Abstract
The site-specific DNA rearrangement process, called V(D)J recombination, creates much of the diversity of immune receptor molecules in the adaptive immune system. Central to this reaction is the organization of the protein-DNA complex containing the proteins RAG1 and RAG2 and their DNA targets. A long-term goal is to appreciate the three-dimensional relationships between the proteins and DNA that allow the assembly of the appropriate reaction intermediates, resulting in concerted cleavage and directed rejoining of the DNA ends. Previous cross-linking approaches have mapped RAG1 contacts on the DNA. RAG1 protein contacts the DNA at the conserved heptamer and nonamer sequences as well as at the coding DNA adjacent to the heptamer. Here we subject RAG1, covalently cross-linked to DNA substrates, to partial cyanogen bromide degradation or trypsin proteolysis in order to map contacts on the protein. We find that coding-sequence contacts occur near the C terminus of RAG1, while contacts made within the recombination signal sequence occur nearer the N terminus of the core region of RAG1. A deletion protein lacking the C-terminal DNA-contacting region is still capable of making the N-terminal contacts. This suggests that the two binding interactions may exist on two separate domains of the protein. A trypsin cleavage pattern of the native protein supports this conclusion. A two-domain model for RAG1 is evaluated with respect to the larger recombination complex.
The adaptive immune system in all vertebrates uses a site-specific rearrangement of DNA to assemble functional T-cell receptor and immunoglobulin genes from the arrays of inactive segments inherited in the germ line. In the process, termed V(D)J recombination, each of the various coding segments, named V, D, and J, is targeted for rearrangement by an adjacent recombination signal sequence (RSS) (reviewed in references 13, 15, and 22). An RSS is composed of a conserved heptamer (CACAGTG) and nonamer (ACAAAAACC) motif, separated by a spacer of either 12 or 23 bp in length. The two sequences of differing length are called the 12RSS and 23RSS, respectively. Within a chromosomal locus, similar segments generally carry RSSs of the same length. A productive rearrangement in cells always occurs between pairs of DNA segments bordered by RSS elements of the two different spacer lengths (the 12/23 rule; 43). Owing to the 12/23 rule, this organization permits a V segment (for example) to join to a D segment but not to a second V segment. The recombination mechanism introduces (in several steps) a double-strand break into the DNA precisely between the RSS and its associated coding region. This process is coordinated at two segments, so the two cleavage events yield an intermediate stage with four available DNA ends. Commonly, pairs of these ends are subsequently joined in a directed manner. The two DNA ends belonging to coding regions are joined to each other to form the coding junction. The two RSS-containing ends are also joined to each other to form the signal junction. These DNA molecules are shepherded through the reaction in large part through the action of proteins which contact the DNA within the RSSs and also at the adjacent coding regions. The assembly of a protein-DNA complex and cleavage of the DNA can be performed in vitro (25, 45) and is dependent primarily on the two proteins RAG1 and RAG2 (31, 37). In addition, the cleavage reaction is aided by DNA bending proteins. Either HMG1 (20, 44) or HMG2 (36) functions in this capacity. Cross-linking strategies indicate that RAG1 makes close contacts at both the nonamer and heptamer in the RSS (28, 41) and additionally to the coding DNA (11, 27). So far the cross-linking approach has been used to map the positions on the DNA where the proteins make contact. In principle, the same strategy can also reveal the contact sites on the protein, as has been demonstrated in other systems (26, 30). In this study we use partial cyanogen bromide degradation and trypsin proteolysis of RAG1 cross-linked to labeled DNA substrates to demonstrate that the coding-end contacts occur near the C terminus of RAG1 while contacts made by the RSS to RAG1 occur nearer the N terminus of the core region. The two contacts appear to be independent of each other in that a deletion protein lacking the C-terminal binding region is still capable of making the N-terminal contacts. This suggests that the two binding interactions may exist on two separate domains of the protein. A trypsin cleavage pattern of the native protein supports this conclusion. A two-domain model for RAG1 is evaluated with respect to the larger recombination complex.
MATERIALS AND METHODS
Proteins.
Baculovirus stocks for MR1 and MR2 (25) were obtained from Martin Gellert (National Institutes of Health). MR1 and MR2 are fusion proteins, each containing an N-terminal maltose binding protein (MBP) followed by the functional core region of mouse RAG1 (residues 384 to 1008) or RAG2 (residues 1 to 387) of the mouse protein sequence, respectively. The C termini carry a polyhistidine tag followed by three tandem copies of the c-myc epitope tag as used previously (34). In this study, MR1 or the deletion construct R1 Δ34–38 were expressed using the baculovirus system. R1 Δ34–38 was constructed by deletion of the coding region in the RAG1 core between the SalI sites introduced into constructs pMS134 and pMS138 (34) and then subcloning into a baculovirus expression vector. Amino acid residues 735 to 963 of the mouse protein are thereby deleted from the core. This protein is expressed without the MBP fusion partner.
When used together, MR1 and MR2 proteins were expressed simultaneously, by coinfection of the SF9 insect cell line. Proteins were harvested after 66 h of infection and purified on nitrilotriacetic acid-agarose (Qiagen) charged with Ni2+ as described previously (45). Fractions containing the fusion proteins were pooled and loaded onto amylose resin (New England Biolabs). The column was washed extensively with buffer A (20 mM Tris-HCl [pH 7.4], 500 mM NaCl, 10 mM 2-mercaptoethanol, 1 mM EDTA) containing 0.2% Tween 20, followed by elution in buffer A plus 10 mM maltose. Protein-containing fractions were pooled and dialyzed against buffer R (25 mM Tris-HCl [pH 8.0], 150 mM KCl, 2 mM dithiothreitol [DTT], 10% glycerol) for 3 h. Aliquots were stored at −80°C.
HMG1 protein was expressed and purified from an Escherichia coli plasmid, pDVG83, constructed by Dik van Gent (Erasmus University, Rotterdam, The Netherlands), as done previously (27). It encodes human HMG1 residues 1 to 163, which span the two HMG boxes, but replaces the acidic C-terminal tail with an additional 13 amino acid residues at the C terminus from the vector.
Oligonucleotides and probe preparation.
The probes used in these studies were a subset of those used previously (27). Oligonucleotide probes were 5′ end labeled with [γ-32P]ATP (NEN) using T4 polynucleotide kinase (Amersham Pharmacia) on the strand carrying the phosphorothioate, annealed to the other strands, and gel purified.
