Abstract
We have evaluated two commercially available kits (AMPLICOR MONITOR [Roche] and NASBA HIV-1 QT or NucliSens HIV-1 QT [Organon Teknika]) and two noncommercial methods for the accurate quantitation of human immunodeficiency virus type 1 (HIV-1) RNA in seminal plasma. The same panels of coded specimens were tested on four separate occasions. Laboratories using the commercial assays employed silica beads to isolate HIV-1 RNA, which removed inhibitory factors sometimes found in seminal plasma. Sensitivities and specificities, respectively, for each assay were as follows: AMPLICOR MONITOR, 100 and 73%; NASBA HIV-1 QT, 84 and 100%; NucliSens HIV-1 QT, 99 and 98%; and noncommercial assays, 91 and 73%. When results from the laboratory that was inexperienced with the silica bead extraction method were excluded from the analysis, specificity for the Roche assay increased to 100%. The commercial assays demonstrated highly reproducible results, with intra-assay standard deviations (measured in log10 RNA copies/milliliter of seminal plasma) ranging from 0.11 to 0.32; those of the noncommercial assays ranged from 0.12 to 0.75. Differences in mean estimated HIV-1 RNA concentrations were ≤0.67 log10 and were greater at low viral loads. Suspension matrices that used blood plasma or seminal plasma did not make a difference in recovery of HIV-1 RNA, which suggested that blood plasma specimens can be used as external controls for seminal plasma assays. More variation in the HIV-1 RNA viral loads was observed in the seminal plasma values than in the blood plasma values when paired specimens from HIV-1-infected men were tested. Quantitation of HIV-1 RNA in seminal plasma can be reliably accomplished using two commercially available assays, and may be incorporated into the evaluations of HIV-1 seropositive men enrolled in clinical studies.
Sexual transmission of human immunodeficiency virus type 1 (HIV-1) is postulated to occur through direct exposure of the oral, vaginal, urethral, or rectal mucosal surfaces to genital secretions (semen and cervical vaginal secretions) from a sexual partner. In 1996 the Centers for Disease Control and Prevention indicated that sexual transmission of HIV-1 was the primary route of HIV-1 infection in the United States in persons aged 25 to 44 years (3). If blood and semen HIV-1 concentrations are predictive of the probability of HIV-1 transmission, then strategies to decrease the viral load below a critical level in both these compartments may ultimately reduce the rate of transmission.
Recovery of infectious HIV-1 from seminal cells has been highly variable (9 to 55%) and also inconsistent within an individual over time in longitudinal studies (11). Seminal plasma infrequently yields infectious virus (10, 18). Early efforts to quantitate virus in seminal plasma used noncommercial reverse transcription (RT)-PCR amplification techniques (1, 9, 12, 14, 17) and employed external standards as the method of quantification. When RT-PCR kits that used internal quantitation standards became commercially available, it became clear that seminal plasma often contained inhibitors of the RT-PCR. This inhibition could be removed by using silica beads to isolate the viral RNA (5, 6, 8).
Multicenter clinical drug trials are now actively studying the effects of different antiretroviral drug regimens on the viral load in blood and the genital tract, including the evaluation of viral load in genital secretions in relation to HIV-1 transmission. We have evaluated the various methods in the processing, isolation, and quantitation of HIV-1 RNA in seminal plasma and developed a standardized method to facilitate the comparison of data generated by studies in the field. This manuscript describes the technical performance characteristics of both commercial and noncommercial HIV-1 RNA assays which have been used to measure seminal plasma HIV-1 RNA and compares blood plasma with seminal plasma as a dilution matrix for the preparation of controls and proficiency panels.
MATERIALS AND METHODS
Quantitative HIV RNA assays.
Two commercially available assays were compared: the Roche AMPLICOR MONITOR assay and the Organon Teknika NASBA HIV-1 QT assay or NucliSens HIV-1 QT assay (hereafter in this work the Organon Teknika assays are referred to as NASBA and NucliSens, respectively). These assays have been described elsewhere (7, 13, 16). Additionally, two laboratories employed noncommercial assays. The Roche MONITOR assay was used according to the manufacturer's instructions except for HIV-1 RNA isolation. Instead, viral RNA was isolated using the silica bead procedure of Boom et al. (2). Previous experiments had demonstrated that factors in some seminal plasma specimens inhibited RT-PCR amplification and that these inhibitors were removed by silica bead extraction (5, 6, 8). The limit of detection of the MONITOR assay is 400 copies/ml.
