Abstract
Accurate quantitation of the number of cells of individual bacterial species in dental plaque samples is needed for understanding the bacterial etiology of periodontitis. Real-time PCR offers a sensitive, efficient, and reliable approach to quantitation. Using the TaqMan system we were able to determine both the amount of Porphyromonas gingivalis and the total number of bacterial cells present in plaque samples. Using species-specific primers and a fluorescent probe, detection of DNA from serial dilutions of P. gingivalis cells was linear over a large range of DNA concentrations (correlation coefficient = 0.96). No difference was observed between P. gingivalis DNA alone and the same DNA mixed with DNA isolated from dental plaque, indicating that P. gingivalis levels can be determined accurately from clinical samples. The total number of cells of all bacterial species was determined using universal primers and a fluorescent probe. Standard curves using four different bacterial species gave similar results (correlation coefficient = 0.86). Levels of both P. gingivalis and total bacteria were determined from a series of human plaque samples. High levels of P. gingivalis were observed in several of the samples from subjects with periodontitis and none of those from healthy subjects. Real-time quantitative PCR provided a sensitive and reliable method for quantitating P. gingivalis. In addition, it allowed the determination of the total number of bacterial cells present in a complex sample so that the percentage of P. gingivalis cells could be determined.
The flora found in chronic periodontitis is a mixture of many bacterial species. While Porphyromonas gingivalis has been strongly implicated in periodontitis, the importance of other species and the relationship of P. gingivalis to these species are not completely understood. Several species have been found preferentially in subjects with disease (1, 4, 6, 7, 12, 14, 15, 17, 19), but no single species, including P. gingivalis, has been shown to be present in all periodontal patients. Also, disease-associated species have often been found in apparently healthy subjects (4, 7). Multiple, interacting species may be typical in the disease process (18). It is also likely that irreversible host tissue destruction occurs only when bacterial levels reach a critical threshold. Accurate quantitation of the number of cells of individual species is needed for understanding the bacterial etiology of periodontitis. Commonly employed sampling methods capture variable numbers of bacteria, so it is important also to determine the total number of bacterial cells in a sample so that the percentage of the total flora accounted for by a particular species can be calculated.
Information on the quantity of putative periodontal pathogens present at sites of disease has previously been collected by immunoassay (1, 14), by cultivation and enumeration (17, 19), and by DNA hybridization (7, 14, 17, 19). These studies have provided many useful data that have pointed to a handful of species as potential pathogens. Real-time PCR with species-specific primers can provide a precise and sensitive method for more accurate quantitation of individual species as well as total bacteria, and will be a useful tool for studies on the etiology of chronic periodontitis. A large number of samples can be accurately measured at one time. Because the assay is not based on an endpoint measurement as in competitive PCR (16), no plateauing effect occurs. The linear range is from as little as 10 to more than 108 cells.
Real-time PCR using the TaqMan system is accomplished by the continuous measurement of products throughout the reaction (8). In addition to primers for PCR, an oligonucleotide probe that hybridizes to the target DNA and is labeled with two fluorescent dyes is included in the PCR. Prior to cleavage, the reporter dye is quenched through fluorescent resonance energy transfer. During primer elongation, the 5′-to-3′ exonuclease activity of the Taq polymerase digests the hybridized probe, and the fluorescent tag is released and dissociated from the quenching dye. The resulting fluorescence can be measured and is proportional to the number of copies of the target sequence.
Real-time PCR offers a sensitive, efficient, and reliable approach to quantitation. Using the TaqMan system we were able to determine both the amount of P. gingivalis and the total number of bacteria present in plaque samples directly without culturing.
MATERIALS AND METHODS
Bacterial strains.
P. gingivalis strain ATCC 53978 and Actinobacillus actinomycetemcomitans strain ATCC 29522 were obtained from the American Type Culture Collection (Manassas, Va.). Escherichia coli was obtained from Life Technologies (Gaithersburg, Md.). A group G streptococcus isolate was provided by Phillip Marucha (Columbus, OH). Bacterial cells were enumerated by visualization in a hemocytometer.
Plaque samples.
