Abstract
The epidermal growth factor receptor (EGF-R) plays an important role in the growth and progression of estrogen receptor-negative human breast cancers. EGF binds with high affinity to the EGF-R and activates a variety of second messenger pathways that affect cellular proliferation. However, the underlying mechanisms involved in the regulation of EGF-R expression in breast cancer cells are yet to be described. Here we show that the EGF-induced upregulation of EGF-R mRNA in two human breast cancer cell lines that overexpress EGF-R (MDA-MB-468 and BT-20) is accompanied by stabilization (>2-fold) of EGF-R mRNA. Transient transfections using a luciferase reporter identified a novel EGF-regulated ∼260-nucleotide (nt) cis-acting element in the 3′ untranslated region (3′-UTR) of EGF-R mRNA. This cis element contains two distinct AU-rich sequences (∼75 nt), EGF-R1A with two AUUUA pentamers and EGF-R2A with two AUUUUUA extended pentamers. Each independently regulated the mRNA stability of the heterologous reporter. Analysis of mutants of the EGF-R2A AU-rich sequence demonstrated a role for the 3′ extended pentamer in regulating basal turnover. RNA gel shift analysis identified cytoplasmic proteins (∼55 to 80 kDa) from breast cancer cells that bound specifically to the EGF-R1A and EGF-R2A cis-acting elements and whose binding activity was rapidly downregulated by EGF and phorbol esters. RNA gel shift analysis of EGF-R2A mutants identified a role for the 3′ extended AU pentamer, but not the 5′ extended pentamer, in binding proteins. These EGF-R mRNA-binding proteins were present in multiple human breast and prostate cancer cell lines. In summary, these data demonstrate a central role for mRNA stabilization in the control of EGF-R gene expression in breast cancer cells. EGF-R mRNA contains a novel complex AU-rich 260-nt cis-acting destabilizing element in the 3′-UTR that is bound by specific and EGF-regulated trans-acting factors. Furthermore, the 3′ extended AU pentamer of EGF-R2A plays a central role in regulating EGF-R mRNA stability and the binding of specific RNA-binding proteins. These findings suggest that regulated RNA-protein interactions involving this novel cis-acting element will be a major determinant of EGF-R mRNA stability.
The epidermal growth factor (EGF) receptor (EGF-R) is a protein tyrosine kinase receptor that is the target for high-affinity ligands, such as EGF and transforming growth factor alpha. EGF-R shares extensive homology with the erbB oncogene product of the avian erythroblastosis virus (12, 17, 46, 64), and overproduction and/or amplification of EGF-R has been detected in several different types of human cancers (12, 39, 40, 46, 47, 63), including breast cancer (20, 21, 30, 53). Extensive studies have shown that breast tumors which overexpress EGF-R are most often estrogen receptor (ER) negative and have a poor prognosis (57, 59). Despite the clinical importance of the EGF-R in human breast cancer, the molecular mechanisms governing the control of EGF-R gene expression in these cells remains to be fully elucidated.
MDA-MB-468 and BT-20 estrogen-unresponsive breast cancer cell lines have similar phenotypes and overexpress EGF-R to similar degrees (1 × 106 to 2 × 106 binding sites per cell) (29, 32). Treatment of MDA-MB-468 cells with EGF increases EGF-R mRNA and protein levels (3, 32), due in part to a moderate transcriptional upregulation (19). However, the vast proportion of the EGF-induced increase in EGF-R mRNA levels in MDA-MB-468 cells cannot be accounted for by transcriptional enhancement. This suggests that posttranscriptional events must play a significant role in the regulation of EGF-R mRNA levels in these cells. Little is known of the regulation of EGF-R expression in BT-20 cells.
The regulation of mRNA decay is a central mechanism in the control of gene expression (51). Specific cis-acting structural RNA motifs can confer instability to mRNAs under appropriate conditions. A major class of these regulatory cis elements comprises adenosine-uridine pentamers (AUUUA) which are termed AU-rich elements (AREs). Shaw and Kamen initially observed that an ARE in the 3′ untranslated region (3′-UTR) of granulocyte-macrophage colony-stimulating factor (GM-CSF) mRNA could stimulate the degradation of the normally stable β-globin mRNA, reducing the half-life from many hours to less than 30 min (56). Similarly, the 3′-UTR of c-fos, which contains a 69-nucleotide (nt) ARE, reduced the stability of β-globin mRNA (8, 9). AREs that function as RNA-destabilizing elements and target the mRNA for rapid degradation in the cytoplasm have been found in numerous mRNAs, including c-myc, junB, nur77, beta interferon, various interleukin (IL-1α, IL-2, and IL-3), and tumor necrosis factor (TNF) mRNAs (8, 9, 10, 13, 24, 31, 51, 56). In vivo, mice lacking AREs in their TNF gene have defective destabilization and translational regulation of TNF mRNA and also develop specific phenotypes, suggesting a potential etiopathological role for the ARE (31).
Typically the ARE contains an AUUUA pentamer, repeated once or several times within the 3′-UTR. It is often found within a U-rich region of the mRNA. Recent work has suggested an ARE containing the nonamer UUAUUUA (U/A) (U/A) is more indicative of rapid destabilization (34, 68). Variations of the nonamer, such as extended pentamers (AUUUUA and AUUUUUA), are present in many complex AREs and have been identified as putative binding targets by a random RNA selection procedure (37). However, little is known of the role of isolated extended AU pentamers in the regulation of specific mRNAs in vivo. This is particularly relevant, as it is the combination of functionally and structurally distinct sequence motifs, such as AU pentamers, nonamers, extended AU pentamers, and U-rich stretches, that determines the ultimate destabilizing ability of each particular ARE.
A family of proteins that bind to AU-rich, and sometimes U-rich, RNAs with high affinity has been characterized by RNA electrophoretic mobility shift assay (REMSA). In many cases, binding of these AU-rich region-binding proteins (AUBPs; 30 to 45 kDa) regulates the turnover of ARE-containing mRNAs (15, 27). One of the best-characterized AUBPs is HuR, a 36-kDa member of the elav family of RRM (RNA recognition motif)-containing RNA-binding proteins (27, 37). HuR binds with high affinity to ARE sequences (27, 37) and plays an active role in the stabilization of specific mRNAs containing AREs, such as GLUT1, c-fos, GM-CSF, plasminogen activator inhibitor type 2, and p21WAF1 mRNAs (18, 22, 26, 27, 38, 44). AUF1 is another well-characterized RNA-binding protein that binds to AREs, in particular that of c-myc (6, 16, 61, 67). AUF1 has been implicated in the regulation of many cytokine and G protein-coupled receptor mRNAs (61) and plays a major role in development (33). Interestingly, the binding of some AUBPs to AREs is regulated by activators of protein kinase C, such as phorbol esters (phorbol 12-myristate 13-acetate [PMA]) (4).
The EGF-R mRNA contains four separate AU-rich sequences in the 3′-UTR. Interestingly, A431 epidermoid cancer cells express both the full-length EGF-R mRNA and a truncated EGF-R mRNA which lacks the 3′-UTR. The truncated EGF-R mRNA transcript is more stable than the full-length transcript (28), suggesting that one or more of the AU-rich sequences in the EGF-R mRNA 3′-UTR may contribute to basal and possibly regulated changes in EGF-R mRNA stability. We used breast cancer cells (MDA-MB-468 and BT-20) to determine the contribution of mRNA stability to the regulation of EGF-R expression. In both cell lines, EGF stabilized EGF-R mRNA >2-fold. Transfection experiments identified a novel 260-nucleotide (nt) AU-rich (66%) cis element in the 3′-UTR of EGF-R mRNA that contained two ∼75-nt AU-rich destabilizing sequences (EGF-R1A and EGF-R2A). trans-acting protein factors (55 to 80 kDa) that targeted the cis element and whose binding affinity was regulated by EGF and phorbol esters were identified. Mutational analysis demonstrated an important role for the extended AU pentamer at the 3′ end of the cis element in regulating EGF-R mRNA stability. These findings demonstrate a central role for mRNA turnover in the regulation of EGF-R expression in breast cancer cells and illustrate that complex cis element RNA-protein interactions contribute to basal and regulated EGF-R mRNA decay.