The Organon Teknika NASBA assay was used for the first panel according to the manufacturer's instructions, with the exception that kit calibrators were diluted 10-fold to increase assay sensitivity. Over the course of the study period, Organon Teknika introduced a more-sensitive RNA assay, NucliSens, which was used for the remaining test panels without modification. The silica bead extraction method of viral RNA is an inherent part of the Organon Teknika assay. The limit of detection is 1,000 copies/ml for the NASBA assay and 400 copies/ml for the NucliSens assay.
Two sites used similar assays that were developed in their own laboratories. Both methods pelleted HIV-1 virions from seminal plasma using centrifugation at ≥14,000 × g for 60 min at 4°C. Pellets were then resuspended in Tri Reagent (4), and viral RNA was extracted with chloroform and isopropanol. The HIV-1 RNA was then subjected to RT-PCR using published methods (14, 15). Briefly, HIV RNA was reverse transcribed to cDNA that was then amplified using Taq polymerase and either SK38 and SK39 primers over 35 cycles (14) or the following primers: Sense, CAATGAGGAAGCTGCAGAATGGGATAG, and Anti-sense, CATCCATCCTATTTGTTCCTGAAGG. Amplicons were detected by hybridization with a 32P-labeled SK19 probe or ATGAGAGAACCAAGGGGAAGTGACATAGCA (15). The RNA copy numbers were estimated by comparison to a standard curve.
Participating laboratories.
Eight laboratories (coded A to H) participated in the evaluation at one or more times. Technologists performing the respective assays had previous experience in the performance of the nucleic acid amplification assays using blood plasma. However, two laboratories (B and E) did not have previous experience with the Boom silica bead method of viral RNA extraction (2) and four laboratories (B, D, E, and F) did not have previous experience with viral RNA measurements from seminal plasma specimens. In the first round of testing, two laboratories (one experienced in testing seminal plasma [A] and one inexperienced [B]) used the Roche MONITOR assay. Similarly, an experienced laboratory (C) and an inexperienced laboratory (D) used the NASBA assay. Two laboratories used their own noncommercial assay (laboratories G and H). For the second and third rounds of testing, two different laboratories (E and F), neither of which had experience with seminal plasma viral RNA determination, assayed the panels using the Organon Teknika assay, replacing laboratory D. Overall, six laboratories tested seminal plasma panel 1 (SP01), seven tested panel 2a (SP2a), five tested panel 2b (SP02b), and five tested panel 3 (SP03).
Composition of test panels.
The Virology Quality Assurance Laboratory of the National Institutes of Health provided three separate blinded panels of specimens for testing (SP01, SP02, and SP03) to each of the participating laboratories. Panels SP01 and SP02 consisted of pooled HIV-negative seminal plasma diluted 1:1 in either Hanks' balanced salt solution (SP01) or phosphate-buffered saline (SP02) into which a known quantity of HIV-1 was added (henceforth referred to as the nominal concentration of HIV-1 RNA). Panel SP01 consisted of six specimens each at nominal concentrations of 103 and 104 HIV RNA copies/ml, five specimens each at 105 and 106 copies/ml, and two negative specimens. Panel SP02 consisted of four negative specimens and five replicates at each of the following nominal concentrations: 103, 104, 105, and 106 HIV RNA copies/ml. The viral stock used was a highly characterized subtype B HIV-1 obtained from pooled culture supernatants from three HIV-1-seropositive donors (19). Virology Quality Assurance Laboratory standards used in each panel were prepared by diluting the same viral stock into HIV-1-seronegative blood plasma. Panel SP02 was tested on two separate occasions approximately 4 months apart, designated SP2a and SP2b. Panel SP03 consisted of triplicate aliquots of paired blood plasma and cell-free seminal plasma from one HIV-1-seronegative and five HIV-1-seropositive donors. Specimens were shipped frozen via overnight express to each of the participating laboratories.
Statistical methods.
