Abstract
Background
Examining immunity-related DNA methylation alterations in blood could help elucidate the role of the immune response in lung cancer etiology and aid in discovering factors that are key to lung cancer development and progression. In a nested, matched case–control study, we estimated methylation-derived NLR (mdNLR) and quantified DNA methylation levels at loci previously linked with circulating concentrations of C-reactive protein (CRP). We examined associations between these measures and lung cancer risk and survival.
Results
Using conditional logistic regression and further adjusting for BMI, batch effects, and a smoking-based methylation score, we observed a 47% increased risk of non-small cell lung cancer (NSCLC) for one standard deviation (SD) increase in mdNLR (n = 150 pairs; OR: 1.47, 95% CI 1.08, 2.02). Using a similar model, the estimated CRP Scores were inversely associated with risk of NSCLC (e.g., Score 1 OR: 0.57, 95% CI: 0.40, 0.81). Using Cox proportional hazards models adjusting for age, sex, smoking status, methylation-predicted pack-years, BMI, batch effect, and stage, we observed a 28% increased risk of dying from lung cancer (n = 145 deaths in 205 cases; HR: 1.28, 95% CI: 1.09, 1.50) for one SD increase in mdNLR.
Conclusions
Our study demonstrates that immunity status measured with DNA methylation markers is associated with lung cancer a decade or more prior to cancer diagnosis. A better understanding of immunity-associated methylation-based biomarkers in lung cancer development could provide insight into critical pathways.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-021-01214-2.
Keywords: Lung cancer, DNA methylation, Methylation-based inflammation measures, C-reactive protein, mdNLR
Background
Lung cancer is the leading cause of cancer death in the USA, projected to account for 21.7% of all cancer deaths in 2021 [1]. A large percentage of lung cancer patients are diagnosed at an advanced stage [2] and five-year relative survival rates for those patients are between 3 and 6% [3]. Thus, early detection remains a key strategy to improve survival. However, the currently recommended strategy for lung cancer screening—low-dose computed tomography (LDCT) for persons 50 to 80 years old with at least a 20 pack-year smoking history and currently smoke or have quit within the past 15 years—is expensive and has a high false positive rate [4, 5]. Modifying the current lung cancer screening strategy by performing risk stratification could help prioritize LDCT screening and optimize secondary prevention. We propose that immune system markers could be incorporated into such risk stratification tools to help identify persons at higher risk of lung cancer to target for screening.
While smoking is the most important risk factor for lung cancer in the population, there is growing evidence that the immune system, in response to or independent of smoking, plays an important role in lung cancer development, acting potentially through the genesis of chronic inflammation [6]. For instance, an aggregated genome-wide association studies (GWASs) analysis of lung cancer risk found a direct causal effect of BMI on small cell lung cancer and an inverse effect on lung adenocarcinoma, suggesting the complexity of the role BMI and chronic inflammation plays in lung cancer subtypes [7]. Furthermore, it is plausible that inflammatory profiles prior to lung cancer diagnosis are associated with lung cancer-specific survival. Markers of systemic inflammation, including elevated levels of C-reactive protein (CRP) and the peripheral blood neutrophil-to-lymphocyte ratio (NLR), also have been identified as robust markers of cancer-associated inflammation [8, 9]. Elevated CRP levels [8], elevated serum levels of pro-inflammatory cytokines [10–12], increased neutrophil counts and decreased lymphocyte counts [13, 14], and polymorphisms in inflammation-related genes [15–18] have been associated with increased lung cancer risk. These inflammatory measures have also been associated with poor survival of lung cancer patients in several retrospective and a few prospective studies [19–21]. In addition, both experimental and epidemiologic studies support a role for chronic inflammation as a hallmark of cancer development and progression [8, 22–25]. We posit that a better understanding of the role of inflammation in lung cancer etiology could be gained by examining DNA methylation alterations in blood that are associated with the systemic immune response.
In the current study, we first predicted peripheral blood leukocyte composition and a neutrophil to lymphocyte index using validated DNA methylation markers (mdNLR), then quantified DNA methylation levels at loci previously linked with circulating concentrations of CRP, and calculated methylation-derived immune cell ratios by using an expanded deconvolution library. We evaluated the associations of these potential markers with lung cancer risk and lung cancer-specific survival. To address this question, we used pre-diagnostic blood samples of cases and controls obtained from the CLUE I/II cohorts. Our analyses controlled for self-reported smoking and methylation-predicted cumulative smoking in order to better focus our examinations on the DNA methylation marks that are informative of the immune response profile [26].
Results
Population characteristics
Characteristics of the 208 lung cancer cases and their 208 matched controls included in this analysis are presented in Table 1. Over 99% of the majority of participants were White. The median time between blood draw and lung cancer diagnosis was 14 years. The median age at blood draw in 1989 was 59 and 57 years in cases and controls, respectively. Overall, 55% of cases and controls were women and 11% were never smokers (Table 1).
Table 1.
Baseline characteristics of lung cancer cases and matched controls nested in CLUE I/II
| Controls | Cases | |||
|---|---|---|---|---|
| All lung cancers | Non-small cell lung cancer | Small cell lung cancer | ||
| N | 208 | 208 | 150 | 29 |
| Median age (range, years) | 56 [28, 81] | 59 [30, 83] | 58 [36, 83] | 53 [30, 71] |
| Median time before diagnosis (range, years) | 14 [0, 29] | 14 [0, 28] | 12 [2, 29] | |
| Sex | ||||
| Male | 95 (45.7%) | 95 (45.7%) | 61 (40.7%) | 16 (55.2%) |
| Female | 113 (54.3%) | 113 (54.3%) | 89 (59.3%) | 13 (44.8%) |
| Race | ||||
| White | 208 (100%) | 205 (98.6%) | 149 (99.3%) | 28 (96.6%) |
| Black | 0 (0%) | 3 (1.4%) | 1 (0.7%) | 1 (3.4%) |
| Cigarette smoking status | ||||
| Never smoker | 22 (10.6%) | 22 (10.6%) | 15 (10.0%) | 1 (3.4%) |
| Ever smoker | 80 (38.5%) | 80 (38.5%) | 56 (37.3%) | 10 (34.5%) |
| Current smoker | 106 (51.0%) | 106 (51.0%) | 79 (52.7%) | 18 (62.1%) |
| Median cigarette smoking Intensity (range, cig/day) | 20 [0, 80] | 20 [0, 80] | 20 [0, 70] | 20 [0, 80] |
| Cigar or pipe smoking | ||||
| Never | 176 (84.6%) | 178 (85.6%) | 132 (88.0%) | 25 (86.2%) |
| Ever | 28 (13.5%) | 26 (12.5%) | 15 (10.0%) | 4 (13.8%) |
| Current | 4 (1.9%) | 4 (1.9%) | 3 (2.0%) | 0 (0%) |
| Median BMI, kg/m2 (range) | 25.8 [18.0, 40.9] | 25.6 [15.5, 42.6] | 25.4 [15.5, 42.6] | 25.6 [18.3, 34.7] |
| mdNLR mean (SD) | 1.74 (0.976) | 1.86 (1.32) | 1.85 (1.19) | 1.65 (1.07) |
| mdNLR median (range) | 1.58 [0.13, 6.42] | 1.59 [0.28, 11.90] | 1.56 [0.28, 9.45] | 1.44 [0.39, 4.81] |
| CRP score 1 mean (SD) | 0.02 (1.01) | − 0.02 (0.99) | − 0.04 (0.97) | 0.47 (1.08) |
| CRP score 1 median (range) | 0.06 [− 2.42, 2.99] | − 0.03 [− 3.26, 2.94] | − 0.03 [− 3.26, 2.94] | 0.41 [− 1.93, 2.16] |
| CRP score 2 mean (SD) | 0.04 (1.03) | − 0.04 (0.98) | − 0.06 (0.95) | 0.36 (1.17) |
| CRP score 2 median (range) | 0.01 [− 3.35, 3.93] | − 0.10 [− 2.68, 3.10] | − 0.11 [− 2.68, 3.10] | 0.24 [− 2.34, 3.10] |
| CRP score 3 mean (SD) | − 0.06 (0.98) | 0.06 (1.02) | 0.09 (1.03) | 0.32 (0.97) |
| CRP score 3 median (range) | − 0.09 [− 2.43, 2.51] | 0.08 [− 2.34, 2.95] | 0.10 [− 2.34, 2.95] | 0.38 [− 2.19, 1.97] |
Methylation-derived mdNLR index, leukocyte proportions, and lung cancer risk
We observed a 47% increased risk of non-small cell lung cancer (NSCLC) for one standard deviation increase in mdNLR (n = 150 pairs; OR: 1.47 [1.08, 2.02]). However, higher mdNLR values were not statistically associated with overall risk of lung cancer in our study. This association was comparable for NSCLC cases diagnosed within 10 years and beyond 10 years after blood draw. No stable associations could be estimated for small cell lung cancer (SCLC). After multiple comparison adjustments, monocyte/lymphocyte ratio showed a borderline significant 65% increased risk of NSCLC for each standard deviation increase (n = 150 pairs; OR: 1.65, adjusted CI: [0.99, 2.76]). In addition, immune cell ratios for CD4/CD8, NLR, B cell/lymphocyte, T cell/lymphocyte, Neu + Mono/lymphocyte, Eos/lymphocyte, CD4nv/lymphocyte, B cell/CD8, CD8/Treg, Bnv/Bmem, CD4nv/CD4mem, CD8nv/CD8mem, and Treg > 0 vs. Treg = 0 were not statistically significantly associated with lung cancer risk overall or by histologic types (Table 2).
