Abstract
The DNA-binding protein Ume6 is required for both repression and activation of meiosis-specific genes, through interaction with the Sin3 corepressor and Rpd3 histone deacetylase and the meiotic activator Ime1. Here we show that fusion of a heterologous activation domain to Ume6 is unable to convert it into a constitutive activator of early meiotic gene transcription, indicating that an additional function is needed to overcome repression at these promoters. Mutations in UME6 allowing the fusion to activate lie in a predicted amphipathic alpha helix and specifically disrupt interaction with Sin3 but not with Teal, an activator of Ty transcription also found to interact with Ume6 in a two-hybrid screen. The mutations cause a loss of repression by Ume6 and precisely identify the Ume6 Sin3-binding domain, which we show interacts with the paired amphipathic helix 2 region of Sin3. Analysis of these mutants indicates that conversion of Ume6 to an activator involves two genetically distinct steps that act to relieve Sin3-mediated repression and provide an activation domain to Ume6. The mutants further demonstrate that premature expression and lack of subsequent rerepression of Ume6-Sin3-regulated genes are not deleterious to meiotic progression and suggest that the essential role of Sin3 in meiosis is independent of Ume6. The model for Ume6 function arising from these studies indicates that Ume6 is similar in many respects to metazoan regulators that utilize Sin3, such as the Myc-Mad-Max system and nuclear hormone receptors, and provides new insights into the control of transcriptional repression and activation by the Ume6-URS1 regulatory complex in yeast.
Saccharomyces cerevisiae adapts to changing environmental conditions by initiating new programs of gene expression that alter the normal progression of mitosis. Depending on cell type and nutritional cues, budding cells switch to a form of invasive pseudohyphal growth, mate to form diploids, stably arrest in stationary phase, or undergo meiosis to form spores (see reference 48, 50, 61, and 91 for reviews). Sporulation typically occurs in MATa/MATα diploids starved for nitrogen and glucose in the absence of a fermentable carbon source and involves the coordinated expression of over 500 genes (13, 63). Analysis of this process in yeast is providing important new insights into common strategies and components that regulate not only meiosis and gametogenesis but also cell proliferation and differentiation in multicellular organisms.
Meiosis-specific genes in yeast have been divided into several classes depending on their time of expression (13, 48, 84). UME6 is one of nine UME genes (UME1 to UME9) whose loss causes unscheduled meiotic expression of early meiotic genes during vegetative growth (76, 77; B. Washburn, unpublished data). Although it was originally identified as a transcriptional repressor of early genes, UME6 has also been shown to function in their meiosis-specific activation (9, 65, 74). UME6 encodes a C6 zinc cluster protein that binds to the URS1 cis-elements found upstream of most early meiotic genes (2, 74, 77) and some middle and late meiotic genes (13, 63, 94). Work from several labs, as well as recent whole-genome analysis, has indicated that many nonmeiotic genes containing URS1 elements are also regulated by UME6 (19, 20, 47, 71, 77, 80; R. Williams, M. Primig, E. A. Winzeler, B. K. Washburn, R. W. Davis, and R. E. Esposito, unpublished data). These genes participate in a wide variety of metabolic pathways, including phospholipid biosynthesis, peroxisomal function, acetyl coenzyme A synthesis, nitrogen metabolism, and heat shock response. UME6 also participates in regulating genes that contain noncanonical URS1 elements, such as PHR1 and other DNA repair genes (79). Although Ume6 is required for normal repression and/or activation of genes in these various pathways, their regulation also usually involves a complex interplay of additional trans-acting and cis-acting factors which integrate the various signals influencing the expression of each particular set of genes.
Repression of meiotic gene expression by UME6 during vegetative growth involves the additional participation of SIN3 (UME4) and RPD3 (UME7) (8, 85, 86, 88; Washburn, unpublished). Aside from UME6, these are the only other UME genes whose loss leads to a nearly complete failure to sporulate (32, 76, 85). Recent studies have demonstrated that Sin3 and Rpd3 are components of a corepressor complex that represses transcription of many genes in yeast as well as higher eukaryotes. Sin3, which can act as a transcriptional repressor when fused to a DNA binding domain (42), contains four paired amphipathic helices thought to be involved in protein-protein interactions (43). It has been shown to interact with Ume6 in vivo and in vitro, indicating that Ume6 acts to recruit the Sin3 corepressor to early meiotic gene promoters (40). Sin3 appears to exert its effect on gene expression, at least in part, by altering chromatin structure through its association with Rpd3 (39, 66). Rpd3 is a histone deacetylase that genetically and physically interacts with Sin3 in yeast (40, 43), and this interaction has been shown to result in the deacetylation of one to two nucleosomes at the Ume6-binding site (41, 66).
Sin3 and Rpd3 homologs also provide a similar repression function in multicellular organisms. In mammalian cells, corepressor complexes containing homologs of Sin3 (mSin3a or mSin3b) and Rpd3 (HDAC1p or HDAC2p) are involved in regulation of transcription by a number of important systems, including the nuclear hormone receptors (reviewed in reference 26) and the Max-interacting members of the Myc/Mad/Max family of basic helix-loop-helix-leucine zipper (bHLHZip) transcription factors (Mad1, Mad3, Mad4, Mxi, and Mnt[Rox] [reviewed in reference 57]). Like Ume6, the nuclear hormone receptors and Myc/Mad/Max families participate in both activation and repression of genes involved in differentiation and development (reviewed in references 14, 31, and 82; see also Discussion).
In yeast meiosis, the conversion of Ume6 from a repressor to an activator of early meiotic gene expression is critical for induction of the early genes and for the progression of meiosis. The switch in Ume6 activity requires the presence of Ime1 (inducer of meiosis), which accumulates to high levels very early in sporulation (9, 65, 74). Ime1 is known to function as an activator of transcription when fused to a DNA binding domain (56, 72) and, like Sin3, has been shown to interact with Ume6 in two-hybrid assays (65). Thus, one function of Ime1 is apparently to provide an activation domain to Ume6, allowing it to be converted to an activator. This interaction is facilitated by phosphorylation of the Ume6 amino terminus by either Rim11 or Mck1 (65, 93).
Taken together, these findings have led to the current view of Ume6 as a multifunctional DNA-binding protein component of transcriptional regulatory complexes, capable of alternately repressing or activating early meiotic transcription through its association with Sin3 or Ime1, respectively. The function and regulation of these interactions are further addressed below. In this study, we observed that fusion of a heterologous activation domain to Ume6 fails to convert it to an activator of early gene expression during vegetative growth. We took advantage of this behavior to isolate mutations in UME6 that allow the fusion to function as an activator. These mutations disrupt interaction with Sin3 in vivo and in vitro and identify the Sin3 binding domain of Ume6, the first target binding sequence for Sin3 defined in yeast. These mutations were used to further investigate the function and regulation of the Sin3-Ume6 interaction during vegetative growth and meiosis.
MATERIALS AND METHODS
Strains.
All yeast strains used in genetic studies are closely related to W303 (81). SFY59 (MATa ade2 ade6 can1-100r his3-11,15 leu2-3,112 trp1-1 ura3-1) and YC105 (MATa ade2 ade6 can1-100r his3-11,15 leu2-3,112 trp1-1 ume6-Δ1 ura3-1) were derived from W303 as previously described (74). YC121 (MATα ade2 can1-100:ADE2:CAN1 his3-11,15 leu2-3,112 trp1-1 ume6-Δ1 ura3-1) was derived from W303 by insertion of the ume6Δ and can1-100:ADE2:CAN1 cassettes as previously described for YC122 (74). REE3574 (MATa ade2 ade6 can1-100r his3-11,15 leu2-3,112 trp1-1 ume6Δ::KanMX ura3-1) was constructed by replacement of the UME6 open reading frame (ORF) in SFY59 with a KanMX6 cassette, using the short flanking PCR-generated homology (87). Isogenic derivatives of this strain containing UME6 (REE4175), ume6-6 (REE4178), ume6-7 (REE4181), ume6-9 (REE4184), ume6-6,9 (REE4187), and ume6-7,9 (REE4190) were produced by transformation with the integrating plasmids pBK100, pBK102, pBK103, pBK104, pBK105, and pBK106, respectively (see below), cut with PacI to select for integration adjacent to ume6Δ::KanMX (64). An isogenic sin3Δ strain (REE4123) was similarly derived by integration of UME6 into REE3575 (MATa ade6 can1-100r his3-11,15 leu2-3,112 trp1-1 sin3Δ::HIS3 ura3-1 ume6Δ::KanMX). REE2276 (MATa ade2 ade6 can1-100r his3-11,15 ime1::URA3 leu2-3,112 trp1-1 ura3-1) was constructed by insertion of an ime1::URA3 disruption into SFY59, and REE3086 (MATα ade2 ade6 can1 his3 his4 leu2 trp1 ura3 ime1::URA3) was derived from outcrosses with W303.
