Abstract
Acanthamoeba causes opportunistic eye infections in humans, which can lead to severe keratitis and may ultimately result in blindness. Current methods for identifying this organism rely on culture and microscopy. In this paper, we describe the isolation of antibody fragments that can be used for the unequivocal identification of Acanthamoeba. A bacteriophage antibody display library was used to isolate antibody fragments that bind specifically to Acanthamoeba. Individual clones were studied by enzyme-linked immunosorbent assay, flow cytometry, and immunofluorescence. Four antibody clones that specifically bind to Acanthamoeba spp. were identified.
Acanthamoeba is a free-living, opportunistic protozoan parasite of humans. Acanthamoeba can cause a fatal meningoencephalitis, but it is most commonly associated with eye infections, typically, Acanthamoeba keratitis associated with contact lens use. The increasing prevalence of this keratitis is thought to be linked to the increased use of contact lenses (8, 10, 15). Acanthamoeba keratitis is usually diagnosed after viral and bacterial causes have been eliminated (1, 5) and, as a result, there is often a significant delay before appropriate treatment is administered. Because of the severity of Acanthamoeba keratitis, a significant loss of visual acuity is common and in many cases total loss of sight in the infected eye occurs (4, 6). Current methods of detection involve culture and microscopic identification (9, 16). These methods are time consuming, laborious, and open to error. The development of a rapid, simple detection method for Acanthamoeba is thus important (2, 14).
Bacteriophage antibody display libraries expressing single-chain Fv antibody fragments have recently been developed as an alternative way of isolating specific antibodies (17). Antibody fragments are generated by the random pairing of large diverse repertoires of variable heavy and light chain genes, derived by PCR from activated or naive human lymphocytes, and cloned for expression of individual specificities on the surface of filamentous bacteriophages. A library contains a vast number of different antibody specificities, varying from 107 to 1012, depending on how the library is constructed. This approach to generating antibodies has the major advantage that epitopes do not have to be immunogenic, i.e., antibodies that recognize native cell surface structures can be isolated. Here, we describe the isolation of antibody fragments that can be used to detect Acanthamoeba immunofluorescence and flow cytometry. These antibodies provide the reagents to establish a specific and rapid detection assay for Acanthamoeba.
MATERIALS AND METHODS
Cell culture.
Pathogenic and nonpathogenic Acanthamoeba species were obtained either from the Culture Collection of Algae and Protozoa or from S. Kilvington (Leicester PHL). Bacteria used in this study were obtained from Hull PHL (Hull Royal Infirmary). All Acanthamoeba spp. and the Hartmanella sp. were maintained in PYG medium (0.75% [wt/vol] proteose peptone, 0.75% [wt/vol] yeast extract [Difco Laboratories, Detroit, Mich.] and 1.5% [wt/vol] glucose) at 30°C, and cultures reached mid-log phase after 7 days. Mid-log-phase cells were used for all experiments and were harvested by centrifugation at 800 × g for 8 min. Cells were resuspended in Page's amoeba saline (PAS) (13) and centrifuged at 800 × g for 8 min; this was repeated twice. Cells used for isolating antibody fragments, enzyme-linked immunosorbent assay (ELISA), or fluorescence-activated cell sorter analysis were fixed in 50% (vol/vol) methanol:PAS. After fixation, cells were washed three times with PAS as described above. Organisms used for immunofluorescence microscopy were harvested by centrifugation and then placed onto slides prior to fixation.
Bacteriophage display library.
The human synthetic ScFv library no. 1 (Nissim Library), a human-derived bacteriophage antibody library expressing a single-chain Fv fragment was obtained from G. Winter (Centre for Protein Engineering, Medical Research Council Centre, Cambridge, United Kingdom) (12). This library consists of a single Vλ3 light chain paired with a bank of in vitro-rearranged VH gene fragments containing a random VH CDR3 of 4 to 12 amino acid residues in length. This library possesses more than 108 specificities.
Preparation of bacteriophage particles.
