Abstract
Fip1 is an essential component of the Saccharomyces cerevisiae polyadenylation machinery and the only protein known to interact directly with poly(A) polymerase (Pap1). Its association with Pap1 inhibits the extension of an oligo(A) primer by limiting access of the RNA substrate to the C-terminal RNA binding domain (C-RBD) of Pap1. We present here the identification of separate functional domains of Fip1. Amino acids 80 to 105 are required for binding to Pap1 and for the inhibition of Pap1 activity. This region is also essential for viability, suggesting that Fip1-mediated repression of Pap1 has a crucial physiological function. Amino acids 206 to 220 of Fip1 are needed for the interaction with the Yth1 subunit of the complex and for specific polyadenylation of the cleaved mRNA precursor. A third domain within amino acids 105 to 206 helps to limit RNA binding at the C-RBD of Pap1. Our data demonstrate that the C terminus of Fip1 is required to relieve the Fip1-mediated repression of Pap1 in specific polyadenylation. In the absence of this domain, Pap1 remains in an inhibited state. These findings show that Fip1 has a crucial regulatory function in the polyadenylation reaction by controlling the activity of poly(A) tail synthesis through multiple interactions within the polyadenylation complex.
Accurate processing of the 3′ end of the primary RNA transcript is an essential step in the mRNA maturation of all eukaryotes. The resulting poly(A) tail has been implicated in numerous aspects of RNA metabolism, including efficiency of mRNA export from the nucleus, message stability, and initiation of translation (6, 11, 24). Mechanistically, polyadenylation consists of a tightly coupled two-step reaction: a site-specific endonucleolytic cleavage of the pre-mRNA, followed by the processive synthesis of a poly(A) tail onto the 3′ end of the upstream cleavage product. This requires the presence of cis-acting signal sequences in the untranslated region of the pre-mRNA as well as trans-acting protein factors (34, 35). The ability to uncouple cleavage and poly(A) addition in vitro (9, 19) has allowed the biochemical identification of factors involved in either one or both steps of the process. In Saccharomyces cerevisiae, cleavage requires cleavage/polyadenylation factor I (CF I) and cleavage factor II (CF II), while tail synthesis requires poly(A) polymerase (PAP; Pap1), CF I, polyadenylation factor I (PF I), and Pab1 (10, 18). A combination of biochemical and genetic approaches has identified almost all the genes involved in this process. This work has revealed a striking degree of conservation from yeast to mammals among most of the protein components required for polyadenylation, despite substantial differences in the signals on the pre-mRNA (34, 35).
As the catalytic subunit, PAP is at the heart of the machinery required for poly(A) addition. There is 47% identity in the amino-terminal 400 amino acids between yeast and mammalian PAP (14, 33) and a remarkable similarity throughout their three-dimensional structures (4, 17). The C termini are divergent but nonetheless display similarities such as the presence of RNA binding domains (RBD) (16, 40). When isolated from the rest of the polyadenylation complex, both yeast and mammalian enzymes synthesize a tail of unregulated length onto any given RNA in vitro, a process referred to as nonspecific polyadenylation (15, 31). Upon association with other factors, PAP specifically uses the appropriate RNA substrate and limits poly(A) synthesis to a tail of the correct length (10, 29), approximately 250 residues in mammals and 50 to 90 residues in yeast. The mechanism by which the other factors confer this specificity to Pap1 involves a network of protein-protein as well as protein-RNA interactions. For example, interactions of mammalian PAP with p160, the largest subunit of cleavage/polyadenylation specificity factor (CPSF), recruits the enzyme to the AAUAAA polyadenylation signal in the pre-mRNA (7) and regulates PAP activity (20).
In yeast, regulation of poly(A) tail synthesis likely involves Fip1, the only protein known to date to interact with Pap1 directly (23, 39). The purification of polyadenylation factors revealed that Fip1 is one of nine subunits that copurifies with PF I activity, the others being Pap1, Pta1, Pfs1, Pfs2, Cft1 (Yhh1), Cft2 (Ydh1), Brr5 (Ysh1), and Yth1 (22). A subset of these components, consisting of Cft1, Cft2, Brr5, and Pta1, was found to provide CF II activity (36). To reflect its involvement in both steps of polyadenylation, this CF II-PF I complex of nine proteins has been renamed cleavage polyadenylation factor (CPF) (21). Interestingly, CPF, when isolated from CF I and Pab1, can nonspecifically synthesize tails onto RNA at a higher rate than recombinant Pap1 (22), indicating that Pap1 is activated in the context of these proteins. The molecular events behind this activation are not clear but probably involve an increased affinity of Pap1 for the RNA mediated through protein-protein and protein-RNA interactions. Fip1 has been proposed to play an important role in this process by tethering Pap1 to CPF and to RNA through its interactions with Pfs2 and Yth1 (2, 3, 21) as well as to CF I through its interaction with Rna14 (23). However, Fip1's role must be more complex. Recombinant Fip1 inhibits nonspecific polyadenylation by binding to a region overlapping the C-terminal RBD (C-RBD) of Pap1, thereby reducing Pap1's affinity for the primer and shifting it from a processive to a distributive mode of poly(A) synthesis (39).
While our knowledge of the architecture of the polyadenylation holoenzyme has increased, we do not understand how interactions among the subunits provide a context that prevents poly(A) synthesis until the completion of cleavage and then allows a limited burst of processive polymerization. To explore this regulation, we identified domains of Fip1 important for protein-protein interactions and investigated in vivo and in vitro the consequences of disrupting these interactions. We show here that amino acids 80 to 105 of Fip1 are necessary for the direct interaction with Pap1. Fip1 lacking this domain fails to support growth and cannot inhibit nonspecific polyadenylation. An additional domain within amino acids 105 to 206, while not required for the interaction with Pap1, contributes to the inhibition of Pap1 activity. We also demonstrate that the 14 amino acids between residues 206 and 220 of Fip1 are required for the interaction with Yth1 and for specific polyadenylation in vitro. Our data indicate that the inhibition of Pap1 by Fip1 is essential for viability and that Fip1 plays a central role in the regulation of Pap1 activity in the polyadenylation complex. They support a model of specific polyadenylation in which Pap1 is kept in an inhibited state by Fip1 but can be activated through a mechanism that requires interactions at the C terminus of Fip1.
MATERIALS AND METHODS
Nucleic acids.
The bacterial expression plasmid pFL11 and the yeast shuttle plasmid pIA34 were kind gifts from Walter Keller and have been described elsewhere (23). Plasmid p314Fip1 was made by subcloning a 1.8-kb ScaI-KpnI fragment from plasmid pIA34, which covers the entire FIP1 gene, into the SacI (blunted) and KpnI sites of pRS314 (26). Plasmids pFL11 and p314Fip1 served as parental vectors for all deletion constructs generated in this study. All deletions of the FIP1 coding sequence were generated by PCR. The restriction sites SacI and BstBI at the 5′ and 3′ ends, respectively, of the Fip1 open reading frame are unique in pFL11 and p314Fip1. For this reason, all primers used for the 5′ ends of the deletion constructs were designed to contain the sequence 5′-CCCGAGCTCC upstream of the annealing sequence; primers used for the 3′ end were designed with 5′-GGGGAACTTCGAATT (sites are underlined). This allowed replacement of the full-length FIP1 coding sequence in pFL11 and p314Fip1 with any deletion made by PCR and maintained the natural stop codon plus the two preceding amino acids. The strategy also preserves the N-terminal Met-Ala-His6 tag in pFL11. However, these residues are not taken into consideration in the nomenclature used throughout the text. The internal deletions fipΔ60-105 and fipΔ80-105 were generated by inverse PCR using the reverse primers 5′-ACTTCTGGCAGTAGCTGGAG and 5′-ACTGTCAGAATCACTATCGTC with the forward primer 5′-GGGCAGTACTGCGACATCTTCAAGCAAAG-3′ and religation of the resulting PCR products. Deletions from both termini were constructed by fusing DraIII-Af1II fragments from either p314F40-327 (1.2 kb) or p314F80-327 (1.1 kb) with the large Af1II-DraIII fragment (5.5 kb) from p314F1-206 or p314F1-220. Full-length, precleaved, and mutated precleaved GAL7 RNAs were made by in vitro transcription with T3 RNA polymerase using linearized plasmids pJC7-1, pJC7-9, and pJC7-10, respectively, as previously described (10, 40).