Plaque samples for the present study were selected from those collected for a previous study of adult subjects with periodontitis and age-matched healthy controls (4). The clinical diagnostic criteria for health and disease have been previously described (4). Only samples identified as containing P. gingivalis in the previous study were analyzed for the present study. These pooled subgingival dental plaque samples were collected with endodontic paper points, and DNA was isolated and frozen for later analysis as previously described (10).
Amplification and quantitation of the rDNA spacer region.
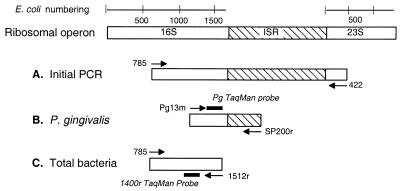
The rRNA operon was the target for the assay using nested, two-step PCR. Figure 1 shows the positions of the oligonucleotides used in this study. The sequence of primers and fluorescent probes is shown in Table 1.
FIG. 1.
Outline of quantitative PCR assay. The target for PCR is the ribosomal operon shown at the top with numbering corresponding to E. coli rDNA. (A) Primers used for initial PCR amplify the entire ISR plus partial 16S and 23S genes from all bacteria. (B) P. gingivalis specific primer PG13m and tRNA primer SP200r were used along with a P. gingivalis-specific probe to quantitate P. gingivalis. (C) Universal eubacterial primers and probe used to determine total amount of bacteria in a sample.
TABLE 1.
Primers and probes used in this study
| Specificity | Primer or probe | Sequence |
|---|---|---|
| P. gingivalis | PG13m | CATCGGTAGTTGCTAACAGTTTTCG |
| PG | ATGACGTCAAATCAGCACGGCCCTTACAT | |
| Ile-tRNA | SP200r | CCGACCTCTACATTATCAG |
| Eubacteria | 785 | GGATTAGATACCCTGGTAGTC |
| 422 | GGAGTATTTAGCCTT | |
| 1512r | TACCTTGTTACGACTT | |
| 1400r | TGACGGGCGGTGTGTACAAGGC |
The first amplification targeted conserved bacterial sequences in the 16S and 23S gene. The universal primers 785 and 422 and the conditions of amplification were as previously described (13).
Second, nested PCRs were used for quantitation of P. gingivalis or total bacteria. Primers and probes for quantitation were selected using Primer Express 1.0 from ABI/Perkin-Elmer (Foster City, Calif.) (11).
The forward primer for quantitation of P. gingivalis, PG13m, is a minor modification of the P. gingivalis specific primer PG13 (9). The modification was made in order to conform to parameters set by the Primer Express software. PG13m corresponds to position 1140 in the 16S gene of P. gingivalis. The reverse primer, SP200r, is specific for the Ile-tRNA in the intergenic spacer region (ISR) of P. gingivalis. This primer pair produces a 900-bp amplicon.
Quantitative PCR was performed on 2 μl of PCR product from the first PCR with 0.5 U of Platinum Taq polymerase (Life Technologies) in a total volume of 25 μl in buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.8; Promega, Madison, Wis.), 5 mM MgCl2 (Promega), a 0.2 mM concentration of each deoxynucleoside triphosphate (Amersham Pharmacia, Piscataway, N.J.), 10 ng of yeast tRNA, a 0.8 μM concentration of each primer (Biosynthesis, Lewisville, Tex.), 100 nM probe (Synthegen, Houston, Tex.), and 60 nM Rox reference dye (Synthegen). Amplification and detection were carried out in optical-grade 96-well plates in an ABI Prism 7700 Sequence Detection System (ABI/PE, Foster City, Calif.) with an initial cycle of 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 52°C for 1 min, and 72°C for 2 min.
All reactions were analyzed by agarose gel electrophoresis to confirm that only one PCR product was synthesized. Even the most closely related species have different sizes of ribosomal intergenic spacer regions (ISRs), therefore the use of primers that flank the ISR makes it possible to distinguish PCR products from other species.