For bacteriophage particle preparation, the library stock or individual bacteriophage clones were added to a culture of Escherichia coli (TG1) grown in 2× TY (0.8% [wt/vol] NaCl, 1.6% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract) supplemented with 100 μg of ampicillin per ml and 1% (wt/vol) glucose. This culture was incubated at 37°C until the absorbance at 600 nm was between 0.4 and 0.5. VCS-M13 helper bacteriophage was then added to the culture, which was incubated for a further 30 min at 37°C without shaking. The culture was centrifuged at 1,500 × g for 10 min, the pellet was resuspended in 2× TY supplemented with 100 μg of ampicillin per ml and 25 μg of kanamycin per ml (to select for bacteriophage-containing clones), and incubated at 30°C overnight. The overnight culture was centrifuged at 10,800 × g for 10 min and the pellet was resuspended in a 1/5 volume of 20% (wt/vol) polyethylene glycol 6000 in 2.5 M NaCl for 1 h at 4°C. After incubation and three washes, the pellet was resuspended in PAS with 15% glycerol and centrifuged at 1,500 × g for 10 min. Finally, the supernatant containing the bacteriophage particles was filtered (pore size, 0.45 μm) prior to storage at −80°C. The bacteriophage particle titer was determined by serial dilution of infected E. coli (TG1) plated out on TYE (1.5% [wt/vol] Bacto-agar, 0.8% NaCl, 1% tryptone, and 5% yeast extract) supplemented with 25 μg of kanamycin per ml.
Use of bacteriophage antibody display library.
To isolate Acanthamoeba-specific antibody fragments the bacteriophage library (2 × 1011 bacteriophages) was added to a suspension of whole fixed cells (2 × 106 cells) which had been blocked by incubation in MPAS (2% [wt/vol] dried milk powder, 1% [wt/vol] bovine serum albumin in PAS) at 37°C for 1 h prior to use. The mixture of bacteriophage particles and Acanthamoeba parasites was incubated at 20°C with gentle shaking for 1 h and then centrifuged at 200 × g for 5 min. The pellet was resuspended in 0.1% (wt/vol) bovine serum albumin in PAS and centrifuged again; this process was repeated 10 times in total, to remove unbound bacteriophage particles. The pellet was resuspended in citric acid (76 mM) and incubated at 20°C with shaking to elute bound bacteriophage particles. The pH of the mixture was then adjusted to 7.0 by adding Tris-HCl (1 M), pH 7.4. This procedure was termed a panning round. The bacteriophage particles selected by this procedure were then amplified as described previously (see “Preparation of bacteriophage particles” above), except that bacteria infected with bacteriophage were spread onto TYE bioassay dishes and incubated at 30°C overnight. The E. coli (TG1) cells containing bacteriophage were scraped from the plate and resuspended in 2× TY containing 15% (wt/vol) glycerol and this suspension, termed the library stock, was stored at −80°C.
To remove nonspecifically binding bacteriophage, a negative panning round against the amoeba Hartmanella sp. was performed as described for Acanthamoeba, except that the supernatant containing unbound bacteriophage particles was retained. Four further negative panning rounds were performed to ensure complete removal of all cross-reactive bacteriophage particles. The supernatant obtained from the final negative panning was then incubated with 2 × 106 fixed and blocked Acanthamoeba cells. In summary, four panning rounds were performed in the following order: a positive pan against Acanthamoeba; a negative pan on Hartmannella followed immediately by a positive pan on Acanthamoeba; and, finally, two positive panning rounds against Acanthamoeba.
PCR.
PCR for CDR3 length was performed to demonstrate the diversity of bacteriophage clones isolated after each panning round (11). Primers used were CDR-FOR (5′ CAG GGT ACC TTG GCC CCA 3′) and CDR-BACK (5′ GTG TAT TAC TGT GCA AGA 3′) (11). PCR fragments amplified directly from bacterial colonies were separated on 5% (wt/vol) Metaphor agarose gel (Flowgen, FMC Bioproducts) stained with ethidium bromide and visualized under UV light.
ELISA.
Single bacterial colonies were picked directly from the bioassay dish and inoculated into 2× TY supplemented with 100 μg of ampicillin per ml and 1% (wt/vol) glucose and grown in 96-well round-bottom plates overnight at 37°C. Glycerol stocks of the overnight incubations were made by adding 15% (wt/vol) glycerol, and they were stored at −80°C until required. To rescue the bacteriophage, 5 μl of the glycerol stock culture was transferred into 2× TY and incubated with shaking at 37°C for 1 h. VCS-M13 helper bacteriophage particles (1 × 109) were added to each well and incubated at 37°C for 30 min without shaking and for 1 h with shaking. The culture was centrifuged at 400 × g for 10 min, and the supernatant was aspirated. The pellet was resuspended in 2× TY supplemented with 100 μg of ampicillin per ml plus 50 μg of kanamycin per ml and incubated overnight at 30°C with shaking (bacteriophage plate). Fixed and MPAS-blocked Acanthamoeba parasites (2 × 105) were added to a conical 96-well plate and centrifuged at 200 × g for 5 min. The supernatant was carefully removed, and the wells were washed with PAS. Bacteriophage plates incubated overnight were centrifuged at 400 × g for 10 min, and the supernatant was used as a source of the bacteriophage clones. To each well of the plate containing immobilized Acanthamoeba, individual bacteriophage clones were added, and the plate was incubated with shaking at 20°C for 1 h and then washed with PAS. Sheep anti-M13–horseradish peroxidase (Pharmacia) (diluted 1:500 in MPAS) was added to each well, and the plates were incubated with shaking at 20°C for 1 h. Wells were washed twice with PAS before 2,2′-azinobis(3-ethylbenzthiazoline sulfonic acid) (ABTS) substrate (Vector Laboratories) was added. The plate was then incubated at 20°C for 30 min in the dark. The supernatant was transferred to a flat-bottom plate and the absorbance (at 405 to 690 nm) was determined.