Yeast strains and culturing conditions.
All yeast strains used in this study are derived from strain PJP22 (MATα leu2-3, 112 ura3-52 trp1 his4 fip1::LEU2 pIA34 [CEN4 URA3 FIP1]) (23) and were obtained by plasmid shuffling. Plasmids of interest were introduced into PJP22 by transformation according to the lithium acetate method (5) and plated on complete medium lacking uracil and tryptophan. Transformants were isolated, grown in liquid culture in the presence of uracil for 24 h, and plated on media containing 5-fluoroorotic acid (FOA). Colonies growing in the presence of FOA were reexamined for their nutritional growth requirements, and the presence of the correct construct was verified by Southern blotting. To compare growth rates of mutants, cells were grown at 22°C overnight in liquid culture, cell densities were standardized by dilution with fresh medium, and equal volumes of serial dilution were spotted on solid medium. The plates were incubated at the temperatures indicated in the figure legends.
Recombinant proteins.
Recombinant Pap1 was expressed using the T7 expression system (27) and purified as described elsewhere (40). The Fip1 truncations contain a His6 tag and were expressed and purified as described for the wild type (39). For most truncations, a one-step affinity purification on Ni-agarose was sufficient to obtain a single band on sodium dodecyl sulfate (SDS)-polyacrylamide gels stained with Coomassie blue. Some truncations required an additional purification step on a 1-ml MonoQ column (Pharmacia) as described for the purification of recombinant full-length Fip1. Recombinant Fip1, like the native protein, migrates at a molecular mass of 50 kDa on SDS-gels despite a calculated molecular mass of 37 kDa. Unusual gel migration was observed for all truncations. Protein concentrations were determined with the Bio-Rad protein concentration kit as described in the manufacturer's protocol, using known concentrations of bovine serum albumin as a standard. Expression of recombinant Yth1 and extract preparation were carried out as described for Pap1. The NP-40 extract obtained after ultracentrifugation was then passed over a DEAE-Sephacel column. Yth1 does not bind to this resin under these conditions and elutes in the flowthrough. While the protein is purified only moderately by this treatment, its stability is greatly enhanced, allowing the use of this fraction in coimmunoprecipitations. A second source of recombinant Yth1 involved the expression of Yth1 as a fusion to glutathione S-transferase (GST) as previously described (37). To release Yth1 from the GST tag, the fusion was incubated with the PreScission protease as specified by the manufacturer (Pharmacia). Both sources of Yth1 were used in nonspecific polyadenylation assays with identical results.
Antibodies and coimmunoprecipitation of recombinant proteins.
Monoclonal antibodies against Pap1 and polyclonal antibodies against Hrp1, Fip1, and Yth1 have been described previously (12, 13, 37, 39). The monoclonal anti-His5 antibody was from Qiagen. Coimmunoprecipitations were carried out as follows. For each immunoprecipitation, 20 μl of a 50% slurry of protein A-agarose (Gibco) was incubated with either 40 μl of monoclonal antibody (tissue culture supernatant) or 0.75 μl of serum in 40 μl of buffer IP-150 (150 mM KCl, 20 mM Tris-Cl [pH 8], 0.1% NP-40) for 90 min at room temperature with slight agitation. The beads were washed three times with ice-cold IP-150 for 5 min each and resuspended in 100 μl of blocking solution (IP-150 containing 10% fetal calf serum). This mixture was rotated for 60 min at 4°C, at which point the recombinant proteins of interest were added (usually 100 to 500 ng). The incubation was continued for 90 min at 4°C and followed by washing the beads three times as before. After the last wash, all liquid was removed and the proteins in the immunoprecipitate were eluted by incubation of the beads with 15 μl of 3× SDS buffer for 10 min at 50°C. The samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting with antibodies against recombinant Fip1, Yth1, and Pap1 and detection by alkaline phosphatase. To avoid heavy cross-reaction of the immunoprecipitating antibody with the secondary antibody during the Western blotting, the primary and secondary antibodies were preincubated as described previously (25). This method significantly reduced the background from the immunoglobulin G G light and heavy chains. Molecular weight markers were from New England Biolabs.
Extract preparation and in vitro 3′-end processing assays.
Protein extracts from yeast were prepared as described elsewhere (12). After the ammonium sulfate precipitation, the protein was resuspended in 300 μl of buffer D (20 mM Tris-HCl [pH 7.9], 50 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 20% glycerol, 2 μM pepstatin A, 0.6 μM leupeptin) and dialyzed twice against 1 liter of the same buffer for 3 h each. The protein concentrations are generally between 15 and 20 mg/ml. Cleavage and polyadenylation assays were carried out as described previously (12, 37), with small modifications. Generally, 32P-labeled GAL7 full-length (GAL7-1) or precleaved (GAL7-9) RNA (200,000 counts [10 nM]) was incubated with 2 μl of yeast cell extract (∼30 μg of protein) at 30°C in a total volume of 12 μl. The buffer in these reactions consisted of 2% polyethylene glycol 8000 (Fisher), 75 mM potassium acetate, 1 mM magnesium acetate, 2 mM ATP, 2 mM phosphocreatine, 2.5 μM tRNA, 1 mM DTT, and 0.4 U of RNasin (Pharmacia). After 20 min, 2.5 μl of stop- solution (2.5% SDS, 5 mg of protease K/ml, 135 mM EDTA) were added, and the incubation continued for an additional 10 min. This treatment was followed by the addition of 15 μl of 50 mM Tris-HCl (pH 7.9) to each sample and extraction with 30 μl of phenol-chloroform-isoamyl alcohol (25:24:1). Then 2.5 μl of the aqueous phase was electrophoresed on a 5% polyacrylamide gel containing 8.3 M urea. The RNA products were visualized with a Molecular Dynamics PhosphorImager. For the complementation of polyadenylation-deficient extracts with recombinant proteins, we used 50 ng of Pap1, 60 ng of Fip1 or any Fip1 truncation, and 2 μl of extract unless indicated otherwise in the figure legends. These amounts of Fip1 and Pap1 correspond to a 2:1 molar ratio of Fip1 to Pap1. Protein components in each sample were preincubated for 2 min at 37°C.
Nonspecific polyadenylation and inhibition by Fip1.
Nonspecific polyadenylation assays were carried out as described previously (39, 40) in a volume of 12 μl containing 20 mM HEPES (pH 7.5), 50 mM KCl, 0.25 mM EDTA, 1 mM MnCl2, 10% glycerol, 0.5 mg of bovine serum albumin/ml, 0.5 mM DTT, 1 μM oligo(A)12, 250 μM ATP, 1 μCi of [α-32P]ATP, and 25 ng of Pap1 at 25°C for the indicated time. Reaction products were analyzed either by Cerenkov counting of acid-precipitable counts or electrophoresis on 18% polyacrylamide–8.3 M urea denaturing gels. In reactions containing Fip1 or any Fip1 truncation, the molar ratio of Fip1 to Pap1 was 2:1. This corresponds to 30 ng of Fip1 per 25 ng of Pap1.
RESULTS
Deletions in FIP1 identify domains important for growth and viability.
Because of its critical role in poly(A) tail synthesis (23) and its direct effect on Pap1 in vitro (39), we set out to determine which regions of Fip1's primary protein structure are important for polyadenylation and for protein-protein interactions. Based on the assumption that a disruption of any crucial interaction should retard growth, we identified deletions within the FIP1 coding region that affect cell viability or cause conditional growth phenotypes. The deletions were generated by PCR, cloned into the yeast shuttle vector pRS314 (26), and introduced into yeast strain PJP22 (23). This strain contains a lethal disruption of the chromosomal copy of FIP1 covered by plasmid pIA34, which carries the wild-type FIP1 gene. None of the fip1 deletions analyzed exert a dominant negative effect on growth in the presence of the wild-type gene. After loss of pIA34 by counterselection with FOA, the growth behavior of nonlethal fip1 mutant strains can be analyzed. Because strains with deletion constructs missing regions important for viability cannot lose pIA34, this approach identifies essential domains as well as important, albeit nonessential, regions.