Table 2.
Association between methylation-predicted immune cell profiles and risk of total lung cancer and NSCLC risk, overall and stratified by time to diagnosis, case–control study nested in the CLUE I/II cohort
| All lung cancersa OR (95% CI) |
Non-small cell lung cancerb OR (95% CI) |
|
|---|---|---|
| mdNLRc | 1.11 (0.89, 1.38) | 1.47 (1.08, 2.02) |
| Time to diagnosis ≤ 10 years | 0.97 (0.69, 1.35) | 1.41 (0.90, 2.20) |
| Time to diagnosis > 10 years | 1.22 (0.91, 1.64) | 1.57 (0.98, 2.49) |
| CD4/CD8 Ratioc | 1.17 (0.80, 1.72) | 1.16 (0.74, 1.82) |
| Time to diagnosis ≤ 10 years | 0.94 (0.56, 1.57) | 0.79 (0.38, 1.66) |
| Time to diagnosis > 10 years | 1.47 (0.82, 2.62) | 1.72 (0.77, 3.85) |
| B cell/lymphocyte ratioc | 1.07 (0.73, 1.57) | 0.94 (0.58, 1.52) |
| Time to diagnosis ≤ 10 years | 1.16 (0.63, 2.14) | 0.91 (0.42, 1.97) |
| Time to diagnosis > 10 years | 0.98 (0.60, 1.59) | 0.93 (0.50, 1.71) |
| T cell/lymphocyte ratioc | 1.06 (0.72, 1.56) | 0.94 (0.60, 1.48) |
| Time to diagnosis ≤ 10 years | 1.11 (0.62, 1.98) | 1.03 (0.49, 2.16) |
| Time to diagnosis > 10 years | 1.05 (0.63, 1.76) | 0.88 (0.48, 1.62) |
| Monocyte/lymphocyte ratioc | 1.15 (0.78, 1.69) | 1.65 (0.99, 2.76) |
| Time to diagnosis ≤ 10 years | 1.18 (0.66, 2.11) | 1.62 (0.75, 3.51) |
| Time to diagnosis > 10 years | 1.16 (0.69, 1.94) | 1.81 (0.86, 3.80) |
| (Neutrophil + monocyte)/lymphocyte ratioc | 1.13 (0.79, 1.61) | 1.46 (0.87, 2.44) |
| Time to diagnosis ≤ 10 years | 0.96 (0.59, 1.56) | 1.32 (0.67, 2.60) |
| Time to diagnosis > 10 years | 1.26 (0.78, 2.04) | 1.62 (0.80, 3.29) |
| Eosinophil/lymphocyte ratioc | 0.94 (0.64, 1.38) | 0.89 (0.53, 1.49) |
| Time to diagnosis ≤ 10 years | 0.81 (0.44, 1.49) | 1.21 (0.48, 3.08) |
| Time to diagnosis > 10 years | 1.01 (0.64, 1.59) | 0.73 (0.36, 1.48) |
| CD4 naïve/lymphocyte ratioc | 1.22 (0.83, 1.80) | 1.36 (0.84, 2.20) |
| Time to diagnosis ≤ 10 years | 1.47 (0.72, 2.99) | 1.70 (0.74, 3.93) |
| Time to diagnosis > 10 years | 1.11 (0.68, 1.80) | 1.19 (0.67, 2.12) |
| B cell/CD8 ratioc | 0.97 (0.68, 1.38) | 1.00 (0.66, 1.52) |
| Time to diagnosis ≤ 10 years | 1.06 (0.61, 1.83) | 1.11 (0.58, 2.11) |
| Time to diagnosis > 10 years | 0.85 (0.45, 1.62) | 0.90 (0.44, 1.83) |
| CD8/treg ratioc | 0.81 (0.55, 1.19) | 0.73 (0.45, 1.18) |
| Time to diagnosis ≤ 10 years | 0.86 (0.47, 1.59) | 0.72 (0.33, 1.56) |
| Time to diagnosis > 10 years | 0.78 (0.45, 1.35) | 0.68 (0.33, 1.38) |
| Treg > 0 versus Treg = 0c | 1.05 (0.74, 1.50) | 1.07 (0.70, 1.63) |
| Time to diagnosis ≤ 10 years | 0.95 (0.53, 1.70) | 1.12 (0.55, 2.27) |
| Time to diagnosis > 10 years | 1.10 (0.70, 1.73) | 1.06 (0.61, 1.83) |
| B cell naïve/B cell memory ratioc | 0.85 (0.58, 1.25) | 0.87 (0.57, 1.32) |
| Time to diagnosis ≤ 10 years | 0.65 (0.24, 1.76) | 0.65 (0.21, 2.01) |
| Time to diagnosis > 10 years | 0.93 (0.56, 1.56) | 0.96 (0.56, 1.66) |
| CD4 naïve/CD4 memory ratioc | 1.04 (0.73, 1.48) | 1.59 (0.81, 3.13) |
| Time to diagnosis ≤ 10 years | 0.98 (0.59, 1.64) | 2.67 (0.65, 11.01) |
| Time to diagnosis > 10 years | 1.18 (0.60, 2.32) | 1.34 (0.58, 3.10) |
| CD8 naïve/CD8 memory ratioc | 0.99 (0.72, 1.37) | 1.01 (0.53, 1.92) |
| Time to diagnosis ≤ 10 years | 0.66 (0.15, 2.81) | 0.74 (0.17, 3.15) |
| Time to diagnosis > 10 years | 1.04 (0.71, 1.53) | 1.73 (0.36, 8.38) |
Bold OR and CI values indicate statistical significance. Underline OR and CI values indicate borderline statistical significance after multiple comparison adjustment
a208 cases and 208 controls matched on age at blood draw, sex, smoking status, and model further adjusting for BMI, surrogate variables for batch effects, and a methylation-predicted pack-years smoked
b150 cases and 150 controls matched on age at blood draw, sex, smoking status, and model further adjusting for BMI, surrogate variables for batch effects, and a methylation-predicted pack-years smoked
cOR results reported per 1 unit SD increase (mdNLR SD: 1.16; CD4/CD8 ratio SD: 2.07; BL ratio SD: 0.0526; TL ratio SD: 0.0802; ML ratio SD: 0.118; NM/L ratio SD: 1.22; EL ratio SD: 0.0722; CD4nv/L ratio SD: 0.0915; B/CD8 ratio SD: 3.07; CD8/Treg ratio SD: 115; Bnv/Bmem ratio SD: 109; CD4nv/CD4mem ratio SD: 0.429; CD8nv/CD8mem ratio SD: 2.05) and OR results reported for Treg > 0 versus 0; Bonferroni-adjusted CI reported for all except mdNLR (a prior hypothesis)
Methylation-derived CRP scores and lung cancer risk
CRP Score 1 was built using 54 CpG sites that were previously associated with inflammatory markers, while CRP Score 2 and 3 were each built with a subset of these 54 CpGs that were putative cell-specific or cell type invariant, respectively. Using data from a previously published pancreatic cancer dataset [27], all three scores were moderately correlated with log CRP and log IL-6 levels (Table 3). In this nested case–control study, we found all three CRP Scores inversely associated with risk of NSCLC after additionally adjusting for methylation-predicted pack-years (n = 150 pairs; Score 1 OR: 0.57 [0.40, 0.81]; Score 2 OR: 0.62 [0.45, 0.84]; Score 3 OR: 0.65 [0.44, 0.95]). We also found statistically significant inverse association between CRP Score 1 and risk of NSCLC among cases diagnosed within 10 years and beyond 10 years, and between CRP Score 2 for NSCLC cases diagnosed within 10 years of blood draw (Table 4). CRP Scores 1, 2, and 3 were not associated with lung cancer risk when taking into account the matching factors and only adjusting for BMI and four surrogate variables for batch effects (n = 208 pairs; Score 1 OR: 0.96 [0.77, 1.21]; Score 2 OR: 0.89 [0.71, 1.11]; Score 3 OR: 1.11 [0.89, 1.40]). However, when additionally adjusting for methylation-predicted pack-years, inverse associations with total lung cancer risk were observed for Score 1 (OR: 0.76 [0.59, 0.99]) and Score 2 (OR: 0.77 [0.61, 0.98]). We also observed a 33% decreased risk of lung cancer for one standard deviation increase in CRP Score 1 (OR: 0.67 [0.47, 0.97]) among those with time to diagnosis over 10 years.
