Highlights
-
•
YTHDC2 is significantly upregulated in human GC tissues, which is linked to poor prognosis.
-
•
Knockout of YTHDC2 inhibits GC cell proliferation, migration and invasion.
-
•
YTHDC2 enhances YAP translation in a m6A-dependent manner.
-
•
YAP transcriptionally activates YTHDC2.
-
•
Knockout of YTHDC2 inhibits GC tumor growth and metastasis in vivo.
Keywords: m6A, YTHDC2, YAP, Translation regulator, Biomarker
Abstract
N6-methyladenosine (m6A) modification is the most prevalent internal modification in eukaryotic mRNA. YTH domain containing 2 (YTHDC2), a m6A binding protein, has recently been identified as a key player in human cancer. However, its contribution to gastric cancer (GC) remains unknown. Herein, we found that YTHDC2 was significantly upregulated in human GC tissues and associated with poor prognosis. CRISPR-Cas9 mediated YTHDC2 knockout notably inhibited GC cell viability, proliferation and invasion. Transcriptome analysis coupled with mechanism experiments revealed that yes-associated protein (YAP), the well-known oncogene, is the target of YTHDC2 in GC cells. Specifically, YTHDC2 recognized m6A-modified YAP mRNA at 5`-UTR, resulting in enhancing the translation efficiency of YAP, without affecting its mRNA level. In turn, YAP/TEAD directly targeted -843∼-831 region on the promoter of YTHDC2 and activated the transcription of YTHDC2, thus forming a positive regulatory loop. Further, using the xenograft tumor model, we found that knockout of YTHDC2 markedly reduced tumor size and lung metastasis nodules in vivo. And high YTHDC2 was strongly positively correlated with high YAP in clinical GC tissues. Collectively, our data demonstrate that YTHDC2 is a novel oncogene in GC, which provides the theoretical basis for the strategy of targeting YTHDC2 for GC patients.
Graphical abstract
Introduction
Gastric cancer (GC) is a malignant tumor originating from gastric mucosa epithelium, it caused about one million new cases and estimated 769 thousand deaths in 2020, ranking fifth and fourth in the global incidence and mortality rate, respectively [1]. Despite its worldwide decline in incidence over the past century, GC remains a major killer across the globe [2]. The etiology of GC is extremely complex, from diet and lifestyle to genetics and ethnicity [3], scientists are still making unremitting efforts to find key regulatory molecules controlling GC progression, which will provide effective prevention strategies and suggestions for clinical diagnosis and treatment.
N6-methyladenosine (m6A) is the most common internal modification in eukaryotic mRNA [4]. It is dynamically installed and removed, and acts as a new layer of mRNA fate, involving in m6A methyltransferase, demethylase and binding protein [5], [6], [7]. m6A binding protein, known as “reader”, is capable of decoding m6A methylation and yielding a series of functional signals, mainly including YTH domain containing protein, IGF2BP and HNRNP families [8,9]. YTH domain can recognize m6A via a conserved aromatic cage [10], thus regulating RNA transcription, splicing, processing, stability and translation [11,12]. YTH domain containing 2 (YTHDC2), the final member of the YTH protein family, has recently been reported as a key regulator in several human diseases [13]. For example, by m6A sequencing and RNA sequencing, Zhou et al. found that YTHDC2 was markedly downregulated in livers of obese mice and nonalcoholic fatty liver patients, it directly bound to lipogenic genes and reduced their mRNA stability [14]. Besides, YTHDC2 was shown as a tumor inhibitor in lung adenocarcinoma via promoting SLC7A11 decay and inducing ferroptosis [15]. Further, emerging evidence suggests that YTHDC2 was also involved in mRNA translation process in a context-dependent way [16,17]. However, to date, the biological role of YTHDC2 in GC remains uncovered.
Therefore, this study aimed to explore the expression, function as well as clinical implication of YTHDC2 in GC. We preliminarily found that YTHDC2 was significantly increased in fresh and paraffin-embedded GC tissues. Further, we investigate the potential of YTHDC2 as a GC marker and its potential mechanisms for promoting the progression of GC.