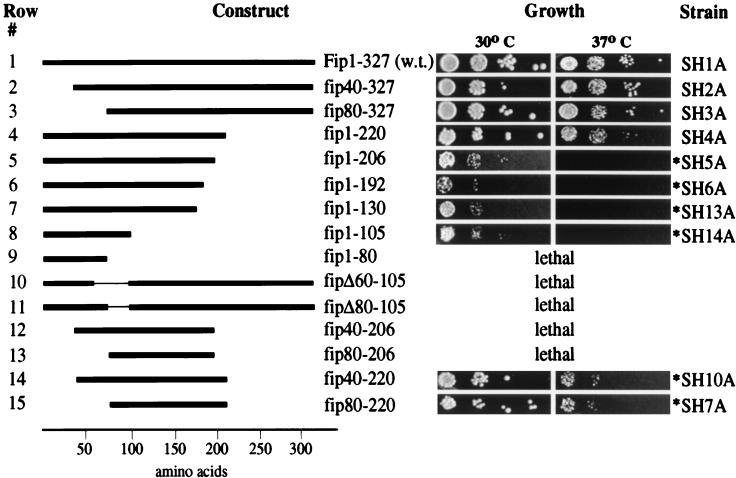
Constructs with deletions of 40 (fip40-327) or 80 (fip80-327) amino acids from the N terminus display no obvious effect on growth behavior when compared to the wild type (Fig. 1, rows 1 to 3). Similarly, the C-terminal 107 amino acids (fip1-220) are dispensable for normal growth (Fig. 1, row 4). The same result was obtained when these mutants were tested for a potential cold sensitivity at 16°C (data not shown). However, a strain with a deletion of the C-terminal 121 amino acids (fip1-206) grows slowly at the permissive temperature of 30°C and is unable to support growth at 37°C (Fig. 1, row 5). The temperature sensitivity persists in strains carrying deletions of the C-terminal 135 (fip1-192), 197 (fip1-130), and 222 (fip1-105) amino acids (Fig. 1, rows 6 to 8). While the slow growth at the permissive temperature is more pronounced in the strain expressing fip1-192, the two strains expressing fip1-130 and fip1-105 curiously grow slightly better under these conditions. It is remarkable that the N-terminal 105 amino acids, which represent only 35% of the total sequence of Fip1, are sufficient to support growth at the permissive temperature. Deletion of an additional 25 amino acids (fip1-80), however, is lethal (Fig. 1, row 9), suggesting that the stretch between amino acids 80 and 105 provides an essential function. This is supported by the finding that internal deletions of 45 (fipΔ60-105) or 25 (fipΔ80-105) amino acids covering this region are unable to rescue a genomic disruption (Fig. 1, rows 10 and 11). Deletions from both termini leaving 180 (fip40-220) or 140 (fip80-220) internal amino acids grow normally at 30°C but display a severely reduced growth rate at 37°C and require an extended incubation period for detection of colonies (Fig. 1, rows 14 and 15). The same deletions from either terminus have no effect individually (Fig. 1, compare rows 14 and 15 with rows 2 to 4). Moreover, in conjunction with a larger C-terminal deletion, which is temperature sensitive on its own (fip1-206), truncation of 40 (fip40-206) or 80 (fip80-206) amino acids from the N terminus is lethal (Fig. 1, rows 12 and 13). Taken together, these results indicate the existence of at least three domains required for normal Fip1 function: an essential domain within amino acids 80 to 105, an important but nonessential domain within amino acids 206 to 220, and two regions within the N-terminal 40 and C-terminal 107 amino acids that are important when missing in combination with each other.
FIG. 1.
Growth behavior of S. cerevisiae carrying deletions in FIP1. Plasmids carrying the indicated constructs of FIP1 were introduced into strain PJP22 by plasmid shuffling as described in Materials and Methods. The viable strains obtained from this approach were grown overnight in liquid culture, and their cell densities were normalized to an optical density of 0.5. An equal volume of serially diluted (by a factor of 10, going from left to right) cell suspensions was spotted on solid medium and incubated at the indicated temperature for up to 120 h. A schematic representation of each FIP1 deletion is shown on the left; the strain carrying the respective construct is indicated on the right. Pictures were recorded 48 h after plating except those marked with an asterisk, which were taken after 100 h. w.t., wild type.
Extracts from fip1 mutants are deficient in polyadenylation and can be complemented with recombinant proteins.
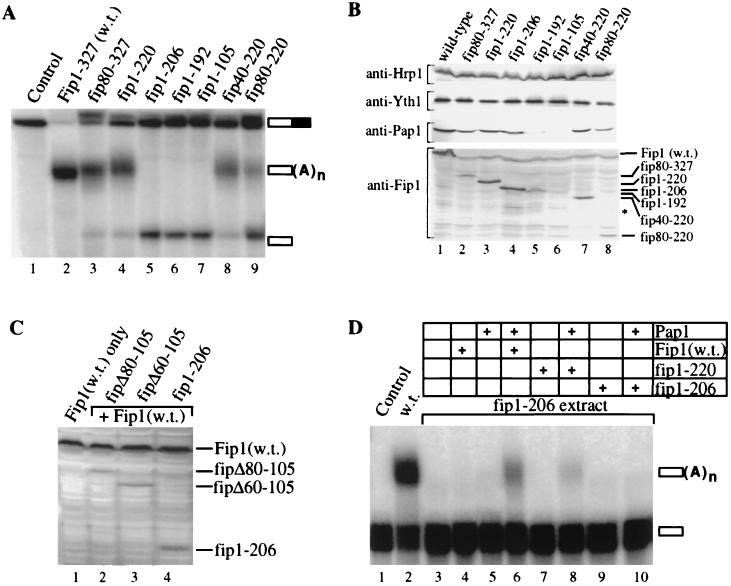
We next prepared cell extracts from viable mutants and analyzed these for the ability to polyadenylate RNA in vitro. To assay for cleavage and poly(A) addition, we incubated 32P-labeled GAL7 mRNA precursor, which contains all signals necessary for processing, with the different extracts and analyzed the products of the reaction by gel electrophoresis. Extract prepared from a strain containing the wild-type FIP1 gene efficiently cleaves and polyadenylates the RNA substrate (Fig. 2A, compare lanes 1 and 2). Unpolyadenylated cleavage product cannot be detected. Extracts from strains expressing fip80-327 or fip1-220 cleave and polyadenylate the GAL7 pre-mRNA, but without the efficiency of wild-type extract, as seen from the accumulation of cleavage product (Fig. 2A, lanes 3 and 4). Thus, although these deletions do not affect growth under the conditions tested, they display a weak polyadenylation defect. Extracts from strains expressing fip40-220 or fip80-220 exhibit a similar or slightly more severe reduction in polyadenylation activity (Fig. 2A, lanes 8 and 9), corresponding with their slow growth at 37°C. Extracts from strains carrying deletion fip1-206, fip1-192, or fip1-105, which fail to grow at 37°C, do not polyadenylate the cleaved product (Fig. 2A, lanes 5 to 7). The finding that all mutants are functional in cleavage supports previous conclusions that Fip1 is not required for this step of the polyadenylation process (23, 36). Interestingly, the polyadenylation defect in vitro is observed at 30°C, a temperature at which all of the conditional mutants are able to grow. This indicates that the extract preparation renders the polyadenylation complex more sensitive to defective Fip1 and suggests that the in vitro assay resembles the more stringent conditions found at elevated temperatures in vivo. All in all, the extent of the polyadenylation defect corresponds reasonably well with the severity of the temperature sensitivity.
FIG. 2.
Deletions in Fip1 cause a deficiency in polyadenylation. (A) In vitro cleavage and polyadenylation of GAL7 RNA using cell extracts from strains carrying deletions in FIP1. Two microliters of cell extract (30 μg of protein) prepared from strains carrying the indicated FIP1 construct was incubated with full-length radiolabeled GAL7 precursor mRNA as described in Materials and Methods. The samples were resolved on a 5% polyacrylamide–8.3 M urea gel and visualized by PhosphorImager scanning. Lane 1 contains unreacted precursor RNA. Migration of the cleaved, uncleaved, and polyadenylated RNA is indicated on the right. (B) Immunodetection of proteins required for polyadenylation in extracts from strains carrying deletions in FIP1. Thirty micrograms of protein from the indicated extract was resolved by SDS-PAGE, transferred to a solid support, and probed with antibodies against Hrp1/Nab4, Pap1, Fip1, and Yth1 as described in Materials and Methods. Migration of truncations of Fip1 is marked on the right. The asterisk denotes the migration of recombinant Fip1-105. (C) Immunodetection of Fip1, fip1-206, fipΔ60-105, and fipΔ80-105 in yeast extracts. Mutations fipΔ60-105, fipΔ80-105, and fip1-206 were expressed in a FIP1 wild-type (w.t.) background. Each lane contains 30 μg of the indicated protein extract. Samples were resolved by SDS-PAGE, transferred to a solid support, and probed with antibodies against Fip1 as described in Materials and Methods. (D) Complementation of a polyadenylation-deficient extract by the addition of recombinant Pap1 and Fip1. Two microliters of cell extract from the strain expressing fip1-206 was incubated with precleaved, radiolabeled GAL7 RNA in the presence of the recombinant protein indicated above each lane (see Materials and Methods for details). The samples were processed and visualized as for panel A. Lane 1 contains unreacted RNA; lane 2 shows the reaction with wild-type extract.