Estimates of HIV-1 RNA concentration were first transformed to the log10 scale. The intra-assay standard deviation (SD) of log-transformed estimates from each panel in each laboratory was estimated from the mean square error for a one-way analysis of variance, with the nominal concentration as the predictor variable. Estimates that were below the limit of detection were set at the limit for each kit. This negatively biased the SDs, but the bias was confined to a few laboratories and was small, given the small number of values below the limit of detection. Estimates were compared among laboratories using regressions of log10 estimated HIV-1 RNA concentration on log10 nominal HIV-1 RNA concentration. Both slopes and intercepts were allowed to vary among laboratories in the initial model for each panel. If no differences among slopes were detected on a panel, then a model with parallel regression lines was assumed. Under this model, differences among laboratories did not vary with nominal concentration and the differences were estimated from the intercepts for the regressions. However, if slopes did differ, then the differences in estimated RNA concentration among laboratories varied with RNA concentration. When this occurred, the regression lines were compared graphically to determine if the differences increased or decreased with concentration, and to estimate the magnitude of interlaboratory variation. A similar regression approach was used to compare data between rounds of testing on panel SP02, which used different nonplasma dilution matrices, within laboratories and to compare estimates from spiked seminal plasma and blood plasma for this panel.
RESULTS
Sensitivity and specificity of assays used for the quantitation of HIV-1 RNA from seminal plasma.
Tables 1 and 2 summarize the performance for each assay and participating laboratory for the first two panels. Six laboratories (A, B, C, D, G, and H) participated in the first round of testing (SP01) (Table 1). One laboratory, inexperienced in the Boom RNA extraction method and using a commercial kit, reported three false-positive results (the two negative seminal plasma specimens and the negative control), all of which had high estimates of viral RNA (6,000 to 11,000 copies/ml). The two laboratories that used the NASBA assay reported false negatives from samples at 103 viral RNA copies/ml. Five of the six false negatives from laboratory H occurred at 103 viral RNA copies/ml, and one occurred at 104 viral RNA copies/ml.
TABLE 1.
Performance characteristics for HIV-1 RNA quantification for the coded seminal plasma samples of panel SP01
| Laboratory | Assay | No. of false positives | No. of false negatives | Intra-assay SD (log10 RNA copies/ml) | Slope | Intercept (log10 RNA copies/ml) | r2 |
|---|---|---|---|---|---|---|---|
| A | Roche | 0 | 0 | 0.16 | 0.91 | 0.674 | 0.98 |
| B | Roche | 3 | 0 | 0.15 | 0.75 | 1.581 | 0.96 |
| C | NASBA | 0 | 4 | 0.20 | 0.94 | 0.399 | 0.96 |
| D | NASBA | 0 | 3 | 0.14 | 0.86 | 0.790 | 0.98 |
| G | Noncommercial | 0 | 0 | 0.12 | 0.89 | 2.146 | 0.98 |
| H | Noncommercial | 0 | 6 | 0.75 | 0.65 | 1.503 | 0.36 |
TABLE 2.
Performance characteristics for HIV-1 RNA quantification for the coded seminal plasma samples of panel SP02a
| Laboratory | Assay | No. of false positives | No. of false negatives | Intra-assay SD(s) (log10 RNA copies/ml) | Slope(s) | Intercept(s) (log10 RNA copies/ml) | r2 |
|---|---|---|---|---|---|---|---|
| A | Roche | 0, 0 | 0, 0 | 0.11, 0.16 | 0.92, 0.96 | 0.30, 0.17 | 0.99, 0.98 |
| B | Roche | 5, 0 | 0, 0 | 0.19, 0.14 | 0.73, 0.94 | 1.47, 0.31 | 0.93, 0.99 |
| C | NucliSens | 0, 0 | 0, 0 | 0.14, 0.14 | 0.93, 1.03 | 0.31, −0.15 | 0.98, 0.99 |
| E | NucliSens | 0, 0 | 0, 0 | 0.21, 0.17 | 0.98, 1.01 | 0.14, −0.10 | 0.97, 0.98 |
| F | NucliSens | 0, 0 | 2, 0 | 0.29, 0.18 | 1.00, 0.94 | −0.07, 0.41 | 0.95, 0.98 |
| G | Noncommercial | 3 | 0 | 0.61 | 0.59 | 2.87 | 0.59 |
Panel SP02 was tested twice in five of the laboratories and once in laboratory G.