In order to quantitate total bacteria, conserved sequences were selected in the 16S gene. The forward primer 785 and the reverse primer 1512r, corresponding to position 1512 of E. coli, target a conserved region at the 3′ end of the 16S gene (2, 20). The universal bacterial TaqMan 1400r probe hybridizes to a conserved position of the 16S gene at 1,400 bp (3). Amplification of total rDNA was carried out in a separate reaction at the same time under the same conditions used for the P. gingivalis-specific amplification, except that the Taq polymerase was pretreated to reduce contaminating bacterial DNA. For quantitative PCR with universal primers, 200 μl of Taq polymerase (5 U per μl) was treated with 1 U of DNase I at 37°C for 2 h. The resulting enzyme preparation was calibrated with untreated Taq polymerase and adjusted for any loss in activity.
Both fluorescent probes were labeled at the 5′ end with the reporter dye 6-carboxyfluorescein (6-FAM) and at the 3′ end with the quencher dye 6-carboxytetramethylrodamine (TAMRA). Data were analyzed using the Sequence Detection System software from ABI.
RESULTS
The number of P. gingivalis cells and the total number of bacterial cells present in a sample were determined by real-time PCR.
Quantitation of P. gingivalis.
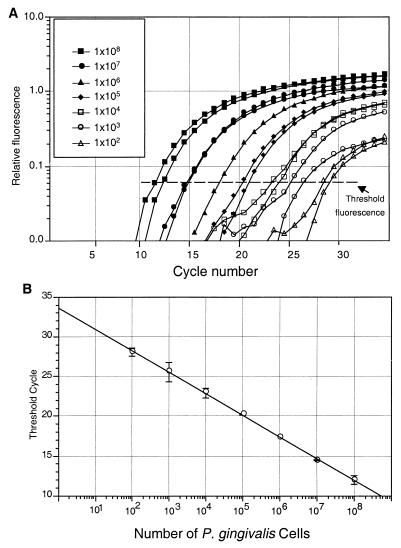
The number of P. gingivalis cells was determined with the TaqMan PCR procedure using a P. gingivalis-specific primer. DNA from known amounts of P. gingivalis was added in serial dilutions from 102 to 108 cells to a series of PCRs. The reactions were carried out in a PE 7700 thermocycler, and the fluorescence was monitored throughout the reaction. The results are shown in Fig. 2A. A standard curve from these data is shown in Fig. 2B. Detection and quantitation were linear over the range of DNA concentrations examined.
FIG. 2.
Amplification of P. gingivalis rDNA. Serial dilutions of genomic DNA from P. gingivalis were used as templates for real-time PCR. (A) The relative fluorescence is the increase in reporter dye intensity relative to the passive internal reference dye. The amount of P. gingivalis DNA in each sample is shown in the key. The threshold fluorescence, or level at which the threshold cycle was determined, is shown. (B) A standard curve was generated from the amplification plot in panel A (correlation coefficient = 0.96). Threshold cycle is the cycle number when the threshold fluorescence is reached. Standard deviations from two measurements are shown as error bars.
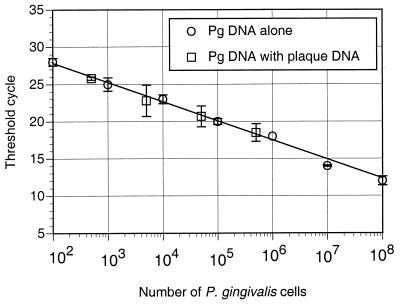
In order to determine if the quantity of P. gingivalis in a complex mixture of bacterial DNA could be determined accurately using the TaqMan assay, known amounts of P. gingivalis DNA were added to DNA isolated from subgingival plaque samples that were negative for P. gingivalis as determined by either agarose gel electrophoresis and ethidium staining or analysis with the TaqMan PCR procedure (data not shown). The fluorescent signal was compared to a standard curve generated from P. gingivalis DNA alone (Fig. 3). Each point represents an average of measurements from three to four independent samples. Similar values were obtained from P. gingivalis genomic DNA alone and the same DNA mixed with DNA from plaque, indicating that P. gingivalis levels can be determined accurately even in the presence of DNA from other species.
FIG. 3.
Serial dilutions of P. gingivalis DNA were added to DNA isolated from subgingival plaque that did not contain P. gingivalis and then quantitated using real-time PCR. Values for known amounts of P. gingivalis DNA alone are also shown. Standard deviations are shown as error bars.