The resulting nicked double-stranded DNA was derivatized with the cross-linker by first resuspending it in 50 μl of buffer TN (10 mM Tris-HCl [pH 7.0], 30 mM NaCl). To this was added 4 μl of 1 M Tris-HCl (pH 7.0), 36 μl of methanol, and 12 μl of azido-phenacyl bromide (Sigma) (10 mM concentration in methanol). The reaction was incubated in the dark for 3 h at room temperature, and then the probe was purified from the reactants by passing it through a G-50 Sepharose spin column (Eppendorf-5 Prime, Inc.) preequilibrated with buffer TN in the dark.
DNA binding and UV cross-linking assays.
Binding was scaled up 50-fold from previous conditions (27) in 25 mM morpholinepropanesulfonic acid (pH 7.0), 5 mM MgCl2, 1 mM DTT, 50 μg of bovine serum albumin (BSA)/ml, and 50 mM KCl. Nonspecific DNA pdIdC · pdIdC (Amersham Pharmacia) was added to a 50-μg/ml final concentration when it was used. RAG1 (and RAG2 when used) protein was added in the range of 2.5 μg each per reaction. When used, HMG1 protein was added at 2.5 μg per reaction. Typically, binding reactions were assembled and incubated in the binding buffer for 10 min on ice. UV exposure for 1 min was performed using a 6-W UV lamp equipped with a 302-nm filter at a distance of 3 cm from the sample. The sample was supplemented with 0.1 volume denaturation cocktail (1% sodium dodecyl sulfate [SDS], 1 M DTT, 10% glycerol) and heated. SDS-polyacrylamide gel electrophoresis (PAGE) was performed on continuous 6% acrylamide gels. The cross-linked protein was located by autoradiography of the wet gel using a Molecular Dynamics PhosphorImager. The desired band was excised and electroeluted using a Bio-Rad electroelution apparatus (model 422).
Cyanogen bromide cleavage.
Protein in SDS buffer following electroelution was concentrated by precipitation with acetone (final concentration, 80%). Cleavage was obtained following an existing protocol (17). The pellet was redissolved in a solution containing 100 μl of 1% SDS and 5 mM triethylamine HCl (pH 9.0). The sample was heat denatured at 95°C for 5 min. Three microliters each of 1 M HCl and 1 M CnBr in acetonitrile was added and incubated at room temperature. This resulted in a final pH of 1.0 for the proteolysis, which is gentler than the pH of 0 at which CnBr digestion is commonly performed when complete digestion is desired. At intervals, 25-μl samples were mixed with stop solution (5% 2-mercaptoethanol, 0.5 M triethylamine [pH 9.0], 0.1% bromphenol blue, 50% glycerol). The samples were analyzed by SDS-PAGE using either SDS-glycine or SDS-tricine buffers as indicated.
Metal chelate chromatography.
For samples analyzed by metal chelate chromatography following CnBr cleavage, the cleavage reaction was halted with an alternate stop buffer lacking 2-mercaptoethanol. To this mixture was added 50 μg of BSA. The solution was precipitated with acetone added to an 80% final concentration. The pellets were redissolved in 100 μl of buffer D (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, 0.1% SDS, 10 mM imidazole) and heated to 95°C for 5 min. One-half of the sample was bound to nitrilotriacetic acid agarose (Qiagen) charged with Ni2+ in a batch at room temperature for 1 h. The beads were washed three times (10 min each) in buffer D and once in 6 M guanidine-HCl for 5 min. Bound peptide was eluted with buffer D supplemented to a 200 mM concentration with imidazole.
Trypsin proteolysis.
One-microgram samples of MR1 were treated with a range of trypsin concentrations in 25 mM Tris (pH 8.0)–150 mM KCl–2 mM DTT–10% glycerol at room temperature for 5 min. The products were precipitated with an equal volume of 20% trichloroacetic acid on ice for 15 min, centrifuged, washed twice with acidified cold acetone, and air dried. The sample was resuspended in SDS sample buffer, boiled for 10 min, and analyzed by SDS-PAGE. Silver staining or transfer to a polyvinylidene difluoride membrane and immunoblotting were performed by standard protocols. Anti-MBP antibody was purchased from New England Biolabs. Anti-c-myc monoclonal antibody was prepared from 9E10 cells obtained from American Type Culture Collection. Peptide sequencing was performed in the molecular biology core facility at the Medical College of Georgia. Protein samples transferred to polyvinylidene difluoride membranes were stained with Coomassie blue R-250 in 40% methanol and 10% acetic acid. The membrane was destained and washed with 50% methanol and air dried, and specific bands were excised and subjected to automated Edman degradation.
Immunoprecipitation of tryptic fragments.
Protein cross-linked to the radioactive probe was digested with trypsin, denatured, and analyzed by SDS-PAGE in Tris-glycine buffer. The radioactive bands were identified by PhosphorImager (Molecular Dynamics), electroeluted as described above, and dialyzed overnight at 4°C into IP buffer (20 mM Tris-HCl [pH 7.4], 0.5 M NaCl, 0.3% NP-40). Mouse immunoglobulin G1 (IgG1) (Sigma) or monoclonal antibody prepared from cell line 9E10 (ATCC) directed against the c-myc epitope tag was added (2 μg) to 2 mg of protein G-agarose (Sigma) in IP buffer and incubated overnight at 4°C. Beads were washed 3 times (10 min per wash) in IP buffer and then combined with the soluble dialyzed protein in a volume of 0.5 ml. Binding continued for 4 hours at 4°C, followed by three washes as described above. The beads were collected, and peptides were released by denaturation at 95°C in 0.1% SDS followed by SDS-PAGE and visualization by PhosphorImager (Molecular Dynamics).
RESULTS
RAG1 alone will bind DNA nonspecifically but forms a more specific complex in the presence of RAG2 (4, 5, 28). The specificity of this complex can be further increased by the addition of HMG1 (3, 27), and previously we have mapped the contacts formed by these three proteins on either 12- or 23RSS probes (27). Within that complex, RAG1 is cross-linked efficiently when the cross-linker is positioned in the nonamer, heptamer, or 2 bp from the heptamer in the coding DNA. In this study we used the same probes and strategy we used previously to cross-link RAG1 to selected probes and then purify the covalent RAG1-DNA product for further analysis.