Two-hybrid analysis was performed in REE3311 (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ GAL80Δ GAL-ADE2 LYS2::GAL-HIS3 met2::GAL7-lacZ rpd3Δ::KanMX6), a derivative of PJ69-4A (38) in which the RPD3 ORF was replaced with KanMX6. BJ2168 (MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407), obtained from E. Jones, was used for production of yeast lysates. Escherichia coli lysates were prepared in strain BL21 (78). All other E. coli plasmid manipulations were performed in strain DH5α or DH10B (Gibco BRL).
Plasmids.
pBK8 carries two SPO13 fusions, SPO13-URA3 and SPO13-lacZ, on a single 2μm TRP1 plasmid. Each fusion contains −847 to +45 of the SPO13 gene and exhibits meiosis-specific regulation similar to that of SPO13. pBK8 was constructed by inserting a BamHI fragment containing SPO13-lacZ (10) into the unique BglII site of pPL128 (provided by R. Surosky, this lab), which contains the SPO13-URA3 fusion from pMS49 (77) inserted into YEp13. The wild-type UME6 plasmid pPL5905 carries UME6 on a centromeric (CEN) LEU2 vector (77). pBK54, a ume6-6 derivative of this plasmid, was produced by swapping the BamHI-StuI and BssHII-SalI UME6 fragments, respectively, from pGAD-ume6-6 into pPL5905. Integrating UME6 and ume6-6 plasmids (pBK100 and pBK102, respectively) were constructed by cloning the SpeI-SalI UME6 fragments from pPL5905 and pBK54, respectively, into pRS305 (69). Integrating ume6-7, ume6-9, ume6-6,9, and ume6-7,9 plasmids (pBK103-106) were derived from pBK100 and pBK102 by swapping appropriate restriction fragments from the GAD-ume6 mutants.
Several transcriptional and translational fusions to full-length Ume6 were constructed for these studies. For construction of the UME6 translational fusions, the start site of UME6 in pPL59095 was altered by site-directed mutagenesis (67) (provided by C. Steber) to contain a unique NcoI site, creating pPL5905-M1P. To construct Gal4 activation domain and Gal4 DNA binding domain fusions (GAD-Ume6 and GBD-Ume6), the NcoI (filled with Klenow enzyme)-SalI UME6 fragment from pPL5905-M1P was inserted into pGADC3 and pGBDUc3 (38) cut with BamHI (filled with Klenow enzyme) and SalI. Two-hybrid studies have previously indicated that it is the amino-terminal end of Ume6 that normally interacts with the meiotic activator Ime1 in meiosis (65), and so the heterologous activation domain in the GAD-Ume6 fusion was positioned as closely as possible to the location of the native activator. Yeast glutathione S-transferase (GST)–UME6 fusions were similarly constructed by inserting the NcoI (filled with Klenow enzyme)-HindIII fragment from pPL5905-M1P into the XbaI (filled with Klenow enzyme)-HindIII site of pEG-KG (62). For all yeast Ume6 fusion plasmids, the junctions were sequenced to verify that they were in frame. All of these fusions (including GAD-UME6) are functional in yeast, as determined by complementation of the SPO13 derepression phenotype of a ume6Δ mutant.
It is important for our studies to note that the ADH1 promoter in pGADc3 (38), used above in the construction of GAD-UME6, is derived from pGAD424 and is truncated and located at a different position in comparison to the ADH1 promoter in pGAD2F (6), used in the construction of the GAD-Ume6 fusions by Rubin-Bejerano et al. (65). The promoter in our study confers a lower level of expression than that of pGAD2F (51), making it suitable for use with proteins that are deleterious when overexpressed. High-level overexpression of Ume6 from a GAL1 promoter is lethal (Washburn, unpublished), and in our strains the constitutive high level of expression of GAD-Ume6 from the full-strength ADH1 promoter on pGAD2F derivatives causes poor viability. In contrast, expression of our GAD-Ume6 fusion under the control of the attenuated ADH1 promoter, like expression of UME6 on a high-copy-number plasmid under the control of its own promoter, causes no ill effects. This difference in expression levels affects some of the interpretation of the results from the two studies (see Discussion). It should also be noted that the Ume6 fusions used in the present study are full length, whereas those used by Rubin-Bejerano et al. lack the first 158 amino acids of Ume6. However, we find that the first 158 amino acid residues Ume6 are not involved in vegetative repression or activation by GAD-UME6 (see Fig. 1B), and this difference therefore has no impact on the present study.
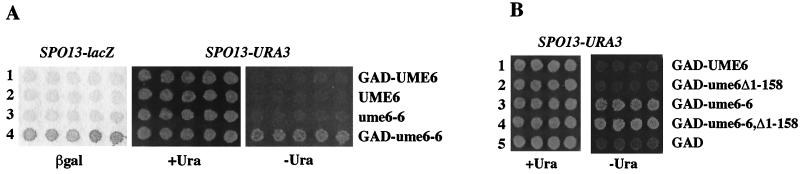
FIG. 1.
Activation of SPO13 expression by wild-type and mutant GAD-Ume6 fusions. A wild-type haploid (SFY59) containing pBK8 (SPO13-URA3 SPO13-lacZ) and UME6 plasmids was replica plated to either Ura− medium or filters on X-Gal medium to detect expression of SPO13. (A) Comparison of SPO13 activation by wild-type (GAD-UME6; lane 1) and mutant (GAD–ume6-6; lane 4) fusions, and wild-type (pPL5905; lane 2) and mutant (pBK54; lane 3) UME6, in the absence of GAD. (B) Activation by GAD-UME6 derivatives lacking the Ime1 binding domain. Fusions contain either full-length UME6 (lane 1) or ume6-6 (lane 3), or derivatives of each (lanes 2 and 4, respectively) lacking the first 158 amino acids of Ume6. Deletions derivatives were created by cutting the parent plasmids with BamHI and religating, thus removing the coding region between the polylinker at the GAD-UME6 junction and UME6 nucleotide +474.
Yeast and E. coli methods.
Growth and sporulation media have been previously described (45). Yeast strains were transformed by the lithium acetate method (25), and E. coli strains were transformed by electroporation (Bio-Rad).
β-Galactosidase assays.
For plate assays, yeast cells were grown on paper filters (3MW; Midwest Scientific) overlaid on agar medium selective for the plasmids. Filters were then frozen in liquid nitrogen and placed on agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (76). After 2 to 36 h, color development was stopped by air drying. For determination of β-galactosidase activity due to activation of SPO13-lacZ expression by GAD-Ume6 derivatives in liquid cultures, at least three independent transformants were grown to an optical density at 600 nm of 0.7 to 0.75 in synthetic dextrose medium and collected by centrifugation. Colorimetric assays of lysates obtained by bead beating were then performed with o-nitrophenyl-β-d-galactopyranoside (ONPG) as previously described (10). For determination of β-galactosidase activity due to derepression of SPO13-lacZ by ume6 mutations, at least two independent transformants were grown on selective agar medium and lysed in Y-PER buffer (Pierce), and activity was determined using the reagents and protocols in the Pierce yeast β-galactosidase activity kit.
Two-hybrid screen and assays.
The two-hybrid strain and plasmids used to identify Ume6 interactors have previously been described (38). The PJ69-4A reporter strain was modified by replacement of the RPD3 ORF with KanMX6 (above), in order to alleviate repression of the reporters by the GAD-UME6 bait. Approximately 2 million transformants for each of the three reading frame Gal4 activation domain fusion libraries were plated to His− medium. His+ transformants (∼1,000) were replica plated to Ade− medium, and His+ Ade+ isolates were then tested for bait dependence. Six bait-dependent clones were obtained. DNA sequencing (Applied Biosystems Inc.) indicated that four isolates encoded Tea1 fusions, and two encoded identical Sin3 fusions. A Sin3 paired amphipathic helix 1 (PAH1) deletion (amino acid residues 238 to 295) was derived from the Sin3 library clone by removal of the internal BspEI fragment. A deletion of the entire Sin3 PAH2 region (residues 290 to 670) was constructed by removal of the internal BsmI-PstI fragment, followed by treatment with T4 polymerase and ligase. The more precise PAH2 deletion (residues 424 to 450) was from plasmid M1285 (90), inserted on a BsmI-CelII fragment. All Sin3 deletion constructs were sequenced to verify intact reading frames.
Ume6 mutagenesis and mapping.
Plasmid DNA was mutagenized by growth in the bacterial mutator strain XL1-Red (Stratagene) and used to transform wild-type yeast (SFY59) containing pBK8 (SPO13-lacZ SPO13-URA3). Ura+ Lac+ transformants were selected as described in Results. Restriction fragment swaps between mutant and wild-type parent plasmids were used to localize the mutations in UME6 responsible for causing activation of SPO13. Of the 10 mutations mapped, 9 localized to a 157-bp EcoRI-StuI fragment encoding amino acids 508 to 560 of Ume6, and one mutation localized to a PvuII-AlwNI fragment encoding amino acids 593 to 659. Sequencing of the indicated fragments indicated that none contained multiple mutations.