Materials and methods
GC tissues
A total of 120 pairs of paraffin-embedded GC and adjacent normal tissues were collected from Department of Pathology, Hainan Provincial People's Hospital to generate tissue microarray (TMA). In addition, 6 paired fresh GC and normal tissues were also obtained from patients who have undergone surgical resection and were subsequently pathologically confirmed to be GC. Each patient provided written informed consent and was followed up. This study was approved by the Medical Ethics Committee of Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University.
RNA extraction and qRT-PCR analysis
Total RNA was extracted using PureLink™ RNA Separation kit (Invitrogen, CA, USA) as per the manufacturer's instructions. Then, cDNA was synthesized with Superscript IV reverse transcriptase (Invitrogen), followed by RNA amplification and quantification using SYBR® GreenER™ qPCR SuperMix (Invitrogen) with ABI PRISM® 7900HT system. 2−ΔΔCt formula was used to calculate gene relative expression.
Western blot
Total protein was collected using RIPA buffer (Thermo Scientific, CA, USA), followed by protein quantification using Pierce™ BCA Protein Quantification Kit (Thermo Scientific). Then, 20 μg protein was used for electrophoresis loading, transfer and blocking. After incubation with primary antibody and corresponding secondary antibody, the blot was developed using SuperSignal™ West Dura Persistence Substrate (Thermo Scientific). The primary antibody used in this study: anti-YTHDC2 (#ab271139, Abcam), anti-E-Cadherin (#14472, CST), anti-N-Cadherin (#13116, CST), anti-YAP (#14074, CST), and anti-Tubulin (#2148, CST).
Immunohistochemistry (IHC)
The paraffin-embedded sections were dewaxed and rehydrated, followed by antigen repair and endogenous peroxidase blockade. Then, the sections were incubated with anti-YTHDC2 (#ab271139, Abcam), anti-YAP (#14074, CST) and anti-Ki-67 (#9449, CST) at 4 °C overnight. After incubation with ready-to-use universal secondary antibody, the sections were stained with DAB solution. The staining scores were determined based on both the intensity and proportion of positive cells in 10 random fields using H-score method as previously described [18].
Cell lines, transfection and CRISPR/Cas9
Two GC cells AGS and HGC-27 were purchased from ATCC, and cultured in DMEM medium supplemented with 10% fetal bovine serum. Cell transfection was conducted using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. To generate stable YTHDC2 knockout cell lines, sgRNA targeting YTHDC2 was designed and inserted into CRISPR/Cas9 lentivirus vector (Hanbio, Shanghai, China). AGS and HGC-27 cells were infected with above lentivirus, and the stable cell lines were generated using puromycin. The knockout efficiency was verified by western blot.
CCK-8, EdU and Transwell assays
For CCK-8 assay, cells were plated onto 96-well plates, and cultured for 24 h, 48 h, 72 h and 96 h. 10μL CCK-8 reagent was added into each well at each point. Then, the absorbance at 450 nm was recorded using a microplate reader. DNA synthesis rate was assessed by EdU staining with Yefluor EdU Imaging Kits (Yeasen Biotechnology, Shanghai, China) based on the manufacturer's protocol. Transwell assay was used to test the migration and invasion of GC cells using Transwell chamber counted with (invasion) or without (migration) matrigel.
mRNA sequencing
Total RNA from control and YTHDC2 knockout cells was extracted and mRNA was enriched with Oligo(dT) magnetic beads, followed by fragmentation and cDNA synthesis and library preparation. The sequencing was conducted using Illumina HiSeq2000 platform system. The differentially expressed mRNAs were set as log2 (fold change) >1 and P-value < 0.05. The raw data are deposited in GEO database (GSE236281).
Luciferase reporter assay
The full-length of wild-type or mutant YAP 5`-UTR was synthesized and inserted into pmirGLO reporter vector (Promega, WI, USA), followed by co-transfection with wild-type YTHDC2 or YTHDC2ΔYTH pcDNA 3.0 expressing plasmid into GC cells using Lipofectamine 3000. For assessing the effect of YAP on YTHDC2 transcription activity, the YTHDC2 promoter with wild-type or mutant YAP/TEAD binding site was synthesized and inserted into pGL3-basic vector (Promega), followed by co-transfection with YAP siRNA (RiboBio, Guangzhou, China) into GC cells. After 48 h of transfection, the luciferase activity was tested using a luciferase reporter commercial kit (Promega, Madison, WI).