Flow cytometry.
Acanthamoeba parasites (1 × 106) were fixed and blocked as described above, mixed with 1 × 1012 bacteriophage particles from the appropriate clone and incubated at 4°C for 1 h. Cells were washed three times with 0.25% (wt/vol) bovine serum albumin in PAS (PAA). The final pellet was resuspended in sheep anti-M13 antibody (10 μg/ml) and incubated at 4°C for 1 h. Cells were washed twice with PAA, resuspended in fluorescein isothiocyanate-conjugated anti-sheep immunoglobulin G (IgG) (10 μg/ml; Vector Laboratories) and incubated at 4°C for 1 h. Cells were washed twice as described above and finally resuspended in PAA and analyzed by flow cytometry. Fluorescence intensity was measured on a FACSCalibur (Becton Dickinson), using an excitation wavelength of 488 nm.
RESULTS
ELISA.
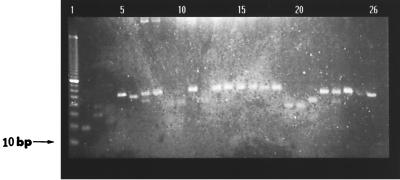
Over 300 clones were analyzed by ELISA. The 10 clones with the highest level of binding to Acanthamoeba palestinensis (the absorbance at between 405 and 690 nm was >0.8) were selected for further study. All these clones showed cross-reactivity with all other Acanthamoeba species studied (the absorbance at between 405 and 690 nm was between 0.8 and 1.1). PCR (Fig. 1) showed the diversity of the selected clones.
FIG. 1.
Diversity in insert sizes of different clones selected from 96-well ELISA plate using CDR-FOR and CDR-BACK primers. Lane 1, 10-bp DNA ladder; lanes 2 to 26, PCR reaction products from 25 individual bacterial colonies selected at random from a TYE plate.
Flow cytometry.
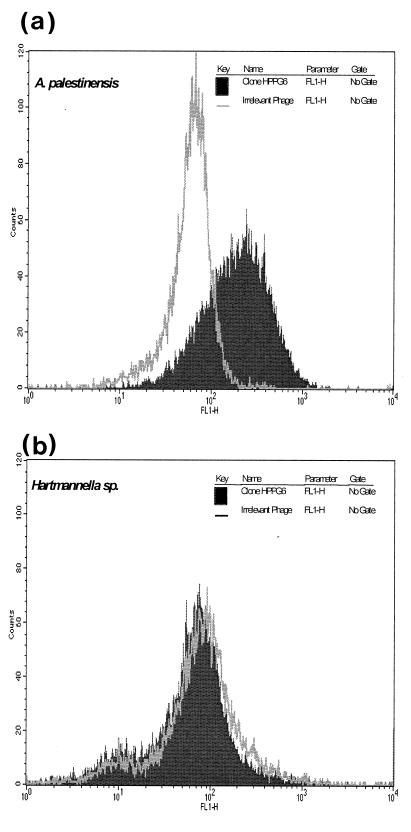
Flow cytometry showed a high level of antibody fragment binding to Acanthamoeba cells with no binding to a range of other cell types (Table 1). Mean channel fluorescence for clone HPPG6 with A. palestinensis was 200 as compared to 48 for cells stained with the negative irrelevant bacteriophage (Fig. 2a). When this was repeated for a Hartmanella sp. (Fig. 2b), no change in mean channel fluorescence could be observed between the negative phage and HPPG6.
TABLE 1.