GST pulldowns.
GST-Ume6 fusion plasmids were constructed by in-frame fusion of the PvuII-EcoRI fragment encoding amino acids 508 to 584 of Ume6 to GST in pGEX-KG (29). Plasmid M1155, containing ADH1-HA (hemagglutinin)-SIN3, was obtained from David Stillman (43). For production of GST-Ume6 fusion proteins in bacteria, E. coli BL21 containing GST-Ume6 fusions (Ume6 amino acid residues 508 to 584) was grown to an optical density at 600 nm of 0.5 at 37°C, and expression of the fusions was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. Cells were collected by centrifugation, and the pellet from 40 ml of cells was resuspended in 500 μl of bacterial lysis buffer (phosphate-buffered saline [PBS] containing 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, and 1.5 μg of leupeptin/ml). Lysates were prepared by sonication. For production of yeast lysates containing HA-tagged Sin3, BJ2168 containing the Adh1-HA-Sin3 plasmid M1155 (43) was harvested at a density of 107 cells/ml. Lysates were prepared by bead beating in 300 μl of yeast lysis buffer (20 mM Tris, 150 mM NaCl, 10 mM MgCl2, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 μM aprotinin, leupeptin [1.5 μg/ml], pepstatin A [3 μg/ml], chymostatin [1 μg/ml]).
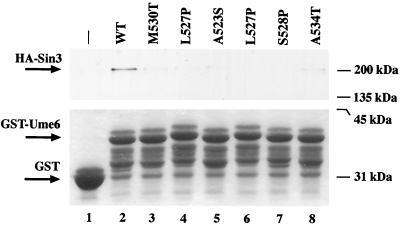
For the GST pulldown experiments, 500 μl of E. coli lysate (above) was mixed with 25 μl (bed volume) of glutathione-agarose (Sigma) for 15 min at 4°C, collected by centrifugation, and washed three times with PBS. A single HA-Sin3 lysate was prepared and divided equally (500 μg each) between the various mutant conjugate pellets and incubated overnight at 4°C. Glutathione-agarose– GST–Ume6–HA-Sin3 complexes were recovered by centrifugation, washed three times with PBS, and eluted by boiling in sodium dodecyl sulfate (SDS) loading buffer for 5 min. One-third of this eluate was electrophoresed on an SDS–10% polyacrylamide gel and electroblotted to a polyvinylidene difluoride membrane (Millipore) in CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] buffer (10 mM CAPS, pH 11.0, 10% methanol). HA-Sin3 was detected with anti-HA-peroxidase antibody conjugate (Boehringer Mannheim Biochemicals) and visualized with enhanced chemiluminescence detection (Amersham). Parallel gels were stained with Coomassie R-250 to verify that equivalent amounts of GST-Ume6 were present in each sample.
RESULTS
Addition of an activation domain to Ume6 is not sufficient to convert it to an activator.
Two-hybrid studies have indicated that Ime1 physically interacts with Ume6, providing a transcriptional activation domain for the induction of early meiotic gene expression (55, 65). If the only requirement for conversion of Ume6 from a repressor to an activator is the addition of an activation domain, one might predict that fusion of a heterologous activation domain to Ume6 would cause it to function as a constitutive activator of SPO13 and other early meiotic genes during vegetative growth (i.e., in the absence of Ime1 induction). The ability of such heterologous activation domains to activate transcription of reporter genes when linked to appropriate DNA-binding proteins is fundamental to the widespread success of yeast two-hybrid systems (6).
To determine whether the addition of an activation domain to Ume6 could promote vegetative expression of SPO13, we fused the Gal4 activation domain (Gal4 amino acids 768 to 881) in frame to the amino-terminal end of full-length Ume6 to produce GAD-UME6. Vegetative SPO13 expression was monitored using SPO13-URA3 and SPO13-lacZ reporter fusions and assayed by growth of ura3-1 strains in the absence of uracil and the development of blue color on X-Gal medium. Wild-type cells, in which expression of SPO13 is repressed, are Ura− and white.
As shown in Fig. 1A, a high-copy-number plasmid expressing the GAD-UME6 fusion from the ADH1 promoter does not cause detectable activation of SPO13 in a wild-type haploid (lane 1) or diploid (not shown), resulting in cells that remain Ura− and white. The failure of the GAD-UME6 fusion to activate gene expression in vegetatively growing cells suggested that Ime1 or some other meiosis-specific component was specifically required for conversion of Ume6 to an activator. Such factors may act to modify and/or interact with specific regions of Ume6 to relieve Sin3-mediated repression or may in some way allow Ume6 to adopt an activating configuration. We took advantage of the failure of GAD-Ume6 to activate transcription by using this fusion as the starting point in a mutant screen designed to identify regions of Ume6 involved in this process.
Isolation of mutations that allow activation by GAD-Ume6.
To identify amino acid residues in Ume6 involved in regulating its conversion to an activator, mutations in GAD-UME6 that led to expression of SPO13 during vegetative growth were sought. These mutants were selected by transforming a wild-type haploid, containing SPO13-URA3 and SPO13-lacZ fusions, with a pool of mutagenized GAD-UME6 plasmid. Ura+ transformants were selected and then screened for β-galactosidase activity to identify mutants exhibiting high-level expression of SPO13; 115 Ura+ Lac+ transformants were obtained from a population of ∼106 transformants. Since the Gal4 activation domain and Ume6 DNA binding domain are located at opposite ends of the GAD-Ume6 protein and are both required for activation of the reporters, this collection of mutants should not include simple loss-of-repression alleles resulting from frameshifts leading to a truncated protein or from mutations that cause a loss of transcription of UME6 or translation of the UME6 message or decreased Ume6 stability.
A typical activating isolate, ume6-6, is shown in Fig. 1A. Unlike the wild-type GAD-UME6 fusion, the GAD–ume6-6 fusion (lane 4) causes vegetatively growing wild-type cells to express SPO13-URA3 at levels sufficient to allow robust growth in the absence of uracil and to express SPO13-lacZ at easily detectable levels. This activation requires the presence of the Gal4 activation domain, since expression of the corresponding Gal4 binding domain (GBD)–ume6-6 fusion (from a promoter identical to that used for the GAD–ume6-6 fusions) fails to cause activation of GAL1-HIS3 or GAL2-ADE2 reporters (see Fig. 4) or SPO13-lacZ (not shown). Similarly, a CEN plasmid carrying UME6 (under the control of its own promoter) into which the ume6-6 mutation was inserted causes no activation (lane 3). Thus, the ume6-6 mutation allows activation by Ume6 only when an activation domain is supplied but does not cause activation itself or uncover a cryptic activation domain in Ume6. Based on the requirement for a linked activation domain, we conclude that the increased SPO13 expression caused by GAD–ume6-6 results from a gain of activation function by the fusion rather than a loss of repression by ume6-6 alone. The participation of the meiotic activator Ime1 is not required for this activation, since deletion of amino acid residues 1 to 158 of Ume6, previously shown to contain the Ime1 binding domain (65), has no effect on activation by GAD-UME6 or GAD–ume6-6 (Fig. 1B, lanes 2 and 4).
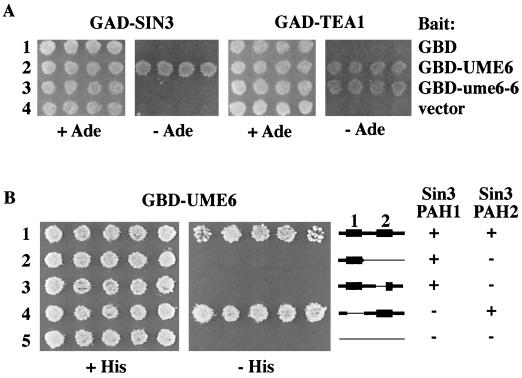
FIG. 4.
Two-hybrid analysis of Ume6 interactions, using an rpd3Δ derivative of PJ69-4A containing GBD-Ume6 baits and either GAD-Sin3 or GAD-Tea1 library clones. (A) Interaction between Ume6 and library clones is shown by growth on Ade− medium resulting from expression of the GAL2-ADE2 reporter. (B) Interaction between GBD-Ume6 with deletion derivatives of GAD-Sin3, shown by growth on His− medium resulting from expression of the GAL1-HIS3 reporter.
