Highlights
-
•
Most patient with pancreatic cancer are treated by chemotherapy.
-
•
Treatments selection are not personalized on the tumor characteristics.
-
•
Signatures predicting chemotherapy efficiency are essential for personalizing treatments.
-
•
An RNA signature of gemcitabine-sensitivity is developed leveraged on the dissimilarities between 2D and 3D in vitro models.
-
•
Combining different in vitro models can help in defining clinically efficient transcriptomic signatures.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) patients are frequently treated by chemotherapy. Even if personalized therapy based on molecular analysis can be performed for some tumors, PDAC regimens selection is still mainly based on patients' performance status and expected efficacy. Therefore, the establishment of molecular predictors of chemotherapeutic efficacy could potentially improve prognosis by tailoring treatments. We have recently developed an RNA-based signature that predicts the efficacy of adjuvant gemcitabine using 38 PDAC primary cell cultures. While demonstrated its efficiency, a significant association with the classical/basal-like PDAC spectrum was observed. We hypothesized that this flaw was due to the basal-like biased phenotype of cellular models used in our strategy. To overcome this limitation, we generated a prospective cohort of 27 consecutive biopsied derived pancreatic organoids (BDPO) and include them in the signature identification strategy. As BDPO's do not have the same biased phenotype as primary cell cultures we expect they can compensate one with each other and cover a broader range of molecular phenotypes. We then obtained an improved signature predicting gemcitabine sensibility that was validated in a cohort of 300 resected PDAC patients that have or have not received adjuvant gemcitabine. We demonstrated a significant association between the improved signature and the overall and disease-free survival in patients predicted as sensitive and treated with adjuvant gemcitabine. We propose then that including BDPO along primary cell cultures represent a powerful strategy that helps to overcome primary cell cultures limitations producing unbiased RNA-based signatures predictive of adjuvant treatments in PDAC.
Introduction
Pancreatic Ductal Adenocarcinoma (PDAC) is a highly fatal malignancy with a 5-year survival below 5% [1]. Approximately 85% of patients present either locally advanced or metastatic stages at the diagnosis time, leaving only a few percent of patients that are eligible for surgery, the only treatment that offers curative potential for PDAC [2]. Neo-adjuvant and adjuvant chemotherapy are administered in the pre- or post-operative setting, respectively in the hope of improving patients’ chances of survival. Therefore, most PDAC patients will receive chemotherapeutic treatments [3,4]. However, despite considerable efforts during the last years to define the best chemotherapy regimen, minor benefits in most cases are observed. In fact, most used regimens have an estimated response rate of 10%−23% for gemcitabine in advanced patients [5], whereas FOLFIRINOX has a positive response of around 20–25% [6]. This low response rate is mostly linked to the high PDAC heterogeneity which is reflected in a very diverse pattern of clinical outcomes and responses to therapies associated with different tumor phenotypes. Predicting the cancerous cells response to the different therapeutic strategies would significantly increase chemotherapy efficiency avoiding treatment resistance and relapse.
Transcriptomic signatures are promising tools based on the analysis of transcript expression levels for selected groups of genes. They are good indicators of the biological behavior of a tumor, hence capable of stratifying patients into different subtypes as the quantification of RNA is the most comprehensive approach to determine the phenotype of a tissue. They have also the advantage of being applicable on clinical grade formalin fixed paraffin embedded samples and has overall good reproducibility and quantification properties. It has been shown that RNA has by far the most predictive potential to predict drug-treatment efficacy [7].
For several decades, scientists have been developing these signatures in order to determine prognostic and theranostic values. Prognostic signatures have already been applied in clinical settings, for example in breast and colon [8,9] cancer, helping clinicians to choose the best therapeutic strategy based on the patient's genomic characteristics. On the other hand, theranostic signatures in PDAC are very challenging mostly due to the difficulty of obtaining reliable pre-clinical models to develop them [10].
In a previous study, we proposed GemPred, an RNA-based signature to predict gemcitabine clinical efficacy [11]. GemPred was developed from 2D primary cell cultures and showed several limitations. GemPred is correlated to the basal-like/classical molecular differentiation axis of PDAC. While it has been shown that basal-like tumors may be more chemoresistant [12,13], it would be possible that the strong signal induced by these major phenotypes could have biased the development of GemPred from 2D primary cell cultures which were shown to be mainly on the basal-like side of the phenotypic transcriptomic spectrum [10]. Initially developed GemPred is limited by the lack of prediction (i.e., statistical interaction) of the disease-free survival rate (DFS). Finally, the initial GemPred is lacking association with known biomarkers (reviewed in [14]) in particular genes involved in gemcitabine metabolism such as hENT1 [15] (also known as SLC29A1, the main transmembrane transporter), CDA [16] (the main catabolic enzyme), and DCK [17] (the main activating enzyme).
A more recent preclinical model that is emerging as a high-fidelity tool in precision medicine are the biopsy-derived pancreatic organoid (BDPOs). They present the advantage of being directly derived from a patient's tumours producing mini-tumours that can be amplified and preserved in 3D cultures in vitro. They also have the advantage of preserving many characteristics of the original tumor as differentiation status and overall patient responses to chemotherapies as was shown in pancreatic and colorectal cancer [18], [19], [20].