Seven laboratories participated in the analysis of panel SP02a (Table 2). One laboratory which was experienced in seminal plasma testing encountered problems with a noncommercial assay, and these data were excluded from the analysis. Another laboratory experienced in seminal plasma testing, using a noncommercial assay, reported three false-positive results, all of which had very high estimates of viral RNA concentration (116,000 to 177,000 copies/ml). An additional previously inexperienced laboratory, using the Roche assay, reported five false positives (the four negative seminal plasma specimens and the negative control), with estimates of viral RNA concentration ranging from 2,874 to 7,267 copies/ml. One inexperienced laboratory using the NucliSens assay could not correctly identify two of the samples with 103 viral RNA copies/ml (false negatives). No false positives or false negatives were observed when five laboratories repeated testing of this panel 4 to 5 months later (SP02b) (Table 2).
Panel SP03 consisted of paired seminal plasma and blood plasma from five HIV-infected men and one uninfected man. One false positive was reported for a semen specimen with a calculated viral load of 200 HIV-1 RNA copies/ml (data not shown). Combining data from all of the panels, the overall sensitivities of the assays for detecting HIV-1 RNA in seminal plasma were as follows: AMPLICOR, 100%; NASBA, 84%; NucliSens, 99%; and noncommercial assays, 91%. Overall assay specificities for HIV-1 RNA were as follows: MONITOR, 73%; NASBA, 100%; NucliSens, 98%; and noncommercial assays, 73%. If the laboratory inexperienced in the Boom extraction procedure was excluded from the analysis, the specificity for the Roche assay increased to 100%.
Intra-assay variation.
Standard deviations for log-transformed estimates from seminal plasma varied widely among panels and assays (Tables 1 and 2 and data not shown for SP03) (range of log10 SDs: 0.11 to 0.75 viral RNA copies/ml). The Roche MONITOR assay was the most consistent, with all log10 SDs being ≤0.19 viral RNA copies/ml. Most of the log10 SDs for the NASBA and NucliSens assays were <0.20 viral RNA copies/ml, but values of 0.29 and 0.32 were also obtained. The highest SDs were obtained from noncommercial assays (range of log10 SDs, 0.12 to 0.75 viral RNA copies/ml).
In general, laboratories that used commercial kits obtained similar estimated values for the specimens, which correlated well with the nominal HIV-1 RNA concentrations for each sample. This is shown by the slopes and intercepts for the calculated regression lines (Tables 1 and 2). Slopes determined by the laboratories that used the commercial assays ranged from 0.73 to 1.03, with 11 of the 14 calculated slopes between 0.91 and 1.03. Intercepts derived from the data from these same laboratories were also comparable, ranging from −0.15 to 1.59, with 12 of the 14 values between −0.15 and 0.79. In contrast, laboratories which used noncommercial assays had more shallow slopes (0.59 to 0.89) and higher intercepts (1.50 to 2.87), indicating that HIV-1 RNA estimates from these laboratories would tend to be higher than the estimates obtained in the five laboratories that used the commercially available kits. In addition, data from the laboratories that used commercial kits were very precise (r2 ≥ 0.93), compared with data from the other two laboratories (r2, 0.36 to 0.98).
Intralaboratory variation.
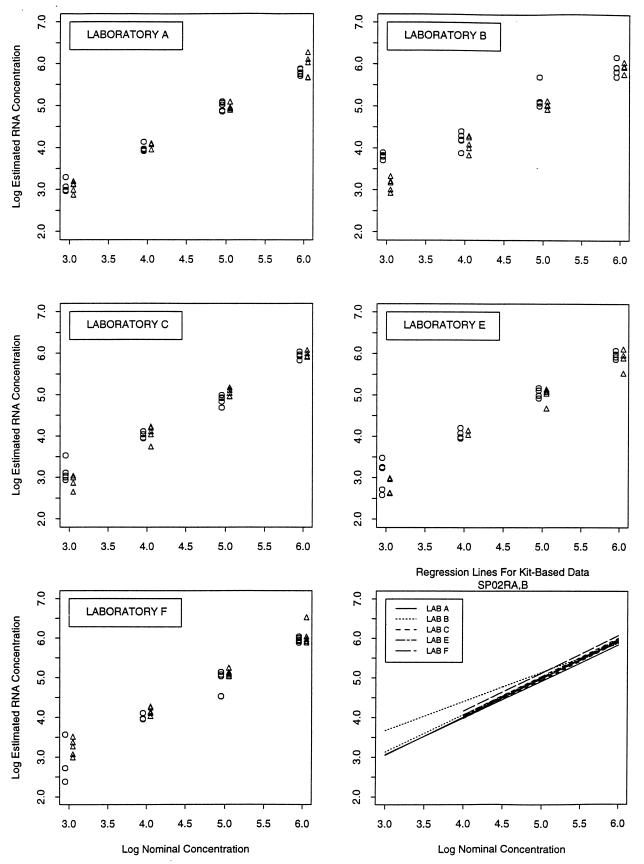
The consistency of performance within each of five of the laboratories that used commercial assay kits was assessed by comparing estimates from the two rounds of testing of panel SP02 that occurred 4 to 5 months apart for both seminal and blood plasma (Table 2). Despite the use of different kit lots between runs within laboratories and the different suspension matrices, the results were highly reproducible (Fig. 1 [panels for laboratories A, B, C, E, and F]). Intra-assay variation was greater for samples with 103 HIV-1 RNA copies/ml than those with higher concentrations, especially in laboratories that used the NucliSens assay (Fig. 1 [panels for laboratories C, E, and F]). Overall there was remarkable consistency in the results observed within laboratories as well as between laboratories (Fig. [bottom right panel]).