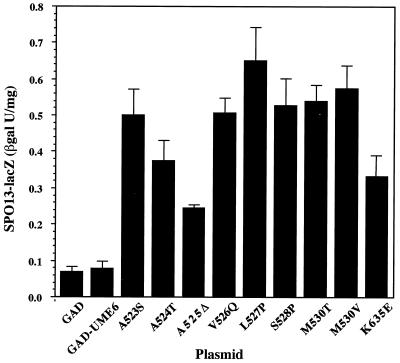
GAD–ume6-6 and nine additional activating GAD-ume6 mutants, picked at random, were chosen for further study. Each of these causes a three- to ninefold increase in SPO13 expression in comparison to wild-type GAD-UME6 in β-galactosidase assays (Fig. 2). Like ume6-6, all of the alleles fail to show detectable activation when fused to the Gal4 DNA binding domain (not shown). To determine whether these GAD-ume6 fusions could also cause activation of other early meiotic genes, GAD–ume6-6 (M530T [see below]) was also tested for vegetative activation of a HOP1-lacZ fusion (pAV79 [21]). Like SPO13, HOP1 contains a URS1 element and its expression is induced in meiosis. In the presence of this reporter, the GAD–ume6-6 fusion causes a greater than 20-fold increase in β-galactosidase activity to 3.5 U, versus 0.12 and 0.17 U for GAD and GAD-UME6, respectively. The effect of the ume6-6 mutation on HOP1 expression is even more dramatic than that seen with SPO13 and indicates that the ability of GAD–ume6-6 to activate early meiotic gene expression is not restricted to SPO13.
FIG. 2.
Activation of SPO13-lacZ expression by mutant GAD-Ume6. ONPG liquid assays of β-galactosidase (βgal) activity in a wild-type haploid (SFY59) containing pBK8 (SPO13-URA3 SPO13-lacZ) and plasmids carrying GAD, GAD-UME6, and various GAD-ume6 mutant fusions (denoted by the amino acid alteration [Fig. 3]) were performed as described in Materials and Methods. The M530T, L527P, S528P, and K635E alleles have been designated ume6-6, ume6-7 ume6-8, and ume6-9, respectively.
The mutations allowing activation cluster in a region of UME6 predicted to encode an amphipathic α helix.
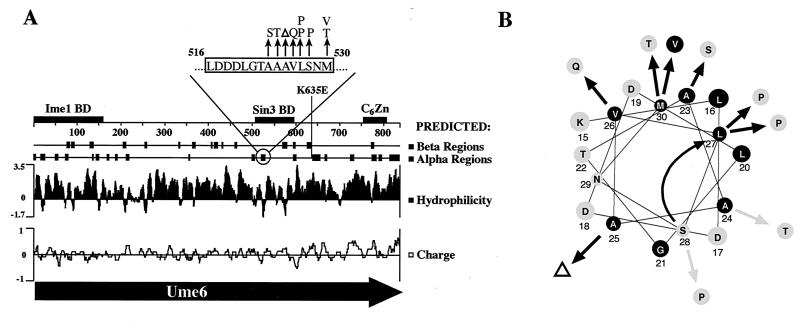
The locations of ume6-6 and the nine other mutations were determined to identify possible functional domains. As shown in Fig. 3A, ume6-6 (M530T) and all but one of the other nine mutations localized to an eight-amino-acid sequence (residues 523 to 530) in the central part of the protein. This sequence is within the only region of Ume6 (residues 508 to 584) previously identified as being sufficient for Sin3 binding and for conferring repression when fused to LexA (40). Sequence analysis indicates that this cluster of mutations lies within a region, residues 516 to 530, that is predicted to adopt an α-helical configuration. This is also one of the few hydrophobic regions of an unusually hydrophilic protein (2). When displayed as a helical wheel plot (Fig. 3B), the hydrophobic residues appear to be distributed in an amphipathic pattern (i.e., one face of the helix is predominantly hydrophobic and the opposite face is predominantly hydrophilic). The nature of the ume6 mutations recovered in this study supports the conclusion that the ability of this region to adopt an amphipathic α-helix configuration is important for repression by Ume6. Except for the M530V substitution, all of the mutations in the predicted α helix either change an amino acid residue on the hydrophobic face to hydrophilic (A523S, A524T, A525Δ, V526Q, and M530T) or introduce a proline (L527P and S528P), which is expected to disrupt the α helix. The unusual deletion of an alanine at position 525, resulting from the loss of a single CAG triplet in a (CAG)4 repeat, is predicted to cause a hydrophilic amino acid (S528) to shift into a position on the hydrophobic face previously occupied by a hydrophobic amino acid (L527).
FIG. 3.
Mutations in UME6 allowing activation by GAD-UME6. (A) Map of UME6 showing locations of mutations and predicted structure. Overall sequence predictions were by the Chou-Fasman method, performed by the Protean sequence analysis program (DNASTAR). The more detailed presentation of the α helix predicted from 516 to 530 (top) is the consensus of four prediction methods (16, 22–24, 52). Mutations are denoted as arrows. Regions known to contain the Ime1 binding domain (BD) (65), Sin3 binding domain (40), and C6Zn DNA binding domain (77) are indicated as boxes. (B) Helical wheel diagram of the predicted α helix (residues 516 to 530 are shown) showing locations of mutations. The curved arrow indicates the shift in position of amino acid 528 resulting from the deletion of amino acid 525.
Intriguingly, in mammalian cells the Sin3 binding domains of Mnt/Rox (35, 59), Mxi (68), and the Mad proteins (5, 36), though different in sequence from Ume6, have also been shown to contain predicted amphipathic helices that are essential for repression. For the Mad1 and Mxi helices, mutation of hydrophobic residues in the helix was shown to result in a loss of binding to Sin3 (5, 18, 68) and in the case of Mad1, to result in a loss of repression (5, 18). It has recently been demonstrated that the Sin3 binding domain in Mad1 does indeed adopt an amphipathic α-helix configuration in vitro and is sufficient for Sin3 recruitment (18). The location of the ume6 mutations within the stretch of amino acids already known to contain the Sin3 interaction domain, as well as the similarity of the predicted secondary structure of the region to that of previously identified Sin3 interaction domains, suggested that these mutations allow the GAD-ume6 fusions to activate transcription by interfering with the interaction of Ume6 with Sin3. This conclusion was confirmed in the studies below.
Mutations in the α helix interfere with the ability of Ume6 to interact with Sin3.
Two-hybrid assays were used to test the ability of Ume6-6 to form stable complexes in vivo. For these experiments, full-length Ume6 was fused to the Gal4 DNA binding domain as bait. The interactions were tested using two Gal4 activation domain fusions that we isolated from a two-hybrid screen designed to identify Ume6 interactors (see Materials and Methods). The first of these interactors is Sin3. The interaction of Sin3 with Ume6 had previously been demonstrated in a pairwise two-hybrid test and with GST pulldown assays (40). Our recovery of Sin3 in this screen provides additional evidence for the interaction of Sin3 and Ume6 in vegetatively growing yeast cells. The second gene isolated from this screen was TEA1. Like UME6, TEA1 encodes a C6 zinc cluster DNA-binding protein. Tea1 has been shown to bind a cis-element in the Ty enhancer and is required to achieve full levels of Ty enhancer-mediated transcription (28). The nature of the relationship between Ume6 and Tea1 is the subject of a separate study (Washburn, unpublished).
As shown in Fig. 4A, the ume6-6 mutation has a dramatic effect on the interaction of Ume6 with Sin3. The wild-type GBD-Ume6 bait (lane 2) exhibits interaction with both Tea1 and Sin3, demonstrated by growth on Ade− medium due to activation of GAL2-ADE2 reporter expression. Introduction of the ume6-6 mutation into GBD-Ume6 (lane 3) abolishes the ability of the GBD-Ume6 fusion protein to interact with GAD-Sin3 in this assay. However, it does not alter the ability of Ume6 to interact with GAD-Tea1, which serves as an internal control indicating that this loss of binding ability by Ume6 is specific to the Sin3 interaction. Neither Sin3 or Tea1 interacts with GBD alone (lane 1), demonstrating that these interactions are with the Ume6 portion of the fusion rather than the region containing the Gal4 C6 zinc cluster DNA binding domain, which has some limited homology to Ume6.
To test additional ume6 mutants and to confirm the effects of the ume6-6 mutation on Sin3 interaction, the ability of Ume6 to bind Sin3 was tested in vitro by GST pulldown assays. For these assays, the region of Ume6 encoding amino acids 508 to 584, previously shown to be sufficient for Sin3 binding (40), was used instead of full-length Ume6. These segments from wild-type and mutant Ume6 derivatives were fused to GST, and the resulting proteins were allowed to bind Sin3 in yeast cell extracts. Complexes were recovered by affinity purification on glutathione-agarose. As shown in Fig. 5, Western blot assays detecting HA epitope-tagged Sin3 indicate that the mutations in UME6 (lanes 3 to 8) do indeed dramatically reduce binding of Ume6 to Sin3 in comparison to wild-type UME6 (lane 2). These results confirm that the mutations in this region interfere with the ability of Ume6 to interact with Sin3 and support the identification of the putative amphipathic α helix as the Sin3 binding domain. The one mutation that lies outside the α helix (ume6-9/K635E) has no detectable effect on Sin3 interaction in two-hybrid assays (not shown), suggesting that it exerts its effect by a different mechanism.
FIG. 5.