In this work, we used 27 patient-derived organoid (BDPO) lineages in 77 transcriptomic-drug correlation assays in addition to the previously assayed primary cell cultures. These orthogonal assays were used to derive an improved version of the GemPred RNA signature predicting sensitivity to gemcitabine.
Materials and methods
Biopsy-derived pancreatic organoids generation
The 77 BDPOs models used in this study were obtained from 27 consecutive endoscopic ultrasound-guided fine-needle aspirations (EUS-FNA) from patients with PDAC. Clinical characteristics of patients cohort is described in Supplementary Table 1). Cultures were established as previously described [21].Briefly, PDAC biopsies were slightly digested with the Tumor Dissociation Kit (Miltenyi Biotec) at 37 °C for 5 min. The pancreatic tissue slurry was transferred into a tissue strainer 100 μm and was placed into 12-well plate coated with 150 μL GFR matrigel (Corning, Boulogne-Billancourt, France). The samples cultured with Pancreatic Organoid Feeding Media (POFM) consisted of Advanced DMEM/F12 supplemented with 10 mM HEPES (Thermo Fisher Scientifics, Courtaboeuf, France); 1 × Glutamax (Thermo-Fisher Scientifics); penicillin/streptomycin (Thermo-Fisher Scientifics); 100 ng/mL Animal-Free Recombinant Human FGF10 (Peprotech, Peprotech, Neuilly-Sur-Seine, France); 50 ng/mL Animal-Free Recombinant Human EGF (Peprotech); 100 ng/mL Recombinant Human Noggin (Biotechne, Bio-Techne, Rennes, France); Wnt3a-conditioned medium (30% v/v); RSPO1-conditioned medium (10% v/v); 10 nM human Gastrin 1 (Sigma-Aldrich Lyon, France) 10 mM Nicotinamide (Sigma Aldrich); 1.25 mM N acetylcysteine (Sigma Aldrich); 1 × B27 (Invitrogen, Villebon sur Yvette, France); 500 nM A83–01 (Tocris, Noyal Châtillon sur Seiche, France); 10.5 μM Y27632 (Tocris). The plates were incubated at 37 °C in a 5% CO2 incubator, and the media were changed every 3 or 4 days. For routine passages BDPOs were disaggregated with accutase (Thermo Fisher Scientific) and re-plated as needed.
Gemcitabine chemograms on BDPO
BDPOs were disaggregated with accutase (Thermo Fisher Scientific), and 1000 cells/well were plated in two 96-well round bottom ultra-low attachment plate (Corning, Costar Ref. CLS7007) with the medium described above. Twenty-four hours later, one plate was used directly for RNA preparation representing the organoids before the treatment (Time 0 transcriptome) and on the other the medium was supplemented with increasing concentrations of gemcitabine and 72 h later cell viability was measured with CellTiter-Glo 3D (Promega) reagent quantified using the plate reader Tristar LB941 (Berthold Technologies). Values were normalized and expressed as the percentage of the control (vehicle), which represent 100% of normalized fluorescence. Twelve increasing concentrations of gemcitabine were used ranging from 0 to 1 mmol/L. Each experiment was repeated in average three times and at least 2 times (27 different models and 77 experiments). Dose response values were processed, normalized and summarized into pharmacological sensitivity scores using Grmetrics [22]. Dose response for 2D primary cell cultures were obtained previously [11]. The dose at 50% of cell viability (IC50) was used as a measure of gemcitabine potency, the efficacy (Eff) was measured by the asymptotic response (Einf in the GRmetrics package) and finally the Area Under the dose-response Curve (AUC) was used as a combined average metric. For 2D cell cultures, the growth-corrected equivalents of these metrics were also used, respectively GrcIC50, GrcEff and GrcAUC.
RNA profiling
Total RNA was extracted from 96.000 BDPOs cells from the time 0 transcriptome plate using RNeasy Mini Kit. RNA libraries were prepared with the kit Illumina TruSeq RNA v2 and run on the Illumina High Seq-2000 for 101 bp paired end reads. Reads were mapped using STAR, gene expression quantified using featureCount with gene-level sumarization and normalized using Upper-Quartile normalization. Ensembl GRCh38 was used as the reference genome. RNA profiles for 2D primary cell cultures were obtained in a previous study [11]. Raw RNA-sequencing counts are available as a figshare dataset (doi: 10.6084/m9.figshare.16955539).