Quantitation of total bacteria.
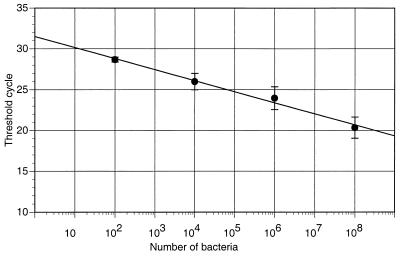
The total number of bacterial cells of any species was determined with the TaqMan PCR procedure using a pair of universal primers and a universal probe. Standard curves using four different bacterial species gave similar results (Fig. 4).
FIG. 4.
Quantitation of total bacteria by real-time PCR. A standard curve was generated for multiple bacterial species (correlation coefficient = 0.86). Each point represents the average value for counted cells from stock cultures of four species, A. actinomycetemcomitans, P. gingivalis, E. coli, and group G streptococcus. Standard deviations are shown as error bars. The same universal primers and probe were used for all amplifications.
Quantitation of clinical samples.
The levels of both P. gingivalis and total bacteria in plaque samples from seven subjects with periodontitis and 11 periodontally healthy subjects were determined (Table 2). To determine the number of P. gingivalis cells present in each sample, the fluorescent signals detected from 2 or 3 serial dilutions in the linear range of the assay were averaged and compared to a standard curve generated with P. gingivalis DNA during the same experiment. The total number of bacteria of all species in each sample was determined with universal primers and a fluorescent probe by averaging 3 or 4 serial dilutions from the linear range. The standard curve for total bacteria was generated from the average of four bacterial species. P. gingivalis levels were expressed as a percentage of total bacteria (Table 2). Strain identification was previously determined by heteroduplex analysis (5) and is reported here for each sample (Table 2).
TABLE 2.
Number of P. gingivalis and total bacterial cells detected in plaque samples
| Group | No. of cells
|
% P. gingivalis | P. gingivalis strain(s) presentc | |
|---|---|---|---|---|
| P. gingivalisa | Total bacteriab | |||
| Periodontal health | 7.30 (0.7) × 1010 | 7.5 (3.2) × 1013 | 0.10 | hHG1691 |
| 1.95 (1.1) × 108 | 1.71 (1.1) × 1013 | 0.00 | hW83 | |
| 7.20 (2.5) × 107 | 2.20 (0.6) × 1014 | 0.00 | hHG1691 | |
| 5.90 (5.2) × 109 | 4.64 (0.5) × 1012 | 0.13 | hA7A1 | |
| 6.25 (4.9) × 106 | 3.40 (1.3) × 1012 | 0.00 | hA7A1 | |
| 4.47 (2.6) × 108 | 4.77 (3.9) × 1014 | 0.00 | h23A4 | |
| 1.75 (0.7) × 107 | 3.00 (2.0) × 1010 | 0.06 | hA7A1 | |
| 2.80 (0.0) × 109 | 1.06 (0.2) × 1012 | 0.26 | hHG1691 | |
| 5.45 (0.7) × 109 | 3.37 (1.6) × 1012 | 0.16 | h381 | |
| 5.21 (1.0) × 108 | 2.77 (1.8) × 1011 | 0.19 | h23A4, hA7A1 | |
| 6.45 (2.5) × 108 | 2.30 (0.4) × 1012 | 0.03 | h381 | |
| Periodontitis | 1.50 (0.5) × 106 | 2.10 (0.7) × 107 | 7.10 | hW83 |
| 3.45 (0.9) × 109 | 4.00 (3.1) × 1013 | 0.00 | h381, hHG1691, hW83 | |
| 8.00 (0.6) × 1010 | 5.90 (3.1) × 1012 | 1.40 | hHG1691 | |
| 2.35 (1.8) × 1011 | 2.14 (0.9) × 1015 | 0.01 | hA50 | |
| 3.45 (1.2) × 1010 | 1.99 (1.4) × 1014 | 0.02 | hHG1691 | |
| 1.19 (0.4) × 1010 | 1.60 (0.7) × 1012 | 0.74 | h49417 | |
| 1.24 (0.4) × 1011 | 1.96 (0.9) × 1012 | 6.32 | hW83 | |
Mean number of cells determined from 2 or 3 serial dilutions found to be in the linear range of the assay (standard deviation).