Table 3.
Correlations between methylation-based CRP scores and circulating log-CRP level, log-IL6 level, peripheral blood leukocyte types, BMI, and smoking score residual among controls only
| Score 1b | Score 2c | Score 3d | |
|---|---|---|---|
| Spearman (p value) | Spearman (p value) | Spearman (p value) | |
| Pancreatic cancer study (controls only) | |||
| Log-CRP levela | 0.284 (4.27e−06) | 0.247 (6.96e−05) | 0.297 (1.56e−06) |
| Log-IL6 levela | 0.213 (7.53e−04) | 0.158 (1.32e−02) | 0.203 (1.33e−03) |
| CLUE I/II (controls only) | |||
| CD4T | 0.425 (1.65e−10) | 0.359 (1.00e−07) | 0.212 (2.13e−03) |
| CD8T | 0.504 (8.78e−15) | 0.461 (2.34e−12) | 0.390 (5.74e−09) |
| NK | 0.062 (3.70e−01) | 0.175 (1.17e−02) | 0.026 (7.09e−01) |
| B cell | 0.103 (1.38e−01) | 0.069 (3.23e−01) | − 0.027 (6.99e−01) |
| Neutrophils | − 0.467 (1.19e−12) | − 0.422 (2.24e−10) | − 0.264 (1.17e−04) |
| Monocytes | − 0.292 (1.92e−05) | − 0.330 (1.17e−06) | − 0.288 (2.38e−05) |
| mdNLR | − 0.488 (7.66e−14) | − 0.449 (1.01e−11) | − 0.281 (3.96e−05) |
| BMI | 0.025 (7.22e−01) | 0.070 (3.12e−01) | − 0.063 (3.68e−01) |
| Methylation-predicted pack-years residuale | 0.475 (4.46e−13) | 0.375 (2.43e−08) | 0.632 (1.42e−24) |
aCorrelations with log-CRP level and log-IL6 level were tested with a pancreatic cancer dataset[43]
bCpG Score 1 is built using 54 CpG sites
cCpG Score 2 is built using the top 10 highly cell-specific CpG sites
dCpG Score 3 is built using the 10 modestly cell-specific CpG sites
eWe calculated a pack-years methylation score to represent pack-years smoked associated methylation alterations. This score correlates with gene expression changes that are affected by smoking
Table 4.
Association between methylation-based CRP scores and risk of total lung cancer and NSCLC risk, overall and stratified by time to diagnosis, CLUE I/II cohort
| All lung cancersa OR (95% CI) |
Non-small cell lung cancerb OR (95% CI) |
|
|---|---|---|
| CRP Score 1c | 0.76 (0.59, 0.99) | 0.57 (0.40, 0.81) |
| Time to diagnosis ≤ 10 years | 0.84 (0.56, 1.26) | 0.57 (0.33, 0.97) |
| Time to diagnosis > 10 years | 0.67 (0.47, 0.97) | 0.53 (0.32, 0.88) |
| CRP Score 2d | 0.77 (0.61, 0.98) | 0.62 (0.45, 0.84) |
| Time to diagnosis ≤ 10 years | 0.79 (0.54, 1.17) | 0.49 (0.28, 0.85) |
| Time to diagnosis > 10 years | 0.73 (0.53, 1.02) | 0.68 (0.46, 1.00) |
| CRP Score 3e | 0.79 (0.58, 1.07) | 0.65 (0.44, 0.95) |
| Time to diagnosis ≤ 10 years | 0.87 (0.53, 1.42) | 0.64 (0.35, 1.18) |
| Time to diagnosis > 10 years | 0.73 (0.49, 1.09) | 0.62 (0.36, 1.05) |
Bold OR and CI values indicate statistical significance
a208 cases and 208 controls matched for age at blood draw, sex, smoking status, and model further adjusting for BMI, surrogate variables for batch effects, and a methylation-predicted pack-years smoked
b150 cases and 150 controls matched for age at blood draw, sex, smoking status, and model further adjusting for BMI, surrogate variables for batch effects, and a methylation-predicted pack-years smoked. The matched pairs were kept within each stratum
cCpG Score 1 is built using 54 CpG sites and OR results reported per 1 unit of SD of CpG Score 1 (SD 1)
dCpG Score 2 is built using the top 10 highly cell-specific CpG sites and OR results reported per 1 unit of SD of CpG Score 2 (SD 1)
eCpG Score 3 is built using the 10 modestly cell-specific CpG sites and OR results reported per 1 unit of SD of CpG Score 3 (SD 1)
Survival analysis
We examined whether the mdNLR, methylation-derived immune cell ratios, and CRP Scores were associated with risk of dying of lung cancer among lung cancer cases (Table 5, Fig. 1).
Table 5.