RNA signature development
The development of the predictive RNA was based on a multiple objective exhaustive search of an optimal linear combination of latent variables. RNA and dose response profiles for 38 primary cell cultures from a previous study [11] were used to extract unsupervised independent components using joint approximation diagonalization of eigen-matrices (JADE), with the number of components k ranging from 2 to 12. For every independent components dimension with fixed k, all combinations of independent components are fitted in a linear regression model as explanatory variables using the dose-response AUC as response variable. In parallel, the same components were projected on the 3D organoids transcriptomes and the same linear models were fitted using the organoid AUC as response variable and the organoid-projected components as explanatory variables. The predicted response was then correlated to all pharmacological dose-response metrics (AUC, IC50, Eff and their growth-corrected counterparts for 2D primary cell cultures). All combinations of independent components were then ranked by their coefficient of determination (R) of 2D-trained and 3D-trained models to 2D and 3D derived pharmacological metrics, including predicting 2D metrics with 3D-trained models, as well as the pearson correlation between the predicted response of 2D- and 3D-trained models in a dataset of resected human primary tumors. The final model, consisting of an independent component space with fixed k and a set of selected components, was identified by selecting the model with the highest rank in all selected metrics. Overall, this approach ensures that the selected model is: consistent in both types of in vitro model, predictive of every pharmacological aspect of gemcitabine (i.e. efficacy and potency), and transferable to primary human tissue (without any training on clinical data). The signatures are available as web application to predict the score and sensitivity to gemcitabine from a transcriptomic profile (app.gebican.fr/pdac-gempred).
Results
Evaluation of gemcitabine-related genes in vitro
Recently we established a transcriptomic signature predictive of gemcitabine sensitivity in PDAC (GemPred) using primary cell cultures and xenografts with concomitant genome-wide RNA profiles and gemcitabine sensitivity analyses. As primary cell cultures have basal-like biased phenotype that can have an impact on the GemPred signature and to further evaluate the gemcitabine-related genes in vitro, data from primary cell culture were re-analyzed and enriched with 27 BDPO 3D models as follows: for 2D primary cell cultures, in vitro sensitivity to gemcitabine was measured using a dose-response assay (chemogram) and summarized for each primary cell culture model in three metrics: i gemcitabine's potency using the dose with 50% reduction in cell viability (IC50), ii gemcitabine's efficacy (Eff) using the asymptotic effect (sometimes referred to as Einf), and iii the area under the dose-response curve (AUC). The proliferation rate could be accurately measured and was used to compute growth-corrected measures (GrcEff, GrcIC50 and GrcAUC). The transcriptome for 2D primary cell cultures were obtained from the cell culture considered as a reference for each patient. For 3D BDPO models and 2D primary cell cultures an average of 3 biological replicates (chemogram) with dose response were calculated. Differently to primary cells cultures, transcriptomes for BDPO were obtained simultaneously with chemograms and for each one of the 3 biological replicates. In this way we obtained 77 different transcriptomes representing very accurately the biological state of the BDPO at the moment when the chemogram is done. The same 3 three metrics used for the cell cultures were calculated for the BDPO's. The relationship between gemcitabine sensitivity and the expression of several genes involved in gemcitabine metabolism was evaluated in these two 2D and 3D in vitro models. Only two genes, CDA and hENT1 (SLC29A1), showed a significant correlation with one of the gemcitabine sensitive metrics both in 2D and 3D models, (Fig. 1).
Fig. 1.
Gene expression correlation with gemcitabine sensitivity. Spearman correlation between gene expression of gemcitabine biomarker response genes and gemcitabine response metrics in 2D primary cell cultures (a) and 3D BDPO (b). Proliferation (Prolif.) is accurately measured only in 2D by the 24 h replication rate. Dose with 50% reduction in cell viability is indicated as IC50, gemcitabine's efficacy as Eff and the area under the dose-response curve as AUC. Grc indicates growth-corrected measures. *: < 0.05; **: < 0.01; ***: < 0.001.
Improvement of gempred signature
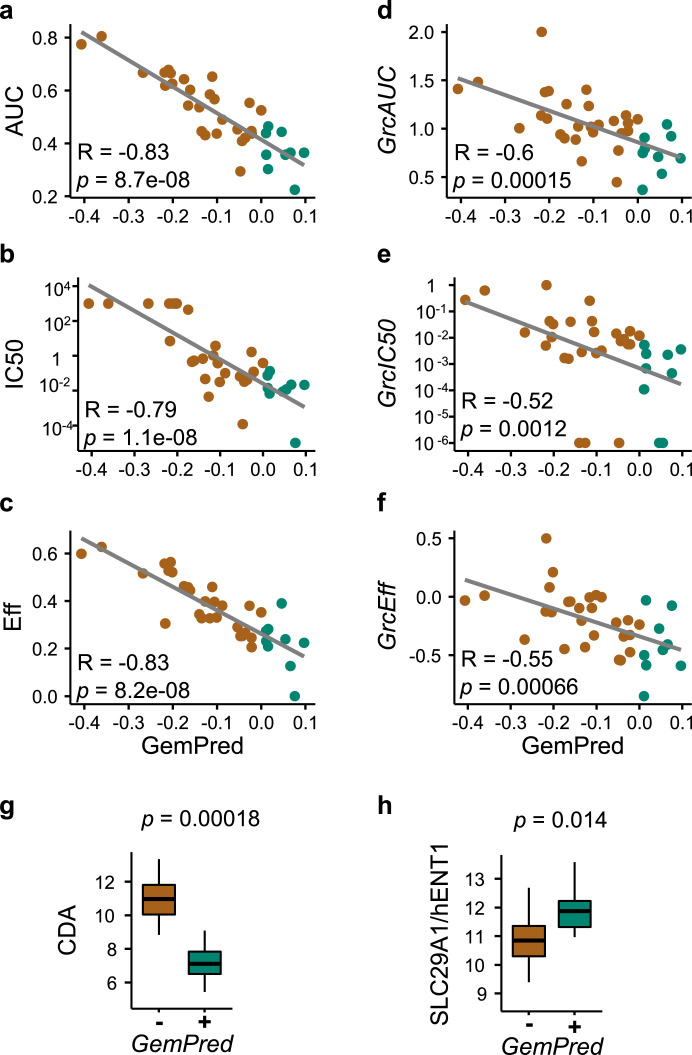
We next sought to use all the transcriptomic-drug correlation assays to improve the GemPred signature which was initially only trained on 2D primary cell cultures. An improved signature was derived from both the 2D and 3D assays using a similar approach as previously described [11]. Briefly, an Independent Component Analysis was applied to extract 2D-derived gemcitabine related components, a multiple-objective exhaustive search was then applied to evaluate linear combinations of components as predictors of gemcitabine sensitivity. The improved GemPred signature scores each sample based on their gene expression profiles and the third quartile was used as a threshold for sensitivity. The GemPred scoring was highly correlated to all 2D metrics of gemcitabine-sensitivity, including potency (IC50), efficacy and AUC, as well as the growth corrected metrics (Fig. 2). GemPred high primary cell cultures had higher hENT1 and lower CDA expression, as expected from higher Gemcitabine sensitivity tumors (Fig. 2 and Supplementary Table 2 showing the top most correlated and anticorrelated genes).