Methylated RNA immunoprecipitation (meRIP) and RIP
The enrichment of m6A and YTHDC2 on YAP 5`-UTR was tested by Magna MeRIP m6A Kit (Millipore, Schwalbach, Germany) and EZ-Magna RIP Kit (Millipore), respectively, according to the manufacturer's instructions, followed by qRT-PCR analysis.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was conducted using SimpleChIP® Plus Enzymatic Chromatin IP Kit (#9004, CST) based on standard protocol. Briefly, protein was cross-linked to DNA with 1% formaldehyde for 10 min, followed by inactivation with glycine for 5 min. Then, DNA fragment was generated using a micrococcal nuclease, followed by incubation with 5 μg anti-YAP (#14074, CST) at 4 °C overnight. The enriched DNA was washed and purified for qPCR analysis.
Nude mice assay
The animal protocols of this study were approved by the Institutional Animal Care and Use Committee (IACUC) of Hainan General Hospital, Hainan Affiliated Hospital of Hainan Medical University. All nude mice were maintained in specific-pathogen-free cages with a standard diet, constant temperature, constant humidity and with standard light/dark cycles. They were subcutaneously and caudal vein injected with YTHDC2 knockout AGS cells, respectively. After 7 weeks, the mice were sacrificed and tumor size and lung metastasis nodules were recorded.
Statistical analysis
The results are the mean±SD of at least three independent experiments carried out in triplicate. Chi-square test and t-test were used for comparison between the two groups. Kaplan-Meier plotter was used to analyze the survival curve of GC patients. P<0.05 was defined as statistically significant. All P-values were two-sided unless otherwise specified.
Results
YTHDC2 is significantly upregulated in human GC tissues
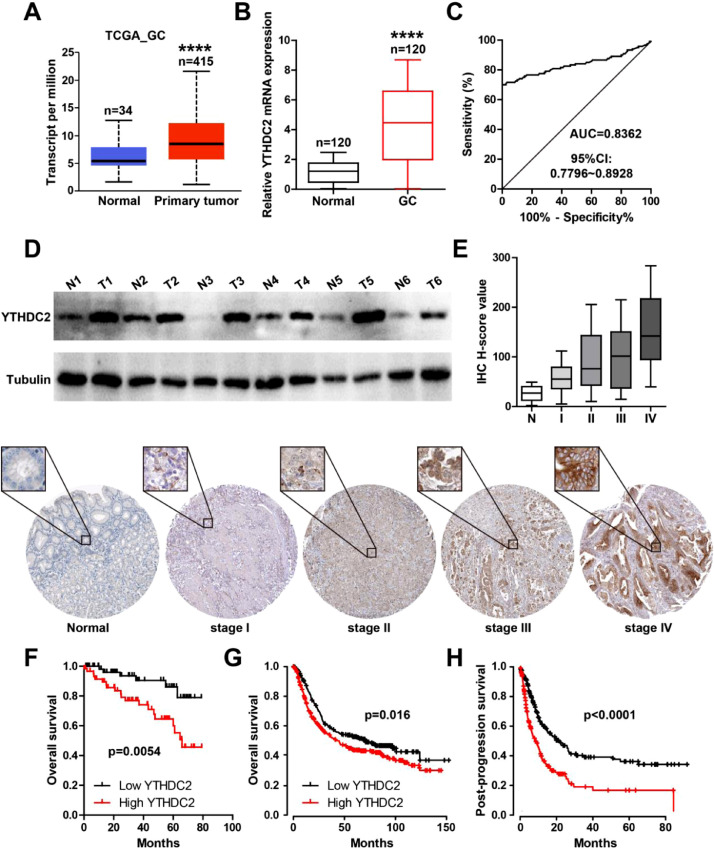
We first analyzed the TCGA database, and found that YTHDC2 mRNA was significantly increased in GC compared to normal tissues (Fig. 1A), which was further verified in our collected 120 paired tissues (Fig. 1B). The AUC value was 0.8362 (95%CI: 0.7796–0.8928) (Fig. 1C). Moreover, the western blot results showed that YTHDC2 protein was notably upregulated in fresh GC tissues in comparison to adjacent normal tissues (Fig. 1D). The IHC staining results in TMA showed that YTHDC2 was highly expressed in GC tissues, and the later the disease stage, the higher the expression (Fig. 1E). As shown in Table 1, high YTHDC2 was positively correlated with larger tumor size, lymph node metastasis and later clinical stage, but not with gender, age and differentiation. Further, GC patients with high YTHDC2 had poorer survival than patients with low YTHDC2 (Fig. 1F). The data from Kaplan-Meier plotter online tool also displayed that YTHDC2 was associated with shorter overall and post-progression survival time (Fig. 1G, H). And Cox multivariate regression analysis showed that TNM stage and YTHDC2 level were independent prognostic indicators for GC overall survival (Table 2).