We next carried out Western analysis to determine whether the polyadenylation deficiency in these extracts is merely the consequence of a decreased stability of the Fip1 truncations. As a control, we examined the extracts for other components of the polyadenylation machinery as well. By immunodetection, the levels of Hrp1/Nab4, a component of CF I, and Yth1, a component of CPF and known interactor with Fip1, remain constant in all mutants (Fig. 2B). The amount of Pap1 is similar in most mutant extracts, but is lower in fip1-192 extract and cannot be detected in fip1-105 extract (Fig. 2B, lanes 5 and 6). There is a similar decline in the levels of truncated Fip1 in these strains. While wild-type Fip1 and most truncations can be easily visualized, the level of fip1-192 is decreased (Fig. 2B, lanes 1 to 5, 7, and 8). We repeatedly failed to detect fip1-105, indicating that this truncation is degraded rapidly in vivo or during the extract preparation (Fig. 2B, lane 6). These findings show that deletions from the C terminus beyond amino acid 206 result in decreased stability of the respective Fip1 derivative. The concurrent decrease in the level of Pap1 suggests a tight regulation of the amount of Pap1 in response to the level of Fip1 in vivo. However, in the temperature-sensitive strain expressing fip1-206, the levels of Pap1 and fip1-206 are similar to those of strains expressing full-length FIP1 and fip1-220 (Fig. 2B, compare lanes 1, 3, and 4). We therefore conclude that the loss of amino acids 206 to 220, and not an instability of Fip1, is responsible for the absence of poly(A) synthesis in extracts from this strain.
The instability of fip1-105 raised the question whether the lethal nature of the constructs fipΔ80-105 and fipΔ60-105 is due to the lack of stability of their gene products. We therefore examined extracts from strains expressing fipΔ80-105 and fipΔ60-105 in a FIP1 wild-type background by immunodetection with anti-Fip1 antibodies (Fig. 2C). The construct fip1-206 was expressed under the same conditions and served as a positive control. Both fipΔ80-105 and fipΔ60-105 can be detected in these extracts, demonstrating that they are expressed and stable (Fig. 2C, lanes 2 and 3). The levels of fipΔ80-105 and fipΔ60-105 are equivalent to the amount of fip1-206 expressed in the FIP1 background (Fig. 2C, lane 4). However, all of the Fip1 derivatives are found at levels lower than that of full-length Fip1, suggesting that the full-length protein may be more stable or expressed more efficiently. The result nonetheless shows that the lethal phenotype associated with the internal deletions fipΔ80-105 and fipΔ60-105 is not due to an inherent instability of their protein products but directly linked to the loss of these amino acids.
To determine whether polyadenylation in vitro can be recovered by supplementing extract with recombinant Fip1, we carried out complementation assays using fip1-206 extract and a synthetic GAL7 RNA precursor that ends at the cleavage site. Wild-type extract polyadenylates most of this precleaved RNA, whereas extract from the mutant strain is not active (Fig. 2D, lanes 2 and 3). Addition of full-length recombinant Fip1 to this extract has no effect (Fig. 2D, lane 4). Since the levels of the Fip1 interactors, Pap1 and Yth1, are normal, this result is unexpected and suggests that the added wild-type Fip1 cannot replace the endogenous fip1-206 in the complex. However, extract activity can be restored with a combination of full-length Fip1 and Pap1 (Fig. 2D, lane 6), suggesting that endogenous fip1-206 and Pap1 are tightly associated with one another. The polyadenylation obtained by the addition of full-length Fip1 and Pap1 is specific because the tail length is regulated correctly and a mutated RNA lacking an essential UA repeat (40) is not processed under the same conditions (data not shown). Addition of the same amount of Pap1 to the extract is not sufficient for activity (Fig. 2D, lane 5), but tripling the amount of recombinant Pap1 results in weak, UA repeat-dependent activity and produces tails of the correct length (data not shown). This finding implies that specific activity does not absolutely require full-length Fip1.
Since the fip1-220 extract is active in polyadenylation, we tested whether recombinant fip1-220 is also sufficient for the complementation of polyadenylation-deficient extracts. Recombinant fip1-220, when mixed with Pap1, indeed restores polyadenylation, although the level of activity is lower than that seen with full-length Fip1 (Fig. 2D, lanes 6 and 8). Recombinant fip1-206 alone or in combination with Pap1 cannot restore poly(A) tail synthesis to the extract (Fig. 2D, lanes 9 and 10).
These data imply that amino acids 206 to 220 of Fip1 are important for the assembly of a productive polyadenylation apparatus in vitro. However, since the fip1-206 construct can support growth at the permissive temperature in vivo, additional interactions that allow a sufficient level of poly(A) tail synthesis must be in place. This is also demonstrated by the finding that addition of excess Pap1 can support a weak level of specific activity.
Identification of Fip1 domains required for interactions with Pap1 and Yth1.
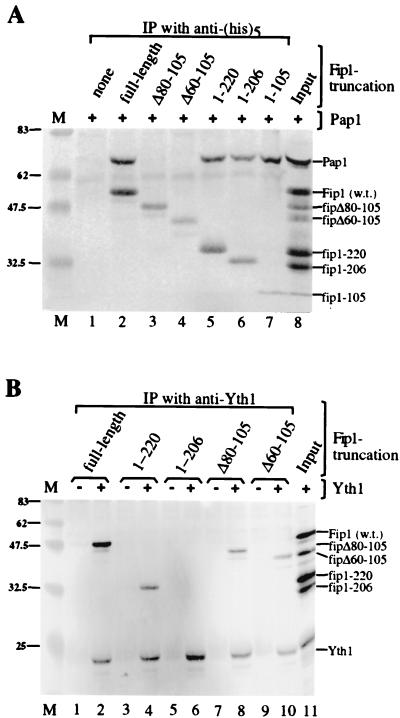
Protein-protein interactions obtained by two-hybrid analysis had previously implicated amino acids 196 to 216 of Fip1 in the association with Pap1 (23). However, the two-hybrid method cannot differentiate between direct and indirect interactions. To investigate whether amino acids 206 to 220 of Fip1 are involved in a direct interaction with Pap1, we examined the ability of purified recombinant Pap1 to coimmunoprecipitate with various Fip1 derivatives. Because the recombinant Fip1 proteins contain a hexahistidine tag, the immunoprecipitations were performed using a commercial monoclonal antibody raised against pentahistidine. For the detection, we used polyclonal antibodies raised against full-length Fip1, which recognize all truncations tested (Fig. 3A, lane 8). Full-length Fip1 and all Fip1 truncations are immunoprecipitated with the anti-His5 antibody (Fig. 3A, lanes 2 to 7), whereas Pap1 on its own is not (Fig. 3A, lane 1). The C-terminal truncations fip1-220, fip1-206, and even fip1-105 are capable of specifically coimmunoprecipitating Pap1 (Fig. 3A, lanes 5 to 7). The levels of Pap1 brought down with these truncations are similar to those seen with full-length Fip1 (Fig. 3A, lane 2). In contrast, Fip1 derivatives lacking amino acids 80 to 105 or 60 to 105 fail to coimmunoprecipitate Pap1 (Fig. 3A, lanes 3 and 4). These results show unambiguously that amino acids 206 to 220 are not required for a direct interaction with Pap1. Instead, the region between amino acids 80 and 105 is necessary to mediate the association with Pap1. Because the deletion of these 25 amino acids is lethal, the result also strongly suggests that the loss of the Pap1-Fip1 interaction cannot be tolerated in vivo. While this analysis does not precisely define the N-terminal boundary of the domain required for Pap1 binding, it is unlikely that it extends much beyond amino acid 80, since cells expressing the construct fip80-327 grow like the wild type and are active in specific polyadenylation.