MATERIALS AND METHODS
Cells and culture conditions.
Cell lines were obtained from American Type Culture Collection (ATCC; Manassas, Va.). MDA-MB-468 (HTB 132) and HT1080 (CCL 121) cells were routinely cultured in Dulbecco's modified Eagle's medium-F12 medium supplemented with 10% fetal calf serum (FCS; Gibco-BRL, Melbourne, Australia). BT-20 (HTB 19), LNCaP (CRL 1740), SK-BR-3 (HTB 30), and MDA-MB-231 (HTB 26) cell lines were grown in RPMI 1640 supplemented with 10% FCS. MCF7 (HTB 22) cells were cultured in RPMI 1640 supplemented with 5% FCS. All cell lines were cultured in the presence of penicillin (100 U/ml) and streptomycin (100 mg/ml). Cells were grown to 70 to 80% confluence and within eight passages of the original stock received from ATCC for all experiments.
Plasmid clones, cDNA probes, and RNA transcripts.
The EGF-R plasmid cDNA contained the entire coding region of EGF-R and the first 131 nt of the 3′-UTR in pBluescript II KS (pBlue) (14). For Northern blots, a 1-kb BglII/HindIII fragment from the EGF-R coding region was random-prime labeled using [32P]dCTP (approximately 3,000 Ci/mmol; Amersham, Sydney, Australia). A 1.1-kb 18S rRNA probe was used as a loading control. BglII, HindIII, and BamHI fragments of EGF-R cDNA were subcloned into the HpaI site of the Rous sarcoma virus-luciferase (RSV-Luc) expression vector (Fig. 1B). Additional sequences in the 3′-UTR were amplified from MDA-MB-468 cell total RNA using reverse transcriptase (RT)-mediated PCR (RT-PCR) and cloned into RSV-Luc (Fig. 1B). The c-fos AU-rich element which generates an unstable mRNA was also cloned into RSV-Luc and used in transfections (68). A plasmid encoding RSV–β-galactosidase (RSV-β-Gal; ATCC) was cotransfected as an internal control. The authenticity of all constructs used for transfections was verified by sequencing.
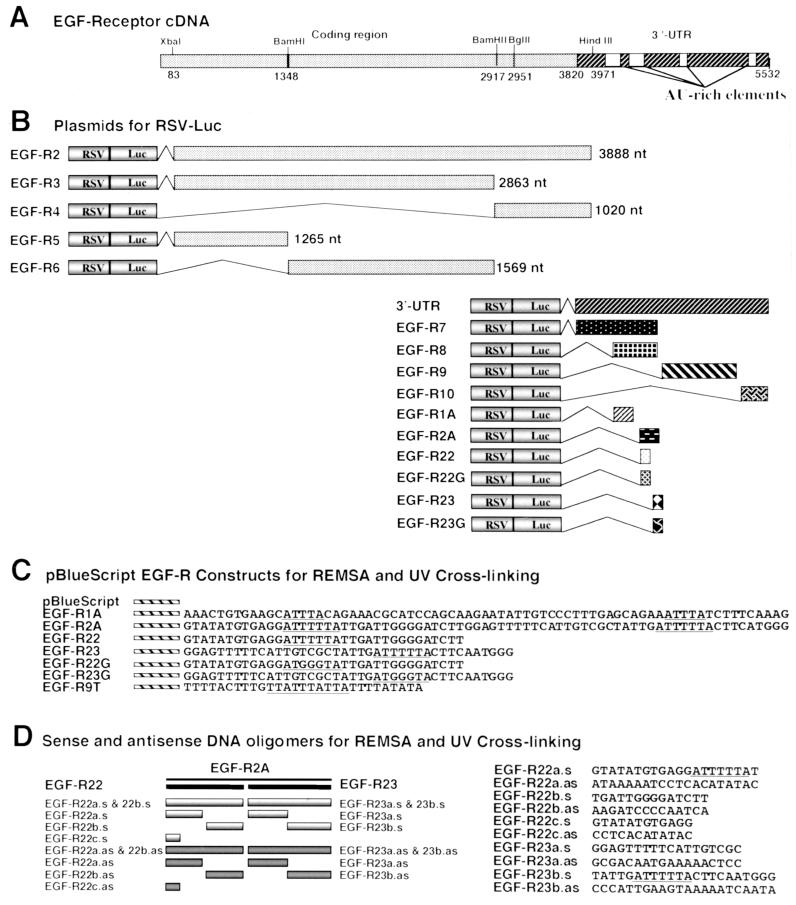
FIG. 1.
Schematic of EGF-R cDNA, RSV-Luc/EGF-R chimeric plasmid constructs, pBlue EGF-R constructs, and DNA oligomers. (A) Schematic representation of the human EGF-R cDNA indicating restriction enzyme sites and AU-rich 3′-UTR sequences. (B) RSV-Luc/EGF-R constructs generated for transfections by subcloning various portions of the EGF-R into the HpaI site in the RSV-Luc expression vector. The length of each EGF-R insert is indicated in nucleotides. EGF-R1A, nt 4017 to 4089; EGF-R2A, nt 4116 to 4190; EGF-R22, nt 4116 to 4153; EGF-R23, nt 4154 to 4190. (C) pBlue EGF-R constructs containing specific sequences of the 3′-UTR of the EGF-R cDNA. The AU-rich elements within the 3′-UTR are underlined. EGF-R1A, EGF-R2A, EGF-R22, and EGF-R23 are clones derived from the same portions of the 3′-UTR as for the RSV-Luc/EGF-R constructs in panel B. EGF-R22G and EGF-R23G are mutant clones in which three thymidine nucleotides in the AU-extended pentamers have been replaced by guanine (underlined). EGF-R9T contains a consensus nonamer AU-rich sequence. (D) Schematic of EGF-R2A showing location and orientation of competitor oligonucleotides. EGF-R22a.s and EGF-R22a.as represent oligonucleotides spanning the 5′ 20 nt of EGF-R22 in the sense and antisense orientations, respectively; EGF-R22b.s and EGF-R22b.as represent oligonucleotides to the remaining 14 nt of EGF-R22; EGF-R22c.s and EGF-R22c.as represent the first 12 nt of EGF-R22 (similarly for EGF-R23a.s, EGF-R23a.as, EGF-R23b.s, and EGF-R23b.as).
pBlue clones containing EGF-R fragments (Fig. 1C) were constructed by subcloning specific sequences of the 3′-UTR of the EGF-R cDNA into the BamHI and HindIII sites of pBlue. To generate riboprobes, all subsequent pBlue/EGF-R plasmids were linearized with HindIII for transcription with T7 RNA polymerase. pBlue was linearized with BamHI to produce a 66-nt transcript. EGF-R1A and EGF-R2A are 74- and 73-nt fragments of the 3′-UTR. EGF-R22 and EGF-R23 are two smaller fragments containing 34 and 40 nt, respectively, of EGF-R2A (Fig. 1C). EGF-R22G and EGF-R23G are equivalent to EGF-R22 and EGF-R23, respectively, but contain mutations in the AU-extended pentamer (Fig. 1C). Smaller sense and antisense DNA oligonucleotides of ∼14 nt were used as unlabeled competitors in REMSA and UV cross-linking (UVXL) assays (Fig. 1D). A consensus nonamer sequence, 9SWT (EGF-R9T) (Fig. 1C), was also used in REMSA. The sequence of each plasmid construct was confirmed by dideoxy sequencing.
Linearized RNA templates were transcribed with T7 RNA polymerase (Gibco) in reactions containing [32P]UTP (37 Ci/mmol; Amersham), as described elsewhere (58), to produce transcripts with a specific activity of approximately 0.5 × 1010 cpm/mg of RNA. Unlabeled RNA transcripts were synthesized as above but with 2.5 mM ribonucleoside triphosphates (rNTPs) and quantified by spectrophotometry.
RNA isolation and northern analysis.
