Abstract
Phage typing and DNA macrorestriction fragment analysis by pulsed-field gel electrophoresis (PFGE) were evaluated for use in the epidemiological subtyping of Escherichia coli serogroup O157 strains isolated in Ontario, Canada. Among 30 strains isolated from patients with sporadic cases of infection, 22 distinct XbaI macrorestriction patterns were identified and 17 strains exhibited unique PFGE patterns. In contrast, phage typing identified only seven different phage types and 17 strains belonged to the same phage type. A total of 25 phage type-macrorestriction pattern combinations were identified among the strains from patients with sporadic cases of infection. PFGE subtyping differentiated between unrelated strains that exhibited the same phage type, and in one group of strains, phage typing differentiated between strains of the same PFGE subtype. Both typing procedures correctly identified outbreak-related isolates as belonging to the same type in four separate outbreaks. Each outbreak strain was characterized by a distinct macrorestriction pattern, while phage typing subdivided the outbreak strains into only three different types. A small percentage of outbreak-related isolates had PFGE patterns that differed slightly (one or two DNA fragment differences) from that of the outbreak strain. On the other hand, each isolate from the same outbreak belonged to the same phage type as that of the outbreak strain. We conclude that phage typing and PFGE fingerprinting represent complementary procedures for the subtyping of E. coli serogroup O157 and that the combined use of these procedures provides optimal discrimination.
Verotoxigenic Escherichia coli serogroup O157 has emerged as an important cause of both sporadic and epidemic disease in North America and other regions of the world. The majority of infections result in mild diarrhea, but more serious manifestations of this infection often result in hemorrhagic colitis and hemolytic-uremic syndrome (10). Most outbreaks and sporadic cases have been associated with the consumption of foods of bovine origin, but in recent years, an increasing number of other foodstuffs have been implicated in the transmission of these pathogenic organisms (4, 5, 14).
Epidemiological investigations of outbreaks caused by E. coli O157 have been greatly assisted by laboratory procedures for the subtyping of isolates. During the last decade, numerous subtyping methodologies have been developed, but phage typing and macrorestriction fragment analysis of DNA by pulsed-field gel electrophoresis (PFGE) have become the most commonly used (16). Despite the successful use of these procedures, several issues, including the heterogeneity and stability of phage types and macrorestriction patterns and the relationships between these epidemiological markers, have yet to be fully resolved (2, 3, 7, 8, 9, 12, 13, 18). Our laboratories have evaluated these procedures for the subtyping of E. coli O157 strains isolated in the province of Ontario, and in this report, we present the results of our investigation.
MATERIALS AND METHODS
Bacteria.
A total of 94 isolates of E. coli O157, all isolated from humans, were analyzed in this study. Thirty of the isolates were isolated from patients with sporadic cases of infection in Ontario during 1995 and were representative of a larger collection of strains. These are referred to as “sporadic isolates.” Sixty-four isolates were associated with four of the largest outbreaks that occurred in Ontario between 1993 and 1996. For the purposes of our study, the term “outbreak strain” refers to a strain designated as typical of the isolates associated with a particular outbreak. Stock cultures of bacteria were maintained in a storage solution consisting of brain heart infusion broth (Difco Laboratories, Detroit, Mich.) and 15% (vol/vol) glycerol at −70°C and were cultivated on blood agar plates at 37°C for 18 h. All cultures were identified as E. coli O157 by using standard laboratory criteria, and E. coli ATCC 43894 was used as a control strain.
Phage typing.
Phage typing was performed by the National Laboratory for Enteric Pathogens, Laboratory Centre for Disease Control, by procedures described previously (1, 11).
DNA macrorestriction fragment analysis.
The incorporation of bacterial cells into agarose plugs and the preparation of genomic DNA were performed by using the following procedure. Bacterial growth from an agar plate was harvested with a sterile cotton swab into 3 ml of SE buffer (75 mM NaCl, 25 mM EDTA [pH 7.5]). The density of the cell suspension was then adjusted to an absorbance of 1.4 at 600 nm with SE buffer, and then 0.5 ml of this suspension was added to 0.5 ml of 2% melted low-melting-point agarose (SeaPlaque GTG Agarose; FMC Bioproducts, Rockland, Maine) at 55°C with gentle mixing. Aliquots of this mixture were added to plug mold slots, and the plugs were allowed to set for 10 min at room temperature. After setting, the plugs were immersed in 2 ml of lysis buffer (1% [wt/vol] N-lauryl sarcosine, 0.5 M EDTA [pH 9.5]) containing 50 μl of 20 mg of proteinase K (Roche Molecular Biochemicals, Laval, Quebec, Canada) per ml. The plugs were incubated for 18 h in a shaking water bath at 50°C. After lysis, the lysis buffer was removed and the plugs were washed four times for 1 h each time with 2 ml of TE buffer (10 mM Tris, 10 mM EDTA [pH 7.5]) at room temperature on an orbital rotator.