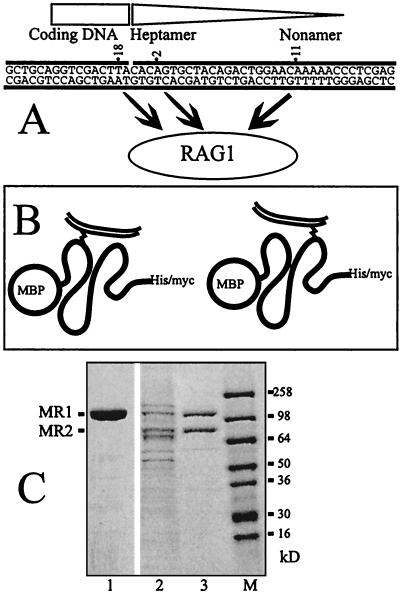
The scheme is shown in Fig. 1. Panel A indicates that various probes can be created containing a photoactivatable cross-linker at one position at a time within the RSS or the coding DNA. The three probes used in this report contain the cross-linkers at the indicated positions and are a subset of those used previously. These probes can be cross-linked to RAG1 under either nonstringent binding conditions (RAG1 alone) or stringent conditions (RAG1, RAG2, HMG1, and dIdC competitor). Panel B shows the logic of a typical experiment. In these experiments we use a fusion protein used previously (5, 25, 45) called MR1. This molecule contains an MBP fusion partner at the N terminus, the enzymatically active core region of RAG1 (residues 386 to 1008 of the mouse protein) and C-terminal polyhistidine and c-myc epitope tags. Folded MR1 is shown cross-linked to radioactively labeled DNA through interactions at two different positions on the protein. These will be mapped by denaturing the protein, cleaving it under partial cleavage conditions, and calculating the position of the cross-link from the sizes of the fragments. Some information can be deduced from the total digests. Most informative are those peptides which still carry the C-terminal peptide tags, since metal chelate chromatography or immunoprecipitation can be used to purify these peptides.
FIG. 1.
(A) The sequences of the three probes used in this analysis are shown, with the site of the cross-linker indicated by the numbers. The region corresponding to the 12RSS is indicated by the triangle, and the coding DNA corresponds to the rectangle. All three probes have been shown in previous work to cross-link to RAG1. (B) The logic of the CnBr mapping procedure. The contact site on the protein can be mapped by partial cleavage and by identifying the shortest peptide that contains both the C-terminal tags and the labeled DNA. The thick line represents folded protein MR1 with the MBP fusion partner at the N terminus. The double thin line is the DNA probe, and the cross-link is the zigzag. (C) The proteins MR1 and MR2 used in this analysis. This panel shows a Coomassie blue-stained SDS-PAGE gel of the proteins. Lane 1 is the purified MR1. Lane 2 shows the copurified MR1 plus MR2 after one column chromatography step, and lane 3 shows them after two column chromatography steps. Lane M shows size markers. This gel was a 4 to 20% gradient gel run in Tris-glycine buffer.
Panel C illustrates the purity of our starting proteins. This panel represents proteins analyzed by SDS-PAGE and stained with Coomassie blue. A silver-stained gel of the MR1 protein is also presented later (see Fig. 7).
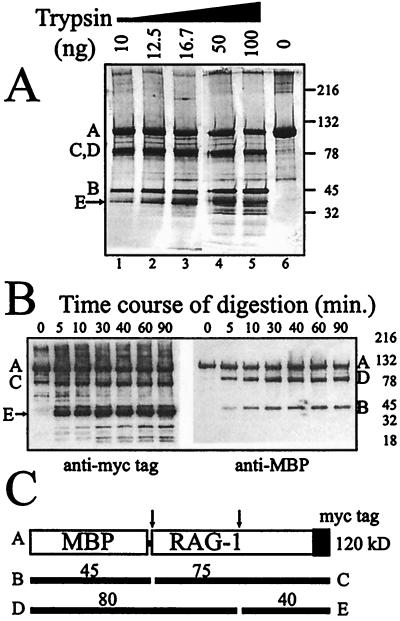
FIG. 7.
Limited tryptic digest of native MR1 suggests a two-domain architecture. (A) Increasing amounts of trypsin were used to digest 1 μg of MR1 protein for 5 min at room temperature. Lane 6 shows the undigested protein. The gel was a 4 to 20% gradient Tris-glycine-SDS-polyacrylamide gel. Other bands indicated by letters are labeled consistently in the other panels. The full-length protein is the top band. The arrow indicates the band for which the N-terminal sequence was determined. (B) Immunoblots of an MR1 digestion time course using a trypsin concentration of 10 ng per μg of MR1. The left panel was developed with the antibody directed against the C-terminal myc epitope tag. The right panel was developed with the anti-MBP antiserum which detects the N-terminal fusion partner. (C) Schematic representation indicating how two preferred cleavage sites produce the digestion pattern. Cleavage products seen in rows A and B are the result of single cleavages at either of these sites.
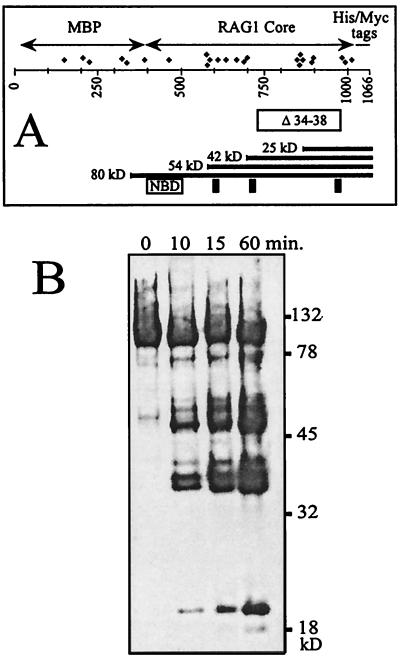
We first wished to determine whether the cyanogen bromide cleavage procedure (17) would provide a useful distribution of fragments. Figure 2A shows a schematic map of the MR1 protein. The panel shows the positions of all the methionines in the protein which are potential targets for cleavage by cyanogen bromide. Also shown is the region of mouse RAG1 (residues 735 to 963 of the mouse peptide) which is deleted in a protein called R1 Δ34–38 (discussed later). Certain landmarks are shown at the bottom of this panel. Mouse RAG1 residues 390 to 460 have been termed the nonamer-binding domain since they are essential for this behavior (13, 39). Residues 600, 708, and 962 of the mouse polypeptide have been identified as essential for catalytic activity (19, 21) and might therefore be expected to also represent close DNA contacts. These are shown as vertical bars. The numbering in this diagram benefits from the coincidence that the MBP protein with its bridging linker (390 amino acids) almost precisely compensates for the 385 amino acids truncated from the N terminus of RAG1. Partial cleavage must generate a nested set of peptides containing the C-terminal tags as well as a complementary set of N-terminal peptides. Figure 2B shows an immunoblot developed with an antibody specific for the c-myc epitope tag. Denatured MR1 protein was allowed to react with CnBr for increasing intervals. The resulting fragments were precipitated, separated by SDS-PAGE, and blotted.