Cells were solubilized in 4 M guanidinium isothiocyanate, and total RNA was isolated by the method of Chomczynski and Sacchi (11). RNA (10 to 15 μg per sample) was size fractionated on a 1% agarose-formaldehyde gel and transferred to a Hybond-N+ membrane (Amersham). RNA was UV cross-linked to the membrane, which was prehybridized for 4 h at 42°C in a buffer containing 50% formamide, 0.75 M NaCl, 0.075 M sodium citrate (pH 7.0), 5× Denhardt's solution, 1% sodium dodecyl sulfate (SDS), and 200 mg of salmon sperm DNA/ml and then hybridized in the same buffer overnight at 42°C with 32P-labeled EGF-R cDNA probe at 106 cpm/ml. The membrane was washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS and then with 0.2× SSC containing 0.1% SDS at 22°C. Membranes were analyzed by autoradiography using Kodak EM-1 film, imaged with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), and quantified using ImageQuant software. In all experiments, an 18S rRNA probe was used for normalization.
mRNA turnover studies.
Cells were grown to 70 to 80% confluence and not treated or treated with EGF (25 ng/ml, 4 nM; Promega, Madison, Wis.) for 8 h, followed by addition of the transcription inhibitor actinomycin D (Act D; 7.5 mg/ml; Sigma, St. Louis, Mo.). Total RNA was isolated from the cells at 0-, 2-, 4-, 8-, and 12-h time intervals after addition of Act D and subjected to Northern analysis as described earlier. EGF-R mRNA half-life was determined by linear regression analysis.
Nuclear run-on transcription assay.
Exponentially growing MDA-MB-468 and BT-20 cells were treated with EGF (25 ng/ml, 4 nM) for 6 to 8 h. Nuclei were isolated as described previously (3), rapidly frozen, and stored at −85°C. The transcription assay was performed as described previously (65). Briefly, for the transcription assay, the nuclei were thawed on ice, resuspended in 100 μl of reaction buffer (10 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 300 mM KCl, 5 mM dithiothreitol, 0.5 mM each ATP, CTP, and GTP, 100 μCi of [32P]UTP [800 Ci/mmol; NEN-Du Pont]), and incubated at 30°C for 30 min. Labeled RNA was isolated and hybridized to nitrocellulose filters onto which 5 μg of EGF-R and 18S cDNAs had been blotted. Filters were washed, analyzed by autoradiography, and quantified using a PhosphorImager.
Immunoblot assay for EGF-R protein.
Cells were treated with EGF (25 ng/ml, 4 nM) for 8 h, harvested, and lysed in ice-cold lysis buffer (1% Triton X-100, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA). After 10 min on ice, the lysate was centrifuged at 750 × g (Eppendorf model 5415C centrifuge) for 10 min at 4°C, after which the supernatant was recovered and stored at −85°C. Total protein concentration of the lysate was determined using the Bio-Rad protein assay; the proteins (5 to 10 μg/lane) were separated by polyacrylamide gel electrophoresis (PAGE) on SDS–6% polyacrylamide gels, and subsequently transferred to nitrocellulose. The membrane was blocked with 5% nonfat dried milk in TBS-T (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) at 22°C for 1 h prior to incubation with an EGF-R polyclonal antibody (1:2,000; Upstate Biotechnology, Lake Placid, N.Y.) for 1 to 2 h at 22°C and horseradish peroxidase-conjugated anti-sheep goat immunoglobulin G (1:2,000; Amersham); the EGF-R protein was visualized by ECL (Amersham) and subjected to autoradiography. The blots were also probed with a human polyclonal actin antibody (1:500; Santa Cruz Biotechnology, Santa Cruz, Calif.) as a control for loading. The 170-kDa EGF-R protein band was quantified using a Kodak DCS-420c digital camera and ImageQuant software.
Transfection, Luc, and β-Gal assays.
MDA-MB-468 cells (70 to 80% confluent) were transfected by electroporation with 10 μg of RSV-Luc or RSV-Luc EGF-R and 6 μg of RSV-β-gal as a control. After electroporation, cells were cultured in medium in the presence or absence of EGF (25 ng/ml) for 6 h prior to lysate extraction and assays for luciferase (Luc) and β-galactosidase (β-Gal) activity. Lysates were prepared by harvesting the cells from the plate in phosphate-buffered saline (PBS). The mixture was centrifuged at 450 × g (Jouan model C3-12 centrifuge) for 5 min, the supernatant was removed, and the cell pellet was resuspended in 250 μl of lysis buffer (125 mM Tris [pH 7.6], 0.5% Triton X-100). The solution was centrifuged at 16,500 × g (Eppendorf model 5415C centrifuge) for 10 min at 4°C, and the supernatant was used in the Luc assay. For each Luc assay, 50 μl of the cell lysate was used with 250 μl of assay buffer (25 mM glycylglycine [pH 7.8], 15 mM MgSO4). Samples in triplicate were analyzed in an automated Berthold luminometer, with 100 μl of luciferin mixture (containing luciferin [50 mg/ml; Promega], 5 mM ATP, and assay buffer) added to each sample. β-Gal activity was determined for each extract as described elsewhere (52).
LightCycler PCR assay.
MDA-MB-468 cells grown to ∼80% confluence were transfected using Fugene (Roche, Indianapolis, Ind.) with RSV-Luc, EGF-R1A, or EGF-R2A plasmid and with β-globin as an internal cotransfection control. Total RNA was isolated from the cells, and 5 μg was reverse transcribed using avian myeloblastosis virus RT (Promega) in a mixture containing 5 mM MgCl2, 1 mM dNTPs, 1 U of RNasin/ml, 10 U of avian myeloblastosis virus RT, and 50 ng of oligo(dT) for 60 min at 42°C. The cDNA was then used in a real-time quantitation assay using a LightCycler (Roche Molecular Dynamics) (62). A total reaction volume of 20 μl contained 2 μl of cDNA from the RT reaction, 2 μl (10 pmol/μl) of Luc antisense and 2 μl (10 pmol/μl) of Luc sense primers, 1 μl of 40 mM dNTPs, 2 μl of 10× PCR buffer, 1 μl of bovine serum albumin (25 mg/ml), 2 μl of 5× Sybr green (Sigma), 1 μl of Taq polymerase (AmpliTaq; 1U/μl; Perkin-Elmer), and 7 μl of water. The Luc primers were 5′ TAC TGG GAC GAA GAC GAA CAC 3′ (sense) and 5′ CAC GCC CGC GTC GAA GAT GTT 3′ (antisense); the β-globin primers were 5′ GAG TCC TTT GGG GAC CTG TCC 3′ (sense) and 5′ GAA GTT CTC AGG ATC CAC GTG 3′ (antisense).
The LightCycler PCR comprised a 2-min denaturation using AmpliTaq followed by 40 cycles of 95°C for 1 s, 60°C for 5 s, and 72°C for 15 s. A melting curve, which is defined as the temperature at which 50% of the DNA becomes single stranded, was determined in all assays. Conditions for the melting curve were 95°C for 1 s and 65°C for 10 s for one cycle. Standards for each LightCycler assay consisted of an RT sample with serial dilutions (1:10) up to 1:1,000.
Preparation of cytoplasmic extracts.
MDA-MB-468 cells were grown to 70 to 80% confluence in 100-mm-diameter culture dishes. Cytoplasmic extracts were prepared as described elsewhere (58). Medium was replenished 12 to 24 h prior to ligand treatment with EGF (25 ng/ml) or PMA (50 ng/ml). Cells were scraped from the culture dishes with chilled PBS and centrifuged for 4 min at 450 × g (Jouan model C3-12 centrifuge) at 4°C; the supernatant was discarded. The cells were washed with cold PBS and centrifuged at 450 × g (Jouan model C3-12 centrifuge) for 4 min. Cell pellets were incubated with cytoplasmic extract buffer (10 mM HEPES, 3 mM MgCl2, 40 mM KCl, 5% glycerol, 0.2% NP-40, 1 mM dithiothreitol), with freshly added protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 2 μg of aprotinin/ml [Roche]), for 20 min and then centrifuged for 2 min at 12,100 × g (Eppendorf model 5415C centrifuge) and 4°C; the supernatant was snap-frozen in liquid nitrogen. Extracts from MDA-MB-231, MCF7, HT1080, SK-BR-3, and LNCaP cells were all processed in a similar manner. The HT1080 fibrosarcoma line required extra steps for cell lysis: heating at 37°C for 5 min followed by snap-freezing in liquid nitrogen for 2 min, repeated five times. Protein concentrations were determined using a Bio-Rad protein assay kit.