Association between immune cell ratios and methylation-based CRP scores and lung cancer-specific mortality among lung cancer cases, CLUE I/II cohort
| All lung cancersa HR (95% CI) |
Non-small cell lung cancerb HR (95% CI) |
|
|---|---|---|
| mdNLRc | 1.28 (1.09, 1.50) | 1.47 (1.20, 1.81) |
| Time to diagnosis ≤ 10 years | 1.34 (1.01, 1.76) | 1.73 (1.19, 2.51) |
| Time to diagnosis > 10 years | 1.20 (0.98, 1.48) | 1.39 (1.05, 1.85) |
| CD4/CD8 ratioc | 1.02 (0.76, 1.36) | 1.07 (0.78, 1.48) |
| Time to diagnosis ≤ 10 years | 1.28 (0.79, 2.07) | 1.29 (0.70, 2.38) |
| Time to diagnosis > 10 years | 0.96 (0.65, 1.41) | 1.03 (0.66, 1.62) |
| B cell/lymphocyte ratioc | 0.97 (0.73, 1.30) | 1.06 (0.74, 1.51) |
| Time to diagnosis ≤ 10 years | 0.87 (0.50, 1.50) | 0.99 (0.52, 1.89) |
| Time to diagnosis > 10 years | 1.11 (0.75, 1.63) | 1.18 (0.75, 1.85) |
| T cell/lymphocyte ratioc | 1.01 (0.78, 1.31) | 0.95 (0.69, 1.31) |
| Time to diagnosis ≤ 10 years | 1.01 (0.62, 1.64) | 0.94 (0.53, 1.68) |
| Time to diagnosis > 10 years | 1.05 (0.69, 1.60) | 0.91 (0.54, 1.52) |
| Monocyte/Lymphocyte Ratioc | 1.16 (0.87, 1.55) | 1.21 (0.85, 1.72) |
| Time to diagnosis ≤ 10 years | 1.45 (0.87, 2.43) | 1.52 (0.80, 2.89) |
| Time to diagnosis > 10 years | 1.25 (0.80, 1.96) | 1.54 (0.84, 2.84) |
| (Neutrophil + monocyte)/lymphocyte ratioc | 1.29 (1.00, 1.67) | 1.48 (1.04, 2.11) |
| Time to diagnosis ≤ 10 years | 1.36 (0.87, 2.13) | 1.76 (0.95, 3.24) |
| Time to diagnosis > 10 years | 1.22 (0.86, 1.74) | 1.41 (0.87, 2.29) |
| Eosinophil/lymphocyte ratioc | 1.01 (0.76, 1.35) | 0.86 (0.60, 1.23) |
| Time to diagnosis ≤ 10 years | 1.55 (0.93, 2.59) | 1.40 (0.74, 2.67) |
| Time to diagnosis > 10 years | 0.98 (0.67, 1.44) | 0.77 (0.49, 1.21) |
| CD4 naïve/lymphocyte ratioc | 0.92 (0.67, 1.27) | 0.95 (0.65, 1.40) |
| Time to diagnosis ≤ 10 years | 0.92 (0.53, 1.59) | 0.73 (0.35, 1.53) |
| Time to diagnosis > 10 years | 0.91 (0.60, 1.38) | 0.99 (0.61, 1.60) |
| B cell/CD8 ratioc | 1.03 (0.82, 1.29) | 1.07 (0.83, 1.38) |
| Time to diagnosis ≤ 10 years | 1.01 (0.69, 1.49) | 1.04 (0.64, 1.69) |
| Time to diagnosis > 10 years | 1.05 (0.71, 1.55) | 1.20 (0.74, 1.95) |
| CD8/treg ratioc | 0.92 (0.67, 1.27) | 0.92 (0.63, 1.35) |
| Time to diagnosis ≤ 10 years | 0.73 (0.42, 1.26) | 0.63 (0.30, 1.32) |
| Time to diagnosis > 10 years | 1.03 (0.66, 1.62) | 1.09 (0.63, 1.88) |
| Treg > 0 versus treg = 0c | 0.96 (0.72, 1.28) | 0.91 (0.64, 1.30) |
| Time to diagnosis ≤ 10 years | 1.27 (0.76, 2.13) | 1.37 (0.70, 2.69) |
| Time to diagnosis > 10 years | 0.85 (0.58, 1.25) | 0.75 (0.45, 1.26) |
| B cell naïve/B cell memory Ratioc | 1.09 (0.84, 1.41) | 1.23 (0.89, 1.70) |
| Time to diagnosis ≤ 10 years | 1.31 (0.86, 1.99) | 1.66 (0.74, 3.71) |
| Time to diagnosis > 10 years | 1.03 (0.75, 1.42) | 1.19 (0.73, 1.93) |
| CD4 naïve/CD4 memory ratioc | 0.97 (0.70, 1.34) | 1.03 (0.70, 1.52) |
| Time to diagnosis ≤ 10 years | 0.91 (0.53, 1.57) | 0.71 (0.32, 1.59) |
| Time to diagnosis > 10 years | 1.00 (0.64, 1.57) | 1.17 (0.70, 1.96) |
| CD8 naïve/CD8 memory ratioc | 0.96 (0.70, 1.32) | 1.10 (0.77, 1.57) |
| Time to diagnosis ≤ 10 years | 1.09 (0.63, 1.88) | 1.19 (0.59, 2.42) |
| Time to diagnosis > 10 years | 0.92 (0.61, 1.40) | 1.05 (0.67, 1.65) |
| CRP Score 1cd | 0.92 (0.67, 1.26) | 0.88 (0.60, 1.30) |
| Time to diagnosis ≤ 10 years | 1.00 (0.57, 1.75) | 1.22 (0.67, 2.22) |
| Time to diagnosis > 10 years | 1.04 (0.67, 1.63) | 0.82 (0.46, 1.46) |
| CRP Score 2cd | 0.86 (0.65, 1.13) | 0.98 (0.69, 1.41) |
| Time to diagnosis ≤ 10 years | 0.76 (0.47, 1.23) | 1.11 (0.61, 2.03) |
| Time to diagnosis > 10 years | 1.10 (0.73, 1.66) | 0.93 (0.55, 1.56) |
| CRP Score 3cd | 1.05 (0.76, 1.47) | 1.07 (0.73, 1.59) |
| Time to diagnosis ≤ 10 years | 1.01 (0.56, 1.81) | 1.01 (0.46, 2.22) |
| Time to diagnosis > 10 years | 1.16 (0.74, 1.83) | 1.22 (0.69, 2.16) |
Bold HR and CI values indicate statistical significance. Underline HR and CI values indicate borderline statistical significance after multiple comparison adjustment
a205 cases (3 cases with person-year = 0 or > 25 years were removed from the analytical dataset) Model adjusted for age at blood draw, sex, smoking status, BMI, surrogate variables for batch effects, and a pack-year-based smoking methylation score. The results for Inflammation Scores additional adjust for cell proportions. BMI was removed from model when stratified by BMI. Group person-year: all lung cancer cases = 421.3; cases with time to diagnosis ≤ 10 years = 104.3; cases with time to diagnosis > 10 years = 316.9; cases with BMI < 25 kg/m2 = 184.3; cases with BMI ≥ 25 kg/m2 = 236.9
b149 cases (1 case with person-year = >25 years were removed from the analytical dataset) Model adjusted for age at blood draw, sex, smoking status, BMI, surrogate variables for batch effects, and methylation-predicted pack-years smoked. The results for Inflammation Scores additional adjust for cell proportions. BMI was removed from model when stratified by BMI. Group person-year for all non-small cell lung cancer (NSCLC) cases = 337.6; NSCLC cases with time to diagnosis ≤ 10 years = 84.4; NSCLC cases with time to diagnosis > 10 years = 253.2; NSCLC cases with BMI < 25 kg/m2 = 141.4; NSCLC cases with BMI ≥ 25 kg/m2 = 196.2
cHR results reported per 1 unit of SD increase among all cases (mdNLR SD: 1.86; CD4/CD8 ratio SD: 2.14; BL ratio SD: 0.0534; TL ratio SD: 0.0844; ML ratio SD: 0.125; NM/L ratio SD: 1.38; EL ratio SD: 0.079; CD4nv/L ratio SD: 0.0920; B/CD8 ratio SD: 2.21; CD8/Treg ratio SD: 112; Bnv/Bmem ratio SD: 81.3; CD4nv/CD4mem ratio SD: 0.335; CD8nv/CD8mem ratio SD: 1.64; CpG Score 1 SD: 0.979; CpG Score 2 SD: 0.945; CpG Score 3 SD: 1.02) and HR results reported for Treg > 0 versus Treg = 0 among all cases; Bonferroni-adjusted CI reported for all except mdNLR and CRP Scores 1–3 (a prior hypotheses)
dCpG Scores 1, 2, and 3 are built using 54 CpG sites, the top 10 highly cell-specific CpG sites, and the 10 modestly cell-specific CpG sites, respectively
Fig. 1.
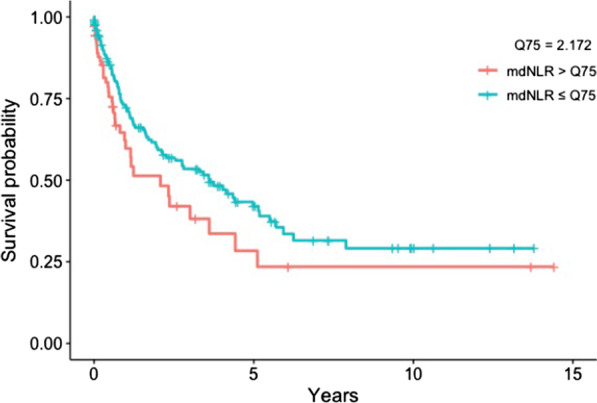
Survival curves for lung cancer-specific mortality among lung cancer cases in the mdNLR high and low groups (> or ≤ 75% quartiles). Plot adjusted for age, sex, smoking status, methylation-predicted pack-years smoked, BMI, stage, and batch effects
We observed a 47% increased risk of dying for one standard deviation of mdNLR for NSCLC cases (n = 149 cases; HR: 1.47 [1.20, 1.81]). Among the NSCLC cases whose mdNLR was from < = 10 years before their diagnosis, we found a 73% increased risk of dying for a one standard deviation increase in mdNLR (HR: 1.73 [1.19, 2.51]). In comparison, the risk of dying for a one standard deviation increase in mdNLR was lower among the NSCLC cases whose mdNLR was from 10 to 25 years prior to diagnosis (HR: 1.39 [1.05, 1.85]). Lastly, we observed a 28% increased risk of dying from lung cancer for one standard deviation increase in mdNLR (n = 205 cases deleted 3 cases with person-year = 0 or > 25 years; HR: 1.28 [1.09, 1.50]).