FIG. 2.
(A) Schematic map of the MR1 protein showing the MBP fusion partner, the RAG1 core region (amino acids 386 to 1008 of the mouse sequence), and the C-terminal tags. Diamonds represent methionines. Below the number line is a box indicating the sequence deleted in the mutant R1 Δ34–38. Selected C-terminal fragments following CnBr cleavage are illustrated with their predicted masses. Other landmarks indicated are the sequence identified as essential for nonamer binding (NBD) and the three acidic residues proposed to participate in formation of the enzymatic active site (mouse residues 600, 708, and 962). (B) Immunoblot of a CnBr digest of MR1 developed with anti-myc tag antibody. A time course of digestion is shown from 0 to 60 min. The gel is 10% acrylamide in Tris-glycine-SDS buffer.
On this blot we do not see the cleavage products expected to be generated from the three methionines closest to the C terminus. These products are 10 kDa or less in size and, if formed, were probably not retained on the membrane during transfer. The smallest visible band, at 20 kDa, is of the appropriate size for peptides generated by cleavage within the cluster of methionines located at positions 850 to 900 on the map of Fig. 1B. We cannot assign a distinct band to every potential cleavage site. It is likely that cleavage efficiency is influenced by surrounding peptide context and that under the limiting conditions used, only the most favored sites are cleaved. It is appreciated, for example, that methionine followed by either serine or threonine is resistant to CnBr (17). We note that under these reaction conditions, the full-length protein remains prominent even at 60 min and smaller fragments increase in abundance over the reaction interval. It is likely that the peptides generated at only 10 min of digestion represent products of single cleavages. If multiple cleavages were occurring, then the smallest observed peptides should accumulate at the expense of the bands of higher molecular mass at later time points. This was not observed.
A cross-linker in the coding region contacts near the C terminus of RAG1.
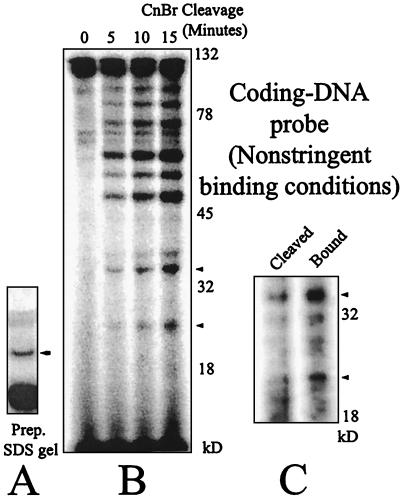
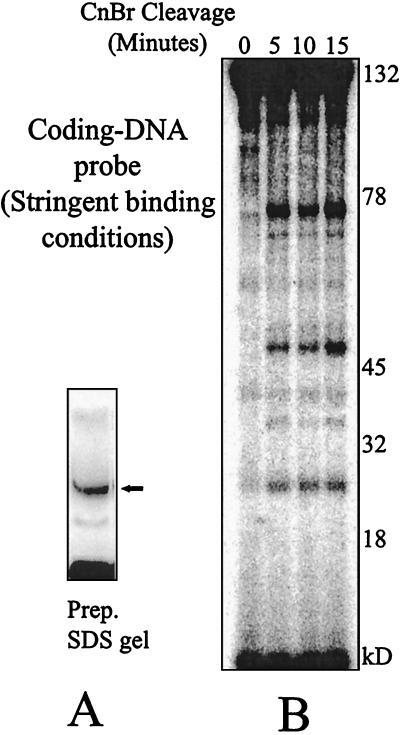
The complex containing the MR1 protein alone (nonstringent conditions) was cross-linked to a radioactive probe (no. 18) in which the cross-linker is situated in the coding region adjacent to a 12RSS. The complex was denatured, and the MR1-DNA adduct was separated from the other components by SDS-PAGE. Figure 3A shows one lane of the preparative gel. The indicated band, which is the MR1 protein covalently linked to the DNA probe, was eluted and subjected to partial CnBr cleavage under the conditions used for Fig. 2B, which are expected to generate single cleavages. This experimental reasoning has been used in other systems (26, 30). The resulting products were resolved on a 10% acrylamide SDS-tricine gel, shown as Fig. 3B. A ladder of bands was generated, with the fastest band appearing at about 25 kDa. The size of this peptide already requires that this represent a C-terminal contact in RAG1. If a peptide is only cleaved once, all the products must represent either N-terminal or C-terminal fragments. In the MR1 construct, cross-links within the N-terminal portion of RAG1 would yield only peptides larger than 45 kDa, since even the smallest N-terminal cleavage peptides would carry the MBP fusion partner while C-terminal peptides would contain large portions of RAG1. MBP by itself does not form contacts with DNA (not shown). We note that a complicated set of higher-molecular-mass peptides is also present, which could represent cross-links to RAG1 at other positions within the protein. We also tested the behavior of the same probe in the context of an adjacent 23RSS with the same result (not shown).
FIG. 3.
(A) Preparative SDS-PAGE gel of the MR1-DNA cross-linked product bound to the coding-end probe (no. 18) under nonstringent conditions (MR1 alone). The band indicated with the arrow was excised and eluted. (B) Partial CnBr cleavage time course of the cross-linked MR1-DNA from panel A. The gel is 10% acrylamide in Tris-tricine-SDS buffer. The persistence of a high fraction of undigested protein at the full length of 120 kDa supports the contention that the cleavage products seen are the result of single cleavage events. (C) Metal chelate chromatography indicates that the smaller peptides shown in panel B carry the polyhistidine tag and are therefore cross-linked near the C terminus of the protein. Cleaved products as in the 15-min time point were divided. Twenty percent was saved and loaded directly in the lane marked Cleaved. The remainder was bound to the metal chelate resin, washed four times, eluted, and loaded in the lane marked Bound. Bands marked with arrowheads appear in both panels B and C. The gel is 12% acrylamide in Tris-tricine-SDS buffer.