GST pulldown assay. HA-Sin3 from yeast extracts was bound to GST (lane 1) or GST fused to amino acids 508 to 584 of wild-type (wt; lane 2) or mutant (lanes 3 to 8) Ume6 as described in Materials and Methods. A single HA-Sin3 lysate was evenly divided among the tubes (500 μg each). Complexes were eluted with boiling loading buffer, separated by SDS-polyacrylamide gel electrophoresis and visualized with anti-HA antibodies (top) to assay Sin3 interaction or Coomassie (bottom) to verify equivalent amounts of GST-Ume6 fusion in each sample. Several presumed GST-Ume6 degradation products uniformly appear (as previously observed [55]) in all of the GST-Ume6 lanes.
Ume6 interacts with the PAH2 region of Sin3.
Having identified the region of Ume6 that interacts with Sin3, it was of interest to identify the corresponding region in Sin3 that interacts with Ume6. Sin3 contains four paired amphipathic helices (PAH1 to PAH4) which have been proposed to be involved in protein-protein interactions (43). Sin3 is now known to interact with several corepressors in higher eukaryotes, through a variety of contact points (1, 3). We found that both of the Ume6-interacting Sin3 clones isolated from the two-hybrid screen encoded fusions to Sin3 amino acid residues 228 to 671 and therefore contain only PAH1 (residues 238 to 285) and PAH2 (residues 426 to 472); PAH3 and PAH4 are predicted to start at residues 680 and 1153, respectively (88). The region of Sin3 required for interaction with Ume6 was further determined by deletion analysis of the GAD-Sin3 clones. These studies showed that removal of residues 238 to 295, encompassing PAH1 in Sin3, has no effect on its ability to interact with Ume6 (Fig. 4B, lane 4). Thus, neither PAH1, PAH3, nor PAH4 in Sin3 is required for interaction with Ume6. That PAH2 is necessary for the interaction was initially suggested by the finding that deletion of residues 290 to 670 abolishes the interaction (Fig. 4B, lane 2). A more precise deletion specifically localized within PAH2 (residues 424 to 450; lane 3) confirmed this conclusion. Among known Sin3-binding proteins, the Max-interacting proteins, which as described above also contain a Sin3 binding domain with some similarity to that of Ume6, have also been shown to interact with PAH2 of yeast Sin3 (42) and mammalian Sin3 homologs (5, 18, 35, 59, 68).
Premature expression of Ume6-Sin3-regulated meiotic genes is not detrimental to subsequent sporulation.
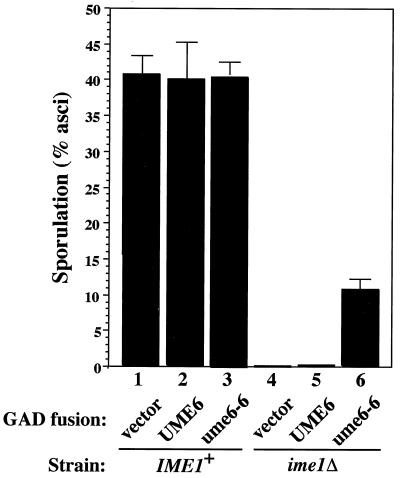
As shown above, the mutant GAD-ume6 fusions cause early meiotic genes to be expressed during vegetative growth, whereas normally the early genes are not induced until several hours after the initiation of meiosis. The three regulators whose loss leads to the highest level of derepressed early meiotic gene expression (ume6, sin3, and rpd3) also confer a severe sporulation defect (32, 76, 85). Until now it has been difficult to determine whether premature expression interfering with meiotic progression is a cause of the Spo defect of these strains and/or whether it due to other meiotic roles of these genes, such as Ume6 function in activation. The properties of the GAD–ume6-6 fusion provided a convenient tool to address this question. As described above, the introduction of a GAD–ume6-6 plasmid into an otherwise wild-type strain (i.e., UME6+ SIN3+ RPD3+) results in vegetative expression of the early genes, similar to the levels observed in sin3 and rpd3 mutants. If premature expression of early meiotic genes is responsible for the sin3 and rpd3 sporulation defects, then dominant overexpression of the early genes conferred by GAD–ume6-6 is expected to inhibit sporulation. As shown in Fig. 6, wild-type diploids containing GAD–ume6-6 (column 3) sporulate as efficiently as strains containing the wild-type GAD-UME6 fusion (column 2) or no fusion at all (column 1). We thus conclude that premature expression of the early genes alone does not interfere with subsequent sporulation and is therefore an unlikely explanation for the inability of sin3 and rpd3 mutants to sporulate.
FIG. 6.
Sporulation of strains containing fusions of the Gal4 activation domain to UME6 or ume6-6. A wild-type diploid (W303a/α; lanes 1 to 3) or ime1Δ diploid (REE2276/REE3086; lanes 4 to 6) was transformed with GAD, GAD-UME6, or GAD–ume6-6 plasmids. Transformants were replica plated to sporulation medium, and asci were counted after 5 days at 37°C.
Activation of early meiotic gene expression by GAD–ume6-6 can partially substitute for Ime1.
Since the GAD–ume6-6 mutant exhibits a dominant gain of vegetative activation function, it seemed plausible that in the absence of Sin3 binding, high-level activation of early gene transcription by the Gal4 activation domain might be able to substitute for Ime1 in sporulation. To test this, an ime1Δ diploid was transformed with GAD fusion plasmids and transformants were tested for the ability to sporulate. As shown in Fig. 6, the GAD–ume6-6 plasmid (column 6) conferred a significant level of sporulation in the absence of Ime1 (∼25% of the wild-type level), in contrast to plasmids containing GAD (column 4) or the wild-type GAD-UME6 fusions (column 5). Thus, the addition of a functional heterologous activation domain to Ume6-6 can partially substitute for the presence of Ime1 in meiosis. Although the sporulation level afforded by GAD–ume6-6 is less than in the wild-type level (columns 1 to 3), this difference may be explained by the poor expression of the ADH1-GAD–ume6-6 fusion in meiosis (65). The failure of a wild-type GAD-UME6 fusion to promote sporulation in an ime1Δ strain implies that the Sin3-Ume6 interaction resulting in repression persists in the absence of Ime1 even after the shift to sporulation medium. Taken together, these data suggest that in wild-type strains, Ime1 may play a dual role in relieving Sin3-mediated repression as well as providing an activation domain (see Discussion).
Mutations in the α helix cause a loss of repression by Ume6.
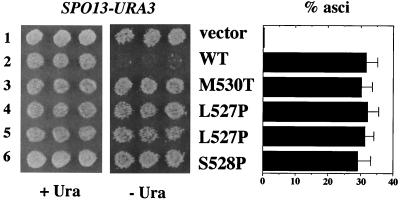
The experiments above indicate that ume6-6 and the other mutations in the α helix allow a GAD-ume6 fusion to activate gene expression by interfering with the ability of Sin3 to bind Ume6. Therefore, these mutants should exhibit a repression defect in the absence of the Gal4 activation domain. This was tested by introducing the mutations into otherwise wild-type UME6 (lacking GAD) on CEN plasmids and transforming a diploid ume6Δ/ume6Δ mutant. As shown in Fig. 7, all of the mutations tested cause a loss of repression by Ume6. Like the uncomplemented ume6Δ mutant containing only vector (lane 1), ume6Δ cells containing ume6-6 and the other mutant ume6 alleles (lanes 3 to 6) are Ura+, indicating that expression of SPO13-URA3 is derepressed.
FIG. 7.
Complementation of a ume6Δ diploid by UME6 plasmids. Vegetative derepression and sporulation in a ume6Δ diploid (YC105/YC121) containing pBK8 and either vector (pRS315; lane 1), wild-type (WT) UME6 plasmid (pPL5905; lane 2), or mutant derivatives of pPL5905 containing the indicated mutations in the α helix (lanes 3 to 6). Lanes 4 and 5 represent two independent isolates of the L527P mutation.
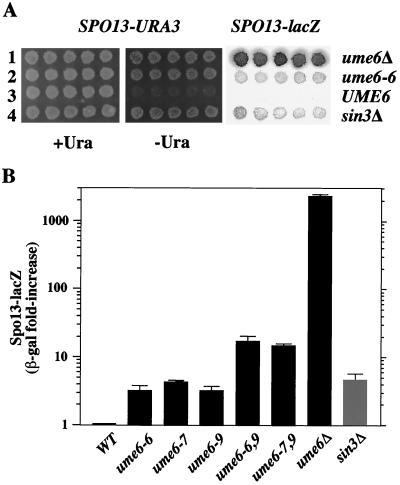
The relative level of derepression resulting from the ume6 mutations cannot be determined using the UME6 and ume6 plasmids described above, since occasional loss of the plasmids in individual cells results in transient derepressed expression of SPO13-lacZ in the nongrowing ume6 cells and therefore an overall low but detectable background of β-galactosidase activity. To determine the degree of derepression conferred by ume6-6, the mutation was integrated into the genome to replace UME6. As shown in Fig. 8A, ume6-6 (lane 2) results in expression of SPO13-URA3 and SPO13-lacZ, in a manner similar to loss of SIN3 (lane 4). As indicated in Fig. 8B, ume6-6, ume6-7, and sin3Δ mutants all exhibit less derepression than a ume6Δ strain, indicating that Ume6 must have a repression function independent of Sin3 (see Discussion). Combination of the mutations within the α helix (ume6-6 or ume6-7) with the one that lies outside the α helix (ume6-9/K635E), creating the double mutations ume6-6,9 and ume6-7,9 results in a strong additive effect on SPO13-lacZ expression. Thus, the K635E mutation may affect the Ume6-dependent Sin3-independent pathway noted above.