Fig. 2.
Improved GemPred signature in 2D primary cell cultures. Association between GemPred score and gemcitabine response metrics including AUC (a), IC50 as a measure of potency (b) and efficacy (c) as well as growth-corrected AUC (d), growth-corrected IC50 (e) and growth-corrected efficacy (f). Spearman's correlation coefficient R and associates p-values are shown (a-f). The association of the GemPred score (fourth quartile) and the gene expression of gemcitabine biomarker genes CDA (g) and hENT1 (h). Green color and GemPred+ indicates gemcitabine sensitivity; brown color and GemPred- indicates gemcitabine resistance.
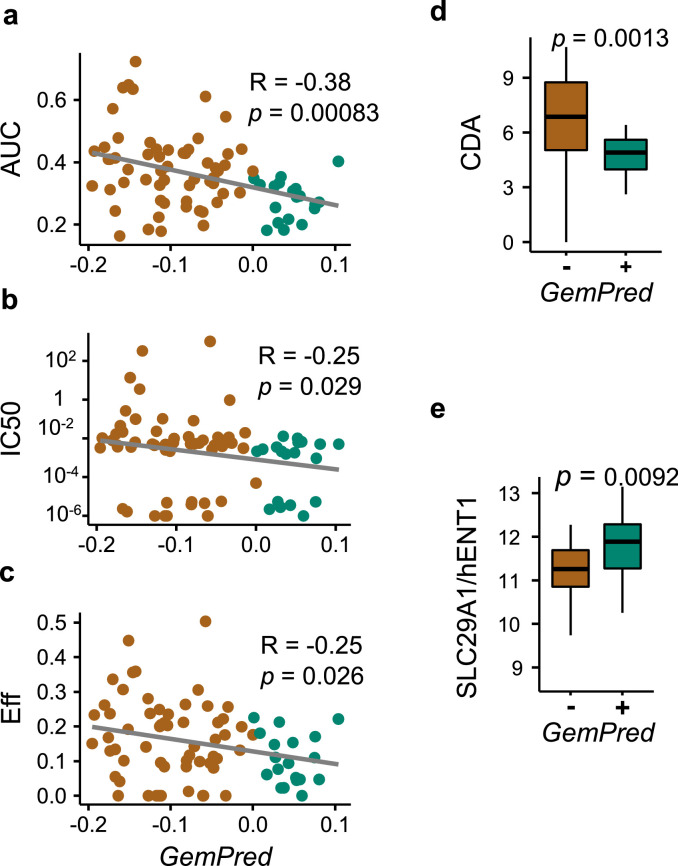
GemPred signature in 3D bdpo cultures
The improved Gempred signature was then applied to the 77 BDPO transcriptomic-drug assays. The GemPred score of BDPO significantly correlated with gemcitabine dose-response metrics, in particular AUC and to a lesser extent to gemcitabine's potency (IC50) and efficacy (Fig. 3b). Growth-corrected measures were not computed for BDPO as growth-rate could not be reliably measured. GemPred+ BDPO over-expressed hENT1 and under-expressed CDA (Fig. 3b).
Fig. 3.
Improved GemPred signature in 3D BDPO lineages. Association between GemPred score and gemcitabine response metrics including AUC (a), IC50 as a measure of potency (b) and efficacy (c). Also shown is the association of the GemPred score (above third quartile) and the gene expression of gemcitabine biomarker genes CDA (d) and hENT1 (e). Green color and GemPred+ indicates gemcitabine sensitivity; brown color and GemPred- indicates gemcitabine resistance.