Immune cell ratios for CD4/CD8, NLR, B cell/lymphocyte, T cell/lymphocyte, Mono/lymphocyte, Eos/lymphocyte, CD4nv/lymphocyte, B cell/CD8, CD8/Treg, Bnv/Bmem, CD4nv/CD4mem, CD8nv/CD8mem, and Treg (> 0 vs = 0) were not associated with lung cancer-specific death, except for a 48% increased risk for one standard deviation increase in Neu + Mono/lymphocyte ratio among the NSCLC cases (HR: 1.48 [1.04, 2.11]) and a borderline significant 29% increased risk of dying from lung cancer (HR: 1.29, adjusted CI: [1.00, 1.67]) for one standard deviation increase in Neu + Mono/lymphocyte ratio after multiple comparison adjustments. Furthermore, the three CRP Scores were not associated with lung cancer-specific death.
Discussion
Our study prospectively assessed predicted immune cell profiles using DNA methylation markers and examined associations between previously identified DNA methylation markers of inflammation and lung cancer risk and survival. Using pre-diagnostic blood samples of lung cancer cases and controls who participated in the CLUE I/II cohorts [23], pre-diagnosis mdNLR was associated with increased risk of NSCLC, and among cases, with total lung cancer and NSCLC lung cancer-specific death. In addition, we built a series of methylation-derived CRP scores to capture individual systemic inflammatory profiles years before lung cancer diagnosis; these scores were inversely associated with risk of lung cancer, especially for NSCLC after adjusting for methylation-predicted pack-years smoked, but not with lung cancer-specific mortality.
Studies on NLR (calculated from measured WBC differentials) and lung cancer risk and survival typically measure pre-treatment NLR at diagnosis or up to 30 days prior to treatment [28–30]. Unlike prior studies, we were able to assess individual systemic inflammation profiles many years prior to diagnosis by using methylation markers of inflammation. Our study is not directly comparable to prior studies since we measured mdNLR using blood samples from subjects with a median of 14 years prior to lung cancer diagnosis. In addition, most cases in our study were diagnosed before the widespread use of immunotherapy. To our knowledge, only one other cohort, the multicenter β-Carotene and Retinol Efficacy Trial (CARET), examined pre-diagnosis mdNLR and lung cancer risk and survival using blood drawn years prior to diagnosis (median 4.7 years) [31, 32]. CARET, a study of heavy smokers, reported a 21% increased risk of lung cancer per one unit increase in mdNLR (OR: 1.21 [1.01, 1.45]), a 30% increased risk of NSCLC for one unit increase in mdNLR (OR: 1.30 [1.03, 1.63], and no association between higher pre-diagnosis mdNLR and risk of developing SCLC (OR: 1.06 [0.77, 1.47]) [31]. Like in CARET, in CLUE I/II we observed a 47% increased risk of NSCLC for a one standard deviation increase in mdNLR (n = 150 pairs; OR: 1.47 [1.08, 2.02]), but in contrast to CARET, we found no statistically significant association for overall lung cancer risk.
CARET researchers recently reported that pre-diagnosis mdNLR was positively associated with increased mortality for SCLC cases, but not for other case types [32]. In comparison, we observed a positive association between pre-diagnosis mdNLR and lung cancer-specific and NSCLC-specific mortality. In the case of SCLC, the number of cases was too limited for us to estimate stable associations (N = 29). Taken together, the CLUE and CARET results suggest that a systemic inflammatory profile marked by elevated NLR could indicate a lesser ability to mount a robust immune response to a developing lung cancer and/or a more favorable environment for cancer progression. Differences in findings between the two studies could stem from differences in study populations. The CARET cohort is exclusively heavy smokers, including a subgroup exposed to asbestos. In comparison, our analysis in the CLUE I/II cohorts included never, ever, and current smokers. Furthermore, our study population had a lower mdNLR in the lung cancer cases (mean 1.86 and SD 1.32) than in CARET (mdNLR mean 2.18 and SD 1.46).
Using a newly expanded deconvolution library, we were able to parse apart the granulocyte subtypes (neutrophils, eosinophils, and basophils) and investigate the balance between naïve and memory cell compartments for lung cancer. Previous research has identified the monocyte/lymphocyte (or lymphocyte/monocyte) ratio as an independent prognostic factor in NSCLC, demonstrating significant association with overall survival in patients with NSCLC [33–35]. In comparison, our exploratory analyses of immune cell ratios suggest that one standard deviation increase in the monocyte/lymphocyte ratio could potentially indicate increased risk of NSCLC after additionally adjusting for methylation-predicted pack-years. In addition, we found an increased risk of dying from lung cancer associated with an increase in Neu + Mono/lymphocyte ratio among the NSCLC cases after multiple comparison adjustments.
We also investigated three CRP Scores that we built from 54 CpG sites that had been strongly associated with CRP in previous studies. We found these methylation-predicted CRP Scores to be moderately correlated with log-CRP and log-IL6 in the controls of a previously published pancreatic cancer dataset [27]. CRP is a systemic marker of chronic inflammation and has been reported as a risk factor for cancer development [36]. Previous studies of pre-diagnostic circulating CRP concentration and lung cancer risk (7 cohorts [10, 11, 19, 37–39] and 3 nested case–control studies [8, 12, 40]) have consistently found a moderate positive association between pre-diagnostic CRP concentrations and lung cancer risk. In our study, CRP Scores were not associated with lung risk when taking into account the matching factors, BMI, and batch effects. However, we observed an inverse association when additionally adjusting for methylation-predicted pack-year. Our results suggest that when strict control of smoking is applied, our CRP Score is likely capturing the unique individual immune response that is not driven by smoking.
Furthermore, these results provide preliminary evidence supporting the hypothesis that systemic inflammation not driven by smoking could have a protective effect on individuals. While smoking is by far the most important risk factor for lung cancer, our DNA methylation-based CRP Scores provide the opportunity to examine inflammatory measures not related to smoking that could play a role in modulating cancer risk years prior to diagnosis. Lastly, our experience with the CRP Scores suggests that measuring methylation-derived inflammatory responses using pre-diagnostic samples provides the opportunity to capture informative individual systemic inflammatory profiles years prior to diagnosis, potentially shedding light on risk factors key to lung cancer development and progression, e.g., underlying genetics, exposure to environmental risk factors, and behavior risk factors.
Like other observational studies, our study included a limited number of NSCLC and SCLC cases. The relatively small sample size of SCLC cases (N = 29) impacted our ability to observe associations for this subtype (SCLC comprises about 15% of lung cancer cases in the USA). In our survival analysis, we adjusted for stage and restricted our analysis to samples whose time between blood draw and date of lung cancer diagnosis was less than 25 years; however, our survival analysis did not have access to post-diagnosis smoking status information. Our study is also limited by a lack of replication dataset and reduced generalizability. (Study population is mainly White and with very few cases in never smokers.) The CRP Scores we built should be investigated in other populations to ensure that what we observed did not arise due to chance.
Conclusions
Our study suggests that elevated pre-diagnosis mdNLR and a lower non-smoking-related systemic inflammatory profile before diagnosis are associated with higher cancer risk and poorer lung cancer-specific survival. These relationships were especially evident for NSCLC. As the most common subtype of lung cancer, most NSCLC cases are diagnosed with locally advanced or metastatic disease. Our prospective results support future evaluation of whether DNA methylation-based inflammatory measures could enhance lung cancer risk stratification to improve targeted lung cancer screening.
Methods
Study Population
This nested case–control study selected cases and controls from individuals who participated and provided blood in both CLUE I and CLUE II [26]. The CLUE I cohort was developed to identify serologic precursors of cancer and was conducted in Washington County, Maryland, in the fall of 1974. A blood sample was collected from 25,620 volunteers at the time of participation [41, 42]. The CLUE II cohort was conducted from May through October 1989. During this time, 32,894 participants donated a blood sample which was collected in tubes containing heparin and kept chilled until centrifuged, aliquoted into plasma, erythrocytes, and buffy coat, and frozen at 70 °C [43]. In CLUE II, the baseline for this study, health information was collected at the time of blood draw, including attained education, cigarette smoking status, cigarette smoking dose, cigar/pipe smoking status, and self-reported weight and height.