The digestion of genomic DNA with the infrequently cutting restriction enzymes XbaI and SfiI (New England Biolabs, Mississauga, Ontario, Canada) was carried out according to the manufacturer's recommendations. Prior to digestion, the DNA plugs were immersed in 200 μl of the appropriate enzyme buffer for 1 h at room temperature on an orbital rotator. High-molecular-weight DNA restriction fragments were separated by using the clamped homogeneous electric field (CHEF) PFGE system (CHEF-DRIII apparatus; Bio-Rad Laboratories, Mississauga, Ontario, Canada). Electrophoresis was performed with 1% agarose gels (Pulsed Field Certified Agarose; Bio-Rad) in 0.5× TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA [pH 8.0]) for 20 h at 14°C. The voltage gradient was 6 V/cm, and the reorientation angle was 120°. For XbaI digests, a linearly ramped pulse time of 5 to 50 s was used, and for SfiI digests, a linearly ramped pulse time of 2 to 20 s was used. Following electrophoresis, the gels were stained with ethidium bromide (0.5 μg/ml) and the macrorestriction fragment patterns were visualized by UV illumination and then photographed.
Sporadic isolates with indistinguishable macrorestriction patterns were considered to belong to the same PFGE subtype. For epidemiologically associated outbreak isolates, the patterns were interpreted according to the criteria proposed by Tenover et al. (17).
RESULTS
In the first phase of the study, the phage types and XbaI macrorestriction patterns of 30 sporadic strains of E. coli O157 were compared. Phage typing subdivided these strains into seven different phage types. Seventeen of the strains exhibited phage type 14, and only two strains had unique phage types. In contrast, a total of 22 different PFGE subtypes were identified among these sporadic strains. Five of the strains displayed macrorestriction pattern E, and 17 strains exhibited unique patterns.
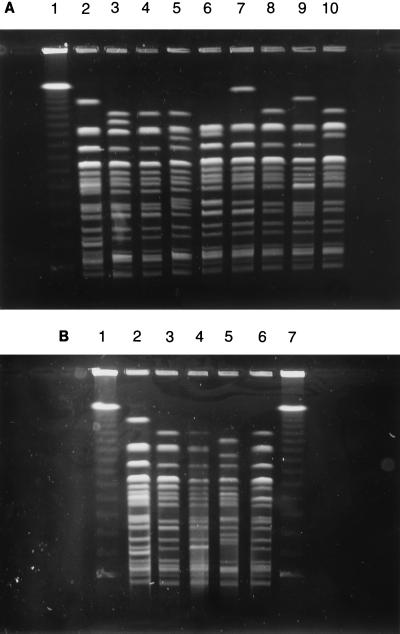
Comparison of individual strains showed that PFGE typing could subdivide strains that belonged to the same phage type. Among the 17 strains of phage type 14, 13 different PFGE subtypes were detected, and representative results are shown in Fig. 1A. In addition, PFGE typing subdivided four phage type 8 strains into four different subtypes and three phage type 2 strains into two different subtypes. Among strains that exhibited the same macrorestriction pattern, phage typing subdivided five strains characterized by pattern E into phage types 1, 8, 14 (two strains), and 32, and further PFGE analysis of these strains with restriction enzyme SfiI revealed genomic differences among some of these strains (data not shown). However, phage typing did not subdivide any other groups of strains that had the same XbaI PFGE pattern. Among the 30 sporadic strains, a total of 25 unique phage type-PFGE subtype combinations were identified.
FIG. 1.
(A) Representative DNA macrorestriction patterns of XbaI-digested genomic DNAs from E. coli O157 phage type 14 strains isolated in Ontario. Lanes: 1, lambda phage concatemer; 2, E. coli ATCC 48394; 3 through 10, sporadic E. coli O157 isolates. (B) DNA macrorestriction patterns of XbaI-digested genomic DNAs from E. coli O157 outbreak strains isolated in Ontario. Lanes: 1 and 7, lambda phage concatemer; 2, E. coli ATCC 48394; 3 through 6, E. coli O157 strains representative of four distinct outbreaks.