REMSA.
Binding reactions were performed as described elsewhere (35, 36, 58) with 5 μg of cytoplasmic extract and 105 cpm of RNA (∼10 to 20 pg). Briefly, following incubation at 22°C for 30 min, 0.3 U of RNase T1 (Roche) was added for 10 min, followed by the addition of heparin (final concentration, 50 mg/ml; Sigma) for 10 min. Samples were subjected to electrophoresis on a 4% native acrylamide gel (acrylamide/bisacrylamide ratio of 36:1), dried, and analyzed with a PhosphorImager followed by autoradiography as described elsewhere (58).
In RNA competition assays, excess (50- to 150-fold) unlabeled sense RNA transcript (e.g., EGF-R or pBlue) or single-base RNA homopolymer [poly(U), poly(C), or poly(A); Amersham Pharmacia Biotech, Sydney, Australia] was preincubated with the extract for 30 min at 22°C prior to incubation with the labeled probe as described above. In other competition assays, sense or antisense unlabeled DNA oligonucleotides were mixed with the 32P-riboprobe at 70°C for 10 min and renatured for 1 h at 22°C; then 5 μg of protein cell extract was added, the mixture was incubated for 30 min, and RNase T1 and heparin were added, as described above. In some assays, antibodies to specific RNA-binding proteins were added (as described in reference 44) in an effort to supershift RNA-protein complexes.
UVXL of RNA-protein complexes.
RNA-protein binding reactions were carried out as described above, using 20 to 30 μg of cytoplasmic extract and 1.5 × 105 cpm of RNA (15 to 30 pg) of 32P-riboprobe (35, 55, 58). Following the addition of heparin, samples were placed on ice in a microtiter tray and UV irradiated for 10 to 15 min, 1 cm below the Stratalinker UV light source (240-nm UV bulb; Stratagene, La Jolla, Calif.). Samples were then incubated with RNase A (100 mg/ml; Roche) at 37°C for 15 min. The samples were boiled for 3 min in SDS sample buffer; RNA-protein complexes were separated by SDS-PAGE on 8 to 10% gels and analyzed by autoradiography. 14C-labeled Rainbow molecular weight markers (Amersham) were used as standards.
Secondary structure prediction of EGF-R mRNA.
Nucleic acid sequence was obtained from GenBank accession number X00588, and stem-loop structure prediction plots were modeled using foldRNA and Squiggles programs (webANGIS) on a Macintosh workstation.
Statistical analysis.
Statistical analysis was performed using General Linear Models. Analysis of variance was performed to assess the significance of treatment (EGF) over different time intervals from Act D chase assays. Where appropriate, means comparisons at specific time points were made using Student's t test. A P value less than or equal to 0.05 was considered statistically significant.
RESULTS
EGF increases EGF-R mRNA and protein levels in MDA-MB-468 and BT-20 cells.
In preliminary experiments, we evaluated the biological potency of EGF by examining levels of phosphotyrosine activation of the EGF-R. Incubation of MDA-MB-468 cells for 15 min with EGF at 12.5 or 25 ng/ml (∼4 nM) induced substantial EGF-R tyrosine phosphorylation (data not shown), so that in subsequent experiments we used either 12.5 or 25 ng of EGF/ml.
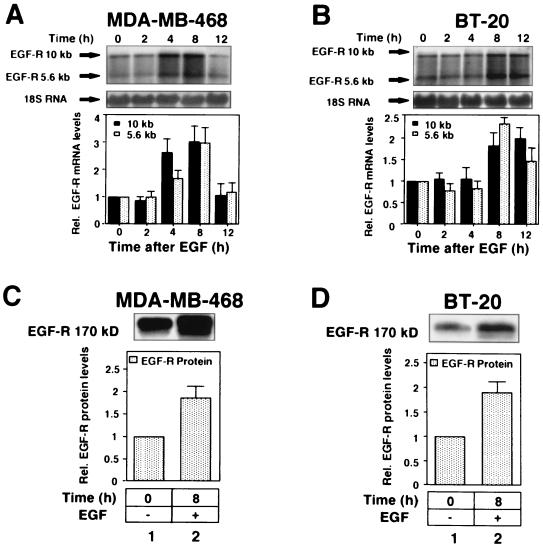
We next evaluated the effect of EGF on endogenous EGF-R mRNA in MDA-MB-468 and BT-20 cells using Northern analysis and an EGF-R-specific cDNA probe. Preliminary experiments determined that there was no difference in response to EGF using either serum-free medium or 10% serum (data not shown); therefore, all subsequent experiments were performed with 10% serum. Two major EGF-R mRNA species of 10 and 5.6 kb were identified in Northern blots (Fig. 2A and B). The 10-kb species was predominant in both cell lines, consistent with findings of previous reports (19, 63). The result in Fig. 2 showed that EGF increased expression of both the 10- and 5.6-kb EGF-R mRNA species in MDA-MB-468 cells ∼2-fold at 4 h but only after 8 h in the BT-20 cells. Interestingly, at 12 h the 10- and 5.6-kb mRNAs were still elevated in the BT-20 cells, whereas the mRNA had returned to basal levels in MDA-MB-468 cells. Cells were also treated with EGF to analyze the effect on EGF-R protein levels. In both cell lines, EGF-induced an increase in EGF-R protein (∼2-fold) (Fig. 2C and D). In contrast, EGF did not increase actin levels, which were used as loading controls (data not shown).
FIG. 2.
EGF upregulates EGF-R mRNA and protein levels in MDA-MB-468 and BT-20 cells. (A and B) Northern blot analysis of total RNA extracted from MDA-MB-468 (A) and BT-20 (B) cells after treatment with EGF (25 ng/ml, ∼4 nM) as indicated. RNA was fractionated on 1% agarose gels, transferred to a nylon membrane, and hybridized to a 32P-labeled EGF-R-specific cDNA probe. Each blot was normalized using a 32P-labeled 18S rRNA probe. Arrows denote EGF-R mRNA species at 10 and 5.6 kb and 18S rRNA. Blots were quantitated using a PhosphorImager and autoradiography. Each value is shown relative to an arbitrary value of 1 at time zero and is representative of at least three separate experiments performed in duplicate. Error bars represent standard errors of the means. Black and grey columns represent 10- and 5.6-kb EGF-R mRNA species, respectively. (C and D) Immunoblot analysis of EGF-R protein levels in MDA-MB-468 (C) and BT-20 (D) cells following treatment with EGF (25 ng/ml) for 8 h. Cells were lysed, and 5 μg (MDA-MB-468 cells) or 10 μg (BT-20 cells) of protein lysate was electrophoresed on SDS–6% polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed for EGF-R protein expression using a sheep polyclonal anti-human EGF-R antibody and ECL detection (see Materials and Methods). The size of the EGF-R protein is ∼170 kDa. The bar graphs depict the quantified changes in EGF-R protein after EGF treatment relative to an actin control (data not shown) and are representative of at least three different experiments performed in duplicate. Error bars represent standard errors of the means. Rel., relative.
EGF stabilizes EGF-R mRNA and increases EGF-R mRNA transcription in MDA-MB-468 and BT-20 cells.