FIG. 3.
Domains of Fip1 involved in protein-protein interactions. (A) Coimmunoprecipitation (IP) of recombinant Pap1 and His-tagged Fip1 truncations using anti-His5 antibody. (B) Coimmunoprecipitation of recombinant Fip1 truncations and Yth1 using anti-Yth1 antibody. The recombinant proteins indicated above the lanes were incubated with antibody coupled to protein A-agarose. Proteins in the precipitate were resolved by SDS-PAGE (12% gel) and visualized by Western detection as described in Materials and Methods. Lane 8 in panel A and lane 11 in panel B contain a mixture of the recombinant proteins loaded directly on the gel and correspond to 25% of the input. fipΔ80-105 is missing in lane 11 in panel B. The mobility of each recombinant protein is indicated on the right; positions of molecular weight markers (lane M) are shown in kilodaltons on the left. w.t., wild type.
Our finding that the Pap1-Fip1 interaction is unaffected by C-terminal truncations in Fip1 implies that the requirement for amino acids 206 to 220 in specific polyadenylation in vitro involves an interaction with a component other than Pap1. Yth1, the yeast homologue of mammalian CPSF 30, is required for poly(A) addition and interacts directly with Fip1 (2). Using antibodies against Yth1, we tested the Fip1 derivatives for the ability to coimmunoprecipitate with recombinant Yth1. Full-length Fip1 is easily visualized in the immunoprecipitate in the presence but not in the absence of Yth1 (Fig. 3B, lanes 1 and 2). Proteins fip1-220, fipΔ80-105, and fipΔ60-105 also specifically coimmunoprecipitate with Yth1 (Fig. 3B, lanes 4, 8, and 10). In contrast, truncation fip1-206 cannot be detected in the immunoprecipitate (Fig. 3B, lane 6), indicating that the 14 amino acids between residues 206 and 220 of Fip1 are required for the interaction with Yth1 in vitro. Thus, the temperature sensitivity and polyadenylation deficiency of the strain expressing fip1-206 are most likely the consequence of a disruption of the Fip1-Yth1 association. However, since our analysis of mutants also indicated the presence of a functional domain in the region C-terminal from amino acid 220, this domain may contribute to the phenotype of fip1-206 as well.
While we cannot rule out the possibility that the loss of binding is due to denaturation of the recombinant truncations, the fact that disrupting the interaction with Pap1 does not have an adverse effect on the interaction with Yth1 and vice versa suggests that the tertiary structure is maintained. Moreover, all truncations of Fip1 are immunoprecipitated by polyclonal antibodies raised against full-length Fip1 (data not shown), indicating that epitopes for the antibodies remain intact.
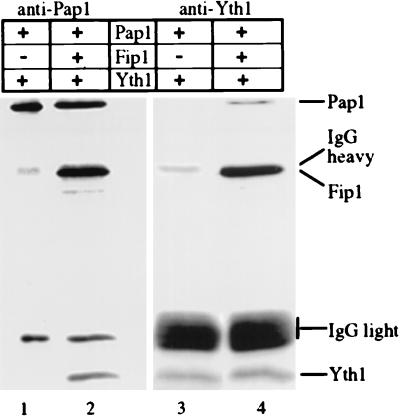
Our results imply that the interaction domains for Pap1 and Yth1 are distinct and that Pap1 can be linked to Yth1 through the interaction of both proteins with Fip1. To demonstrate this directly, we carried out immunoprecipitations of recombinant proteins using antibodies against either Yth1 or Pap1. Coimmunoprecipitation of Pap1 and Yth1 with either antibody is strictly dependent on the presence of Fip1 (Fig. 4, lanes 2 and 4), as no such interaction can be observed in its absence (Fig. 3, lanes 1 and 3). Thus, the three proteins can form a ternary complex with Fip1 functioning as a bridge between Pap1 and Yth1.
FIG. 4.
Fip1 interacts simultaneously with Pap1 and Yth1. The indicated recombinant proteins were subjected to immunoprecipitation with antibody against Pap1 (lanes 1 and 2) or Yth1 (lanes 3 and 4) coupled to protein A beads as described in Materials and Methods. After washing the beads, the proteins in the precipitates were eluted, resolved by SDS-PAGE (12% gel), and analyzed by immunoblotting. The migration of the recombinant proteins and the immunoglobulin G (IgG) heavy and light chains are indicated on the right.
Amino acids 80 to 105 of Fip1 are required for the inhibition of nonspecific polyadenylation.
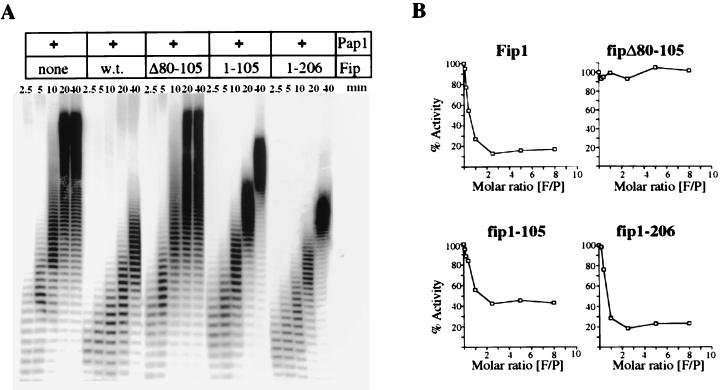
We had previously shown that the association with Fip1 impairs the processivity of Pap1 in nonspecific polyadenylation assays by limiting access of the primer to the RBD-C of Pap1 (39). To determine whether our recombinant Fip1 deletions are capable of this inhibitory activity, we tested them in nonspecific assays using Pap1, oligo(A)12, and α-32P-labeled ATP. Samples were removed at different time points, and the products were resolved on an 18% polyacrylamide gel (Fig. 5A). Pap1 on its own processively synthesizes a long tail onto the primer. In the presence of wild-type Fip1, the overall activity is decreased and the mode of poly(A) synthesis becomes more distributive, as was shown previously. No effect on poly(A) synthesis is observed when fipΔ80-105 replaces full-length Fip1 in the reaction, demonstrating that the inhibitory effect of Fip1 requires the interaction between Pap1 and Fip1. Inclusion of fip1-105 or of fip1-206 in the reaction produces a pattern similar to that observed with wild-type Fip1. The tails synthesized are shorter, and the mode of poly(A) addition becomes distributive. However, fip1-105 does not inhibit Pap1 as effectively as full-length Fip1.
FIG. 5.
Inhibition of nonspecific polyadenylation by Fip1 truncations. (A) Twenty-five nanograms of Pap1 was incubated at 25°C with 1 μM oligo(A)12 and labeled ATP in the absence or presence of 30 ng of wild-type (w.t.) Fip1 or the indicated Fip1 truncations as described in Materials and Methods. Samples were removed at the indicated time points and resolved by electrophoresis on 18% polyacrylamide–8.3 M urea gels. (B) Reactions were carried out as for panel A except that the amounts of Fip1 or the indicated Fip1 derivatives were varied and the entire reaction was terminated after 10 min by addition of 100 μl of trichloroacetic acid. Acid-precipitable counts were collected by filter binding, quantitated by scintillation counting, and expressed as a percentage of the activity obtained with Pap1 in the absence of Fip1. The resulting values were plotted as a function of the Fip1/Pap1 (F/P) molar ratio.
To quantitate the inhibition of nonspecific polyadenylation by the different Fip1 derivatives, were terminated polyadenylation reactions after 10 min and determined the activity by scintillation counting of acid-precipitable radioactivity. This value was expressed as a percentage of the Pap1 activity without Fip1, and the results were plotted as a function of the Fip1/Pap1 molar ratio (Fig. 5B). At a primer concentration of 1 μM, wild-type Fip1 reduces the activity to 18% of the activity of Pap1 alone, as previously demonstrated. Increasing the Fip1/Pap1 molar ratio beyond 2:1 does not cause further inhibition. In the presence of fipΔ80-105, polyadenylation proceeds uninhibited. Even an eightfold molar excess of fipΔ80-105 does not inhibit activity, suggesting that binding is completely abolished. In the presence of fip1-105 or fip1-206, the activity of Pap1 at 1 μM oligo(A)12 is reduced to 45 or 23%, respectively. The ability to associate with Pap1 is not affected in either C-terminal truncation since both, like full-length Fip1, reach close to maximal inhibition at a 1:1 molar ratio of Fip1 to Pap1. Since the inhibition of Pap1 by Fip1 can be competed by increasing the primer concentration, we also determined the Michaelis-Menten constant Km of Pap1 for oligo(A)12 in the presence of fip1-105. This value is 5.3 μM in comparison to 10 μM for Pap1 in the presence of full-length Fip1. The Km of Pap1 alone is 0.5 μM.