In the next phase of the study, we analyzed isolates associated with four separate food-borne outbreaks of E. coli O157 infection that occurred in Ontario between 1993 and 1996. Phage typing revealed that the isolates associated with outbreaks 1 and 3 belonged to phage type 2, while the isolates associated with outbreaks 2 and 4 belonged to phage types 10 and 4, respectively. PFGE analysis showed that each group of outbreak isolates was characterized by a distinct outbreak pattern, despite marked similarities between these patterns (Fig. 1B). Two of the patterns identified among the group of outbreak strains were indistinguishable from two patterns identified among the group of sporadic strains. The patterns of all individual isolates from outbreaks 1 and 4 were identical to those of the outbreak strains. In contrast, 1 of 14 isolates associated with an outbreak in a hospital and 4 of 26 isolates from a day care center outbreak exhibited patterns that differed slightly (one or two fragment differences) from those of the outbreak strains (data not shown).
DISCUSSION
The results of our investigation have provided new insights into the degree of diversity and the stability of phage types and PFGE macrorestriction patterns and the relationships between these epidemiological markers among E. coli O157 strains isolated in Ontario. At present, more than 80 phage types are recognized by the phage typing scheme, and this procedure represents the only internationally standardized subtyping method with universally accepted interpretative criteria for these organisms. In recent years, DNA macrorestriction analysis by PFGE has increasingly been used for the molecular subtyping of a wide range of bacterial and fungal pathogens, and it is now considered the “gold standard” for the molecular subtyping of many of these organisms. For E. coli serogroup O157, the usefulness of PFGE fingerprinting during outbreak investigations has been demonstrated previously, and in addition, the standardization of PFGE analysis in public health laboratories in the United States has been achieved recently (2, 3, 14, 15, 18). In our laboratory, these two procedures are used for the subtyping of isolates suspected to be part of an outbreak.
The results of our study confirmed earlier investigations that found that the macrorestriction patterns of epidemiologically unrelated E. coli O157 strains have a high degree of similarity due to the relatively limited genetic diversity within this serogroup (3, 6, 18). Nevertheless, on the basis of the pattern interpretative criteria used, 22 distinct patterns were identified among 30 strains isolated from patients with sporadic cases of infection in Ontario, and a total of 17 strains exhibited unique patterns. In contrast, phage typing of the same group of isolates resulted in the identification of only seven different phage types, and over one-half of the strains exhibited the same phage type. Therefore, as in previous studies, our results provided clear evidence for the superior discriminatory capability of PFGE analysis compared to that of phage typing for unrelated strains (2, 3, 12). A total of 25 phage type-PFGE subtype combinations were identified among the sporadic isolates, suggesting that the combined use of these procedures could identify more subtypes than the use of either methodology alone. These results also provided evidence for considerable genetic heterogeneity among E. coli serogroup O157 strains isolated in Ontario.
Our findings also demonstrated that PFGE subtyping could discriminate between strains that exhibited the same phage type. Thirteen distinct DNA macrorestriction patterns were identified among a total of 17 phage type 14 strains. This phage type represents the most commonly isolated E. coli O157 phage type in Canada. For one group of strains, the results showed that phage typing could subdivide unrelated strains with the same XbaI PFGE pattern into phage types 1, 8, 14, and 32, and the use of an additional restriction enzyme for PFGE indicated that there were genomic differences between some of these strains. These results suggest the presence of distinct genotypes among E. coli O157 isolates beyond that revealed by PFGE analysis with XbaI, and they provide evidence for the view that more than one restriction enzyme should be used routinely for analysis of isolates of this serogroup. These results also contradict a previous suggestion regarding the dissociation between an isolate's phage type and its genetic background (12). Interestingly, Izumiya et al. (8) recently reported that a group of E. coli O157 strains, isolated in Japan, with similar XbaI PFGE fingerprints could be subdivided into phage types 1, 4, 8, 14, and 32.
By analyzing isolates associated with four separate outbreaks in Ontario, we confirmed previous studies which reported that both phage typing and PFGE fingerprinting could correctly identify epidemiologically related E. coli O157 isolates as belonging to the same type (2, 3). PFGE analysis showed that each outbreak strain was characterized by a distinct genomic fingerprint. In contrast, phage typing differentiated the four outbreak strains into only three different phage types. In addition, our results showed that each isolate associated with the same outbreak exhibited the same phage type as that of the outbreak strain. On the other hand, a small minority of isolates had macrorestriction patterns that differed slightly (one or two DNA fragment differences) from that of the outbreak strain in two of the outbreaks, but these isolates would be considered closely related and probably part of the outbreak by using previously published PFGE interpretative criteria (17). Previous investigations have also shown that the PFGE patterns of isolates isolated from different patients and associated with the same outbreak and those of sequential isolates isolated from the same patient can vary slightly (7, 9, 16).