Incident lung cancer cases were ascertained from linkage to the Washington County cancer registry (before 1992 to the present) and the Maryland Cancer Registry (since 1992 when it began to the present). We ascertained 241 incident lung cancer cases who participated in CLUE I and were diagnosed after the day of blood draw in CLUE II through January 2018. Cases were characterized with respect to histology. We used incidence density sampling to select one control matched to each case on age, sex, smoking status and intensity (cig/day), and cigar/pipe smoking status. Death from lung cancer as the underlying cause was obtained from death certificates. The Institutional Review Board at the Johns Hopkins Bloomberg School of Public Health and the Tufts University Health Sciences Campus Institutional Review Board approved this study.
DNA methylation measurements
Extracted DNA was bisulfite-treated using the EZ DNA Methylation Kit (Zymo), and DNA methylation was measured with the 850 K Illumina Infinium MethylationEPIC BeadChip Arrays (Illumina, Inc., CA, USA). All samples and all array experiments were performed blinded to case–control status. Details on DNA methylation measurements, data preprocessing processing, and quality control assessment/screening are provided in the Additional file 1. The 850 K methylation microarray has been validated from a biological and technical standpoint. Reproducibility of results from 850 K Illumina array has been previously shown to be very high (r = 0.997) [44]. DNA volume and quality were sufficient for 208 of the cases and 222 controls totaling 208 matched pairs.
Estimation of peripheral blood leukocyte composition
Peripheral blood leukocyte subtypes proportions, including myeloid lineage sub-types [neutrophils (Neu), eosinophils (Eos), basophils (Bas), and monocytes (Mono)] and lymphoid lineage subtypes [B lymphocytes naïve (Bnv), B lymphocytes memory (Bmem), T helper lymphocytes naïve (CD4nv), T helper lymphocytes memory (CD4mem), T regulatory cells (Treg), T cytotoxic lymphocytes naïve (CD8nv), T cytotoxic lymphocytes memory (CD8mem), and natural killer lymphocytes (NK)], were estimated using a newly expanded reference-based deconvolution library EPIC IDOL-Ext [45]. This library used the IDOL methodology [46] to optimize the currently available six-cell reference library [47] in order to deconvolve the proportions of 12 leukocyte subtypes in peripheral blood. This EPIC IDOL-Ext library (Bioconductor package FlowSorted.BloodExtended.EPIC) was validated using flow cytometry gold standard data and substantiated by including publicly available data from > 100,000 samples [45].
Methylation-Derived Neutrophil Lymphocyte Ratio (mdNLR)
The peripheral blood neutrophil-to-lymphocyte ratio (NLR) is a cytological marker of both inflammation and poor outcomes in cancer patients [48–52]. We used a DNA methylation-derived NLR (mdNLR) index to predict the common clinical NLR parameter using a previously described approach [9]. This index is based on normal isolated leukocyte reference DNA methylation libraries and established reference-based cell mixture deconvolution algorithms [9, 53].
Inflammation-associated CpG score
We used 54 CpG sites that have been strongly associated with C-reactive protein (CRP) [54, 55] to build three CRP Scores. We selected these 54 CpGs (remaining 4 were not on the 850 K array that we used) from the 58 CpGs identified by Ligthart and colleagues [54] for their association with serum CRP level (listed in Table 3) using 450 K DNA methylation data. Forty-five of these 58 CpG sites were validated to have the same direction of protein–methylation associations by Myte et al. [55]. These CpGs, while identified based on their CRP association, have also been shown to be associated with other inflammatory mediators [54–56]. To compute CRP Score 1, we multiplied the beta value at each selected CpG site with the effect size estimates reported by Ligthart et al. These estimated beta coefficients represented the change in DNA methylation per one unit increase in log CRP. In the CRP Score 1 formula, we weighted the beta coefficients estimated by Ligthart et al. with their corresponding standard errors.
Bij is the beta value for the ith participant at the jth CpG site. ∆j is the beta coefficients reported by Ligthart et al. for the jth CpG site. SEj is the SE reported by Ligthart et al. for the jth CpG site.
Since most of the estimated beta coefficients are negative, CRP Score 1 ranged between − 0.059 and -0.026 in these participants. A score closer to zero indicated higher CRP levels. Based on CRP Score 1, we computed two additional CRP Scores, one cell (leukocyte)-type invariant (CRP Score 2) and one cell-specific (CRP Score 3). Among the 54 inflammation (CRP)-associated CpGs, we identified putative cell-type invariant and cell-specific CpGs by conducting ANOVA using the dataset described in Salas and Koestler et al. [47] and publicly available on the Gene Expression Omnibus (GSE110555). The dataset used for this ANOVA consisted of EPIC methylation data profiled in purified leukocyte cell population isolated from different healthy adults. Specifically, methylation signatures were available for CD4 + T cells, CD8 + T cells, NK cells, B cells, monocytes, and neutrophils. One-way ANOVA models were fit independently to each of the 54 CRP-associated CpGs treating methylation as the dependent variable and cell type as the independent variable. We tested the null hypothesis that the mean methylation beta-value is the same across the cell types. The F-statistic, corresponding p value, and maximum absolute pairwise difference in the mean methylation beta value across cell types were calculated for each of the 54 CpGs. We then selected subgroups of CpG sites that had the top 10 smallest or top 10 largest F-statistic value to build the two additional CRP Scores. CRP Score 2 consists of putative cell-specific CpGs with high F-statistics, e.g., those exhibiting a difference in mean methylation beta-values between at least two of the six cell types. CRP Score 3 is made of cell-type invariant CpGs with low F-statistics, e.g., CpGs for which there did not appear to be a substantial difference in mean methylation beta-values across the normal six leukocyte subtypes. Score 2 ranged between − 0.0002 and 0.0046, while Score 3 ranged between − 0.025 and − 0.016. In the regression analyses, we used a standardized version of CRP Scores 1, 2, and 3 (mean = 0, sd = 1) for easier interpretation of results and allowing us to compare the results for each of the scores.
Statistical analyses
All statistical analyses were performed in R (version 3.5.1). We estimated mdNLR as described above, used an independent pancreatic cancer dataset [27] to estimate the correlation between estimated values of CRP Scores 1–3 with the log CRP and log IL-6 levels, and tested a series of a priori hypotheses concerning the mdNLR and CRP Scores. In addition, we also conducted exploratory analyses to generate novel hypotheses regarding the role of methylation-derived leukocyte proportions in lung cancer. Immune cell ratios (e.g., CD4/CD8, Neu/lymphocyte, B cell/lymphocyte, T cell/lymphocyte, Mono/lymphocyte, Neu + Mono/lymphocyte, Eos/lymphocyte, CD4nv/lymphocyte, B cell/CD8, CD8/Treg, Bnv/Bmem, CD4nv/CD4mem, and CD8nv/CDmem) were calculated for each sample by taking the ratio of its predicted cell proportions described above and tested as continuous variables. The presence of Treg was tested as a dichotomous variable. Given the need for multiple comparison adjustment, Bonferroni adjustment (family-wise error rate = 0.0013) was conducted for all exploratory analyses.
We used conditional logistic regression to examine the association between DNA methylation-based inflammatory measures (CRP Scores 1–3 and continuous mdNLR) and lung cancer risk. Models were fit with age, sex, and smoking status (never, former, current) as matching factors and were adjusted for potential confounding factors, including body mass index (BMI), batch effect, and previously described methylation-predicted pack-years smoked [57]. These analyses did not additionally adjust for methylation-derived cell proportions given how these proportions correlated with methylation-based inflammatory measures (Table 3). We repeated these analyses by lung cancer histology (NSCLC, SCLC), length of time between blood draw and diagnosis (< = 10, > 10 years), and BMI (< 25, ≥ 25 kg/m2).
Among the lung cancer cases, we examined the association between these same pre-diagnostic DNA methylation-based inflammatory measures (CRP Scores 1–3 and continuous mdNLR) and risk of lung cancer-specific death using a series of multivariable Cox proportional hazard regression adjusting for age, gender, smoking status, BMI, stage at diagnosis (three strata: stage 1 & 2, stage 3 & 4, and missing), cell proportion, batch effects, and methylation-predicted pack-years smoked. The proportional hazards assumption was checked by conducting global tests of correlating the set of scaled Schoenfeld residuals with time for each covariate. We excluded three lung cancer cases whose date of diagnosis and date of death were the same, or whose time between blood draw and date of lung cancer diagnosis was longer than 25 years. Cases were followed until their date of death from lung cancer, death from another cause, or the end of follow up in 2018, whichever came first.