If our reasoning is correct, we anticipate that each of the C-terminal bands would also contain the polyhistidine tag and could be affinity purified using metal chelate chromatography. One technical difficulty was the interfering effect of the SDS already present in the peptide mixture, which would prevent binding to the column. Total elimination of the SDS was likely to render the peptides insoluble. We reduced this effect by adding a large excess of BSA to the peptide mixture following cleavage to provide a sink for most of the SDS. At moderate efficiency, the resulting peptides were retained on the affinity resin as shown in Fig. 3C. The bands present in the cleaved sample at 25 and 35 kDa were retained on the affinity matrix through four cycles of washing and eluted with imidazole (lane labeled “Bound”), supporting their identification as C-terminal peptides. This result, however, can only be evaluated qualitatively. An independent proof will be presented later.
We repeated this experiment under more stringent binding conditions, with MR1 as well as a RAG2 derivative (MR2), HMG1, and nonspecific competitor dIdC cross-linked to the same labeled probe as above. The complex was denatured, and the MR1-DNA adduct was separated from the other components by SDS-PAGE (Fig. 4A). That band was eluted and subjected to partial CnBr cleavage. The resulting products were resolved on a 10% acrylamide SDS-tricine gel, shown as Fig. 4B. The labeled cleavage products contain the same band at 25 kDa as that seen in Fig. 3B but fewer bands higher in the gel. We will comment on this point later. Cleaved products from this analysis were also subjected to metal chelate affinity chromatography as described above with the same qualitative result. Several bands were retained at moderate efficiency on the resin and eluted with imidazole (not shown).
FIG. 4.
(A). Preparative SDS-PAGE gel of the MR1-DNA cross-linked product bound to the coding-end probe (no. 18) formed under stringent binding conditions using MR2, HMG1, and dIdC. The band indicated with the arrow was excised and eluted. (B) Partial CnBr cleavage time course of MR1 prepared for panel A. The size of the fastest-migrating band is consistent only with a C-terminal contact site. The gel is 10% acrylamide in Tris-tricine-SDS buffer.
Heptamer and nonamer probes cross-link near the N terminus.
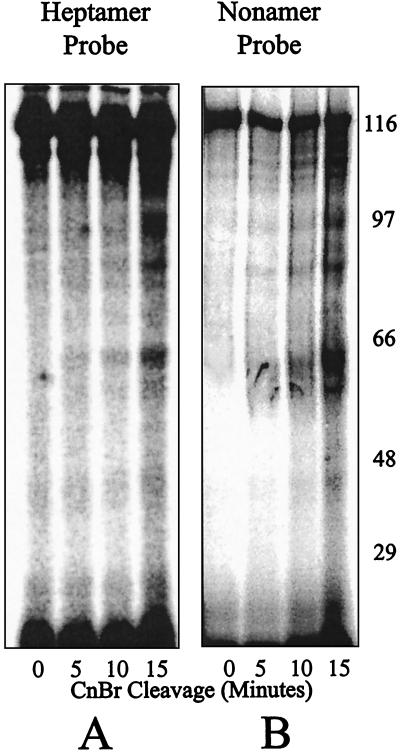
The cross-linking to MR1 under stringent binding conditions identical to those used for Fig. 4B was repeated with additional probes containing the cross-linking moiety within the heptamer or nonamer. Cross-linked MR1 was gel purified, cleaved with CnBr, and analyzed as before. The result using one of each of these probes (no. 2 and 11 in Fig. 1A) is shown in Fig. 5. Other probes were also tested, with similar results (not shown). The digestion products obtained with these two probes appear similar to each other and different from the pattern obtained with the previous coding region probe. The smallest reliable band in each case appears at about 65 kDa. This suggests that the heptamer and nonamer cross-linking sites are close to each other along the protein primary sequence (within the low resolution of this analysis) and occur at a different site in the protein from the contact made by the coding region probe. This site could well be within the part of the protein already considered a DNA binding region, the nonamer binding domain. A second DNA contact site in the protein would also be consistent with the differences at higher masses shown between Fig. 3B and 4B. Under the less stringent binding conditions, the probe seemed to be labeling two different positions in RAG1, resulting in a greater number of bands and the more complicated pattern of peptides shown in Fig. 3B. Furthermore, the molecular mass of this band is also consistent with a DNA binding site located nearer the N terminus of the RAG1 core region. We have attempted (unsuccessfully) to purify the 65-kDa bands using metal chelate chromatography (not shown), suggesting that this band could represent an N-terminal peptide no longer retaining the polyhistidine tag. A 65-kDa peptide including the 42-kDa MBP fusion would locate this RAG1 cross-link within the 20 kDa at the N-terminal side of the core region.
FIG. 5.
Time course of CnBr digestion of cross-linked MR1 to probes with the cross-linker positioned in the heptamer (A) (probe 2) or nonamer (B) (probe 11) under the same stringent binding conditions of Fig. 4. The distribution of labeled peptides to only above 45 kDa indicates that cross-linking occurred nearer the N terminus of the core region. The gels are 10% acrylamide in Tris-tricine-SDS buffer.
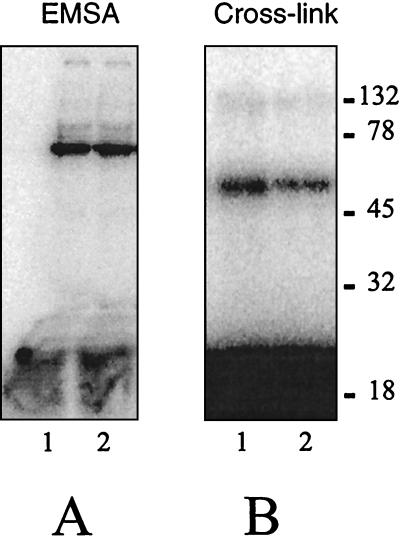
Consistent with this interpretation is the behavior of the RAG1 deletion construct called R1 Δ34–38 (Fig. 2A). The C-terminal region spanning the contact to the coding DNA (Fig. 3B and 4B) has been removed in this protein. Nevertheless, this protein is capable of binding DNA, which indicates that the N-terminal DNA binding region is able to bind in the absence of the C-terminal region. An electrophoretic mobility shift assay (EMSA) and cross-linking data using the heptamer or nonamer probe are presented in Fig. 6. The stability of the complex under EMSA conditions is roughly comparable to that of the MR1 protein, but additional experiments will be necessary to characterize the behavior of the deletion mutant. Specificity of binding is not addressed in this analysis, which was performed under the nonstringent conditions used for Fig. 3.
FIG. 6.
Behavior of RAG1 deletion construct R1 Δ34–38. (A) EMSA showing the deleted RAG1 protein alone binding under native conditions to the heptamer and nonamer probes (lanes 1 and 2, respectively). (B) Following cross-linking, the resulting covalent complex was resolved on this 8% acrylamide Tris-glycine-SDS gel.