In summary, these findings show that amino acids 80 to 105 of Fip1 are required for the inhibition and that a region between amino acids 105 and 206 contributes to preventing RNA from binding at the C-RBD of Pap1.
Interactions at the C terminus of Fip1 are necessary to overcome the inhibition of Pap1 in specific polyadenylation.
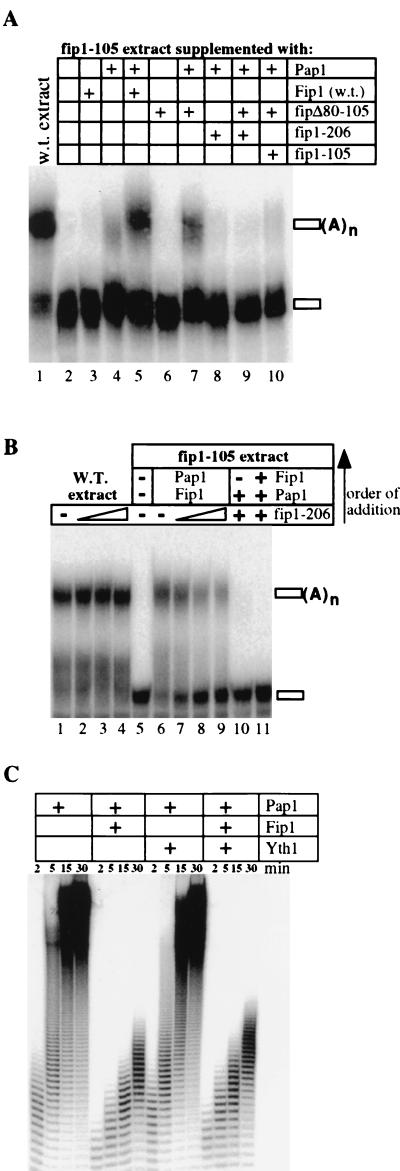
For Pap1 to function efficiently in specific polyadenylation, there must be a mechanism to release Fip1's negative effect on Pap1 activity in the context of the entire polyadenylation machinery. We have shown that recombinant fip1-206 inhibits nonspecific Pap1 activity with an efficiency similar to that of wild-type Fip1. It is therefore possible that the polyadenylation deficiency of fip1-206 extract is due to an inability to reverse the inhibiting effect of Fip1. To test this hypothesis, we used extract from a strain expressing fip1-105. Since this extract is depleted of Pap1 and Fip1 but contains normal levels of other polyadenylation components (Fig. 2B), one can study the effects of recombinant Fip1 derivatives on specific polyadenylation. Hence, it allows the examination of lethal Fip1 derivatives, such as fipΔ80-105, or of combinations of Fip1 derivatives. The fip1-105 extract alone is not active for polyadenylation (Fig. 6A, lane 2), and this deficiency cannot be alleviated by the addition of recombinant full-length Fip1 or fipΔ80-105 (Fig. 6A, lanes 3 and 6). Supplementing the fip1-105 extract with Pap1 supports a weak level of polyadenylation (Fig. 6A, lane 4). This discovery, like the observation that excess Pap1 weakly restores specific activity to fip1-206 extract, indicates that Pap1 can be directed to the rest of the complex in the absence of Fip1 and weakly polyadenylate the RNA in vitro. Addition of fipΔ80-105 alongside Pap1 stimulates the polyadenylation activity despite this construct's inability to interact directly with Pap1 (Fig. 6A, lane 7). While the activity does not reach the level of complementation obtained with Pap1 and full-length Fip1, it is reproducibly higher than that observed with Pap1 on its own (Fig. 6A, compare lanes 4 and 5 with lane 7). In contrast, inclusion of fip1-206 with Pap1 in the reaction does not stimulate the poly(A) addition and instead suppresses the poly(A) synthesis seen with Pap1 added on its own (Fig. 6A, lane 8). Moreover, the repression of polyadenylation by fip1-206 is dominant over the stimulatory effect of fipΔ80-105 (Fig. 6A, lane 9). These results suggests that fip1-206 locks Pap1 in an inhibited state. A similar effect is observed with recombinant fip1-105 in extract supplemented with Pap1 and fipΔ80-105 (Fig. 6A, lane 10). However, in agreement with the reduced ability of fip1-105 to inhibit nonspecific polyadenylation, a weak level of poly(A) synthesis can still be detected in this reaction. All observed activity is specific with regard to the substrate, because it cannot be obtained with a mutant RNA that lacks an essential UA repeat (data not shown).
FIG. 6.
Fip1-mediated inhibition of Pap1 activity can be relieved in the polyadenylation complex. (A) Effects of recombinant Fip1 derivatives in specific polyadenylation using fip1-105 extract. The recombinant proteins indicated above the lanes were incubated with labeled precleaved GAL7 RNA, ATP, and 2 μl of extract from a strain expressing fip1-105, which contains no detectable amounts of Pap1 or Fip1-105. Reactions were carried out and processed as described in Materials and Methods. Lane 1 contains wild-type (w.t.) extract as a positive control. (B) Binding of fip1-206 to Pap1 prevents specific polyadenylation. The recombinant proteins indicated above the lanes were incubated with labeled precleaved GAL7 RNA, ATP, and 2 μl of extract from a wild-type strain (lanes 1 to 4) or from a strain expressing fip1-105 (lanes 5 to 11). The reactions were supplemented with recombinant proteins as follows: 100, 400, and 800 ng of fip1-206 (lanes 2 to 4, respectively); 75 ng of Pap1 and 100 ng of Fip1 (lane 6); 100 ng of Fip1, 75 ng of Pap1, and 100, 400, or 800 ng of fip1-206 (lanes 7 to 9, respectively) with fip1-206 and Fip1 mixed before the addition of Pap1; 75 ng of Pap1 and 100 ng of Fip1-206 (lane 10); 100 ng of fip1-206, 75 ng of Pap1, and 100 ng of Fip1 (lane 11), assembled in that order. The amounts of Fip1 and fip1-206 are always in molar excess of Pap1; extract was always added last. The order of addition is also indicated by the arrow on the right. (C) Yth1 does not relieve Fip1-mediated inhibition of nonspecific polyadenylation. Pap1 (25 ng) was incubated at 25°C with 1 μM oligo(A)12 and labeled ATP either in the absence or in the presence of 30 ng of Fip1, 50 ng of Yth1, or both as described in Materials and Methods. Samples were removed at the indicated time points and resolved by electrophoresis on 18% polyacrylamide–8.3 M urea gels.
To analyze in more detail an inhibiting effect of fip1-206 in specific polyadenylation, we carried out specific polyadenylation assays with wild-type extract in the presence of fip1-206. Wild-type extract on its own efficiently polyadenylates the precleaved GAL7 RNA (Fig. 6B, lane 1). No inhibition of polyadenylation can be observed by including increasing amounts of fip1-206 in the reaction with wild-type extract (Fig. 6B, lanes 2 to 4). Thus, like the failure to complement fip1-206 extract with recombinant wild-type Fip1 (Fig. 2D), exogenous fip1-206 cannot inhibit polyadenylation in wild-type extract. This result again suggests a tight complex between Pap1 and Fip1. To address whether fip1-206 can inhibit specific polyadenylation, we used recombinant Pap1, Fip1, and fip1-206 with fip1-105 extract. Since this extract requires exogenous Pap1 and Fip1 for activity, the recombinant proteins can be assembled in the desired order of addition. As shown before, the fip1-105 extract on its own is not active (Fig. 6B, lane 5). Addition of a combination of Fip1 and Pap1 to the extract efficiently restores polyadenylation (Fig. 6B, lane 6). This complementation is progressively inhibited when Pap1 is added to a mix consisting of Fip1 and increasing amounts of fip1-206 (Fig. 6B, lanes 6 to 8). The degree of inhibition increases with the fip1-206/Fip1 ratio and is most obvious when comparing the unreacted GAL7 RNA. We next wanted to test the effect of preincubating Pap1 with fip1-206 before adding wild-type Fip1 to the mixture. Supplementing fip1-105 extract with Pap1 and Fip1-206 alone cannot restore polyadenylation (Fig. 6B, lane 10). Supplementing the extract with a mixture containing Pap1, Fip1, and fip1-206, in which Pap1 and fip1-206 had been preincubated for 5 min before the addition of full-length Fip1, also fails to restore polyadenylation (Fig. 6B, lanes 11). These data indicate that Pap1 binds tightly to either Fip1 or fip1-206 and remains associated with its binding partner under these reaction conditions. Since fip1-206 suppresses the specific activity restored to fip1-105 extract by the addition of Pap1 alone or Pap1 in combination with fipΔ80-105 (Fig. 6A, lanes 8 and 9), these results strongly suggest that fip1-206 traps Pap1 in the inhibited state.