FIG. 8.
Derepression of SPO13 expression in chromosomal ume mutants. Strains containing UME6, ume6Δ::KanMX, ume6-6, ume6-7, ume6-9, ume6-6,9 and ume6-7,9, or sin3Δ were constructed as described in Materials and Methods. (A) Plate assays of SPO13-URA3 and SPO13-lacZ expression. (B) Quantitative β-galactosidase (β-gal) assays of SPO13-lacZ expression of cells grown on plates, normalized to wild-type levels.
The Ume6-Sin3 interaction is not required for sporulation.
sin3 mutants were previously shown to be Spo−, and it was initially proposed that their failure to sporulate might be due to premature expression of the early genes (76). The results described above indicate that this explanation is unlikely to be correct. Since both sin3 and rpd3 mutants show a failure to rerepress the early genes later in meiosis (32), an alternative hypothesis is that the Spo− defect is due to the absence of repression, which may be necessary for meiotic progression. The process of restoring the early meiotic genes to the repressed state is expected to require binding of Sin3 to Ume6, presumably using the same mechanisms as employed for vegetative repression. Another possibility is that Sin3, like Ume6, is involved in activation of meiotic gene expression as well as repression. If so, this activation pathway may also involve interaction with Ume6.
To determine whether disrupting the interaction of Sin3 with Ume6 causes a Spo− phenotype, ume6Δ diploids transformed with plasmids containing wild-type UME6, ume6-6 (M530T), ume6-7 (L527P), or ume6-8 (S528P) were sporulated. As shown in Fig. 7, strains containing these ume6 α-helix mutants as the only available source of Ume6 (columns 3 to 6) sporulated normally. Since the disruption of Sin3 interaction has no effect on sporulation, these results suggest that the essential role of Sin3 in meiosis is independent of its interaction with Ume6. The ability of these mutants to sporulate also confirms the previous conclusion that premature expression of the early genes does not inhibit sporulation. Efficient binding of Sin3 to Ume6 is thus dispensable for sporulation.
DISCUSSION
This study addresses the mechanism by which Ume6 switches from a repressor to an activator of meiosis-specific genes and the role of Sin3-mediated repression in the regulation of meiosis by Ume6. Prior studies have shown that Ume6 binds to URS1 elements in the promoters of early meiotic genes (2, 77) and that UME6 and URS1 are both required for repression as well as activation (8, 9, 74, 77). Ume6-mediated activation is known to be dependent on interaction with Ime1 (65), which does not bind to DNA on its own but exhibits activation function when fused to LexA (72) or the Gal4 DNA- binding domain (56). It has therefore been proposed that one role of Ime1 is to provide an activation domain to the Ume6 complex (9, 65, 74). This model gained support from the finding that a GAD-UME6 fusion was able to cause Ime1-independent vegetative activation of the early meiotic gene HOP1 (65). We constructed a similar GAD-UME6 fusion in the course of our two-hybrid studies, using a weaker ADH1 promoter variant to alleviate the poor viability resulting from high-level overexpression of GAD-Ume6 from the stronger ADH1 promoter construct (see Materials and Methods) and found that it did not activate transcription of HOP1 or SPO13. For this fusion the repression activity provided by the Sin3 binding domain appeared to be dominant and in that respect similar to Sin3 interaction domains in other proteins (4, 30, 35, 42) as well as to direct fusions of activation domains to yeast Sin3, which fail to activate transcription (44). The only critical difference between the constructs in the two studies appears to be the promoters used to express the GAD-Ume6 fusions. The possibility that variations in the GAD-Ume6 protein sequence are responsible was excluded by demonstrating that the coding region from the wild-type Ume6 fusion used in this study can activate expression of SPO13 and HOP1 when moved into the expression plasmid used by Rubin-Bejerano et al. (not shown). We further determined that neither the strains or lacZ reporters used in the two studies can account for the different results, since the GAD-Ume6 fusion used in the previous study (65) activates the same reporter fusions and strains that fail to be activated by the GAD-Ume6 fusion used in this study (not shown). The analysis of ume6 mutants in this study indicates that the higher level of GAD-UME6 expression from the stronger ADH1 promoter variant used by Rubin-Bejerano et al. may exceed the steady-state levels of Sin3 available to bind to Ume6, thereby allowing activation.
Our finding that fusion of the Gal4 activation domain to Ume6 fails to activate early meiotic genes (e.g., SPO13 and HOP1) indicated that an additional function that cannot be substituted for by Gal4 is required. Here we provide evidence indicating that two genetically separable steps—relief of Sin3-mediated repression and addition of an activation domain—are involved in conversion of Ume6 to an activator. When ume6 mutations that disrupt Sin3-Ume6 interaction are introduced, GAD-Ume6 can function as a transcriptional activator of meiosis-specific genes. Furthermore, GAD-Ume6 under these conditions can substitute for the presence of Ime1 in meiosis.
The Sin3 binding domain in Ume6.
Prior two-hybrid and GST pulldown assays localized the Sin3 binding domain in Ume6 to the region between amino acid residues 508 and 584 (40). The mutant analysis described in this study has now allowed the domain to be precisely mapped to sequences within a predicted amphipathic helix encoded by amino acid residues 515 to 530. Although Sin3 was first identified in yeast (75, 89) and has mammalian homologs and binding partners that are currently the subject of extensive investigation, this is the first time a Sin3 interaction site in a yeast protein has been defined. Here we show that the Ume6 amphipathic helix that binds Sin3 is similar in structure, but not sequence, to the amino-terminal Sin3 binding domains of Mad1 and other mammalian Max-interacting bHLHZip proteins (reviewed in reference 57). These proteins also resemble Ume6 in that their repression domains interact with Sin3 PAH2 and are dominant with respect to linked activators (42, 57). At present, Sin3 PAH2 is also thought to interact with as many as six other yeast proteins (STB1 to STB6) (44), but none of these proteins appear to contain strong homologies to the Sin3 binding domain in Ume6. Therefore, if any of them interact with Sin3 PAH2 by a direct physical contact, they may do so by a mechanism that differs from that used by Ume6. Intriguingly, one yeast protein that shows significant homology to the Ume6 Sin3 binding domain is the DNA-binding protein Sum1. Six of eight consecutive amino acids within a predicted α helix in Sum1 are identical to the critical eight-amino-acid Sin3 binding sequence (residues 523 to 530) in the Ume6 α helix. Both Sum1 and Sin3 have been implicated in DNA silencing (12, 83), and both were identified as regulators of middle gene expression (32, 94). The possibility that Sum1, like Ume6, recruits Sin3 to repress transcription is currently being investigated. In higher eukaryotes, aside from the Max-interacting proteins, most of the proteins that appear to interact directly with Sin3 (including the Krüppel-like transcriptional repressors Laz3 [BCL6] and PLZF, the corepressors NCoR and SMRT, Sin3-associated polypeptides SAP18 and SAP30, and Rpd3 histone deacetylase homologs) do so through regions other than Sin3 PAH2 (15, 17, 49, 92, 95). Although the precise sequences through which most of these proteins make contacts with Sin3 are not yet known, current indications suggest that multiple mechanisms are involved.
Previous studies showed that amino acid residues 508 to 594 in Ume6 comprise the only region that exhibits significant interaction with Sin3 in vitro and that it is necessary and sufficient for Sin3 binding (40). The present analysis indicates that all but one of the mutations in UME6 that allow activation by the GAD-Ume6 fusion fall within this region. The one exception is located at residue 635, over 100 amino acids away from the predicted α helix. Since the mutation appears to have no effect on Ume6-Sin3 interaction, it may identify another repression site that is Sin3 independent (see below). For example, it may interfere with recruitment of the Isw2 chromatin remodeling complex, which has recently been shown to interact with Ume6 and repress transcription independently of Sin3 (27). Since the mutation alters a lysine residue and such residues in transcription factors are potential sites of ubiquitination (33) and acetylation (46), it may also prevent a crucial regulatory posttranslational modification or increase the stability of the protein.
What is the role of Ime1 in conversion of Ume6 to an activator?