Fig. 1.
YTHDC2 is upregulated in GC. A. YTHDC2 mRNA level in TCGA_GC database. B, C. qRT-PCR analysis of YTHDC2 mRNA expression in 120 paired GC and normal tissues, followed by ROC curve analysis. D. Western blot assay detecting YTHDC2 protein level in 6 paired fresh GC and normal tissues. E. IHC staining of YTHDC2 in GC tissues with different TNM stage. F-H. The survival curve of GC patients in out cohort and in K-M database based on YTHDC2 level. ⁎⁎⁎⁎P<0.0001.
Table 1.
Correlation between YTHDC2 expression and clinicopathological features in GC patients (n = 120).
| Parameters | All cases | YTHDC2 expression |
p value | |
|---|---|---|---|---|
| Low (n = 60) | High (n = 60) | |||
| Gender | ||||
| Male | 86 | 44 | 42 | 0.685 |
| Female | 34 | 16 | 18 | |
| Age (years) | ||||
| ≤ 60 | 59 | 33 | 26 | 0.201 |
| > 60 | 61 | 27 | 34 | |
| Tumor size | ||||
| ≤ 5 | 70 | 43 | 27 | 0.003 |
| > 5 | 50 | 17 | 33 | |
| Lymph node metastasis | ||||
| No | 62 | 42 | 20 | <0.001 |
| Yes | 58 | 18 | 40 | |
| TNM stage | ||||
| I-II | 68 | 46 | 22 | <0.001 |
| III-IV | 52 | 14 | 38 | |
| Differentiation grade | ||||
| Well/moderate | 74 | 40 | 34 | 0.260 |
| Poor | 46 | 20 | 26 | |
Table 2.
Uni- and multivariate analysis of prognostic predictors in GC patients (n = 120).
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95%CI) | P value | HR (95%CI) | p value | |
| Gender (male) | 1.038 (0.522–1.258) | 0.631 | ||
| Age (>60) | 1.075 (0.527–1.869) | 0.736 | ||
| Tumor size (>5) | 2.452 (1.263–4.587) | 0.005 | 1.328 (0.458–1.985) | 0.236 |
| Lymph node metastasis (yes) | 2.313 (1.521–3.816) | 0.009 | 1.072 (0.887–4.231) | 0.153 |
| TNM stage (III-IV) | 3.724 (1.558–5.368) | <0.001 | 2.308 (1.289–6.384) | 0.032 |
| Differentiation (poor) | 1.661 (0.438–2.651) | 0.337 | ||
| YTHDC2 (high) | 4.685 (2.347–9.035) | <0.001 | 3.867 (2.511–8.698) | 0.004 |
Knockout of YTHDC2 inhibits GC cell proliferation, migration and invasion
We established YTHDC2 knockout AGS and HGC-27 cell lines using CRISPR-Cas9 gene editing, which was verified by western blot assay (Fig. 2A). The CCK-8 results showed that cell viability was significantly decreased after YTHDC2 knockout (Fig. 2B). And less DNA synthesis was observed in YTHDC2-depleted AGS and HGC-27 cells compared to normal cells (Fig. 2C). Transwell assay showed that YTHDC2 knockout resulted in a drastic weakening in cell migration and invasion (Fig. 2D, E). Further, compared with control cells, YTHDC2 knockout cells become round in shape (Fig. 2F), implying that YTHDC2 may be correlated with epithelial-mesenchymal transition (EMT). As expected, the epidermal marker E-cadherin was markedly increased, while the stromal marker N-cadherin was decreased in YTHDC2 knockout AGS and HGC-27 cells (Fig. 2G).
Fig. 2.