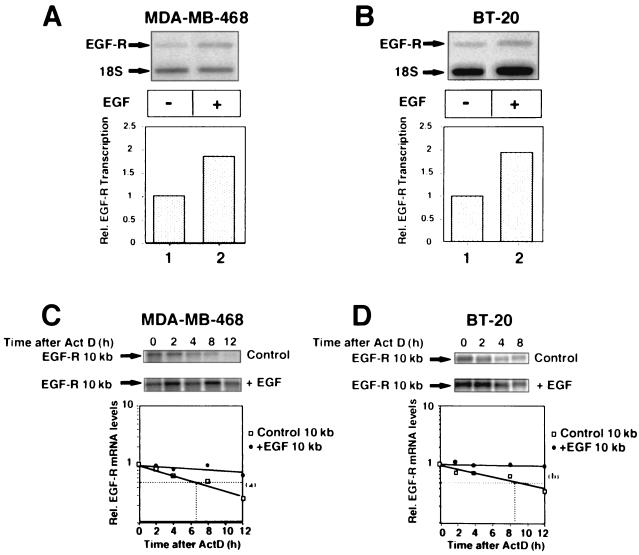
The contribution of posttranscriptional events to the regulation of EGF-R mRNA expression in breast cancer cells has not previously been addressed. Act D chase studies were used to determine the half-life of EGF-R mRNA in each cell line (Fig. 3C and D). The basal half-lives of EGF-R mRNA were ∼6.5 and 8.5 h in MDA-MB-468 and BT-20 cells, respectively. In both cell lines, EGF stabilized EGF-R mRNA, prolonging the half-life to greater than 14 h (>2-fold). Nuclear run-on assays demonstrated that EGF upregulated transcriptional activity ≤2-fold in each cell line (Fig. 3A and B). Thus, the increase in EGF-R mRNA (∼3-fold) induced by EGF in each cell line resulted from a combined increase in mRNA stability and transcriptional upregulation, with the former appearing predominant.
FIG. 3.
EGF increases EGF-R gene transcription and stabilizes EGF-R mRNA in MDA-MB-468 and BT-20 cells. (A and B) Transcriptional analysis of MDA-MB-468 (A) and BT-20 (B) cells after treatment with EGF (25 ng/ml) for 6 h. EGF-R transcription rates were determined in isolated nuclei by run-on transcription assays, analyzed by autoradiography, and quantitated using a PhosphorImager. Results were normalized relative to 18S RNA transcription levels and are representative of two separate experiments performed in duplicate. (C and D) Act D chase studies in MDA-MB-468 (C) and BT-20 (D) cells. Cells were grown to 70 to 80% confluence, treated with EGF (25 ng/ml) for 8 h, and then chased with 7.5 μg of Act D/ml. Total RNA was extracted from the cells at 0, 2, 4, 8, and 12 h after Act D treatment and analyzed by Northern blotting using a 32P-labeled cDNA EGF-R-specific probe as in Fig. 2. The 10-kb species of EGF-R mRNA was normalized against 18S RNA (image not shown) and quantitated using a PhosphorImager. Half-lives were determined by linear regression analysis. The graph at the bottom shows composite data from three separate experiments performed in duplicate. Rel., relative. Means comparisons: (a) P < 0.05; (b) P < 0.02.
Identification of cis-acting elements in EGF-R mRNA 3′-UTR.
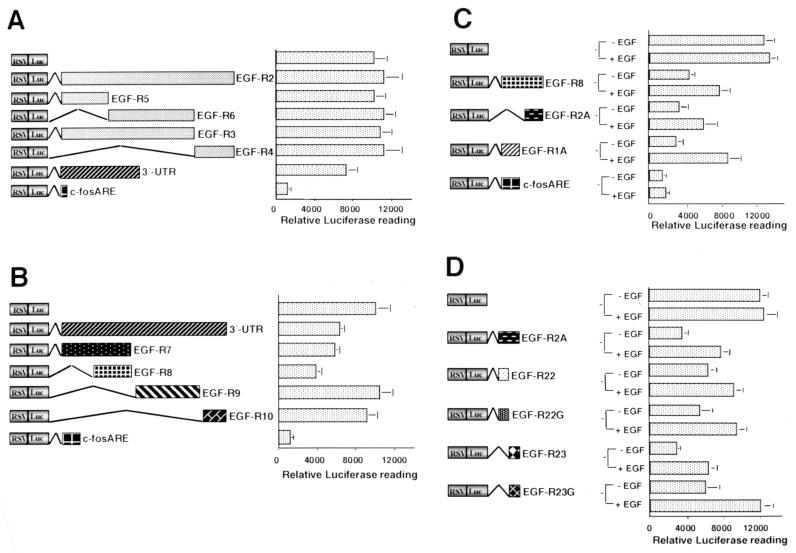
To localize the cis element(s) contributing to the regulation of EGF-R mRNA stability, we developed a transient transfection system in MDA-MB-468 breast cancer cells. A set of plasmids containing different portions of the EGF-R mRNA inserted downstream of the Luc coding region in RSV-Luc was used (Fig. 1B). Initially we examined the change in reporter activity induced by various portions of the EGF-R mRNA. Interestingly, basal Luc activity was modified with the RSV-Luc/3′-UTR construct to ∼60 to 70% of control levels (Fig. 4A). However, no effect was seen with any of the constructs that contained 5′-UTR or coding region sequences of EGF-R mRNA. This included sequences containing the 5′ end of the EGF-R mRNA (EGF-R2) and several smaller clones from within that sequence, EGF-R3, -4, -5, and -6 (Fig. 4A). Reporter activity of the RSV-Luc/c-fos ARE plasmid was reduced to ∼10% of the basal level. As the c-fos ARE sequence is known to have a short half-life in other systems (25), this observation was consistent with it conferring rapid decay and shortening the mRNA half-life of the Luc reporter. Taken together, these results suggested that the 3′-UTR was the major contributor to the regulation of EGF-R mRNA turnover.
FIG. 4.
The 3′-UTR of EGF-R mRNA contains an EGF-regulated 260-nt ARE that regulates Luc activity in MDA-MB-468 cells. (A and B) MDA-MB-468 cells were transfected by electroporation with 10 μg of one of several RSV-Luc constructs, either RSV-Luc or an RSV-Luc/EGF-R hybrid (Fig. 1B), or c-fos ARE and 6 μg of RSV-β-Gal. Cells were harvested 24 h later, and the lysate was analyzed in Luc and β-Gal assays (see Materials and Methods). The luciferase results were normalized to the β-Gal reading and plotted. The graphs are representative of at least five separate experiments performed in triplicate. Error bars denote standard deviation. (C and D) Analysis of the AU-rich sequences within the 260-nt EGF-R mRNA cis-acting element. MDA-MB-468 cells were transfected as for panels A and B, EGF (25 ng/ml) was added to the cells 24 h posttransfection, cells were harvested 8 h later, and analysis was performed as described above.
The EGF-R mRNA 3′-UTR contains several AU-rich regions, two of which are contained within a 260-nt sequence at the 5′ end (Fig. 1A). The most 5′ region contains two AU pentamers (nt 4128 to 4134 and 4172 to 4179), and immediately 3′ is a region containing two extended AU pentamers (AUUUUUA) located at nt 4029 to 4035 and 4079 to 4085. Two other pentamers were identified further 3′ at nt 5059 and 5349. Several constructs were generated to include each of the four AREs (Fig. 1B). When transfected into MDA-MB-468 cells, EGF-R7 and EGF-R8 significantly reduced Luc activity (to ∼55 to 60% and ∼30 to 40%, respectively, of basal) (Fig. 4B), while EGF-R9 and EGF-R10 had no effect. EGF-R8 was subsequently analyzed in more detail, as it comprised a 260-nt sequence containing the AU-rich regions described above.
Constructs containing the EGF-R1A and EGF-R2A AU-rich regions (Fig. 1B) were then examined. Each AU-rich sequence reduced Luc activity to ∼30% of basal levels, (Fig. 4C), whereas the c-fos ARE sequence reduced Luc activity to 10% of control levels (Fig. 4C). The EGF-R2A mRNA sequence was studied in greater detail because of a more stable stem-loop structure (see Fig. 8B and C) and because little is known about the role of extended pentamers in the regulation of mRNA stability. We dissected EGF-R2A into two smaller constructs, EGF-R22 and EGF-R23 (Fig. 1B), each of which contained a single extended AUUUUUA pentamer. EGF-R22 reduced Luc reporter activity to a level ∼50% of the control levels but still twofold higher than the level for EGF-R2A. However, EGF-R23 had a more significant effect and reduced reporter activity below the level of EGF-R2A (Fig. 4D). To examine the role of the extended pentamers in each of these regions, we generated two mutants, EGF-R22G and EGF-R23G, in which the AUUUUUA sequence was replaced with AUGGGUA (Fig. 1B). Plasmid EGF-R22G regulated basal reporter activity similarly to EGF-R22 (Fig. 4D). However, basal reporter activity was twofold higher with the EGF-R23G mutant (Fig. 4D), suggesting that the EGF-R23 extended AU pentamer was involved in regulating basal EGF-R mRNA turnover.