FIG. 1.
Results obtained from repeated testing of SP02 in five laboratories that used the commercial HIV-1 RNA assay. (A to E) Scatter plots of log10 RNA copies/milliliter from repeated testing of panel SP02. Circles represent SP02a results, and triangles represent SP02b results. The panel in the lower right corner shows regression lines generated from all five laboratories from both rounds of testing.
Effect of type of body fluid matrix on the measurement of HIV-1 RNA.
We compared estimates of numbers of HIV-1 RNA copies per milliliter obtained from the spiked seminal plasma samples and the spiked blood plasma controls to determine if differences in composition of the suspension matrix affected estimates of HIV-1 RNA concentration (panel SP02). Neither the slopes nor the intercepts differed significantly between spiked seminal plasma samples and spiked blood plasma controls in four of the five laboratories that used the commercial assays (laboratories A, C, E, and F). The fitted regression lines for the semen samples and plasma standards were essentially superimposable, indicating that the estimates from the two were almost identical. The eight regression lines derived from laboratories A, C, E, and F were also extremely precise (r2 > 0.95). RNA recovery from the blood plasma samples was poor in laboratory B, resulting in a flatter slope and higher intercept than those obtained from seminal plasma. However, the great similarity of the paired regression lines in four of the laboratories indicates that the suspension matrix did not make a difference at least when the Boom RNA isolation method was used.
Variation within specimen type.
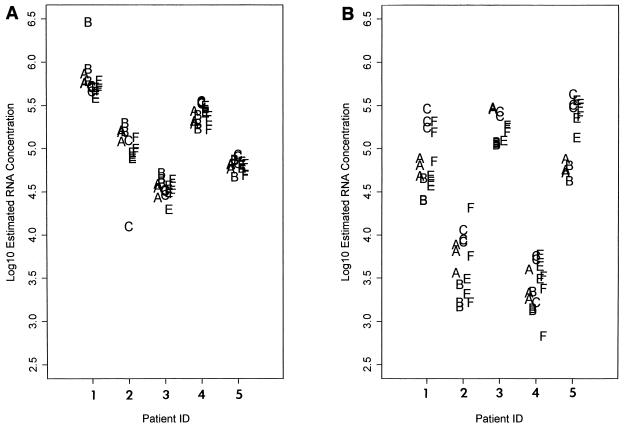
In panel SP03 paired blood plasma and seminal plasma from HIV-1-seropositive donors were evaluated using the two commercial assays. Results were consistent between assays and among laboratories (Fig. 2). Slightly more variation in copy number (average 0.05 log10 SD) was observed when replicates of seminal plasma (Fig. 2B) were tested than was observed with blood plasma (Fig. 2A). The median SD (measured in log10 viral RNA copies/milliliter) for blood plasma was 0.08 for the MONITOR assay and 0.06 for the NucliSens assay. In contrast, the median SD for seminal plasma was 0.12 for the MONITOR assay and 0.10 for the NucliSens assay. None of these differences were statistically significant.
FIG. 2.
HIV-1 RNA results from five HIV-seropositive patients (patients, 1, 2, 3, 4, and 5). Blood (A) and seminal plasma (B) were tested in triplicate. Each letter represents an individual HIV-1 RNA result from each of five laboratories (laboratories A, B, C, E, and F). In many cases results were overlapping and cannot be visualized.