YTHDC2 knockout inhibits GC cell malignant phenotype. A. Western blot verifying the knockout of YTHDC2 in AGS and HGC-27 cells. B-E. CCK-8, EdU and Transwell assays testing the effect of YTHDC2 knockout on GC cell viability, DNA synthesis rate and invasion/migration, respectively. F. The change of cell morphology after YTHDC2 knockout. G. Western blot detecting E-cad and N-cad levels after YTHDC2 knockout. ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
YTHDC2 enhances YAP translation in a m6A-dependent manner
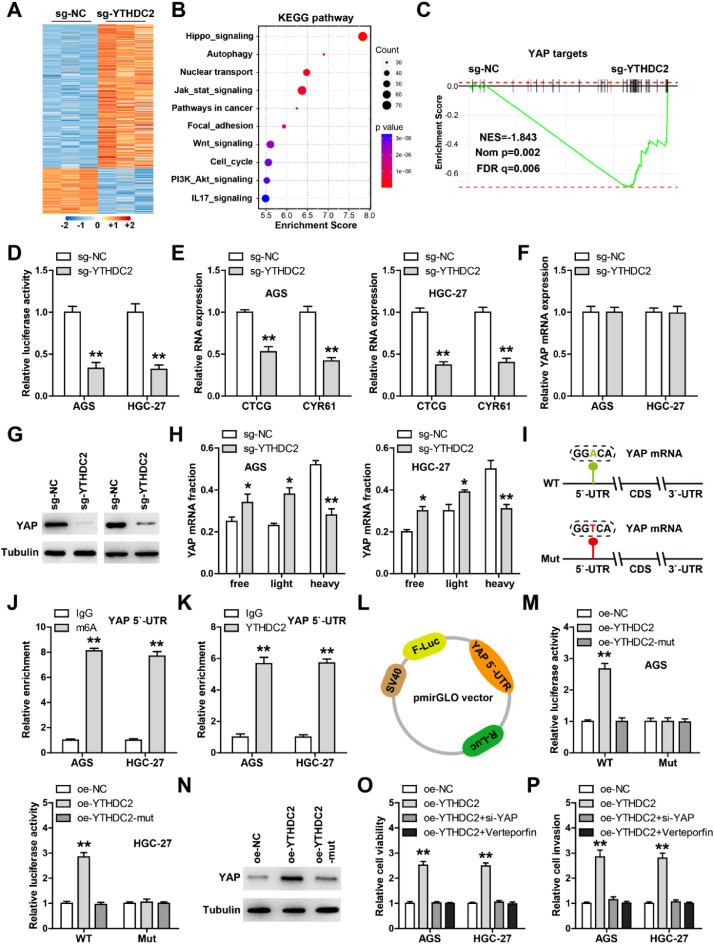
To identify the downstream targets of YTHDC2, we performed mRNA sequencing in YTHDC2 knockout cells (Fig. 3A). A number of genes were altered after YTHDC2 knockout, involving some cancer-related signal pathways, especially Hippo signaling (Fig. 3B). GSEA analysis showed that YTHDC2 was significantly correlated with YAP targets (Fig. 3C), then we conducted luciferase reporter assay, the results showed that YTHDC2 knockout dramatically reduced the YAP/TEAD activity in both AGS and HGC-27 cells (Fig. 3D). Besides, the well-known YAP targets CTCG and CYR61 were decreased after YTHDC2 knockout (Fig. 3E). Importantly, knockout of YTHDC2 markedly reduced YAP protein level, but did not affect its mRNA (Fig. 3F, G). In light of the key regulation on mRNA translation of YTHDC2, we inferred that YTHDC2 affected YAP mRNA translation. As expected, YTHDC2 knockout reduced the association of YAP mRNA with heavy polysome fractions, accompanied by increased association with free and light fractions (Fig. 3H). Further, only one m6A motif was found on YAP mRNA 5`-UTR, and YTHDC2 and m6A were abundantly enriched on this site (Fig. 3J, K). We then mutated this motif and conducted luciferase reporter assay, the results showed that overexpression of wild-type YTHDC2, but not YTH domain deletion, significantly enhanced YAP 5`-UTR translation efficiency, however this effect disappeared after mutation of m6A motif (Fig. 3L, M). Consistently, YAP protein level was significantly increased in wild-type YTHDC2-overexpressing cells, but not in YTHDC2 mutant cells (Fig. 3N). Functionally, YTHDC2 overexpression increased GC cell viability and invasion, which was effectively blocked by YAP siRNA or verteporfin, a small molecule compound inhibiting the interaction between YAP and TEAD (Fig. 3O and P). And inhibition of YTHDC2 or YAP expression could significantly repress the malignant phenotype of GC cells, especially when two genes were interfered with simultaneously (Figure S1).