GemPred signature predictive of adjuvant gemcitabine efficacy
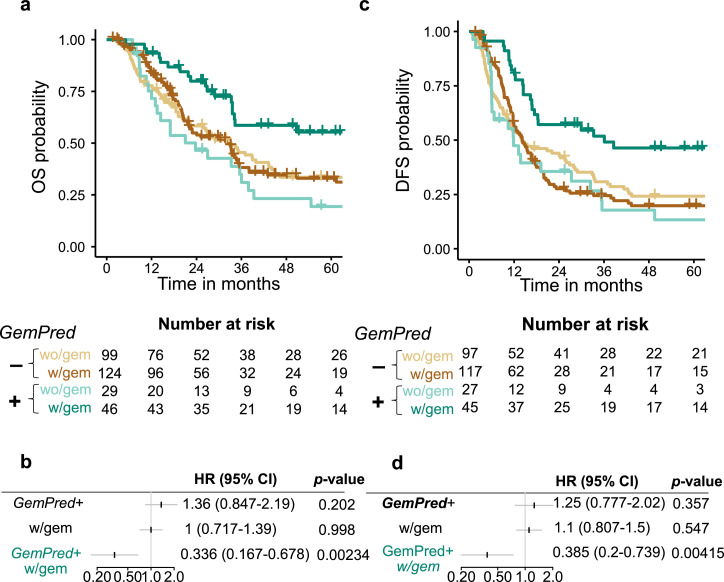
The improved GemPred signature was applied to previously generated transcriptomic profiles [11] of 385 resected PDAC from patients that had (n = 130, 43.3%) or had not (n = 170, 56.7%) received adjuvant gemcitabine. GemPred patients classified as GemPred- or GemPred+ corresponding, respectively to patients predicted as resistant or sensitive to gemcitabine. GemPred+ patients had a greater Overall Survival (OS) only if they had received adjuvant gemcitabine (Fig. 4a) with a significant interaction between adjuvant gemcitabine and Gempred+ (HR = 0.336, 95% Confidence interval (CI) [0.167, 0.678], p = 0.00234). GemPred+ patients having received adjuvant gemcitabine had a 5-year survival rate of 55.4% (95% CI [41.6%, 73.6%]) against 19.4% (95% CI [8.91%, 42.2%]) for those that did not receive gemcitabine while GemPred- patients had a 33% (95% CI [24.6%, 44.1%]) and 33.5% (95% CI [25%, 45%]) whether having, respectively received adjuvant gemcitabine or not. GemPred+ patients also had a significantly longer disease-free survival (DFS) specifically when receiving adjuvant gemcitabine (Fig. 4c) as shown by a significant statistical interaction between GemPred+ and adjuvant gemcitabine (Fig. 4d, HR = 0.385, 95% CI [0.2, 0.739]). The 5-year DFS rate was 46.4% (95% CI [33.4%, 64.3%]) for GemPred+ patients that had received adjuvant gemcitabine, with a median survival of 36.1 months (95% CI [17.4, unattained]). GemPred+ remained significantly associated with OS and DFS among patients having received adjuvant gemcitabine in a multivariate analysis including tumor stage T, N status, tumor differentiation and tumor size (Fig. 5). GemPred had no significant association with OS or DFS in patients that had not received adjuvant gemcitabine.
Fig. 4.
Association of the improved GemPred signature with survival. a. Overall Survival (OS) Kaplan-meier curves stratified by GemPred status and by adjuvant treatment (with vs without gemcitabine). b. OS cox regression model including GemPred, adjuvant gemcitabine and their interaction. c. Disease-free survival (DFS) Kaplan-meier curves stratified by GemPred status and by adjuvant treatment (with vs without gemcitabine). d. DFS cox regression model including GemPred, adjuvant gemcitabine and their interaction. w/gem: With adjuvant gemcitabine. wo/gem: without adjuvant gemcitabine.
Fig. 5.
Multivariate survival models. Multivariate Cox regression model of OS for patients that have (a) or have not received (b) adjuvant gemcitabine. Multivariate Cox regression model of DFS for patients that have (c) or have not received (d) adjuvant gemcitabine. Diff.: Differentiation.
Improvement of gempred signature
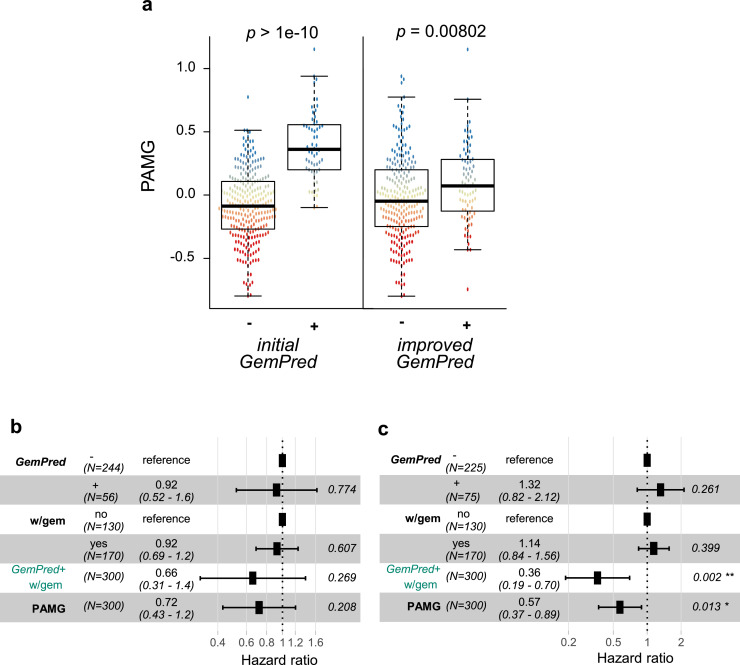
The main issue with the initial GemPred signature is its statistical association with the classical/basal-like phenotypic spectrum. Fig. 6a shows the association between the initial GemPred signature and the here proposed improved GemPred signature, showing a weaker association between the improved GemPred signature and the Pancreatic Adenocarcinoma Molecular Gradient (PAMG), an RNA-based signature quantifying the level of differentiation on the classical/basal-like spectrum (higher being more classical) [12]. This reflects on the predictive value of the GemPred signature. Fig. 6b and C, respectively show the statistical interaction between the GemPred signature (6b: initial GemPred, 6c: improved GemPred) in multivariate models including the PAMG. These results show that only the improved GemPred was significantly predictive of adjuvant gemcitabine, independently of the PAMG.