DISCUSSION
HIV-1 RNA quantitation assays were originally developed to assess viral RNA levels in blood plasma. Some of these assays have also been used, sometimes with modification, to measure viral load in seminal plasma. We evaluated several of the previously published methods for quantifying HIV-1 RNA from seminal plasma (5, 6, 14, 15) and found remarkable consistency in HIV-1 RNA quantification in seminal plasma among laboratories, even for those laboratories that had little prior experience working with semen.
Our data indicate that the commercial assays provide more sensitive and consistent results when used to assess viral load in the seminal plasma than do noncommercial assays. These observations may be a consequence of the internal standards used by both of the commercial assay kits, which can signal the presence of a problem with inhibition of viral nucleic acid amplification. In addition, reagents for commercial kits are prepared in large, well-characterized lots. All laboratories in this study that used a commercial HIV-1 RNA assay kit also used the silica procedure for viral RNA extraction, while those laboratories that used noncommercial assays did not. Previous studies have indicated that seminal plasma frequently contains inhibitors of RT-PCR and that the silica bead isolation extraction procedure (2) can remove these inhibitors (5, 6, 8).
Although all of the laboratories which participated in these evaluations were very experienced in performing HIV RNA assays, laboratories B and E had never used the silica bead RNA isolation procedure and laboratories B, D, E, and F had never worked with seminal plasma. In particular, laboratories A and B prepared their own silica beads and reagents, while laboratories C, D, E, and F used commercially available reagents which were part of the Organon Teknika HIV RNA kits. Laboratory B experienced the most problems, obtaining several false positives in the first two panels tested. By the third round of testing (panel SP02b), more-consistent results were obtained. These results demonstrate that inexperience in just one step in an assay can affect laboratory performance and also indicate the value of quality assurance program evaluations. Laboratories initiating studies that require silica bead extraction techniques might be advised to use the commercially available reagents.
There appeared to be no effect of the suspension matrix on the ability to estimate HIV-1 RNA levels when the Boom silica RNA extraction method and a commercial HIV-1 RNA quantification kit were used. This is an important practical consideration, as it suggests that the controls that were provided by the manufacturer with each assay kit can be reliably used in assays in which seminal plasma is the test specimen. We did observe somewhat more variation when replicates of seminal plasma were tested than when blood plasma was assayed (Fig. 2). The reasons for this are unclear but may be related to the intrinsic complex biological nature of seminal plasma and the fact that it is composed of fluids from several reproductive compartments. Part of the increased variation observed among the seminal plasma samples may also be due to the generally lower copy numbers found in the seminal plasma compared to blood plasma.
In summary, the NucliSens and AMPLICOR MONITOR kits were found to quantify HIV-1 RNA in seminal plasma reliably and comparably after isolation of viral RNA using the silica bead procedure of Boom (2). This suggests that data derived from one of the commercial assays could be easily compared with data from the other, as might occur in different clinical trials or natural history studies, and that data sets could perhaps be pooled to achieve larger numbers and possibly more statistical power for analysis purposes. The accurate and reliable quantification of HIV-1 viral load in the seminal plasma by these methods is fundamental to the evaluation of therapeutic drug and vaccine candidates aimed at the reduction of viral load in the semen and the subsequent reduction in HIV-1 transmission.
ACKNOWLEDGMENTS
We acknowledge the expert technical assistance of Joan Dragavon, Michelle Jack, Reggie Sampoleo, Jody Schock, and Colleen Starkey. We also thank Suzanne Granger for data analysis and graphics. Michael Cronin of Organon Teknika provided NASBA and NucliSens kits used by some of the participating laboratories.
Financial support was provided by the following grants and contracts: AI-27664, AI-30731, and AI27757 (to R.W.C.) and AI-25868 and DK-49381 (to S.A.F.).