Fig. 3.
YTHDC2 enhances YAP translation. A-C, mRNA sequencing in YTHDC2 knockout cells, followed by KEGG and GSEA enrichment analysis. D. Luciferase reporter assay testing the effect of YTHDC2 on YAP transcription activity. E, F. qRT-PCR analysis of CTCG, CYR61 and YAP levels in YTHDC2 knockout on GC cells. G. Western blot testing YAP protein expression after YTHDC2 knockout. H. qRT-PCR analysis of YAP mRNA level in different polysome fraction. I. The wild-type and mutant m6A sites on YAP mRNA 5`-UTR. J, K. RIP assay in GC cells using anti-m6A and -YTHDC2 antibodies, followed by qRT-PCR analysis. L, M. Luciferase reporter assay testing the effect of YTHDC2 on YAP 5`-UTR translation. N. Western blot detecting YAP protein levels after YTHDC2 overexpression. O, P. CCK-8 and Transwell assays testing cell viability and invasion in YTHDC2-overexpressing GC cells transfected with YAP siRNA or treated with Verteporfin. *P<0.05, ⁎⁎P<0.01.
YTHDC2 is a target of YAP
Of note, YTHDC2 mRNA and protein levels were significantly reduced after silencing of YAP (Fig. 4A,B), implying that YTHDC2 may be transcriptionally regulated by YAP. Three YAP/TEAD binding sites were found on YTHDC2 promoter (Fig. 4C), the luciferase reporter assay showed that knockdown of YAP significantly reduced YTHDC2 promoter activity, while only mutation of site 2 could effectively block above effect (Fig. 4D), suggesting that site 2 is critical for YAP-mediated regulation of YTHDC2. Further ChIP assay revealed that YAP indeed directly bound to site 2 on YTHDC2 promoter (Fig. 4E,F). The results of IHC staining showed that YAP protein was significantly overexpressed in GC tissues (Fig. 4G), and 86.7% of GC tissues with YAP high expression showed high expression of YTHDC2 (Fig. 4H).
Fig. 4.
YAP activates YTHDC2. A, B. Detection of YTHDC2 mRNA and protein levels after YAP silencing. C. Three YAP/TEAD binding sites on YTHDC2 promoter. D. The effect of YAP knockdown on wild-type or mutant YTHDC2 promoter activity. E, F. ChIP assay using anti-YAP antibody, followed by PCR analysis. G, H. IHC staining of YAP in TMA, and its correlation with YTHDC2. ⁎⁎P<0.01, ⁎⁎⁎⁎P<0.0001.
Knockout of YTHDC2 inhibits GC tumor growth and metastasis
Lastly, we tested the in vivo effect of YTHDC2, as shown in Fig. 5A-C, tumor volume and weight in YTHDC2 knockout group were significantly less than those in control group. Moreover, IHC staining showed that YTHDC2 protein level were almost undetectable in YTHDC2 knockout tissues, and Ki-67 (a proliferation marker) and YAP were substantially decreased by YTHDC2 knockout (Fig. 5D). Besides, the expression levels of CTCG and CYR61 were reduced in YTHDC2 knockout tissues, however YAP mRNA remained unchanged (Fig. 5E). The lung metastasis model was established, and the results showed that YTHDC2 knockout led to a significant reduction in the number of metastatic nodule (Fig. 5F).
Fig. 5.