Tryptic digests support a two-domain architecture of RAG1.
The fact that the N-terminal region of the RAG1 core can bind DNA in the absence of the C-terminal contact region suggests that these may function as independent protein domains. We tested whether the native MR1 protein could be divided into domains by limited cleavage with trypsin under physiologic conditions. Figure 7A shows the resulting pattern of cleavage products in an experiment with increasing amounts of digestion. The starting protein is shown in lane 6, and four major bands are detected in this silver-stained gel even at the lowest concentration of trypsin (lane 1). The identities of the bands are resolved by the immunoblot results shown in panel B. A diagrammatic representation of the protein with two predominant trypsin cleavage sites that explains the data is shown in panel C. At 120 kDa (band A) is the full-length MR1 protein. At about 78 kDa is actually a cluster of products, one of which (band C) represents the removal of the MBP protein which is expected to fold into a separate domain. The cleaved MBP protein itself is detected at about 45 kDa (bands marked B) in the silver stain and in the immunoblot of panel B visualized with antiserum specific for MBP. The 40-kDa band (labeled E) is a C-terminal peptide which contains the c-myc epitope as confirmed in the immunoblot of panel B (left panel). We isolated this bottom band in a sufficient quantity for peptide sequence determination by means of Edman degradation. The six N-terminal residues of this peptide were EVEGLE, and they correctly follow an arginine residue (amino acid 713 of the mouse RAG1 protein) as expected for a tryptic cleavage product. Cleavage of MR1 at this site is expected to yield a 40.5-kDa C-terminal peptide. If the starting protein had been cleaved only at this site, one would expect to find the N-terminal peptide (D) as an 80-kDa product which still retains the MBP fusion partner. This is also present in the blot shown in panel B (right panel). Additional characterization of the domain architecture is in progress.
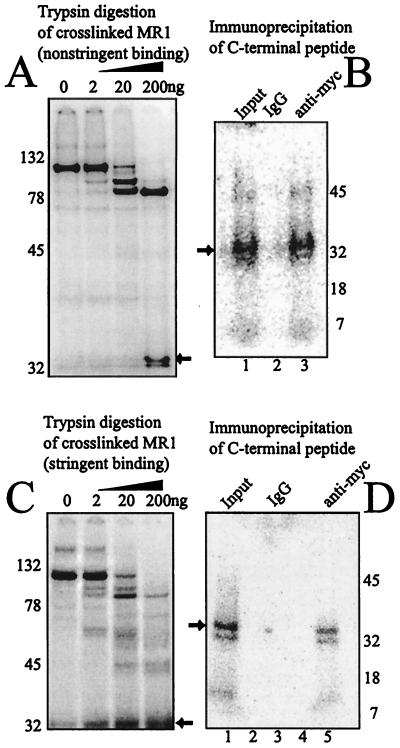
Since we were able to cause the native MR1 protein to be digested into N-terminal and C-terminal fragments with trypsin, we repeated the cross-linking experiment using enzymatic proteolysis rather than CnBr cleavage to demonstrate the specific cross-linking to the C-terminal peptide. Figure 8 shows the results of two experiments that differ in their binding conditions. For panel A, MR1 protein alone was bound and cross-linked to the coding-end probe as for Fig. 3. The protein was digested with increasing quantities of trypsin under physiologic conditions. The labeled products were resolved by SDS-PAGE. We confirmed that the small peptide represented the C terminus of the protein by eluting that product and demonstrating (panel B) that it could be specifically immunoprecipitated with the antibody directed against the myc tag. Under these nonstringent binding conditions, cross-linking to the N-terminal domain was also observed.
FIG. 8.
Tryptic digest of MR1 cross-linked to the labeled coding-DNA probe. (A) When bound and cross-linked under nonstringent conditions, two peptides retain the label when digested to the greatest degree. Lane 1 shows the full-length MR1 protein cross-linked to the probe. Progressive digestion with trypsin generates two predominant labeled products. The smaller peptide (arrow) was eluted. (B) The eluted peptide (arrow) was immunoprecipitated using nonspecific mouse IgG (lane 2) or antibody directed against the C-terminal myc tag (lane 3). Only the specific antibody resulted in precipitation of peptide cross-linked to the labeled probe. (C) Cross-linking was performed using specific binding conditions, as for previous figures. The trypsin cleavage products now show labeled DNA primarily on the smaller peptide (arrow). (D) The indicated peptide from panel C (arrow) was eluted and subjected to immunoprecipitation. Lane 1 shows a portion of the eluted peptide. Lane 3 is the precipitate with nonspecific mouse IgG, and lane 2 is the last wash step prior to collection of the precipitate. Lane 5 is the precipitate formed by antibody directed against the C-terminal myc tag. Lane 4 is the last wash step prior to collection of the precipitate. As was done for panel B, the peptide was immunoprecipitated only with the antibody directed against the C-terminal myc tag.
A parallel experiment was conducted employing the stringent binding conditions previously used for Fig. 4. Cross-linking in the presence of the MR2 and HMG1 proteins prior to trypsin digestion reduced the fraction of the coding-end probe that remained associated with the N-terminal domain (panel C). Again, the identity of the small product was confirmed as a C-terminal peptide by elution and specific immunoprecipitation by the antibody directed towards the myc tag.
DISCUSSION
We report here several observations pertaining to the architecture of RAG1 and its association with DNA. Previous studies have used cross-linking strategies to map which nucleotides formed close contacts with the proteins (11, 27, 28, 41). Our own work has used two different cross-linking techniques. Iodine-substituted nucleotide analogs can cross-link to proteins that make base contacts in the major groove. There are additional constraints using this strategy. Only direct base contacts are detectable, since iodine has a van der Waals radius of 2.15 Å and preferentially forms cross-links only with aromatic amino acids (48). In contrast, the azido-phenacyl cross-linker used in this study and previously (27) is positioned on the phosphate backbone and has a longer radius of 11 Å (7). This adduct extends a distance equal to half the diameter of the DNA helix and will cross-link any amino acid in its vicinity. We find these techniques complementary in their advantages. Using these techniques we previously found close contacts to RAG1 formed with cross-linkers positioned in the nonamer, the heptamer, and the nearby coding DNA. In the present study we turn the question around and ask where these contacts occur on the protein. We find that these cross-linkers show two distinct behaviors with respect to mapping their sites of interaction with RAG1. The cross-linker located in the coding DNA bound under stringent conditions (in the presence of a RAG2 derivative, an HMG1 derivative, and competitor dIdC) primarily to a C-terminal location in RAG1 (Fig. 4B and 8C). The CnBr digestion pattern appears to locate this site between the methionine residues 889 and 974. This region includes the critical acidic residue E962 that has been proposed to represent part of the enzymatic active site (19, 21). Our data are consistent with this model in that a residue which may contribute to the enzymatic active site would likely approach the cleavage site on the DNA.