Taken together, our results indicate that phage typing and PFGE fingerprinting represent complementary techniques for the subtyping of E. coli serogroup O157, and they provide further evidence for the view that the combined use of these procedures provides optimal discrimination.
ACKNOWLEDGMENTS
We thank the staff of the Public Health Laboratories of Ontario for providing the strains used in this study, and we appreciate the assistance of N. denHollander, V. Brunins, and S. Lombardi.
REFERENCES
- 1.Ahmed R, Bopp A, Borczyk A, Kasatiya S. Phage-typing scheme for Escherichia coli serotype O157:H7. J Infect Dis. 1987;155:806–809. doi: 10.1093/infdis/155.4.806. [DOI] [PubMed] [Google Scholar]
- 2.Allison L, Stirrat A, Thomson-Carter F M. Genetic heterogeneity of Escherichia coli O157:H7 in Scotland and its utility in strain subtyping. Eur J Clin Microbiol Infect Dis. 1998;17:844–848. doi: 10.1007/s100960050204. [DOI] [PubMed] [Google Scholar]
- 3.Barrett T J, Lior H, Green J H, Khakhria R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besser R E, Lett S M, Weber J T, Doyle M P, Barrett T J, Wells J G, Griffin P M. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA. 1993;269:2217–2220. [PubMed] [Google Scholar]
- 5.Bielaszewska M, Janda J, Blahova K, Minarikova H, Jikova E, Karmali M A, Laubova J, Sikulova J, Preston M A, Khakhria R, Karch H, Klazasrova H, Nyc O. Human Escherichia coli O157:H7 infection associated with the consumption of unpasteurized goat's milk. Epidemiol Infect. 1997;119:299–305. doi: 10.1017/s0950268897008297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohm H, Karch H. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30:2169–2172. doi: 10.1128/jcm.30.8.2169-2172.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gouveia S, Proctor M E, Lee M S, Luchansky J B, Kaspar C W. Genomic comparison and Shiga toxin production among Escherichia coli O157:H7 isolates from a day care center outbreak and sporadic cases in southeastern Wisconsin. J Clin Microbiol. 1998;36:727–733. doi: 10.1128/jcm.36.3.727-733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumiya H, Masuda T, Ahmed R, Khakhria R, Wada A, Terajima J, Itoh K, Johnson W M, Konuma H, Shinagawa K, Tamura K, Watanabe H. Combined use of bacteriophage typing and pulsed-field gel electrophoresis in the epidemiological analysis of Japanese isolates of enterohemorrhagic Escherichia coli O157:H7. Microbiol Immunol. 1998;42:515–519. doi: 10.1111/j.1348-0421.1998.tb02318.x. [DOI] [PubMed] [Google Scholar]
- 9.Karch H, Russman H, Schmidt H, Schwarzkopf A, Heesemann J. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J Clin Microbiol. 1995;33:1602–1605. doi: 10.1128/jcm.33.6.1602-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karmali M A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khakhria R, Duck D, Lior H. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol Infect. 1990;105:511–520. doi: 10.1017/s0950268800048135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause U, Thomson-Carter F M, Pennington T H. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J Clin Microbiol. 1996;34:959–961. doi: 10.1128/jcm.34.4.959-961.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin I E, Tyler S D, Tyler K D, Khakhria R, Johnson W M. Evaluation of ribotyping as epidemiologic tool for typing Escherichia coli serogroup O157 isolates. J Clin Microbiol. 1996;34:720–723. doi: 10.1128/jcm.34.3.720-723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preston M, Borczyk A, Davidson R, McGeer A, Bertoli J, Harris S, Thususka J, Goldman C, Green K, Low D, Proctor P, Johnson W, Khakhria R. Hospital outbreak of Escherichia coli O157:H7 associated with a rare phage type—Ontario. Can Commun Dis Rep. 1997;23:33–37. [PubMed] [Google Scholar]
- 15.Stephenson J. New approaches to detecting and curtailing foodborne microbial infections. JAMA. 1997;277:1337–1340. doi: 10.1001/jama.277.17.1337. [DOI] [PubMed] [Google Scholar]
- 16.Strockbine N A, Wells J G, Bopp C A, Barrett T J. Overview of detection and subtyping methods. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C.: American Society for Microbiology; 1998. pp. 331–356. [Google Scholar]
- 17.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willshaw G A, Smith H R, Cheasty T, Wall P G, Rowe B. Vero cytotoxin-producing Escherichia coli O157 outbreaks in England and Wales, 1995: phenotypic methods and genotypic subtyping. Emerg Infect Dis. 1997;3:561–565. doi: 10.3201/eid0304.970422. [DOI] [PMC free article] [PubMed] [Google Scholar]