YTHDC2 knockout inhibits GC cell growth and metastasis in vivo. A-C. Tumor volume and weight in control and YTHDC2 knockout groups. D. IHC staining of Ki-67, YTHDC2 and YAP in control and YTHDC2 knockout tissues. E. qRT-PCR analysis of YAP, CTCG and CYR61 levels in control and YTHDC2 knockout tissues. F. The number of lung metastasis nodules in control and YTHDC2 knockout groups. ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
Discussion
In this study, we characterized the role of m6A reader protein YTHDC2 in GC, we found that YTHDC2 is a previously unappreciated oncogene in GC. YTHDC2 promoted GC cell growth, migration and invasion both in vitro and in vivo. The mechanism investigation revealed that YAP was the target of YTHDC2, in detail, YTHDC2 recognized the m6A modification on YAP mRNA 5`-UTR, and potentiated YAP translation in a m6A-dependent manner, resulting in increased yield of oncogenic YAP protein. In turn, YAP was able to directly bind to YTHDC2 promoter and enhanced YTHDC2 transcription (Fig. 6). Therefore, the positive feedback loop between YTHDC2 and YAP was formed, amplifying their cancer-promoting effects. In addition, the regulatory axis of YTHDC2/YAP was also found in human GC tissues and xenograft tumor model. Moreover, YTHDC2 was identified as a promising independent prognostic biomarker for GC patients. Altogether, our data provide evidence for the link between m6A and GC progression, meanwhile uncover a novel target for GC prognosis and treatment.
Fig. 6.
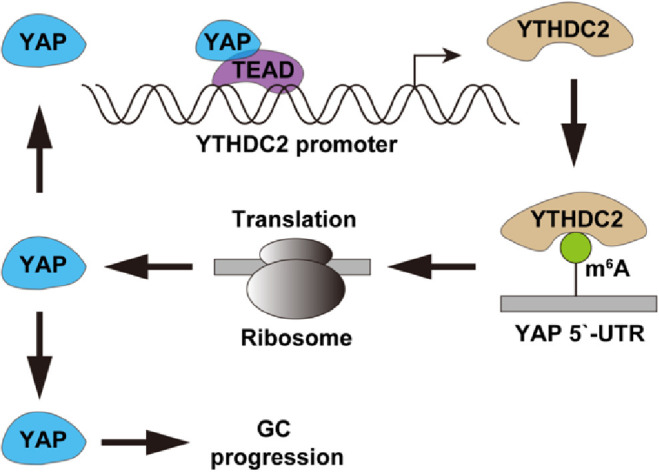
The proposed model of YTHDC2 promoting GC progression via regulation of oncogenic YAP translation.
Hippo signaling pathway, composed of conserved kinases, regulates downstream gene expression by regulating the phosphorylation and nuclear translocation of YAP, and participates in the proliferation, differentiation and migration of various cells [19,20]. After entering the nucleus, YAP binds to TEAD as a transcriptional co-activator to promote cancer cell growth, EMT and distant metastasis [21]. Available evidence shows that YAP protein is abnormally expressed in various human cancers and is emerging as a new target for cancer intervention [22]. However, it is yet undetermined why YAP protein is frequently activated in human malignancies in which the Hippo pathway is still active. Herein, we found that YAP mRNA translation process was affected by YTHDC2, specifically, YTHDC2 increased YAP translation via binding to m6A site on YAP mRNA 5`-UTR, while deletion of YTH domain could not produce the above effect, suggesting that YTHDC2 regulates and activates YAP in a m6A-dependent manner. Hence, our findings provide new insights for Hippo-independent activation of YAP, and further studies are needed to clarify the specific mechanisms by which YTHDC2 promotes YAP translation at 5`-UTR, which may be related to the recruitment of some translation regulatory factors. In addition, spite some studies have shown that YTHDC2 plays a role by affecting mRNA stability, YAP mRNA was not changed after YTHDC2 knockout in GC cells, hinting that YTHDC2 functions in a context-dependent way.
YAP, as a key transcriptional co-activator, regulates gene expression at the transcriptional level via forming the YAP/TEAD dimer [23,24]. A number of oncogenes have been identified as targets of YAP/TEAD, such as CTCG, CYR61 and AXL [25]. In this study, we found that YTHDC2 mRNA expression was significantly decreased after YAP silencing, implying that YTHDC2 may be controlled by YAP. Further, three TEAD targeting sites were found on YTHDC2 promoter, and mutation of site 2 (−843∼−831) blocked the transcriptional activation of YAP on YTHDC2, revealing that YTHDC2 is a novel target of YAP. The newly identified YTHDC2/YAP feedback loop amplifies the mutual carcinogenic effect, and whether this network is also present in other malignancies is worth further investigation.