Mean determined from 3 to 4 serial dilutions found to be in the linear range of the assay (standard deviation).
Data from Griffen et al. (5).
DISCUSSION
Real-time PCR offers the ability to determine the absolute and relative amounts of P. gingivalis in a mixed sample without culturing the sample. With previous methods it has been especially difficult to determine the total number of bacteria collected in a sample. The ability to compare the number of P. gingivalis cells in a sample to the total number of bacteria in the sample makes it possible to adjust for variation in sampling and allows comparisons to be made between samples. Estimates obtained by cultivation offer less sensitivity for P. gingivalis, and are less accurate for total bacteria, since many of the species found in the periodontal pocket are difficult to culture.
Although the TaqMan quantitative PCR method requires expensive instrumentation, it does offer advantages over previous approaches to quantitative PCR. It is more accurate and reliable over a larger range than other methods that rely on endpoint PCR, such as competitive PCR. Also, large numbers of samples can be more easily processed using the TaqMan method because fewer reactions are required to measure each sample.
Total bacteria were determined using universal primers that hybridized to all bacteria. Standard curves generated with DNA from four different species of bacteria were used to quantitate total bacteria present in a plaque sample. Although the data shown in Table 2 were determined by averaging the values from four species, all species gave similar results, and any one species could be used for generating a standard curve for total bacteria. Because there are small shifts in the fluorescent signal from experiment to experiment, it is essential to generate a new standard curve for each set of measurements.
The number of target sequences (rDNAs) may vary among bacterial species in number of copies per genome as well as with the growth phase of the cell. It was assumed that the average number of rDNAs per bacterial cell was similar in all plaque samples. No difference was observed among the species tested, and no attempt was made to compensate for differences in rDNA copy number.
Because Taq polymerase preparations contain some bacterial genomic DNA that is not removed during the purification process, most preparations add approximately 105 copies of bacterial rDNA template to the PCR mixture (data not shown). Therefore, DNA in samples in which the template concentration is not substantially higher than this cannot be accurately measured by real-time PCR with universal bacterial primers. Treating Taq polymerase with DNase I reduces the contaminating template to approximately 103 copies. If preparations of Taq polymerase with little or no contaminating DNA become available, it will be possible to accurately measure lower concentrations of template. The first amplification of the clinical samples was carried out with untreated Taq polymerase. Most samples contained at least 1011 to 1012 cells in the entire sample. One-fifth of the sample was used for the reaction, so that the total number of templates added to the reaction was usually 2 × 1010 or more. Therefore, the contaminant DNA would account for approximately 0.005% of the total DNA measured. The sample with the lowest number of bacteria contained 2 × 107 cells, so the most that contaminating template accounted for in any sample was 2.5% of the total bacterial templates. Treated Taq polymerase was used for the second amplification to minimize detection of extraneous template. PCR with species-specific primers is not affected by contaminating DNA.
The TaqMan method was linear for samples containing from fewer than 100 cells to more than 108 cells. The nested (two-step) PCR procedure used to quantitate P. gingivalis amplifies all bacterial rDNA sequences in the first step. If more than 108 cells of P. gingivalis are present in the reaction, amplification of P. gingivalis-specific DNA may not proceed exponentially in the first PCR. This would not allow an accurate quantitation during the second PCR. Samples that contained more than 108 cells were diluted to obtain accurate measurements.
Quantitation of P. gingivalis from a mixture of bacteria relies on the specificity of the PCR. The PG13m primer used in this study is specific for P. gingivalis. A search of GenBank showed no other matches. Because the target sequence contains part of the ISR that varies in size even among closely related species, amplification of DNA from other bacteria would have resulted in the production of additional bands. PCR products made during quantitation were analyzed by agarose gel electrophoresis, and no size variants were observed.