In contrast, probes with the cross-linker positioned in the heptamer or nonamer also cross-link to RAG1 but do so nearer the N terminus of the core region (Fig. 5). These studies are less resolving than those performed with the coding region probe. We are not able at this time to determine precisely where these contacts occur in the protein. However, we interpret the 65-kDa band to be an N-terminal fragment of MR1, since we were unable to purify it by metal chelate chromatography. Based on this assumption and the size of the MBP fusion partner (43 kDa), it appears that both the heptamer probe and the nonamer probe make contacts within the N-terminal 20 kDa of RAG1 protein in the construct. More than one contact is possible, and the fact that we cannot distinguish a difference in the pattern obtained with the nonamer probe from that obtained with the heptamer probe does not prove that these contacts form at identical sites. Nevertheless, both of these probes make contact with the protein predominantly near the N terminus of the core region within the resolution of this assay. DNA binding has been previously associated with this region (39), and deletion of most of the C-terminal half of the core region in the protein designated R1 Δ34–38 (Fig. 2A) still permits DNA binding by the remaining peptide (Fig. 6).
Our original intent was to use metal chelate chromatography to purify the C-terminal peptides in a quantitative manner. We had difficulty for at least two technical reasons, rendering this analysis only qualitative. First, SDS was used during the CnBr cleavage reaction and could not be completely removed prior to chromatography. It is likely to interfere with binding to some degree. We also came to appreciate that the divalent heavy metal ions used in this chromatography procedure are capable of severing the thiophosphate linkage that joins the cross-linker to the DNA probe, thereby reducing the yield of retained products. Mercury ions have specifically been used to break the linkage with the phosphate, but most heavy metals will act similarly (7). The validity of the chromatography results was confirmed by independent immunoprecipitation, shown in Fig. 8.
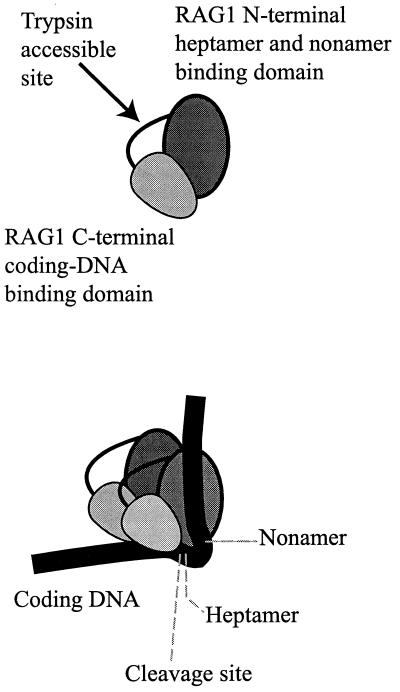
Since two different contact sites were found on the protein, we questioned whether these might reside on two separate domains. A tryptic digestion of the native MR1 protein supports this contention. Our data from experiments using limited proteolysis with trypsin (Fig. 7) and the successful peptide sequencing by Edman degradation of the C-terminal tryptic peptide demonstrate a solvent-accessible site at RAG1 residue R713 (of the full-length mouse protein). We do not yet have insight into the relationship of the C-terminal domain with the rest of the protein. Efforts to extend these data by additional studies of these potential domains are in progress. We also note that this finding is made in the context of a functional but engineered truncated derivative of RAG1. A model demonstrating the simplest organization of RAG1 is presented as Fig. 9. The trypsin-accessible site is presented as a peptide loop. RAG1 exists in solution as a dimer, and a simple representation of the RAG1 dimer bound to one DNA is shown (Fig. 9, bottom). The key features in this model are the location of binding by both the heptamer and the nonamer to the N-terminal part of the RAG1 core, while the coding DNA contact is associated with the C-terminal portion. There is no evidence that these contacts occur within the same monomer of RAG1. Other structures consistent with the data might have one monomer of RAG1 contacting the nonamer while the second binds the heptamer and binding to the coding end independent of other interactions. Further interactions with other proteins in the complex (notably RAG2) or the many possible cis-trans configurations between the DNA and the protein domains in a synaptic complex are not addressed here. The loop that we illustrate in the spacer DNA within the RSS is consistent with our previous cross-linking data, which showed minimal contact between RAG1 or RAG2 and the DNA within the spacer region (27, 28). In these studies the 23RSS showed additional internal binding by HMG1 in the spacer, which suggests sharp DNA bends interpretable as a loop. The loop would be larger in the 23RSS without disturbing the interactions with the heptamer, nonamer, or coding end. However, ethylation protection data (11, 41, 42) suggest that the spacer region is not solvent accessible along one face. Further analysis may reconcile these observations. Evidence for distortion of the DNA helix and bending near the heptamer has been presented in several studies (3, 8, 27, 32, 42).
FIG. 9.
Renditions of a RAG1 core region monomer and dimer bound to a DNA molecule, accounting for the observation presented here. The monomer (top) is drawn with a trypsin-accessible site as a protein loop dividing the protein into two domains. The dimer (bottom) is configured in one of many possible relationships. The model with one DNA indicates that the coding DNA interacts with the C-terminal domain of RAG1, while the heptamer and nonamer interact with the N-terminal domain. These contacts need not be limited to cis configurations on one monomer, as drawn purely for simplicity. The relationships with other proteins present in a bound complex are also unknown.
A similarity in structure between the RAG proteins and bacterial transposases is a plausible expectation based on the similarity of enzymology. The common features of three acid residues forming an essential metal binding motif have been found. In this regard, it is worth noting that the crystal structure of the Tn5 transposase on DNA has recently been reported (9). In this determination, each monomer of the transposase makes both N- and C-terminal contacts with DNA.