Our analysis of GAD-Ume6 fusions has indicated that the conversion of Ume6 to an activator requires two steps that are genetically separable: (i) relief of Sin3-mediated repression and (ii) introduction of an activation domain. We propose that for wild-type Ume6, Ime1 is essential for both processes. With regard to the derepression step, there are two lines of evidence that support the idea that Sin3 remains present and functional in the nucleus when Ume6 is actively promoting transcription, and that it must be inactivated or physically removed from Ume6 by an Ime1-dependent mechanism for activation to occur. First, wild-type GAD-UME6 is unable to promote sporulation if Ime1 is absent but can do so if the Sin3 binding site is disrupted (Fig. 6). This indicates that the Sin3 protein is normally still present during meiosis and that if an activation domain is added to Ume6, Ime1 is dispensable only if repression is also removed (i.e., ume6-6). Second, it is well established that vegetative expression of Ime1 under control of the GAL1 promoter can cause activation of early meiotic genes in vegetative cells, when Sin3 normally functions in repression of transcription (73).
Two models for how Ime1 relieves Sin3 repression are that Ime1 either prevents Sin3 binding to Ume6 or interferes with Sin3 function in the complex. It has been hypothesized that due to its highly hydrophilic nature, the Ume6 protein is rather unstructured on its own and that interaction with other proteins such as Ime1 may have a dramatic impact on its structure (2). Thus, the addition of Ime1 may result in a change in the conformation of Ume6 and thereby directly eliminate Sin3 repression by either mechanism. Alternatively, Ime1 may act indirectly by promoting the synthesis or activity of other proteins that bind to and/or modify (e.g., phosphorylate) Ume6 and/or Sin3, thereby disrupting their interaction or function. At present, it is not clear whether Sin3 remains in the complex following Ime1 induction. Moreover, it is also not known whether indirect effects such as phosphorylation play a role in derepression. For example, while Ume6 is known to be phosphorylated (by Rim11 and/or Mck1 [93]), this phosphorylation does not appear to require Ime1 in vivo or in vitro (55, 93) and is thus unlikely to be involved in the Ime1-dependent derepression step.
The second function of Ime1, providing an activation domain to Ume6, has previously been described (9, 65, 74) and is supported by this work. We have found that this activation function is required in addition to derepression, since in the absence of either Ime1 or a heterologous activation domain, Ume6 cannot activate even when Sin3 binding is disrupted (e.g., in ume6-6 and GBD–ume6-6). This finding, together with the observation that GAD–ume6-6 can partially complement an ime1Δ in meiosis, provides strong evidence that Ime1 does indeed provide an activation domain as proposed (9, 65, 74).
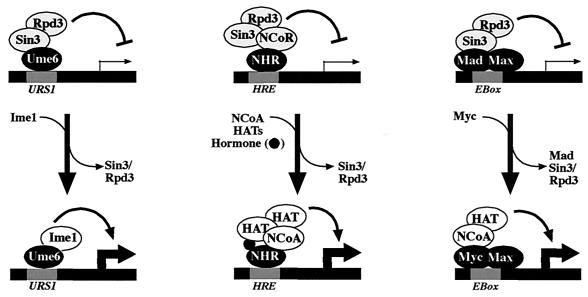
Based on these findings, the conversion of Ume6 from a repressor to an activator by a two-step process dependent on Ime1 is similar to regulation of metazoan Sin3-binding proteins that function in activation and repression, such as the nuclear hormone receptors (e.g., retinoic acid and thyroid hormone receptors) and the Myc/Mad/Max pathway (Fig. 9). For the thyroid hormone and retinoic acid receptors, ligand binding has been shown to result in a conformational change in the receptor and trigger a release of the histone deacetylase corepressor complex, which is replaced by a coactivator complex (53). For Myc/Mad/Max, a change in heterodimer partner occurs to replace the corepressor-binding Mad family protein with the coactivator-binding Myc protein (reviewed in references 31 and 82). In the case of Myc and the nuclear hormone receptors, the switch from repression to activation involves the recruitment of one or more histone acetylases (58). A role for the histone acetylase Gcn5 at the early meiotic promoter IME2 has been similarly demonstrated (11), although it is not known whether Gcn5 interacts with Ime1. Finally, certain oncogenic mutations in the thyroid hormone receptor (v-ErbA), as well as retinoic acid fusions (to PML and PLZF) implicated in the development of acute promyelocytic leukemia, retain the ability to recruit the Sin3 corepressor complex but cannot be converted to activators by physiological levels of hormone, thus blocking differentiation (7, 54). An unusual allele of ume6 identified as rim16-12 (9) behaves analogously. The point mutation (T99N) in rim16-12 interferes with Ime1 binding and conversion of Ume6 to an activator, thereby causing Ume6 to become a constitutive repressor of early meiotic genes, blocking meiotic development (9).
FIG. 9.
Transcriptional regulation by Ume6, nuclear hormone receptors, and Myc/Mad/Max. DNA-binding proteins (Ume6, nuclear hormone receptors [NHR], or Max heterodimer) in conjunction with cis-acting DNA elements (e.g., URS1, hormone response element [HRE], or E box, as indicated), repress transcription through interaction with a corepressor complex containing Sin3, Rpd3, and additional proteins (not all are shown). In each system, the switch from repression to activation requires that the Sin3 corepressor complex be replaced by an activation complex. In meiosis, this involves the binding of Ime1. For NHR, this involves binding of hormone and a number of potential coactivators (not all are shown) including histone acetylases (HAT) (see references 26, 31, and 82 for reviews). For Max, it involves heterodimerization with Myc, which possesses a transactivation domain that may act through a variety of additional proteins (14, 31, 58).
It should be noted that there are also a number of nonmeiotic genes in yeast that require Ume6 for their repression and/or activation (e.g., INO2 and PHR1 [37, 79]). Since Ime1 is not implicated in the regulation of these genes, there may be additional, as yet unidentified Ume6 partners similar to Ime1 that relieve Sin3-mediated repression. For example, ARGR1 and ARGRII, two activators of nitrogen-regulated genes recently shown to interact with Ume6 in two-hybrid assays (60), may act in this way. Likewise, Tea1, a transcriptional activator of Ty enhancer-mediated transcription that interacts with Ume6 (this study), may have a similar function.
UME6-dependent and UME6-independent roles of SIN3.
In the absence of the Gal4 activation domain, mutations in the potential amphipathic α helix (e.g., ume6-6 [M530T], ume6-7 [L527P], and ume6-8 [S528P]) result in a partial loss of repression to a level similar to a deletion of SIN3, as expected for Ume6 mutants that have lost the ability to interact with Sin3. Since the level of derepression that occurs in sin3Δ and the ume6 Sin3-binding-domain mutants is less than occurs in ume6Δ or URS1 mutants, we have concluded that Ume6 must have a repression function at early meiotic promoters that is independent of Sin3. A similar Sin3-independent repression function is seen at the nonmeiotic INO1 promoter, which shows a much higher level of derepression when URS1 or Ume6 is mutated than when SIN3 is deleted (20, 70). The repression conferred by Ume6 in the absence of Sin3 may result from either a Sin3-independent repression site in Ume6 (e.g., perhaps defined by ume6-9/K635E, the derepressing mutation that lay outside the α helix) or simply to the binding of Ume6 to URS1, excluding binding of other activators to URS1 or nearby sequences (21, 80). Ume6-dependent Sin3-independent repression probably does not involve Sin3-independent recruitment of Rpd3, since activity of Rpd3 at URS1 has been shown to require the presence of Sin3 (40, 85). Recent studies suggest that a likely mechanism for Sin3-independent repression of SPO13 by Ume6 may involve the Isw-2 chromatin remodeling complex, which has now been shown to bind to Ume6 and repress transcription independently of Sin3 (27).
Mutations in SIN3 and RPD3 confer a severe sporulation defect, causing arrest in pachytene after premeiotic S phase prior to the onset of middle gene expression (32; A. Helms and R. E. Esposito, unpublished data). Here we provide evidence that this sporulation defect is not a result of the premature or derepressed expression of early meiosis-specific genes that occurs in these mutants in the absence of IME1 induction. This is demonstrated by the sporulation proficiency of ume6-6, ume6-7, and ume6-8 Sin3-binding-domain mutants, which exhibit derepression similar to that of sin3Δ and rpd3Δ, and by the sporulation proficiency of wild-type diploids constitutively overexpressing the early genes due to the presence of GAD–ume6-6. The ability of the Ume6 Sin3-binding-domain mutants to sporulate at wild-type levels also suggests that repression of early meiotic gene expression is not essential for proper progression. These results indicate that the essential role of Sin3 in sporulation may be independent of its interaction with Ume6, which thus far is the only known DNA-binding partner for Sin3 in yeast. The effects of deletions of individual Sin3 PAH on sporulation support this view. Deletion of Sin3 PAH3 has the most severe effect on sporulation (90), rather than deletion of PAH2, which we now know to interact with Ume6. Since mammalian Rpd3 homologs bind Sin3 near PAH3 (92), recruitment of deacetylase activity by Sin3 and repression through proteins other than Ume6 may play a critical, as yet undefined role in sporulation. Two-hybrid studies have identified other potential DNA-binding proteins besides Ume6 that can interact with Sin3 (44). Determination of the targets of these potential transcription factors, together with recent whole-genome analysis of transcription in sin3Δ mutants (34), may allow the essential meiotic role of SIN3 to be identified.