The total number of bacteria collected in each plaque sample varied by a few orders of magnitude (Table 2), demonstrating the variability of the sampling technique. This points to the importance of considering relative rather than absolute numbers of a single species in a mixed sample. High levels of P. gingivalis were observed in several of the samples from subjects with periodontitis and none of the healthy subjects. This is consistent with previously reported qualitative data indicating that P. gingivalis is a factor in many but not all cases of periodontitis (4). Samples contained pooled plaque from each tooth, including those without evidence of disease, so P. gingivalis levels are average values for the entire dentition and may be much higher at sites of disease activity. Further study is needed to determine this. It is interesting that the two samples containing the highest levels of P. gingivalis both contained heteroduplex type hW83, the strain group most strongly associated with periodontitis (5).
In summary, real-time quantitative PCR provided a sensitive and accurate method for measuring the amount of P. gingivalis in plaque samples. In addition, it allowed the determination of the total number of bacterial cells present in a complex sample so that the percentage of P. gingivalis could be determined.
ACKNOWLEDGMENTS
We gratefully acknowledge Michael Caligiuri and Philip Johnson for use of their fluorescent thermocyclers and Zulma Sanchez for technical assistance.
This work was supported by NIH grant DE10467.
REFERENCES
- 1.Alpagot T, Wolff L F, Smith Q T, Tran S D. Risk indicators for periodontal disease in a racially diverse urban population. J Clin Periodontol. 1996;23:982–988. doi: 10.1111/j.1600-051x.1996.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 2.Fry N K, Fredrickson J K, Fishbain S, Wagner M, Stahl D A. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl Environ Microbiol. 1997;63:1498–1504. doi: 10.1128/aem.63.4.1498-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffen A L, Becker M R, Lyons S R, Moeschberger M L, Leys E J. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffen A L, Lyons S R, Becker M R, Moeschberger M L, Leys E J. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol. 1999;37:4028–4033. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossi S G, Zambon J J, Ho A W, Koch G, Dunford R G, Machtei E E, Norderyd O M, Genco R J. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 7.Haffajee A D, Cugini M A, Tanner A, Pollack R P, Smith C, Kent R L, Jr, Socransky S S. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 8.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 9.Lamell C W, Griffen A L, McClellan D L, Leys E J. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J Clin Microbiol. 2000;38:1196–1199. doi: 10.1128/jcm.38.3.1196-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leys E J, Griffen A L, Strong S J, Fuerst P A. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J Clin Microbiol. 1994;32:1288–1294. doi: 10.1128/jcm.32.5.1288-1294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 12.Machtei E E, Dunford R, Hausmann E, Grossi S G, Powell J, Cummins D, Zambon J J, Genco R J. Longitudinal study of prognostic factors in established periodontitis patients. J Clin Periodontol. 1997;24:102–109. doi: 10.1111/j.1600-051x.1997.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 13.McClellan D L, Griffen A L, Leys E J. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1995;34:2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melvin W L, Assad D A, Miller G A, Gher M E, Simonson L, York A K. Comparison of DNA probe and ELISA microbial analysis methods and their association with adult periodontitis. J Periodontol. 1994;65:576–582. doi: 10.1902/jop.1994.65.6.576. [DOI] [PubMed] [Google Scholar]
- 15.Preus H R, Anerud A, Boysen H, Dunford R G, Zambon J J, Loe H. The natural history of periodontal disease. The correlation of selected microbiological parameters with disease severity in Sri Lankan tea workers. J Clin Periodontol. 1995;22:674–678. doi: 10.1111/j.1600-051x.1995.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 16.Rupf S, Merte K, Eschrich K. Quantification of bacteria in oral samples by competitive polymerase chain reaction. J Dent Res. 1999;78:850–856. doi: 10.1177/00220345990780040501. [DOI] [PubMed] [Google Scholar]
- 17.Schlegel-Bregenzer B, Persson R E, Lukehart S, Braham P, Oswald T, Persson G R. Clinical and microbiological findings in elderly subjects with gingivitis or periodontitis. J Clin Periodontol. 1998;25:897–907. doi: 10.1111/j.1600-051x.1998.tb02388.x. [DOI] [PubMed] [Google Scholar]
- 18.Socransky S S, Haffajee A D, Cugini M A, Smith C, Kent R L., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 19.Tanner A, Maiden M F, Macuch P J, Murray L L, Kent R L., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 20.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]