FIG. 8.
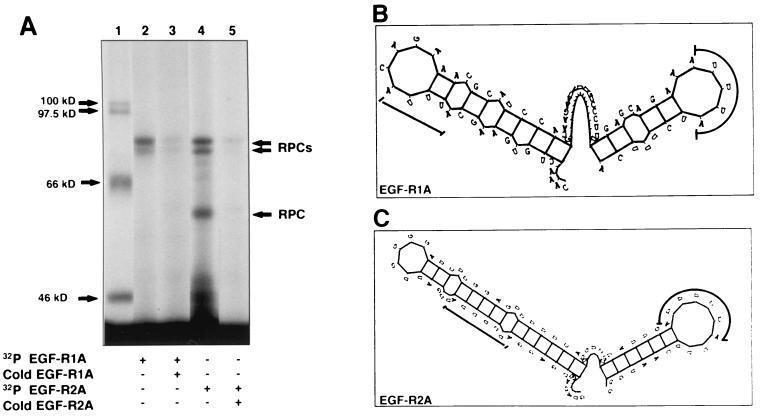
EGF-R mRNA is a target for multiple RNA-binding proteins. (A) Extract from MDA-MB-468 cells was incubated with either 32P-labeled EGF-R1A (lanes 2 and 3) or EGF-R2A (lanes 4 and 5), followed by addition of RNase T1 and heparin. The mixture was then UV cross-linked for 15 min (see Materials and Methods) before digestion with RNase A, SDS-PAGE (7.5% gel), and analysis by PhosphorImager. In lanes 3 and 5, ∼100-fold excess unlabeled (cold) RNA was added (EGF-R1A and EGF-R2A, respectively). 14C-labeled size markers are indicated in lane 1. (B and C) Stem-loop secondary structure plots of EGF-R1A (B) and EGF-R2A (C), using the Genetics Computer Group Squiggles program (see Materials and Methods). The ΔE values for EGF-R1A and EGF-R2A were −6.4 and −9.2 kJ mol−1, respectively. Locations of the AU pentamers (B) and AU-extended pentamers (C) are indicated by the thick lines.
In view of the EGF-induced stabilization of EGF-R mRNA in MDA-MB-468 cells, we next determined the effect of EGF on reporter activity in the presence of the various EGF-R constructs. EGF treatment upregulated Luc activity in cells transfected with EGF-R1A and EGF-R2A (∼2- to 3-fold), although the upregulation was more pronounced with EGF-R1A (Fig. 4C). In contrast, there was no EGF regulation of Luc activity in cells transfected with the c-fos ARE (Fig. 4C). EGF also upregulated reporter activity in cells transfected with EGF-R22 and EGF-R23, the effect being more pronounced for EGF-R23 (Fig. 4D). Interestingly, EGF upregulated EGF-R22G and EGF-R23G to the same degree as EGF-R22 and EGF-R23, respectively (∼2-fold) (Fig. 4D). Taken together, these results suggest that EGF-R1A and EGF-R2A can independently regulate basal and growth factor-stimulated EGF-R mRNA stability in MDA-MB-468 cells, but neither of the extended AU pentamers is required for EGF-induced reporter upregulation.
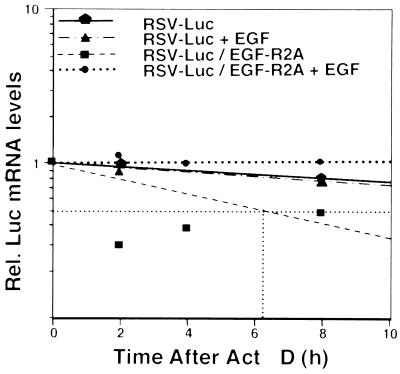
To verify that the Luc assay was representative of changes in Luc mRNA turnover, we performed several additional experiments. First, we demonstrated with nuclear run-on assays that the transcriptional activity of Luc mRNA did not change in cells transfected with RSV-Luc/EGF-R2A compared to RSV-Luc alone (data not shown). Second, we performed a parallel set of transfections to determine the Luc mRNA decay rate using Act D chase, RT-PCR, and a LightCycler. We found that the half-life of RSV-Luc mRNA was long (>14 h) (Fig. 5). However, the half-life of RSV-Luc/EGF-2A mRNA was significantly shortened to ∼6 h, and EGF increased the half-life to well over 14 h while having no effect on the vector alone (Fig. 5). Similar EGF-responsive data were obtained for RSV-Luc/EGF-R1A in this system (∼2-fold change with addition of EGF) (data not shown). Taken together, these data verify the reporter assays above and confirm that the EGF-R1A and EGF-R2A regions act as cis elements within the EGF-R mRNA.
FIG. 5.
The EGF-R2A mRNA sequence acts as a cis destabilizing and EGF-regulated element. MDA-MB-468 cells were transfected with pRSV-Luc or pRSV-Luc/EGF-R2A in triplicate. After treatment with EGF (25 ng/ml) for 6 h, the cells were treated with Act D (7.5 μg/ml), and RNA was extracted at 0, 2, 4, and 8 h followed by RT-PCR for Luc mRNA using a LightCycler (see Materials and Methods). Samples were normalized using a cotransfected control of β-globin. Half-lives were determined by linear regression analysis. The graph is representative of data from four independent experiments in duplicate. Rel., relative.
MDA-MB-468 cells contain cytoplasmic proteins that bind specifically to the EGF-R mRNA cis element.
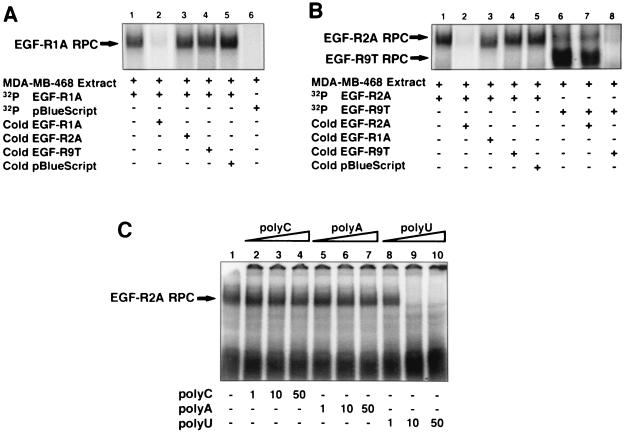
To investigate whether the 260-nt cis-acting EGF-R mRNA element was a target for RNA-binding proteins, we generated riboprobes to the EGF-R1A and EGF-R2A sequences (Fig. 1C, EGF-R1A and EGF-R2A, respectively) for use in REMSA. Cytoplasmic proteins were identified from MDA-MB-468 cells that bound specifically to the EGF-R1A and EGF-R2A riboprobes (Fig. 6A and B). Addition of ∼100-fold excess unlabeled pBlue competitor RNA did not diminish the formation of the RNA-protein complex (Fig. 6A and B, lanes 5). However, addition of excess unlabeled EGF-R1A (Fig. 6A, lane 2) or EGF-R2A (Fig. 6B, lane 2) virtually abolished RNA-protein complex formation, confirming binding specificity for these transcripts. Addition of excess unlabeled EGF-R2A had no significant effect on binding of the EGF-R1A complex (Fig. 6A, lane 3). Similarly, excess unlabeled EGF-R1A had only a marginal effect on the EGF-R2A complex (Fig. 6B, lane 3). No RNA-protein complex was observed with 32P-pBlue riboprobe (Fig. 6A, lane 6).
FIG. 6.
