by stabilization of the U7 snRNP on the HDE. The stabilization of the U7 snRNP may be achieved by direct interaction of the SLBP-RNA complex with one of the proteins of the U7 snRNP. However, it is possible that there is a yet unknown component of the processing machinery which interacts with both SLBP and U7 snRNP and provides a bridging link between the two processing factors, resulting in stable association of the U7 snRNP with the pre-mRNA. One approach to identifying this unknown component may be to screen for proteins that interact with the SLBP/SL complex using a modification of the yeast two-hybrid system. These studies are in progress.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM29832 to W.F.M.
We thank Francesca Perez for help in constructing some of the clones and Xiaocui Yang for technical assistance. We are grateful to Tom Ingledue for the Xenopus SLBP constructs and members of the Marzluff laboratory for helpful discussions.
Z.D. and J.A.E. contributed equally to this work.
REFERENCES
- 1.Calnan B J, Biancalana S, Hudson D, Frankel A D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 2.Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel M L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J. 1988;7:801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominski Z, Marzluff W F. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 4.Dominski Z, Sumerel J, Hanson R J, Marzluff W F. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- 5.Dominski Z, Zheng L-X, Sanchez R, Marzluff W F. The stem-loop binding protein facilitates 3′ end formation by stabilizing U7 snRNP binding to the histone pre-mRNA. Mol Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galli G, Hofstetter H, Stunnenberg H G, Birnstiel M L. Biochemical complementation with RNA in the Xenopus oocyte: a small RNA is required for the generation of 3′ histone mRNA termini. Cell. 1983;34:823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- 7.Gallie D R, Lewis N J, Marzluff W F. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gick O, Krämer A, Keller W, Birnstiel M L. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gick O, Krämer A, Vasserot A, Birnstiel M L. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris M E, Böhni R, Schneiderman M H, Ramamurthy L, Schümperli D, Marzluff W F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingledue T C, Dominski Z, Sanchez R, Erkmann J A, Marzluff W F. Dual role for the RNA-binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA. 2000;6:1635–1648. doi: 10.1017/s1355838200000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krieg P A, Melton D A. Formation of the 3′ end of histone mRNA by post-transcriptional processing. Nature. 1984;308:203–206. doi: 10.1038/308203a0. [DOI] [PubMed] [Google Scholar]
- 13.Martin F, Michel F, Zenklusen D, Müller B, Schümperli D. Positive and negative mutant selection in the human histone hairpin-binding protein using the yeast three-hybrid system. Nucleic Acids Res. 2000;28:1594–1603. doi: 10.1093/nar/28.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marzluff W F. Histone 3′ ends: essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 16.Marzluff W F, Whitfield M L, Dominski Z, Wang Z-F. Identification of the protein that interacts with the 3′ end of histone mRNA. In: Richter J D, editor. mRNA formation and function. New York, N.Y: Academic Press; 1997. pp. 163–193. [Google Scholar]
- 17.Melin L, Soldati D, Mital R, Streit A, Schümperli D. Biochemical demonstration of complex formation of histone pre- mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mowry K L, Steitz J A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 19.Pandey N B, Williams A S, Sun J-H, Brown V D, Bond U, Marzluff W F. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol Cell Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith H O, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel M L. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci USA. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spycher C, Streit A, Stefanovic B, Albrecht D, Koning T H W, Schümperli D. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefanovic B, Hackl W, Lührmann R, Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Streit A, Koning T W, Soldati D, Melin L, Schümperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan E, Santiago C, Parker E D, Dominski Z, Yang X, Lanzotti D J, Ingledue T C, Marzluff W F, Duronio R J. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–181. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J-H, Pilch D R, Marzluff W F. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 1992;20:6057–6066. doi: 10.1093/nar/20.22.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 27.Vasserot A P, Schaufele F J, Birnstiel M L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci USA. 1989;86:4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z-F, Ingledue T C, Dominski Z, Sanchez R, Marzluff W F. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol. 1999;19:835–845. doi: 10.1128/mcb.19.1.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z-F, Whitfield M L, Ingledue T I, Dominski Z, Marzluff W F. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 30.Whitfield M L, Zheng L-X, Baldwin A, Ohta T, Hurt M M, Marzluff W F. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams A S, Ingledue T C, Kay B K, Marzluff W F. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S P, Romfo C M, Nilsen T W, Green M R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 33.Zorio D A R, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]