ACKNOWLEDGMENTS
We thank members of the Esposito lab for helpful discussions and Y. Kassir, D. Stillman and A. Vershon for providing plasmids.
This work was supported by National Service Research Award F32GM15400 to B.K.W. and National Institutes of Health research grant GM29182 to R.E.E.
REFERENCES
- 1.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S F, Steber C M, Esposito R E, Coleman J E. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 1995;4:1832–1843. doi: 10.1002/pro.5560040918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 4.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 6.Bartel P L, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 7.Bauer A, Mikulits W, Lagger G, Stengl G, Brosch G, Beug H. The thyroid hormone receptor functions as a ligand-operated developmental switch between proliferation and differentiation of erythroid progenitors. EMBO J. 1998;17:4291–4303. doi: 10.1093/emboj/17.15.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowdish K S, Mitchell A P. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowdish K S, Yuan H E, Mitchell A P. Positive control of yeast meiotic genes by the negative regulator UME6. Mol Cell Biol. 1995;15:2955–2961. doi: 10.1128/mcb.15.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckingham L E, Wang H T, Elder R T, McCarroll R M, Slater M R, Esposito R E. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:9406–9410. doi: 10.1073/pnas.87.23.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgess S M, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci USA. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi M H, Shore D. SUM1–1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol Cell Biol. 1996;16:4281–4294. doi: 10.1128/mcb.16.8.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 14.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David G, Alland L, Hong S H, Wong C W, DePinho R A, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 16.Deleage G, Roux B. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 17.Dhordain P, Lin R J, Quief S, Lantoine D, Kerckaert J, Evans R M, Albagli O. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eilers A L, Billin A N, Liu J, Ayer D E. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J Biol Chem. 1999;274:32750–32756. doi: 10.1074/jbc.274.46.32750. [DOI] [PubMed] [Google Scholar]
- 19.Einerhand A W, Kos W, Smart W C, Kal A J, Tabak H F, Cooper T G. The upstream region of the FOX3 gene encoding peroxisomal 3-oxoacyl-coenzyme A thiolase in Saccharomyces cerevisiae contains ABF1- and replication protein A-binding sites that participate in its regulation by glucose repression. Mol Cell Biol. 1995;15:3405–3414. doi: 10.1128/mcb.15.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkhaimi M, Kaadige M R, Kamath D, Jackson J C, Biliran H, Jr, Lopes J M. Combinatorial regulation of phospholipid biosynthetic gene expression by the UME6, SIN3 and RPD3 genes. Nucleic Acids Res. 2000;28:3160–3167. doi: 10.1093/nar/28.16.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gailus-Durner V, Xie J, Chintamaneni C, Vershon A K. Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol Cell Biol. 1996;16:2777–2786. doi: 10.1128/mcb.16.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 23.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 24.Gibrat J F, Garnier J, Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 25.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 26.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 27.Goldmark J P, Fazzio T G, Estep P W, Church G M, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 28.Gray W M, Fassler J S. Isolation and analysis of the yeast TEA1 gene, which encodes a zinc cluster Ty enhancer-binding protein. Mol Cell Biol. 1996;16:347–358. doi: 10.1128/mcb.16.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 30.Harper S E, Qiu Y, Sharp P A. Sin3 corepressor function in Myc-induced transcription and transformation. Proc Natl Acad Sci USA. 1996;93:8536–8540. doi: 10.1073/pnas.93.16.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassig C A, Schreiber S L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 32.Hepworth S R, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 34.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, Kidd M J, King A M, Meyer M R, Slade D, Lum P Y, Stepaniants S B, Shoemaker D D, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend S H. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 35.Hurlin P J, Queva C, Eisenman R N. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 36.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson J C, Lopes J M. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 1996;24:1322–1329. doi: 10.1093/nar/24.7.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 41.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasten M M, Ayer D E, Stillman D J. SIN3-dependent transcriptional repression by interaction with the Mad1 DNA-binding protein. Mol Cell Biol. 1996;16:4215–4221. doi: 10.1128/mcb.16.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasten M M, Stillman D J. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol Gen Genet. 1997;256:376–386. doi: 10.1007/s004380050581. [DOI] [PubMed] [Google Scholar]
- 45.Klapholz S, Esposito R E. Recombination and chromosome segregation during the single division meiosis in SPO12–1 and SPO13–1 diploids. Genetics. 1980;96:589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kratzer S, Schuller H J. Transcriptional control of the yeast acetyl-CoA synthetase gene, ACS1, by the positive regulators CAT8 and ADR1 and the pleiotropic repressor UME6. Mol Microbiol. 1997;26:631–641. doi: 10.1046/j.1365-2958.1997.5611937.x. [DOI] [PubMed] [Google Scholar]
- 48.Kupiec M, Byers B, Esposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 49.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, Rosenfeld M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 50.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 51.Legrain P, Dokhelar M C, Transy C. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levin J M, Robson B, Garnier J. An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 53.Lin B C, Hong S H, Krig S, Yoh S M, Privalsky M L. A conformational switch in nuclear hormone receptors is involved in coupling hormone binding to corepressor release. Mol Cell Biol. 1997;17:6131–6138. doi: 10.1128/mcb.17.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin R J, Egan D A, Evans R M. Molecular genetics of acute promyelocytic leukemia. Trends Genet. 1999;15:179–184. doi: 10.1016/s0168-9525(99)01710-2. [DOI] [PubMed] [Google Scholar]
- 55.Malathi K, Xiao Y, Mitchell A P. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandel S, Robzyk K, Kassir Y. IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev Genet. 1994;15:139–147. doi: 10.1002/dvg.1020150204. [DOI] [PubMed] [Google Scholar]
- 57.McArthur G A, Laherty C D, Queva C, Hurlin P J, Loo L, James L, Grandori C, Gallant P, Shiio Y, Hokanson W C, Bush A C, Cheng P F, Lawrence Q A, Pulverer B, Koskinen P J, Foley K P, Ayer D E, Eisenman R N. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harbor Symp Quant Biol. 1998;63:423–433. doi: 10.1101/sqb.1998.63.423. [DOI] [PubMed] [Google Scholar]
- 58.McMahon S B, Wood M A, Cole M D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro C L, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Messenguy F, Vierendeels F, Scherens B, Dubois E. In Saccharomyces cerevisiae, expression of arginine catabolic genes CAR1 and CAR2 in response to exogenous nitrogen availability is mediated by the Ume6 (CargRI)-Sin3 (CargRII)-Rpd3 (CargRIII) complex. J Bacteriol. 2000;182:3158–3164. doi: 10.1128/jb.182.11.3158-3164.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 63.Primig M, Williams R M, Winzeler E A, Tevzadze G G, Conway A R, Hwang S Y, Davis R W, Esposito R E. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 64.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 65.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 67.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 68.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 69.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slekar K H, Henry S A. SIN3 works through two different promoter elements to regulate INO1 gene expression in yeast. Nucleic Acids Res. 1995;23:1964–1969. doi: 10.1093/nar/23.11.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smart W C, Coffman J A, Cooper T G. Combinatorial regulation of the Saccharomyces cerevisiae CAR1 (arginase) promoter in response to multiple environmental signals. Mol Cell Biol. 1996;16:5876–5887. doi: 10.1128/mcb.16.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith H E, Driscoll S E, Sia R A, Yuan H E, Mitchell A P. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics. 1993;133:775–784. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith H E, Su S S, Neigeborn L, Driscoll S E, Mitchell A P. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steber C M, Esposito R E. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc Natl Acad Sci USA. 1995;92:12490–12494. doi: 10.1073/pnas.92.26.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sternberg P W, Stern M J, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 76.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 78.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 79.Sweet D H, Jang Y K, Sancar G B. Role of UME6 in transcriptional regulation of a DNA repair gene in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6223–6235. doi: 10.1128/mcb.17.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szent-Gyorgyi C. A bipartite operator interacts with a heat shock element to mediate early meiotic induction of Saccharomyces cerevisiae HSP82. Mol Cell Biol. 1995;15:6754–6769. doi: 10.1128/mcb.15.12.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 82.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 83.Vannier D, Balderes D, Shore D. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics. 1996;144:1343–1353. doi: 10.1093/genetics/144.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vershon A K, Pierce M. Transcriptional regulation of meiosis in yeast. Curr Opin Cell Biol. 2000;12:334–339. doi: 10.1016/s0955-0674(00)00104-6. [DOI] [PubMed] [Google Scholar]
- 85.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 88.Wang H, Clark I, Nicholson P R, Herskowitz I, Stillman D J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang H, Nicholson P R, Stillman D J. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol Cell Biol. 1990;10:1743–1753. doi: 10.1128/mcb.10.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang H, Stillman D J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Werner-Washburne M, Braun E L, Crawford M E, Peck V M. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 92.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 93.Xiao Y, Mitchell A P. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol Cell Biol. 2000;20:5447–5453. doi: 10.1128/mcb.20.15.5447-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon A K. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]