Supplementary Information
Additional file 1. Supplementary Methods.
Acknowledgements
Cancer data were provided by the Maryland Cancer Registry, Center for Cancer Prevention and Control, Maryland Department of Health, with funding from the State of Maryland and the Maryland Cigarette Restitution Fund. The collection and availability of cancer registry data are also supported by the Cooperative Agreement NU58DP006333, funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
Authors' contributions
DSM, KTK, and EAP designed the study, obtained funding, and acquired the data. JL assisted with identifying the study samples and preparation of dataset. LAS assisted with cell deconvolution. DSM supervised all research activities. MR and NZ conducted the statistical analyses. NZ drafted the manuscript. DSM, EAP, KTK, and DCK interpreted the data and provided critical revisions of the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by 2018 American Association for Cancer Research (AACR)-Johnson and Johnson Lung Cancer Innovation Science (18-90-52-MICH). Note: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Availability of data and materials
The datasets generated during the current study are available from the corresponding author on reasonable request and will be deposited into dbGaP by publication.
Declarations
Ethics approval and consent to participate
The Institutional Review Board at the Johns Hopkins Bloomberg School of Public Health and the Tufts University Health Sciences Campus Institutional Review Board approved this study.
Consent for publication
Not applicable.
Competing interests
Dr. Kelsey is a founder and scientific advisor for Cellintec, which had no role in this research.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.ACS. Cancer Facts & Figures 2021. Atlanta: American Cancer Society, Inc. 2021.
- 2.Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383(7):640–649. doi: 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, et al. SEER cancer statistics review, 1975–2017. Bethesda, MD: National Cancer Institute; 2020. p. 12. [Google Scholar]
- 4.Fabrikant MS, Wisnivesky JP, Marron T, Taioli E, Veluswamy RR. Benefits and challenges of lung cancer screening in older adults. Clin Ther. 2018;40(4):526–534. doi: 10.1016/j.clinthera.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Force UPST. Screening for lung cancer: US preventive services task force recommendation statement. JAMA. 2021;325(10):962–970. doi: 10.1001/jama.2021.1117. [DOI] [PubMed] [Google Scholar]
- 6.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12(10):584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, Liu G, Hung RJ, Haycock PC, Aldrich MC, Andrew AS, et al. Causal relationships between body mass index, smoking and lung cancer: univariable and multivariable Mendelian randomization. Int J Cancer. 2021;148(5):1077–1086. doi: 10.1002/ijc.33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi AK, Caporaso NE, Katki HA, Wong H-L, Chatterjee N, Pine SR, et al. C-reactive protein and risk of lung cancer. J Clin Oncol. 2010;28(16):2719. doi: 10.1200/JCO.2009.27.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koestler DC, Usset J, Christensen BC, Marsit CJ, Karagas MR, Kelsey KT, et al. DNA methylation-derived neutrophil-to-lymphocyte ratio: an epigenetic tool to explore cancer inflammation and outcomes. Cancer Epidemiol Prev Biomark. 2017;26(3):328–338. doi: 10.1158/1055-9965.EPI-16-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Il'yasova D, Colbert LH, Harris TB, Newman AB, Bauer DC, Satterfield S, et al. Circulating levels of inflammatory markers and cancer risk in the health aging and body composition cohort. Cancer Epidemiol Prev Biomark. 2005;14(10):2413–2418. doi: 10.1158/1055-9965.EPI-05-0316. [DOI] [PubMed] [Google Scholar]
- 11.Siemes C, Visser LE, Coebergh J-WW, Splinter TA, Witteman JC, Uitterlinden AG, et al. C-reactive protein levels, variation in the C-reactive protein gene, and cancer risk: the Rotterdam Study. J Clin Oncol. 2006;24(33):5216–5222. doi: 10.1200/JCO.2006.07.1381. [DOI] [PubMed] [Google Scholar]
- 12.Trichopoulos D, Psaltopoulou T, Orfanos P, Trichopoulou A, Boffetta P. Plasma C-reactive protein and risk of cancer: a prospective study from Greece. Cancer Epidemiol Prev Biomark. 2006;15(2):381–384. doi: 10.1158/1055-9965.EPI-05-0626. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie GJ, Charles KA, Roxburgh CS, Horgan PG, McMillan DC, Clarke SJ. The systemic inflammation-based neutrophil–lymphocyte ratio: experience in patients with cancer. Crit Rev Oncol Hematol. 2013;88(1):218–230. doi: 10.1016/j.critrevonc.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Templeton AJ, McNamara MG, Šeruga B, Vera-Badillo FE, Aneja P, Ocaña A, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106(6):dju124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 15.Campa D, Zienolddiny S, Maggini V, Skaug V, Haugen A, Canzian F. Association of a common polymorphism in the cyclooxygenase 2 gene with risk of non-small cell lung cancer. Carcinogenesis. 2004;25(2):229–235. doi: 10.1093/carcin/bgh008. [DOI] [PubMed] [Google Scholar]
- 16.Engels EA, Wu X, Gu J, Dong Q, Liu J, Spitz MR. Systematic evaluation of genetic variants in the inflammation pathway and risk of lung cancer. Can Res. 2007;67(13):6520–6527. doi: 10.1158/0008-5472.CAN-07-0370. [DOI] [PubMed] [Google Scholar]
- 17.Seifart C, Plagens A, Dempfle A, Clostermann U, Vogelmeier C, von Wichert P, et al. TNF-α, TNF-β, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21(3):157–165. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shih C-M, Lee Y-L, Chiou H-L, Hsu W-F, Chen W-E, Chou M-C, et al. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer. 2005;50(3):291–297. doi: 10.1016/j.lungcan.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Allin KH, Bojesen SE, Nordestgaard BG. Baseline C-reactive protein is associated with incident cancer and survival in patients with cancer. J Clin Oncol. 2009;27(13):2217–2224. doi: 10.1200/JCO.2008.19.8440. [DOI] [PubMed] [Google Scholar]
- 20.Heikkilä K, Ebrahim S, Lawlor DA. A systematic review of the association between circulating concentrations of C reactive protein and cancer. J Epidemiol Community Health. 2007;61(9):824–833. doi: 10.1136/jech.2006.051292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu Z, Sun F, Zhou J, Li L, Shapiro SD, Xiao G. Interleukin-6 prevents the initiation but enhances the progression of lung cancer. Cancer Res. 2015;75(16):3209–3215. doi: 10.1158/0008-5472.CAN-14-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballaz S, Mulshine JL. The potential contributions of chronic inflammation to lung carcinogenesis. Clin Lung Cancer. 2003;5(1):46–62. doi: 10.3816/CLC.2003.n.021. [DOI] [PubMed] [Google Scholar]
- 23.Bauer AK, Dwyer-Nield LD, Keil K, Koski K, Malkinson AM. Butylated hydroxytoluene (BHT) induction of pulmonary inflammation: a role in tumor promotion. Exp Lung Res. 2001;27(3):197–216. doi: 10.1080/019021401300053948. [DOI] [PubMed] [Google Scholar]
- 24.Engels EA. Inflammation in the development of lung cancer: epidemiological evidence. Expert Rev Anticancer Ther. 2008;8(4):605–615. doi: 10.1586/14737140.8.4.605. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Genkinger JM, Platz EA, Hoffman SC, Comstock GW, Helzlsouer KJ. Fruit, vegetable, and antioxidant intake and all-cause, cancer, and cardiovascular disease mortality in a community-dwelling population in Washington County, Maryland. Am J Epidemiol. 2004;160(12):1223–1233. doi: 10.1093/aje/kwh339. [DOI] [PubMed] [Google Scholar]