In considering the normal pathway of V(D)J recombination, one must appreciate the high degree of control that RAG proteins exert over several stages in the reaction. They provide the specificity to recognize the two types of RSS (12RSS and 23RSS) and yet can distinguish between them to enforce the 12/23 rule (though exactly how is not yet understood) (10, 20, 46, 47). They cleave the two DNA targets very precisely adjacent to the heptamer (25, 45) and seem to also open the hairpin intermediates that exist transiently on the coding ends formed by the cleavage reaction (6, 38). The RAG proteins persist in their association with the various DNA ends and seem essential for the joining reactions (2, 18, 28, 33). It seems important that the RAG proteins retain their hold on all the DNA ends, else these ends might diffuse from the proximity of their future partner. In fact, it is the case that all four DNA ends must be fairly close in space. Occasional “open and shut” reactions can be detected in which a coding end is rejoined to the same signal end from which it had been recently cleaved and hybrid junctions occur in which a coding end is joined to the opposite signal end (23, 24). Commonly, however, the two coding ends are joined to each other and similarly for the two signal ends. How might the recombination reaction bias the choice of partners for the joining reaction? It seems likely that a persistent complex associated with these ends undergoes a spatial change in configuration or isomerization while never releasing hold of the DNA. Preceding the isomerization, a cleaved coding end would be immediately adjacent to the RSS from which it has just been liberated. After isomerization, we propose that the two coding ends are rotated to favor their eventual coupling and to disfavor joining to either signal end. Similarly, the two signal ends would come to occupy positions that favor their joining to each other. Critical to this progression is the ability of the RAG proteins to contact not only the RSS but also the coding DNA in order to maintain the complex after cleavage. In addition, the contacts cannot be in the context of a single rigid DNA binding site. Flexibility is required for this isomerization proposal. We have previously recognized that a tetramer composed of two RAG1 plus two RAG2 molecules forms a functional unit that can form prior to any interaction with DNA (5). We speculated that the synaptic complex could be comprised of one or two of these units. If a single binding site existed per the RAG1 molecule, it might not be possible to satisfy the constraints of separate binding to coding ends and signal ends for both cleavage sites within a single tetramer. This question remains open at this time. However, the finding of this report that a single RAG1 molecule is capable of making DNA contacts at two separate sites within the protein, and that these may be associated with separate functional protein domains, would remove one constraint from the single-tetramer model. It is also attractive that the coding end may be bound in a separate functional domain from the RSS. This could more easily accommodate the structural reorganization required by the proposed isomerization step.
The architecture of the assembled constituents may affect the efficiency with which the biologically important coding junctions are assembled. Literature exists demonstrating that the sequence of the coding end can alter the efficiency of the recombination reaction (16). Two mechanisms may contribute to this observation. The terminal dinucleotide of the coding end adjacent to the heptamer contacts RAG1 (27) and has been shown to influence the efficiency of recombination using RAG1 mutants (35). One effect could be upon the stability of the bound complex. Some evidence exists, however, that the two steps of the cleavage reaction may be separately influenced by sequence context (49). In addition, coding-end sequence may also influence the efficiency of the subsequent processing of the hairpin (1, 29) and the joining steps (12, 14, 40).
ACKNOWLEDGMENTS
The oligonucleotide synthesis and peptide sequencing were performed in our Molecular Biology Core Facility at the Medical College of Georgia. Valuable encouragement and advice was provided by our colleague William Dynan.
This work was funded by NIH grant AI41711. M.S. is a Scholar of the Leukemia & Lymphoma Society.
REFERENCES
- 1.Agard E A, Lewis S M. Postcleavage sequence specificity in V(D)J recombination. Mol Cell Biol. 2000;20:5032–5040. doi: 10.1128/mcb.20.14.5032-5040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 3.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi M E, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailin T, Mo X, Sadofsky M J. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 7.Burgin A B, Pace N R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990;9:4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuomo C A, Mundy C L, Oettinger M A. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies D R, Goryshin I Y, Reznikoff W S, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 10.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in-vitro obeying the 12/23-rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 11.Eastman Q M, Villey I J, Schatz D G. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezekiel U R, Sun T, Bozek G, Storb U. The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol Cell Biol. 1997;17:4191–4197. doi: 10.1128/mcb.17.7.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fugmann S D, Lee A I, Shockett P E, Villey I J, Schatz D G. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 14.Gauss G H, Lieber M R. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 16.Gerstein R M, Lieber M R. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 17.Grachev M A, Lukhtanov E A, Mustaev A A, Zaychikov E F, Abdukayumov M N, Rabinov I V, Richter V I, Skoblov Y S, Chistyakov P G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989;180:577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- 18.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 19.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D R, Oettinger M A. Functional analysis of coordinated cleavage in V(D)J recombination. Mol Cell Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landree M A, Wibbenmeyer J A, Roth D B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 23.Lewis S M, Hesse J E. Cutting and closing without recombination in V(D)J joining. EMBO J. 1991;10:3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis S M, Hesse J E, Mizuuchi K, Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- 25.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 26.McMahan S A, Burgess R R. Mapping protease susceptibility sites on the Escherichia coli transcription factor sigma70. Biochemistry. 1999;38:12424–12431. doi: 10.1021/bi990824x. [DOI] [PubMed] [Google Scholar]
- 27.Mo X, Bailin T, Noggle S, Sadofsky M J. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 2000;28:1228–1236. doi: 10.1093/nar/28.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo X, Bailin T, Sadofsky M J. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J Biol Chem. 1999;274:7025–7032. doi: 10.1074/jbc.274.11.7025. [DOI] [PubMed] [Google Scholar]
- 29.Nadel B, Feeney A J. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol. 1997;17:3768–3778. doi: 10.1128/mcb.17.7.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Spatial organization of transcription elongation complex in Escherichia coli. Science. 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
- 31.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 32.Ramsden D A, McBlane J F, van Gent D C, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsden D A, Paull T T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 34.Sadofsky M J, Hesse J E, McBlane J F, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadofsky M J, Hesse J E, van Gent D C, Gellert M. RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 36.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 38.Shockett P E, Schatz D G. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–4166. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spanopoulou E, Zaitseva F, Wang F, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 40.Sun T, Ezekiel U R, Erskine L, Agulo R, Bozek G, Roth D, Storb U. Signal joint formation is inhibited in murine scid preB cells and fibroblasts in substrates with homopolymeric coding ends. Mol Immunol. 1999;36:551–558. doi: 10.1016/s0161-5890(99)00053-x. [DOI] [PubMed] [Google Scholar]
- 41.Swanson P C, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanson P C, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 43.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 44.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 46.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23-rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 47.West R B, Lieber M R. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol Cell Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willis M C, Hicke B J, Uhlenbeck O C, Cech T R, Koch T H. Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science. 1993;262:1255–1257. doi: 10.1126/science.7694369. [DOI] [PubMed] [Google Scholar]
- 49.Yu K, Lieber M R. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol Cell Biol. 1999;19:8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]