Reactivity of clone HPPG6 as determined by flow cytometry
| Organism, isolate type, or cell type (n) | Reactivity |
|---|---|
| Acanthamoeba palestinensis | + |
| Acanthamoeba castellanii | + |
| Acanthamoeba polyphaga | + |
| Acanthamoeba astronyxis | + |
| Acanthamoeba griffinii | + |
| Acanthamoeba sp. (clinical isolate) | + |
| Acanthamoeba sp. (clinical isolate) | + |
| Environmental isolates (7) | + |
| Hartmanella sp.a | − |
| Candida albicansa | − |
| Escherichia colia | − |
| Haemophilus influenzaea | − |
| Nisseria sp.a | − |
| Pseudomonas aeruginosaa | − |
| Klebsiella aerogenesa | − |
| Monocytesb | − |
| Lymphocytesb | − |
| Neutrophilsb | − |
This organism is a common cause of eye infection.
This cell type may be found on eye swabs.
FIG. 2.
Flow cytometric analysis of clone HPPG6 against A. palestinensis (A) and Hartmanella sp. (B). Parasites (1 × 106) were incubated with bacteriophage clone HPPG6 or the irrelevant anti-NIP control (1 × 1012) for 1 h at 4°C. Cells were then stained with a polyclonal sheep anti-M13 and fluorescein isothiocyanate-conjugated donkey anti-sheep IgG (10 μg/ml). Samples were analyzed on a FACSCalibur flow cytometer.
Indirect immunofluorescence.
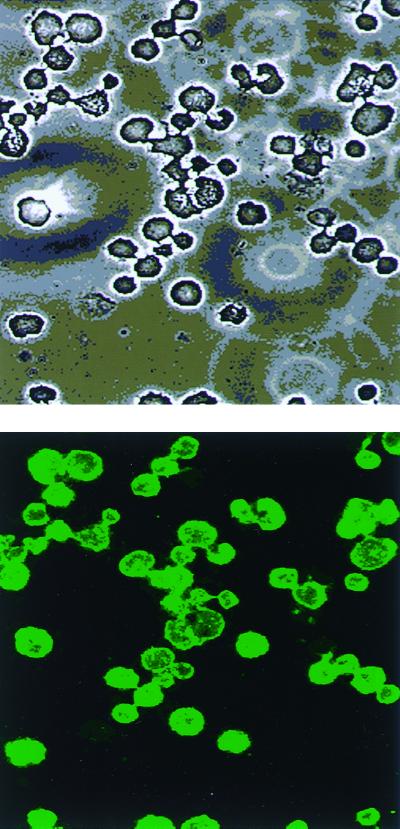
The selected antibody fragments were also assessed by indirect immunofluorescence microscopy and yielded results similar to those obtained by flow cytometry. Figure 3 shows A. palestinensis stained with HPPG6. It can be seen from the photomicrograph that this antibody binds uniformly, with a high intensity, to all cells.
FIG. 3.
(A) Under a light microscope. (B) Indirect immunofluorescence reactivities of clone HPPG 6 with A. palestinensis. Magnification, ×400.
DISCUSSION
In this study we have shown that antibody fragments showing a high level of specificity for Acanthamoeba spp. can be isolated from a naive bacteriophage display library. This is the first time that bacteriophage antibody display technology has been used in the development of an antibody for the identification of a microorganism. Previously, similar technology has been used successfully to isolate a melanoma-specific antibody, but this work required extensive screening of more than 1,700 clones before a useful reagent was identified (3). It is significant that we have been able to isolate four clones with the desired specificities from an initial screen of only 300 clones. This suggests that bacteriophage antibody display technology is ideally suited for the isolation of novel antibodies for use as research and diagnostic tools in clinical microbiology.
Current methods for the rapid identification of Acanthamoeba involve staining preparations with Giemsa, calcofluor white, methylene blue, or acridine orange. Culturing is also widely used in clinical laboratories for the reliable identification of Acanthamoeba. However, accurate diagnosis and interpretation of results using these techniques requires a strong clinical suspicion of amoebic infection and highly trained personnel. Other groups have utilized PCR as a rapid detection method for Acanthamoeba (14), but the specificity and sensitivity of this technique in a clinical environment have not been tested. Rabbit polyclonal antisera have also been used for the detection of Acanthamoeba keratitis and suspected Acanthamoeba meningoencephalitis. However, these reagents showed considerable cross-reactivity with other cell types, and, once again, the sensitivity of the assay has been questioned (7).