The fact that inhibition of specific polyadenylation cannot be observed with full-length Fip1 indicates that the repressive effect can be released in the context of the polyadenylation complex and that this requires interactions at the C-terminal 121 amino acids of Fip1. To test whether the Fip1-Yth1 interaction itself is sufficient to relieve the Fip1-mediated repression of Pap1, we included recombinant Yth1 in nonspecific polyadenylation assays and analyzed the reaction products by gel electrophoresis (Fig. 6C). The results show that the activity of Pap1 and Fip1 is unchanged in the presence of Yth1. Thus, the Fip1-Yth1 interaction is not sufficient to release the repression of Pap1 by Fip1 in this assay.
In summary, a low level of specific polyadenylation can be observed in vitro when Pap1 is added to extracts lacking Fip1. This activity is suppressed in the presence of fip1-206, which binds to Pap1 but not to Yth1. Interactions of Fip1 with components of the polyadenylation complex other than Pap1 have an additional activating effect. The low level of specific polyadenylation observed with Pap1 and fip1-105 extract is stimulated in the presence of fipΔ80-105. Since fipΔ80-105 does not bind to Pap1 directly, the stimulatory effect must be transmitted to Pap1 through interactions of fipΔ80-105 with other subunits of the polyadenylation complex. Thus, these interactions regulate the level of Pap1 activity in specific polyadenylation both through releasing the Fip1-mediated inhibition as well as through a direct activation.
DISCUSSION
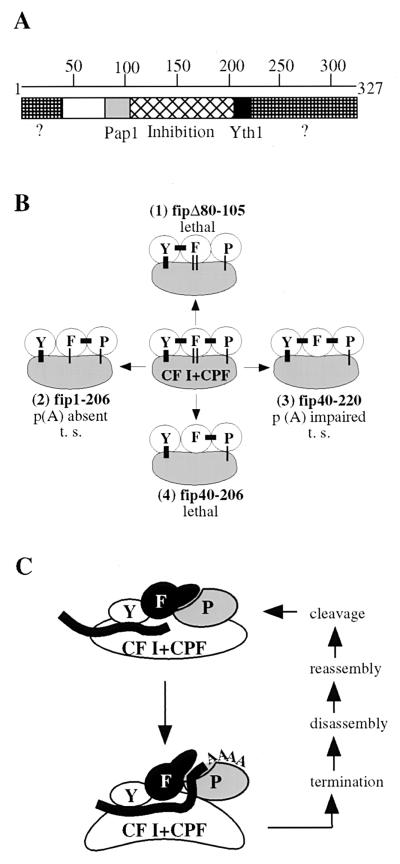
We have previously shown that Fip1 inhibits nonspecific polyadenylation by interfering with binding of RNA to the C-RBD of Pap1 (39). Here we present the identification of domains in Fip1 that are involved in separate protein-protein interactions and the inhibition of Pap1 activity. Moreover, we demonstrate that these interactions are crucial in the control of Pap1 activity during the polyadenylation process. The data for the most important deletions of Fip1 are summarized in Table 1. Our analysis reveals the domain structure of Fip1 illustrated in Fig. 7A. Amino acids 80 to 105 are necessary for the direct Pap1-Fip1 association and essential for viability. A region within amino acids 105 to 206 is important for full inhibition of Pap1. This domain is not required for binding to Pap1 but contributes to limiting access of the RNA substrate to the C-RBD of Pap1. The interaction with Yth1 requires the presence of amino acids 206 to 220. C-terminal deletions beyond amino acid 220 result in temperature sensitivity and a polyadenylation defect. Our analysis also indicates the presence of important regions within the N-terminal 40 and the C-terminal 107 amino acids (Fig. 7A, marked with question marks). The functions of these regions are not known. They may be involved in interactions with other components of the polyadenylation complex.
TABLE 1.
Effects of FIP1 deletionsa
| Fip derivative | Growth at:
|
Interaction
|
Protein level in extract
|
Specific polyadenylation in extract | |||
|---|---|---|---|---|---|---|---|
| 30°C | 37°C | P | Y | P | F | ||
| Fip1 (wild type) | ++ | ++ | + | + | + | + | +++ |
| fip40-327 | ++ | ++ | |||||
| fip80-327 | ++ | ++ | + | + | ++ | ||
| fip1-220 | ++ | ++ | + | + | + | + | ++ |
| fip1-206 | + | − | + | − | + | + | − |
| fip1-105 | + | − | + | − | − | − | − |
| fipΔ80-105 | − | − | − | + | (+) | ||
| fipΔ60-105 | − | − | − | + | (+) | ||
| fip40-206 | − | − | |||||
| fip80-206 | − | − | |||||
| fip40-220 | ++ | + | + | + | ++ | ||
| fip80-220 | ++ | + | + | + | + | ||
P, Pap1; Y, Yth1; F, Fip1 or derivative; (+), tested in a FIP1 background.
FIG. 7.
Fip1 regulates polyadenylation. (A) Domain structure of Fip1. The question marks indicate regions of Fip1 containing domains of unknown functions. The Pap1 interaction domain could extend further toward the N terminus. (B) Deletions of interaction domains of Fip1 lead to defects in polyadenylation or lethality. F, Fip1; Y, Yth1; P, Pap1. Bars indicate known associations. Thick bars represent interactions in which both partners are known; thin bars indicate that one interaction partner has not been identified. The thin double bars linking Fip1 and CF I/CPF represent the associations involving the C and N termini of Fip1. t. s. (temperature sensitive) refers to growth at 37°C; p(A) (polyadenylation) refers to activity obtained with extract from a strain carrying the corresponding fip1 construct. See text for details. (C) Model of the regulatory role of Fip1 during the polyadenylation of cleaved RNA. Multiple interactions between Fip1 and other components of the polyadenylation machinery release the inhibition of Pap1 after cleavage and activate specific polyadenylation. See text for details.
Our data demonstrates that the defects in growth, viability, and polyadenylation caused by elimination of different interactions vary in severity. This is summarized in Fig. 7B, which shows the connections of Fip1 and its associated partners within the complex. While the loss of the Pap1-Fip1 interaction is lethal (Fig. 7B, complex 1), cells with deletions of the Yth1 interaction domain, including the region of unknown function within the C-terminal 107 amino acids, remain viable despite a defect in polyadenylation (Fig. 7B, complex 2). Concurrent truncation of the N-terminal 40 and C-terminal 107 amino acids results in mild effects on growth and polyadenylation (Fig. 7B, complex 3). Good candidates for proteins interacting with these regions are Rna14 of CF I and Pfs2 of CPF, since both proteins have been shown to bind weakly to Fip1 (21, 23). We have repeatedly failed to reproduce these interactions in coimmunoprecipitations with recombinant proteins (data not shown), but their weaker nature may place them outside the detection limits of our assay. The fact that extract from a strain carrying fip40-220 is functional in specific polyadenylation indicates that the contributions of these regions are dispensable for activity. However, the additional loss of the interaction with Yth1 causes lethality (Fig. 7B, complex 4). This implies a functional overlap for these domains and indicates that multiple contacts of Fip1, and especially the interaction with Yth1, contribute to the overall stability of the active polyadenylation complex. These results are in agreement with recent findings by Barabino et al. (3), which show that a mutated Yth1 deficient in binding Fip1 cannot tightly tether a Pap1-Fip1 subcomplex to the polyadenylation machinery. The corresponding yth1-1 mutant strain displays temperature sensitivity and a defect in specific polyadenylation similar to the phenotype of the strain carrying fip1-206.