- 27.Michaud DS, Ruan M, Koestler DC, Alonso L, Molina-Montes E, Pei D, et al. DNA Methylation–derived immune cell profiles, CpG markers of inflammation, and pancreatic cancer risk. Cancer Epidemiol Prev Biomark. 2020;29(8):1577–1585. doi: 10.1158/1055-9965.EPI-20-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohinai Z, Bonanno L, Aksarin A, Pavan A, Megyesfalvi Z, Santa B, et al. Neutrophil–lymphocyte ratio is prognostic in early stage resected small-cell lung cancer. PeerJ. 2019;7:e7232. doi: 10.7717/peerj.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galvano A, Peri M, Guarini AA, Castiglia M, Grassadonia A, De Tursi M, et al. Analysis of systemic inflammatory biomarkers in neuroendocrine carcinomas of the lung: prognostic and predictive significance of NLR, LDH, ALI, and LIPI score. Ther Adv Med Oncol. 2020;12:1758835920942378. doi: 10.1177/1758835920942378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozyurek BA, Ozdemirel TS, Ozden SB, Erdogan Y, Kaplan B, Kaplan T. Prognostic value of the neutrophil to lymphocyte ratio (NLR) in lung cancer cases. Asian Pac J Cancer Prev. 2017;18(5):1417. doi: 10.22034/APJCP.2017.18.5.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grieshober L, Graw S, Barnett MJ, Thornquist MD, Goodman GE, Chen C, et al. Methylation-derived neutrophil-to-lymphocyte ratio and lung cancer risk in heavy smokers. Cancer Prev Res. 2018;11(11):727–734. doi: 10.1158/1940-6207.CAPR-18-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grieshober L, Graw S, Barnett MJ, Goodman GE, Chen C, Koestler DC, et al. Pre-diagnosis neutrophil-to-lymphocyte ratio and mortality in individuals who develop lung cancer. Res Sq 2021. [DOI] [PMC free article] [PubMed]
- 33.Mandaliya H, Jones M, Oldmeadow C, Nordman II. Prognostic biomarkers in stage IV non-small cell lung cancer (NSCLC): neutrophil to lymphocyte ratio (NLR), lymphocyte to monocyte ratio (LMR), platelet to lymphocyte ratio (PLR) and advanced lung cancer inflammation index (ALI) Transl Lung Cancer Res. 2019;8(6):886. doi: 10.21037/tlcr.2019.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu P, Shen H, Wang G, Zhang P, Liu Q, Du J. Prognostic significance of systemic inflammation-based lymphocyte-monocyte ratio in patients with lung cancer: based on a large cohort study. PLoS ONE. 2014;9(10):e108062. doi: 10.1371/journal.pone.0108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Wu J, Zhang F, Ying L, Chen Y. Prognostic significance of pre-operative monocyte-to-lymphocyte ratio in lung cancer patients undergoing radical surgery. Lab Med. 2018;49(2):e29–e39. doi: 10.1093/labmed/lmx069. [DOI] [PubMed] [Google Scholar]
- 36.Tsilidis KK, Branchini C, Guallar E, Helzlsouer KJ, Erlinger TP, Platz EA. C-reactive protein and colorectal cancer risk: a systematic review of prospective studies. Int J Cancer. 2008;123(5):1133–1140. doi: 10.1002/ijc.23606. [DOI] [PubMed] [Google Scholar]
- 37.Heikkilä K, Harris R, Lowe G, Rumley A, Yarnell J, Gallacher J, et al. Associations of circulating C-reactive protein and interleukin-6 with cancer risk: findings from two prospective cohorts and a meta-analysis. Cancer Causes Control. 2009;20(1):15–26. doi: 10.1007/s10552-008-9212-z. [DOI] [PubMed] [Google Scholar]
- 38.dos Santos SI, De Stavola BL, Pizzi C, Meade TW. Circulating levels of coagulation and inflammation markers and cancer risks: individual participant analysis of data from three long-term cohorts. Int J Epidemiol. 2010;39(3):699–709. doi: 10.1093/ije/dyq012. [DOI] [PubMed] [Google Scholar]
- 39.Van Hemelrijck M, Holmberg L, Garmo H, Hammar N, Walldius G, Binda E, et al. Association between levels of C-reactive protein and leukocytes and cancer: three repeated measurements in the Swedish AMORIS study. Cancer Epidemiol Prev Biomark. 2011;20(3):428–437. doi: 10.1158/1055-9965.EPI-10-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki K, Ito Y, Wakai K, Kawado M, Hashimoto S, Seki N, et al. Serum heat shock protein 70 levels and lung cancer risk: a case-control study nested in a large cohort study. Cancer Epidemiol Prev Biomark. 2006;15(9):1733–1737. doi: 10.1158/1055-9965.EPI-06-0005. [DOI] [PubMed] [Google Scholar]
- 41.Braun MM, Helzlsouer KJ, Hollis BW, Comstock GW. Colon cancer and serum vitamin D metabolite levels 10–17 years prior to diagnosis. Am J Epidemiol. 1995;142(6):608. doi: 10.1093/oxfordjournals.aje.a117682. [DOI] [PubMed] [Google Scholar]
- 42.Comstock G, Helzlsouer KJ, Bush TL. Prediagnostic serum levels of carotenoids and vitamin E as related to subsequent cancer in Washington County, Maryland. Am J Clin Nutr. 1991;53(1):260S–S264. doi: 10.1093/ajcn/53.1.260S. [DOI] [PubMed] [Google Scholar]
- 43.Kakourou A, Koutsioumpa C, Lopez DS, Hoffman-Bolton J, Bradwin G, Rifai N, et al. Interleukin-6 and risk of colorectal cancer: results from the CLUE II cohort and a meta-analysis of prospective studies. Cancer Causes Control. 2015;26(10):1449–1460. doi: 10.1007/s10552-015-0641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8(3):389–399. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salas LA, Zhang Z, Koestler DC, Butler RA, Hansen HM, Molinaro AM, et al. Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling. bioRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 46.Koestler DC, Jones MJ, Usset J, Christensen BC, Butler RA, Kobor MS, et al. Improving cell mixture deconvolution by identifying optimal DNA methylation libraries (IDOL) BMC Bioinform. 2016;17(1):120. doi: 10.1186/s12859-016-0943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KT, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19(1):1–14. doi: 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Liu P, Xu Y, Zhang W, Tong L, Guo Z, et al. Preoperative neutrophil-to-lymphocyte ratio predicts response to first-line platinum-based chemotherapy and prognosis in serous ovarian cancer. Cancer Chemother Pharmacol. 2015;75(2):255–262. doi: 10.1007/s00280-014-2622-6. [DOI] [PubMed] [Google Scholar]
- 49.Salim DK, Mutlu H, Eryılmaz MK, Salim O, Musri FY, Tural D, et al. Neutrophil to lymphocyte ratio is an independent prognostic factor in patients with recurrent or metastatic head and neck squamous cell cancer. Mol Clin Oncol. 2015;3(4):839–842. doi: 10.3892/mco.2015.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y, Wang H, Yan A, Wang H, Li X, Liu J, et al. Pretreatment neutrophil to lymphocyte ratio in determining the prognosis of head and neck cancer: a meta-analysis. BMC Cancer. 2018;18(1):1–9. doi: 10.1186/s12885-018-4230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ethier J-L, Desautels DN, Templeton AJ, Oza A, Amir E, Lheureux S. Is the neutrophil-to-lymphocyte ratio prognostic of survival outcomes in gynecologic cancers? A systematic review and meta-analysis. Gynecol Oncol. 2017;145(3):584–594. doi: 10.1016/j.ygyno.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 52.Diem S, Schmid S, Krapf M, Flatz L, Born D, Jochum W, et al. Neutrophil-to-Lymphocyte ratio (NLR) and Platelet-to-Lymphocyte ratio (PLR) as prognostic markers in patients with non-small cell lung cancer (NSCLC) treated with nivolumab. Lung Cancer. 2017;111:176–181. doi: 10.1016/j.lungcan.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 53.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13(1):1–16. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17(1):1–15. doi: 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myte R, Sundkvist A, Van Guelpen B, Harlid S. Circulating levels of inflammatory markers and DNA methylation, an analysis of repeated samples from a population based cohort. Epigenetics. 2019;14(7):649–659. doi: 10.1080/15592294.2019.1603962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahsan M, Ek WE, Rask-Andersen M, Karlsson T, Lind-Thomsen A, Enroth S, et al. The relative contribution of DNA methylation and genetic variants on protein biomarkers for human diseases. PLoS Genet. 2017;13(9):e1007005. doi: 10.1371/journal.pgen.1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugden K, Hannon EJ, Arseneault L, Belsky DW, Broadbent JM, Corcoran DL, et al. Establishing a generalized polyepigenetic biomarker for tobacco smoking. Transl Psychiatry. 2019;9(1):1–12. doi: 10.1038/s41398-019-0430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Methods.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request and will be deposited into dbGaP by publication.