The need for a simple and rapid method for the specific detection of Acanthamoeba has become more urgent as Acanthamoeba keratitis becomes a more significant causative agent of eye keratitis due to greater contact lens use and is increasingly associated with meningoencephalitis in immunocompromised individuals. We believe that the clones isolated in this study will form the basis of a rapid and unequivocal assay for the detection of Acanthamoeba. Thus, bacteriophage antibody display libraries are potentially useful and powerful tools that allow the rapid generation of antibody reagents for use in diagnostic assays.
REFERENCES
- 1.Aitken D, Hay J, Kinnear M C, Lee R C. Amebic keratitis in a wearer of disposable contact lenses due to a mixed Vahlkampfia and Hartmannella infection. Opthalmology. 1996;103:485–494. doi: 10.1016/s0161-6420(96)30667-2. [DOI] [PubMed] [Google Scholar]
- 2.Bacon S A, John K G, Ficker A L, Matheson M M, Wright P. Acanthamoeba keratitis: the value of early diagnosis. Opthalmology. 1993;100:1238–1243. doi: 10.1016/s0161-6420(93)31499-5. [DOI] [PubMed] [Google Scholar]
- 3.Cai X, Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumour cells: Selection of specific antibodies from single-chain Fv fusion phage libraries. Proc Natl Acad Sci USA. 1995;92:6537–6541. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chynn W E, Lopez A M, Langston P D, Talamo H J. Acanthamoeba keratitis. Opthalmology. 1995;102:1369–1373. doi: 10.1016/s0161-6420(95)30862-7. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J E, Buchanan W H, Laughrea A P, Adams P C, Galentine G P, Folberg P, Arentsen J J, Laibson R P. Diagnosis and management of Acanthamoeba keratitis. Am J Opthalmol. 1985;100:389–395. doi: 10.1016/0002-9394(85)90499-4. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J E, Fultan C J, Hoffman J C, Rapuano J C, Laibson R P. Trends in contact lens-associated corneal ulcers. Cornea. 1996;15:566–570. [PubMed] [Google Scholar]
- 7.Flores B M, Garcia C A, Stamm W E, Torian B E. Differentiation of Naegleria fowleri from Acanthamoeba species by using monoclonal antibodies and flow cytometry. J Clin Micro. 1990;28:1999–2005. doi: 10.1128/jcm.28.9.1999-2005.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grey B T, Ray T M, Sherwan F, Rose R P. Acanthamoeba, bacterial and fungal contamination of contact lens storage cases. Br J Opthalmol. 1995;79:601–605. doi: 10.1136/bjo.79.6.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John J K, Steven W, Parrish M C, Williams E T, Robinson D R, Denis M. Examination of hydrophilic contact lenses with light microscopy to aid in the diagnosis of Acanthamoeba keratitis. Am J Opthalmol. 1989;108:329–331. doi: 10.1016/0002-9394(89)90129-3. [DOI] [PubMed] [Google Scholar]
- 10.Ma P, Visvesvara G S, Martinez A J, Theodore F H, Daggett P M, Sawyer T K. Naegleria and Acanthamoeba infections. Rev Infect Dis. 1991;13(Suppl. 5):369–372. doi: 10.1093/clinids/12.3.490. [DOI] [PubMed] [Google Scholar]
- 11.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty A D, Griffiths A, Winter G. Human antibodies from bacteriophage display libraries. J Mol Biol. 1991;222:581. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 12.Nissim A, Hoogenboom R H, Tomlinson M I, Flynn G, Midgley C, Lane D, Winter G. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 1994;13:692–698. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page F C. A new key to the fresh water and soil Gymnamoebae with instructions for culture. Ambleside, Cumbria, United Kingdom: Freshwater Biological Association, Ferry House; 1988. [Google Scholar]
- 14.Vodkin M H, Howe K D, Visvesvara S G, Mclaughlin L G. Identification of Acanthamoeba at the generic and specific levels using the polymerase chain reaction. J Protozool. 1992;39:378–385. doi: 10.1111/j.1550-7408.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 15.Wilhelmus K R. The increasing importance of Acanthamoeba. Rev Infect Dis. 1991;13(Suppl. 5):367–446. doi: 10.1093/clind/13.supplement_5.s367. [DOI] [PubMed] [Google Scholar]
- 16.Winchester K, Mathers D W, Sutphin E J, Daley E T. Diagnosis of Acanthamoeba keratitis in vivo with confocal microscopy. Cornea. 1995;14:10–17. [PubMed] [Google Scholar]
- 17.Winter G, Griffiths D A, Hawkins E R, Hoogenboom R H. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–456. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]