Fig. 6.
Initial versus improved GemPred signature. a. Association between GemPred classification (left: initial GemPred; right: improved GemPred) and Pancreatic Adenocarcinoma Molecular Gradient (PAMG). Testing statistical interaction between adjuvant gemcitabine and initial GemPred (b) or improved GemPred (c) in a multivariate model including PAMG. w/gem: With adjuvant gemcitabine.
Discussion
In this study, we used 2D primary cell cultures and 3D organoids as in vitro models of PDAC. We then combined their genome-wide RNA profiles with the pharmacological evaluation of their sensitivity to gemcitabine to derive a generalizable RNA signature predictive of gemcitabine efficacy in patients receiving adjuvant gemcitabine.
2D and 3D in vitro models of PDAC have broadly different characteristics, the main one being a very different development success rate. While primary cell cultures have an approximately 30% success rate, 3D organoids from human PDAC can be virtually derived from all patients in expert centers [23].
Organoid cultures have acquired a significant interest in translational studies and recent efforts provide evidences that the medium used to provide chemogram and even more RNAseq data is an important parameter to consider. In particular, the influence of WNT seems to have un important impact [24]. We did not test different medium composition as the medium used in this work was satisfactory to improve the GemPred signature. In the future this controversial point needs to be carefully considered. Organoids used in this work are all low passages (between 4 and 5). Clonal evolution of more replicative cells can modify the phenotypic characteristics of the cultured organoids. But once clonally established within the first set of passages, we found as others [25] that a dominant clone is established in the culture of organoids. Even though pancreatic cancer organoids showed in our experience a certain level of variability between one experiment and its biological replicate. As we need a maximum of precision in the transcriptomic characterization associated to the drug response we did an RNA sequencing at the same time of each chemogram replicate.
Phenotypically, 2D PDAC cells tend to present a more basal-like molecular phenotype than 3D organoids which are mostly classical. In agreement with this we observed for 3D organoids globally a more chemosensitive phenotype than in 2D cell cultures. This could be associated to a more basal-like phenotype of cell cultures. These differences make the two types of models difficult to compare for a general description of the disease yet may bring a unique description of the full phenotypic spectrum in integrative study. In particular, the identification of a chemotherapeutic response phenotype, of which there is no reliable evidence of its association with the classical/basal-like spectrum, is greatly improved by the integration of such different models. The approach used here focuses on uncovering an RNA-based gemcitabine response phenotype, independently of the background molecular phenotype or any other specifics of each type of in vitro cell culture model. Overall, this work is based on the hypothesis that the integration of orthogonal cell culture models is crucial to the development of predictive signatures.
The initial GemPred signature demonstrated a significant predictive value of the sensitivity of adjuvant gemcitabine. This initial signature, however, was associated with the general molecular phenotype, along the classical/basal-like spectrum. This association introduced a potential bias whereby the molecular phenotype may hinder the predictive value of the signature. This bias was likely introduced by the use of 2D primary cell cultures, which were shown to mainly present a basal-like phenotype. This study aimed at correcting this bias by integrating 2D models with 3D organoids having a more classical phenotype thus covering a broader range of molecular phenotype. In addition, the multi-objective search aimed at extracting a signature that was significantly associated with several pharmacological characteristics, including gemcitabine potency and efficacy. Overall, the approach proposed in this study leveraged on the dissimilarities between 2D and 3D models to develop a pharmacologically comprehensive RNA signature predictive of gemcitabine efficacy.
The usefulness of transcriptomic predictive signatures aiming to determine the efficacy of drugs in patient care is undeniable. However, their development is still in its early stages. Even if there is a considerable promise of their usefulness and effectiveness it is still necessary to validate their full potential as a guiding line of prospective interventional clinical trials. Another important point is that in a previous work we shown that sequencing RNA from fine needle aspirates, including for metastatic patients, is feasible and that transcriptomic signatures are relevant in these biological samples [12]. Even if the GemPred signature was developed specifically on epithelial cancerous cells, it is also informative on more complex samples such as surgical biopsies that have abundant stroma suggesting its relevance on clinical diagnostic biopsies.