REFERENCES
- 1.Anderson D J, O'Brien T R, Politch J A, Martinez A, Seage G R, Padian N, Horsburgh C R, Mayer K H. Effects of disease stage and zidovudine therapy on the detection of human immunodeficiency virus type 1 in semen. JAMA. 1992;267:2769–2774. [PubMed] [Google Scholar]
- 2.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Update: trends in AIDS incidence—United States. Morbid Mortal Weekly Rep. 1996;46:861–867. [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Coombs R W, Speck C E, Hughes J P, Lee W, Sampoleo R, Ross S O, Dragavon J, Peterson G, Hooton T M, Collier A C, Corey L, Koutsky L, Krieger J N. Association between culturable human immunodeficiency virus type 1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: evidence for compartmentalization of HIV-1 between semen and blood. J Infect Dis. 1998;177:320–330. doi: 10.1086/514213. [DOI] [PubMed] [Google Scholar]
- 6.Dyer J R, Gilliam B L, Eron J J, Grosso L, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in cell free seminal plasma: comparison of NASBA and Amplicor reverse transcription-PCR amplification and correlation with quantitative culture. J Virol Methods. 1996;60:161–170. doi: 10.1016/0166-0934(96)02063-0. [DOI] [PubMed] [Google Scholar]
- 7.Dyer J R, Pilcher C D, Shepard R, Schock J, Eron J J, Fiscus S A. Comparison of NucliSens and Roche Monitor assays for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1999;37:447–449. doi: 10.1128/jcm.37.2.447-449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta P, Mellors J, Kingsley L, Riddler S, Singh M K, Schreiber S, Cronin M, Rinaldo C R. High viral load in semen of human immunodeficiency virus type 1-infected men at all stages of disease and its reduction by therapy with protease and nonnucleoside reverse transcriptase inhibitors. J Virol. 1997;71:6271–6275. doi: 10.1128/jvi.71.8.6271-6275.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamed K A, Winters M A, Holodniy M, Katzenstein D A, Merigan T C. Detection of human immunodeficiency virus type 1 in semen: effects of disease stage and nucleoside therapy. J Infect Dis. 1993;167:798–802. doi: 10.1093/infdis/167.4.798. [DOI] [PubMed] [Google Scholar]
- 10.Krieger J N, Coombs R W, Collier A C, Ross S O, Chaloupka K, Cummings D K, Murphy V L, Corey L. Recovery of human immunodeficiency virus type 1 from semen: minimal impact of stage of infection and current antiviral chemotherapy. J Infect Dis. 1991;163:386–388. doi: 10.1093/infdis/163.2.386. [DOI] [PubMed] [Google Scholar]
- 11.Krieger J N, Coombs R W, Collier A C, Ross S O, Speck C, Corey L. Seminal shedding of human immunodeficiency virus type 1 and human cytomegalovirus: evidence for different immunologic controls. J Infect Dis. 1995;171:1018–1022. doi: 10.1093/infdis/171.4.1018. [DOI] [PubMed] [Google Scholar]
- 12.Mermin J H, Holodniy M, Katzenstein D A, Merigan T C. Detection of human immunodeficiency virus DNA and RNA in semen by the polymerase chain reaction. J Infect Dis. 1991;164:769–772. doi: 10.1093/infdis/164.4.769. [DOI] [PubMed] [Google Scholar]
- 13.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasheed S, Li Z, Xu D. Human immunodeficiency virus load. Quantitative assessment in semen from seropositive individuals and in spiked seminal plasma. J Reproduct Med. 1995;40:747–757. [PubMed] [Google Scholar]
- 15.Vahey M, Wong M T. A quantitative liquid hybridization polymerase chain reaction methodology employing storage phosphor technology. In: Diffenbach C W, Dveksler G S, editors. The Cold Spring Harbor Laboratory manual for PCR. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1995. pp. 313–318. [Google Scholar]
- 16.van Gemen B, Kievits T, Schukkink R, van Strijp D, Malek L T, Sooknanan R, Huisman H G, Lens P. Quantification of HIV-1 RNA in plasma using NASBA during HIV-1 primary infection. J Virol Methods. 1993;43:177–187. doi: 10.1016/0166-0934(93)90075-3. [DOI] [PubMed] [Google Scholar]
- 17.Van Voorhis B J, Martinez A, Mayer K, Anderson D J. Detection of human immunodeficiency virus type 1 in semen from seropositive men using culture and polymerase chain reaction deoxyribonucleic acid amplification techniques. Fertil Steril. 1991;55:588–594. [PubMed] [Google Scholar]
- 18.Vernazza P L, Eron J J, Cohen M S, van der Horst C M, Troiani L, Fiscus S A. Detection and biological characterization of infectious HIV-1 in semen of seropositive men. AIDS. 1994;8:1325–1329. doi: 10.1097/00002030-199409000-00017. [DOI] [PubMed] [Google Scholar]
- 19.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]