Based on the ability to interact simultaneously with Pap1 and Rna14 (23), a major role of Fip1 was thought to be the recruitment of Pap1 to the polyadenylation machinery. We find, however, that Pap1 weakly, but specifically, polyadenylates RNA when added to extracts depleted of Fip1. Thus, Pap1 is able to associate with the polyadenylation complex on its own. This is not surprising given the fact that the N-terminal 18 amino acids of Pap1 are necessary for a productive interaction with the processing complex (40) yet are clearly not required for the direct interaction with Fip1 (39). Consistent with this, in the three-dimensional structure of Pap1, the N-terminal region is located far away on an opposite face from the Fip1 interaction domain (4).
The repression of Pap1 by Fip1 cannot be detected in a fully functional polyadenylation complex, because in this context the polyadenylation machinery can release the inhibition to allow specific and processive poly(A) tail synthesis. However, we show that the specific polyadenylation observed with fip1-105 extracts and recombinant Pap1 is suppressed in the presence of fip1-206. The finding suggests that the Pap1-Fip1 interaction, in addition to stabilizing the association of Pap1 with the complex, is important for the regulation of Pap1 activity. Our data imply that in extracts from strains expressing fip1-206, Pap1 is trapped in an inhibited state. This conclusion is supported by findings of Preker et al., which show that mRNAs from temperature-sensitive mutants with premature stop codons at amino acids 217 or 197 of Fip1 have short poly(A) tails (23).
Our data demonstrate that Yth1 is one of the proteins interacting in the region deleted in fip1-206. However, the fact that the association between Fip1 and Yth1 is by itself not sufficient to relieve Fip1-mediated inhibition of Pap1 in nonspecific assays suggests that additional interactions in the polyadenylation complex are required for the release of inhibition. Although the extent and nature of these interactions are not known, they likely involve components interacting at the C terminus of Fip1 and one or more component interacting with Yth1.
Interactions at the C terminus of Fip1 also activate polyadenylation by a mechanism that does not involve the release of Fip1-mediated inhibition. This is indicated by the fact that fipΔ80-105 stimulates polyadenylation when added to fip1-105 extract supplemented with Pap1. Since fipΔ80-105 neither binds nor inhibits Pap1, its stimulatory effect on polyadenylation must be the result of its interactions with other components of the polyadenylation complex.
Based on these results, we propose the model of polyadenylation shown in Fig. 7C. Fip1 is tightly associated with Pap1 and inhibits its intrinsic activity by limiting access of RNA to the CRBD. Upon completion of cleavage, a burst of processive poly(A) synthesis occurs. The events leading to this activation of polyadenylation require interactions at the C-terminal domain of Fip1, which most likely stabilize the association of the Pap1-Fip1 subcomplex with the polyadenylation machinery. In this postcleavage complex, activation of Pap1 activity occurs in two ways. First, the inhibition of Pap1 by Fip1 is released, most likely through a conformational change, causing a repositioning of the inhibition domain of Fip1, which allows the C-RBD of Pap1 to engage RNA. Second, and probably simultaneously with the release of inhibition, the interactions of Fip1 also provide a direct stimulatory effect by an unknown mechanism. Subsequent steps leading to termination are poorly understood but include the action of Pab1 (1, 12, 18). This restores the Fip1-mediated inhibition, the complex disassembles, and a new round of processing can take place. In this model, Fip1 is the central regulator of the catalytic subunit of the polyadenylation machinery and tightly controls Pap1's state of activity through a network of interactions.
A model involving Pap1 inhibition is appealing because the cell must have a mechanism to avoid the polyadenylation of RNAs that are not subject to this modification. Since Pap1 on its own does not discriminate much between RNA substrates (15), another important function of Fip1 may be the prevention of nondiscriminating Pap1 activity. Overexpression of PAP in chicken cells interferes with cell growth, indicating that it is extremely important for the cell to avoid an increase of deregulated PAP activity (38). Our observations that Pap1 levels in cells decrease in strains with unstable Fip1 truncations and that the loss of the Pap1-Fip1 interaction causes lethality and an inability to inhibit nonspecific polyadenylation support this hypothesis. In contrast, reduced levels of PAP in chicken cells are tolerated well (38).
Such an important regulatory function for Fip1 raises the question why there seems to be no mammalian homologue. However, p160, the homologue of yeast Cft1 (28) and largest subunit of CPSF, inhibits PAP in nonspecific polyadenylation, perhaps in a fashion similar to Fip1 (20). Thus, a mechanism involving repression of PAP activity in mammals could be achieved through different interactions within the complex. The involvement of PAP in cleavage (8) and the stimulation of processivity by PAB II (30, 32), a subunit with no homologue in yeast, also point to differences in the organization and activation of the mammalian polyadenylation machinery. Nonetheless, the basic principle of this mechanism appears to be preserved.
Our study demonstrates how multiple protein contacts act in synchrony to regulate the synthesis of the poly(A) tail. Many details remain to be elucidated. For instance, the signals for the release of repression and concurrent activation are not known. This likely involves a change in the complex triggered by the cleavage step. It is tempting to speculate that Yth1, with its RNA-binding ability (2), its involvement in cleavage (3), and its direct interaction with Fip1 (2) plays the primary role in relaying this message to Pap1. Other questions concern the role of Fip1 during termination and whether the Pap1-Fip1 subcomplex is associated with the machinery during cleavage. Future research is needed to examine these problems and other molecular events of this process.
ACKNOWLEDGMENTS
We thank Pascal Preker and Walter Keller for the yeast strain PJP22 and plasmids pFL11 and pIA34. We are grateful to Kimberly Sparks for critically reading the manuscript and all members of the Moore laboratory for helpful discussions.
This research was supported by NIH grant GM57218 to CLM.
REFERENCES
- 1.Amrani N, Minet M, LeGouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barabino S M, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homologue are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 3.Barabino S M, Ohnacker M, Keller W. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 2000;19:3778–3787. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bard J, Zhelkovsky A M, Earnest T N, Helmling S, Moore C L, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- 5.Becker D M, Guarente L G R. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:183–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 6.Beelman C, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 7.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–19776. [PubMed] [Google Scholar]
- 9.Butler J S, Platt T. RNA processing generates the mature 3′ ends of yeast CYC1 mRNA in vitro. Science. 1988;242:1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Moore C L. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Carmichael G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler M, Henry M, Gross S, Shen E, Zhao J, Silver P, Moore C. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler M, Zhelkovsky A M, Skvorak A, Moore C L. Monoclonal antibodies to yeast poly(A) polymerase (PAP) provide evidence for association of PAP with cleavage factor I. Biochemistry. 1995;34:1750–1759. doi: 10.1021/bi00005a032. [DOI] [PubMed] [Google Scholar]
- 14.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from Saccharomyces cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 15.Lingner J, Radtke I, Wahle E, Keller W. Purification and characterization of poly(A) polymerase from Saccharomyces cerevisiae. J Biol Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- 16.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 17.Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minvielle-Sebastia L, Preker P, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Nat Acad Sci. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore C, Sharp P. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985;41:845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- 20.Murthy K G K, Manley J L. The 160 kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 21.Ohnacker M, Barabino S M, Preker P J, Keller W. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end processing complex. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preker P, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 24.Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz H. Suppression of irrelevant signals in immunoblots by preconjugation of primary antibodies. BioTechniques. 1997;23:1007–1008. doi: 10.2144/97236bm07. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 28.Stumpf G, Domdey H. Dependence of yeast pre-mRNA 3′-end processing on Cft1: a sequence homologue of the mammalian AAUAAA binding factor. Science. 1996;274:1517–1520. doi: 10.1126/science.274.5292.1517. [DOI] [PubMed] [Google Scholar]
- 29.Takagaki Y, Ryner L C, Manley J L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA processing and polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 30.Wahle E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- 31.Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 32.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 33.Wahle E, Martin G, Schiltz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahle E, Ruegsegger U. 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Kessler M, Moore C. Cleavage factor II of S. cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J Biol Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 37.Zhao J, Kessler M, Helmling S, O'Connor J P, Moore C L. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition reactions of mRNA 3′ end formation. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao W, Manley J L. Deregulation of poly(A) polymerase interferes with cell growth. Mol Cell Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhelkovsky A, Helmling S, Moore C. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol Cell Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhelkovsky A M, Kessler M M, Moore C L. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. J Biol Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]