Proteins from MDA-MB-468 cells bind specifically to EGF-R mRNA. (A) MDA-MB-468 cytoplasmic extract (∼5 μg) was incubated with either 32P-labeled EGF-R1A (lanes 1 to 5) or pBlue riboprobe (lane 6), and REMSA was performed as described in Materials and Methods. In lanes 2 to 5, ∼100-fold excess unlabeled (cold) competitor RNA was added to the labeled EGF-R1A probe for 10 min at 22°C before addition of extract and REMSA. Arrow denotes RNA-protein complex (RPC). (B) MDA-MB-468 extract (∼5 μg) was incubated with either 32P-labeled EGF-R2A (lanes 1 to 5) or EGF-R9T (lanes 6 to 8), and REMSA was performed as above. In lanes 2 to 5, 7, and 8, ∼100-fold excess of various unlabeled competitor RNA was added to the labeled probe prior to the extract. Arrows denote RNA-protein complexes (RPC). (C) MDA-MB-468 extract was incubated with labeled EGF-R2A in the absence (lane 1) or presence of increasing concentrations (1, 10, and 50 ng) of unlabeled homopolymers RNA, poly(C) (lanes 2 to 4), poly(A) (lanes 5 to 7), and poly(U) (lanes 8 to 10). Arrow denotes RNA-protein complex (RPC). The data are representative of at least three individual experiments.
Addition of excess unlabeled EGF-R9T, which contains a consensus nonamer sequence (Fig. 1C), had little effect on either the EGF-R1A (Fig. 6A, lane 4) or EGF-R2A (Fig. 6B, lane 4) RNA-protein complex. With the labeled EGF-R9T riboprobe, we observed a large, faster-migrating complex (Fig. 6B, lane 6) that was abolished with excess unlabeled EGF-R9T (Fig. 6B, lane 8). Addition of excess unlabeled EGF-R2A also marginally reduced complex formation (Fig. 6B, lane 7). Taken together, these data suggest that the proteins binding to EGF-R2A were not high-affinity AU-rich nonamer-binding proteins. Unlabeled competitor homopolymer sequences [poly(U), poly(C) and poly(A)] were also used to explore RNA-protein complex specificity. Excess poly(C) and poly(A) did not compete for binding (Fig. 6C, lanes 2 to 4 and 5 to 7, respectively). However, excess poly(U) (10 and 50 ng) abrogated binding completely (Fig. 6C, lanes 9 and 10). This suggested that the U-rich sequences in EGF-R2A were likely to be important for trans-acting factor binding.
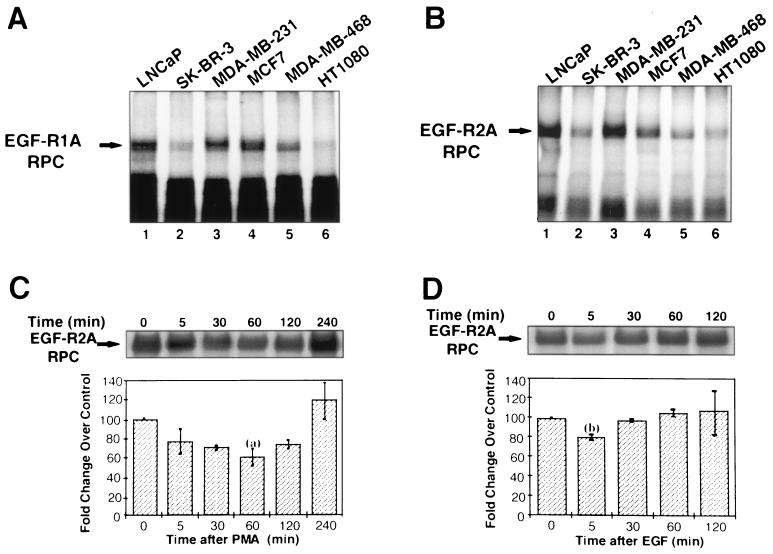
Tissue distribution of these EGF-R mRNA-binding proteins was examined using extracts from a variety of breast cancer (SK-BR-3, MCF7, and MDA-MB-231), prostate cancer (LNCaP), and fibrosarcoma (HT1080) cell lines. EGF-R mRNA-binding proteins were detected in each cell line with each of the EGF-R1A and EGF-R2A riboprobes (Fig. 7A and B, respectively). However, there was marked variation in binding activity to each riboprobe in the different cell lines. Binding activity was not simply related to the level of EGF-R expressed in each cell line, as binding was lower in MDA-MB-468 cells, which have far more EGF-Rs (106/cell) than do LNCaP and MCF7 cells (∼105 and 104/cell, respectively) (21).
FIG. 7.
EGF-R mRNA-binding proteins are present in a variety of cancer cells and are regulated by PMA and EGF. (A and B) REMSA using 32P-labeled EGF-R1A (A) or EGF-R2A (B) and extracts from a variety of human cancer cells, including prostate (LNCaP), breast (SK-BR-3, MDA-MB-231, MCF7, and MDA-MB-468), and fibrosarcoma (HT1080) cells. Arrows denote RNA-protein complexes (RPC). (C and D) REMSA using 32P-labeled EGF-R2A and extracts isolated from MDA-MB-468 cells treated with 100 nM PMA (C) or 4 nM EGF (D) at the time intervals shown. The top panel shows the REMSA; the lower graph depicts the level of binding relative to a control at each time point and normalized to an arbitrary value at 100% at time zero. The data are representative of four experiments performed in duplicate. Error bars represent standard errors of the means. Means comparisons: (a) P < 0.01; (b) P < 0.05.
The binding activity of AUBPs may be regulated by phorbol esters and other activators of cell signaling pathways (5). To determine whether the binding of these proteins to the EGF-R2A region of EGF-R mRNA could be regulated, we generated extracts from MDA-MB-468 cells that were incubated in the presence or absence of EGF and PMA. Both EGF and PMA rapidly decreased RNA-protein complex formation by 20 to 30% within 5 min (Fig. 7C and D, respectively). For PMA, this downregulation persisted for at least 2 h. In contrast, after EGF treatment, RNA-protein complex formation returned to basal levels within 30 min (Fig. 7D).
UVXL analysis using MDA-MB-468 cell extracts demonstrated specific RNA-protein complexes with both EGF-R1A and EGF-R2A riboprobes (Fig. 8A). Several RNA-protein complexes were detected between ∼55 and 80 kDa with the EGF-R2A probe but only the bands at ∼80 kDa were observed with the EGF-R1A riboprobe, suggesting different proteins targeting each sequence (Fig. 8A, lanes 2 and 4, respectively). These distinct RNA-protein profiles were in turn different from that of the EGF-R9T probe, which produced only RNA-protein complexes of lower molecular weight (data not shown). Addition of excess unlabeled EGF-R1A or EGF-R2A mRNA diminished the complex (Fig. 8A, lane 3 or 5, respectively), but excess unlabeled pBlue vector alone had no effect (data not shown). Using MDA-MB-468 cell extracts and REMSAs, we were unable to supershift the EGF-R mRNA-protein complex with antibodies to either HuR or AUF1 (data not shown).
Examination of the predicted secondary structure of the cis element revealed that EGF-R2A formed a stem-loop structure with an energy (ΔE) of −9.2 kJ mol−1 (Fig. 8C). The extended pentamers resided on the 5′ stem and 3′ loop. In contrast, EGF-R1A was less stable (ΔE = −6.4 kJ mol−1) (Fig. 8B).
Mutations within EGF-R2A radically alter RNA protein binding.