PDAC is probably the best candidate for the application of such signatures in patient care mostly due to its aggressive nature and late diagnosis. In fact, RNA-sequencing requires the same logistics, equipment and time as DNA NGS assay, which has been previously used to stratify patient treatment in metastatic pancreatic cancer in clinical trials [26], and are also entering routine tests in some expert centers. Retrospective analyses have shown that the delay from diagnosis (imaging) to treatment in advanced (i.e. metastatic) diseases was around 29 days (median) [27], which makes RNA-sequencing compatible with clinical timeline in this disease. However, this requires an effective organization similar to other NGS tools.
While we are unable to prove that GemPred signature will perform well in multi-agent combination strategies based on gemcitabine with the cohorts used in this study, it may be expected that GemPred+ patients will not only benefit from gemcitabine alone but potentially from gemcitabine-based regimens in general. This is expected from the fact that most multi-agent regimens lack synergy and rely on populational effect [28]. Finally, GemPred+ patient receiving adjuvant gemcitabine have similar three-year survival rate (76.1%, CI: 95% 62.8–92.2) than patients with adjuvant mFOLFIRINOX in the PRODIGE24 trial (63.4%, [6]), suggesting that a biologically-informed first-line single-agent treatment selection may have sufficient efficacy with lower adverse effects than poly-chemotherapy regimens with high toxicities. We are convinced that applying the right treatment from the beginning is imperative, highlighting the clinical importance of chemosensitivity predictive signatures. In this context transcriptomic signatures should be performed routinely during diagnostic biopsy.
Disclosure
RN, JI and NJD have a pending patent entitled ‘Evaluation of the efficiency of an anticancer compound for a PDAC patient’ filed 23 January 2020 (European patent application number EP20305052.1). All other authors have declared no conflicts of interest.
Compliance with ethical standards
In vivo models were derived from patients included under the PaCaOmics clinical trial (ClinicalTrials.gov: NCT01692873) after approval by the Paoli-Calmettes hospital ethics committee and following patient informed consent.
Patient cohorts of resected PDAC patients was approved by the institutional review board (2010/01NICB IRB:00,003,835).
CRediT authorship contribution statement
R. Nicolle: Visualization, Formal analysis. O. Gayet: Formal analysis. M. Bigonnet: Formal analysis. J. Roques: Formal analysis. B. Chanez: Data curation. F. Puleo: Data curation. J. Augustin: Data curation. J.F. Emile: Data curation. M. Svrcek: Data curation. T. Arsenijevic: Data curation. P. Hammel: Data curation. V. Rebours: Data curation. M. Giovannini: Data curation. P. Grandval: Data curation. L. Dahan: Data curation. V. Moutardier: Data curation. E. Mitry: Data curation. J.L. Van Laethem: Data curation. J.B. Bachet: Data curation. J. Cros: . J. Iovanna: Visualization. N.J. Dusetti: Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was supported by INCa; Canceropole PACA; DGOS (labellisation SIRIC); Amidex Foundation; Fondation de France; and the Institut national de la santé et de la recherche médicale (INSERM) (no grant number). This work is part of the national program Cartes d'Identité des Tumeurs (CIT) funded and developed by the Ligue Nationale Contre le Cancer.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101315.
Appendix. Supplementary materials
References
- 1.Bengtsson A., Andersson R., Ansari D. The actual 5-year survivors of pancreatic ductal adenocarcinoma based on real-world data. Sci. Rep. 2020;10(1):16425. doi: 10.1038/s41598-020-73525-y. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395(10242):2008–2020. doi: 10.1016/S0140-6736(20)30974-0. Jun. [DOI] [PubMed] [Google Scholar]
- 3.Oettle H., Post S., Neuhaus P., Gellert K., Langrehr J., Ridwelski K., et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297(3):267. doi: 10.1001/jama.297.3.267. Jan 17. [DOI] [PubMed] [Google Scholar]
- 4.Oettle H., Neuhaus P., Hochhaus A., Hartmann J.T., Gellert K., Ridwelski K., et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310(14):1473. doi: 10.1001/jama.2013.279201. Oct 9. [DOI] [PubMed] [Google Scholar]
- 5.Burris H.A., Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R., et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. JCO. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. Jun. [DOI] [PubMed] [Google Scholar]
- 6.Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. May 12. [DOI] [PubMed] [Google Scholar]
- 7.Aben N., Vis D.J., Michaut M., Wessels L.F.A. TANDEM: a two-stage approach to maximize interpretability of drug response models based on multiple molecular data types. Bioinformatics. 2016;32(17):i413–i420. doi: 10.1093/bioinformatics/btw449. Sep 1. [DOI] [PubMed] [Google Scholar]
- 8.Angell H.K., Bruni D., Barrett J.C., Herbst R., Galon J. The Immunoscore: colon cancer and beyond. Clin. Cancer Res. 2020;26(2):332–339. doi: 10.1158/1078-0432.CCR-18-1851. Jan 15. [DOI] [PubMed] [Google Scholar]