Taken together, our data for the first time demonstrate that YTHDC2, a m6A reader protein, is a novel oncogene in human GC. YTHDC2 enhances YAP translation and activate oncogenic YAP signaling in a m6A-dependent manner, suggesting that m6A is closely linked to human cancer progression.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT author statement
Wei Yuan, Shiqiang Chen, Bo Li: Conceptualization, Methodology, Software, Writing - original draft
Xiaoyu Han, Bo Meng: Visualization, Investigation.
Yongping Zou: Software, Validation
Shunwu Chang: Writing - review & editing, Supervision
Declaration of Competing Interest
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101308.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 3.Petryszyn P., Chapelle N., Matysiak-Budnik T. Gastric cancer: where are we heading? Dig. Dis. 2020;38:280–285. doi: 10.1159/000506509. [DOI] [PubMed] [Google Scholar]
- 4.Zhang W., Qian Y., Jia G. The detection and functions of RNA modification m(6)A based on m(6)A writers and erasers. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao G., Li H.B., Yin Z., Flavell R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6 doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Shi X., Huang T., Zhao X., Chen W., Gu N., Zhang R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48:6251–6264. doi: 10.1093/nar/gkaa347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W., Chen T.Q., Fang K., Zeng Z.C., Ye H., Chen Y.Q. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J. Hematol. Oncol. 2021;14:117. doi: 10.1186/s13045-021-01129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai X.Y., Shi L., Li Z., Yang H.Y., Wei J.F., Ding Q. Main N6-methyladenosine readers: YTH family proteins in cancers. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.635329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao S., Sun H., Xu C. YTH Domain: a family of N(6)-methyladenosine (m(6)A) readers. Genom. Proteom. Bioinform. 2018;16:99–107. doi: 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi R., Ying S., Li Y., Zhu L., Wang X., Jin H. Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics. Cell Death Dis. 2021;12:346. doi: 10.1038/s41419-021-03625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Li G., Li Q., Zhang Q., Zhuo L., Chen X., Zhai B., Sui X., Chen K., Xie T. The roles and mechanisms of YTH domain-containing proteins in cancer development and progression. Am. J. Cancer Res. 2020;10:1068–1084. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou B., Liu C., Xu L., Yuan Y., Zhao J., Zhao W., Chen Y., Qiu J., Meng M., Zheng Y., et al. N(6) -methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology. 2021;73:91–103. doi: 10.1002/hep.31220. [DOI] [PubMed] [Google Scholar]
- 15.Ma L., Chen T., Zhang X., Miao Y., Tian X., Yu K., Xu X., Niu Y., Guo S., Zhang C., et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J., et al. YTHDC2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim G.W., Siddiqui A. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2022024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunshine J.C., Nguyen P.L., Kaunitz G.J., Cottrell T.R., Berry S., Esandrio J., Xu H., Ogurtsova A., Bleich K.B., Cornish T.C., et al. PD-L1 expression in melanoma: a quantitative immunohistochemical antibody comparison. Clin. Cancer Res. 2017;23:4938–4944. doi: 10.1158/1078-0432.CCR-16-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y., Pan D. The hippo signaling pathway in development and disease. Dev. Cell. 2019;50:264–282. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z., Guan K.L. Hippo signaling in embryogenesis and development. Trends Biochem. Sci. 2021;46:51–63. doi: 10.1016/j.tibs.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pocaterra A., Romani P., Dupont S. YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 2020:133. doi: 10.1242/jcs.230425. [DOI] [PubMed] [Google Scholar]
- 22.Reggiani F., Gobbi G., Ciarrocchi A., Sancisi V. YAP and TAZ are not identical twins. Trends Biochem. Sci. 2021;46:154–168. doi: 10.1016/j.tibs.2020.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Crawford J.J., Bronner S.M., Zbieg J.R. Hippo pathway inhibition by blocking the YAP/TAZ-TEAD interface: a patent review. Expert Opin. Ther. Pat. 2018;28:867–873. doi: 10.1080/13543776.2018.1549226. [DOI] [PubMed] [Google Scholar]
- 24.Currey L., Thor S., Piper M. TEAD family transcription factors in development and disease. Development. 2021:148. doi: 10.1242/dev.196675. [DOI] [PubMed] [Google Scholar]
- 25.Totaro A., Panciera T., Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018;20:888–899. doi: 10.1038/s41556-018-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.