- 9.Vieira A.F., Schmitt F. An update on breast cancer multigene prognostic tests – emergent clinical biomarkers. Front. Med. 2018;5:248. doi: 10.3389/fmed.2018.00248. Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoare O., Fraunhoffer N., Elkaoutari A., Gayet O., Bigonnet M., Roques J., et al. Exploring the complementarity of pancreatic ductal adenocarcinoma preclinical models. Cancers. 2021;13(10):2473. doi: 10.3390/cancers13102473. (Basel)May 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolle R., Gayet O., Duconseil P., Vanbrugghe C., Roques J., Bigonnet M., et al. A transcriptomic signature to predict adjuvant gemcitabine sensitivity in pancreatic adenocarcinoma. Ann. Oncol. 2021;32(2):250–260. doi: 10.1016/j.annonc.2020.10.601. Feb. [DOI] [PubMed] [Google Scholar]
- 12.Nicolle R., Blum Y., Duconseil P., Vanbrugghe C., Brandone N., Poizat F., et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102858. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rashid N.U., Peng X.L., Jin C., Moffitt R.A., Volmar K.E., Belt B.A., et al. Purity independent subtyping of tumors (PurIST), a clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clin. Cancer Res. 2019:1078. doi: 10.1158/1078-0432.CCR-19-1467. Nov 21-0432CCR-19–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciccolini J., Serdjebi C., Peters G.J., Giovannetti E. Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: an EORTC-PAMM perspective. Cancer Chemother. Pharmacol. 2016;78(1):1–12. doi: 10.1007/s00280-016-3003-0. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordh S. hENT1 expression is predictive of gemcitabine outcome in pancreatic cancer: a systematic review. World J. Gastroenterol. 2014;20(26):8482. doi: 10.3748/wjg.v20.i26.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjånes T.K., Jordheim L.P., Schjøtt J., Kamceva T., Cros-Perrial E., Langer A., et al. Intracellular cytidine deaminase regulates gemcitabine metabolism in pancreatic cancer cell lines. Drug Metab. Dispos. 2020;48(3):153–158. doi: 10.1124/dmd.119.089334. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohhashi S., Ohuchida K., Mizumoto K., Fujita H., Egami T., Yu J., et al. Down-regulation of deoxycytidine kinase enhances acquired resistance to gemcitabine in pancreatic cancer. Anticancer Res. 2008;28(4B):2205–2212. Aug. [PubMed] [Google Scholar]
- 18.Driehuis E., van Hoeck A., Moore K., Kolders S., Francies H.E., Gulersonmez M.C., et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. U.S.A. 2019;116(52):26580–26590. doi: 10.1073/pnas.1911273116. Dec 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ooft S.N., Weeber F., Dijkstra K.K., McLean C.M., Kaing S., van Werkhoven E., et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11(513):eaay2574. doi: 10.1126/scitranslmed.aay2574. Oct 9. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson F.M., Nabet B., Raghavan S., Liu Y., Leggett A.L., Kuljanin M., et al. Discovery of a selective inhibitor of doublecortin like kinase 1. Nat. Chem. Biol. 2020;16(6):635–643. doi: 10.1038/s41589-020-0506-0. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraunhoffer N.A., Abuelafia A.M., Bigonnet M., Gayet O., Roques J., Telle E., et al. Evidencing a pancreatic ductal adenocarcinoma subpopulation sensitive to the proteasome inhibitor Carfilzomib. Clin. Cancer Res. 2020;26(20):5506–5519. doi: 10.1158/1078-0432.CCR-20-1232. Oct 15. [DOI] [PubMed] [Google Scholar]
- 22.Hafner M., Niepel M., Chung M., Sorger P.K. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods. 2016;13(6):521–527. doi: 10.1038/nmeth.3853. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boj S.F., Hwang C.I., Baker L.A., Chio I.I.C., Engle D.D., Corbo V., et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):324–338. doi: 10.1016/j.cell.2014.12.021. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L., Bockorny B., Paul I., Akshinthala D., Frappart P.O., Gandarilla O., et al. PDX-derived organoids model in vivo drug response and secrete biomarkers. JCI Insight. 2020;5(21) doi: 10.1172/jci.insight.135544. Nov 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seppälä T.T., Zimmerman J.W., Sereni E., Plenker D., Suri R., Rozich N., et al. Patient-derived organoid pharmacotyping is a clinically tractable strategy for precision medicine in pancreatic cancer. Ann. Surg. 2020;272(3):427–435. doi: 10.1097/SLA.0000000000004200. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pishvaian M.J., Blais E.M., Brody J.R., Lyons E., DeArbeloa P., Hendifar A., et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the know your tumor registry trial. Lancet Oncol. 2020;21(4):508–518. doi: 10.1016/S1470-2045(20)30074-7. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruger S., Schirle K., Haas M., Crispin A., Schirra J., Mayerle J., et al. Prolonged time to treatment initiation in advanced pancreatic cancer patients has no major effect on treatment outcome: a retrospective cohort study controlled for lead time bias and waiting time paradox. J. Cancer Res. Clin. Oncol. 2020;146(2):391–399. doi: 10.1007/s00432-019-03061-4. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer A.C., Sorger P.K. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell. 2017;171(7):1678–1691. doi: 10.1016/j.cell.2017.11.009. Dec.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.