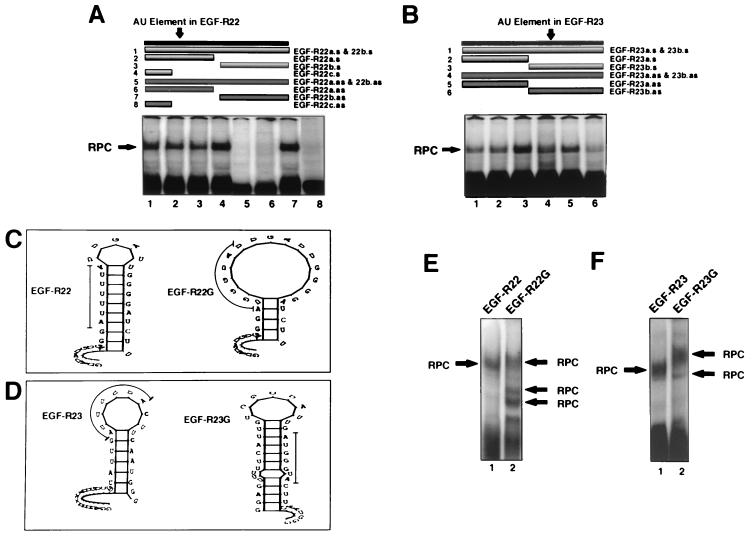
A set of short sense and antisense DNA oligomers (Fig. 1D) was used to identify which half of EGF-R2A was critical for binding. The predominant binding site for EGF-R22 resided in the 5′ end of the sequence within which the AU-extended pentamer was located (Fig. 9A, lanes 5 and 6 versus lane 7). Because of the marked effect of antisense DNA oligomer 6 with EGF-R22, we generated oligomers to the 5′ 12 nt of EGF-R22 (oligomers 4 and 8), which did not include the AU-rich sequence. We found that sequence antisense to the 12-nt region abrogated binding completely, while its sense counterpart had no effect (Fig. 9A, lanes 8 and 4, respectively). A similar but less striking pattern was observed for EGF-R23: a major reduction in binding was observed when the 3′-end antisense oligomer was used (Fig. 9B, compare lanes 3 and 6).
FIG. 9.
Analysis of the EGF-R mRNA binding motif using DNA oligomers and AU-extended pentamer mutants. (A and B) REMSA using MDA-MB-468 extracts and 32P-labeled EGF-R22 (A) or EGF-R23 (B). Before addition of cell extract, a sense or antisense DNA oligomer was incubated with the labeled probe (1:1 molar ratio) for 10 min at 22°C. In panel A, sense oligomers were added to samples in lanes 1 to 4, and antisense oligomers were added to samples in lanes 5 to 8. In panel B, sense oligomers are in lanes 1 to 3, and antisense oligomers are in lanes 4 to 6. The upper portion of each panel illustrates the length and location of each oligomer relative to the labeled probe. The oligomer, probe, and extract mixture was treated as above for REMSA and analyzed by PhosphorImager. The sequence and orientation of each oligonucleotide and probe are shown in Fig. 1D. (C and D) Stem-loop secondary structure plots of EGF-R22 and EGF-R23 and their respective triple-G AU-extended pentamer mutations. The thin line denotes the location of the mutation. The ΔE values for EGF-R22, EGF-R22G, EGF-R23, and EGF-R23G were −3.5, 0.7, −4.0, and 5.7 kJ mol−1, respectively. (E and F) MDA-MB-468 extract was incubated with either 32P-labeled EGF-R22 or EGF-R22G (E) or EGF-R23 or EGF-R23G (F). REMSA was performed as for Fig. 6 and analyzed by PhosphorImager. RPC, RNA-protein complex.
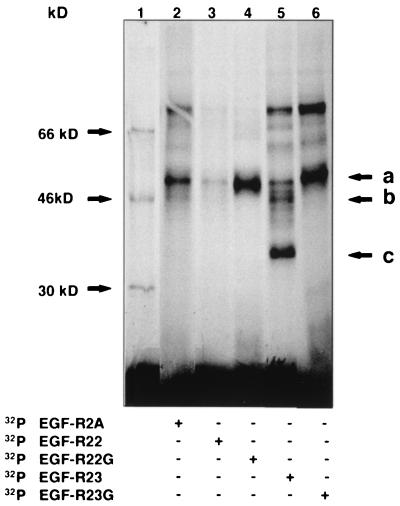
From our transfection studies with the RSV-Luc/EGF-R22G and EGF-R23G mutants, we had established that the 5′ extended pentamer in EGF-R2A was unlikely to be involved in regulating mRNA turnover (Fig. 4D). However, our data suggested an important role for the 3′ extended pentamer, contained within EGF-R23, in regulating basal reporter activity. To investigate whether the transfection data correlated with alteration in the binding of RNA-protein complexes, we generated riboprobes containing the same mutants for REMSA (with the middle 3 U's replaced with G's [Fig. 1C, EGF-R22G and EGF-R23G, respectively]). The predicted secondary structure of EGF-R22G was dramatically different from that of EGF-R22 (Fig. 9C). In contrast, the structural integrity of EGF-R23 was predominately maintained in EGF-R23G (Fig. 9D). Using MDA-MB-468 extracts and the EGF-R22 probe, a single RNA-protein complex was observed (Fig. 9E, lane 1). However, with the mutant EGF-R22G probe, total binding to the probe was increased, but with a different profile of RNA-protein complexes (Fig. 9E, lane 2). A different pattern was observed for the EGF-R23G mutant, where two slower-migrating, less intense RNA-protein complexes were observed (Fig. 9F, lane 2). We used UVXL to further define the effects of these mutants on each RNA-protein complex. In contrast to the full-length EGF-R2A riboprobe, EGF-R22 bound only one predominant protein (∼55 kDa [Fig. 10, lane 3, arrow a]). Interestingly, the binding of this protein to EGF-R22G was markedly increased (Fig. 10, lane 4). Labeled EGF-R23 bound proteins with a similar pattern to EGF-R2A, but with additional RNA-protein complexes at ∼35 and 45 kDa (Fig. 10, lane 5, arrows b and c, respectively). Binding activities of the ∼55- and 80-kDa proteins increased with the mutant EGF-R23G riboprobe, while the RNA-protein complexes at ∼35 and 45 kDa were abolished (Fig. 10, lane 6).
FIG. 10.
UVXL analysis of EGF-R mRNA mutants. MDA-MB-468 extract was incubated with one of several different 32P-labeled probes (Fig. 1C), UV cross-linked, resolved by SDS-PAGE on an 8.5% gel, and analyzed by PhosphorImager. Lane 1 shows 14C-labeled molecular mass standards. Arrows a, b, and c denote specific RNA-protein complexes discussed in the text.
These data, together with the data from Fig. 9A, suggest that the 12-nt sequence upstream of the extended AU pentamer in EGF-R22, rather than the extended AU pentamer itself, is critical for binding. Furthermore, mutations in the AU pentamer predict a structure that increases rather than decreases binding. The findings for the EGF-R23 probe are significantly different, as they suggest an important role for the AU-extended pentamer in binding, because specific RNA-protein complexes were abolished with the introduction of the mutation. However, the extended pentamer clearly does not comprise the entire binding motif, as several higher-Mr RNA-protein complexes were still evident with the mutant probe. Furthermore, it can be deduced that the predominant protein binding to EGF-R2A occurs at the 3′ end within the EGF-R23 sequence. These data support the transfection data which implicate the 3′ AU-extended pentamer in the regulation of EGF-R mRNA turnover.
DISCUSSION
These data establish the importance of mRNA stability in the regulation of EGF-R gene expression by EGF in ER-negative breast cancer cell lines that overexpress EGF-R. In both MDA-MB-468 and BT-20 breast cancer cells, the EGF-induced stabilization of EGF-R mRNA (>2-fold) was accompanied by a ∼2-fold increase in total EGF-R protein. Taken together with previous studies in which we reported EGF-induced stabilization of EGF-R mRNA in epidermoid KB cancer cells (∼2-fold) (45) and in human prostate cancer cells (LNCaP and DU-145; ∼2-fold) (39), it is apparent that this effect on EGF-R mRNA stability is shared across many different cancer cell lines and is of primary importance in the regulation of EGF-R gene expression.
Despite increasing evidence for tight control of EGF-R expression at the level of mRNA decay, little is known of the molecular mechanisms involved. In particular, there have been no attempts to locate and characterize a cis-acting element(s) within the EGF-R mRNA or the trans-acting protein(s) that binds these regions. The 3′-UTR of the EGF-R mRNA contains four major AU-rich regions, two within a ∼260-nt sequence (EGF-R8) at the 5′ end and two smaller regions further toward the 3′ end. Our transfection studies indicated that the 5′ 260-nt AU-rich (66%) region contains a complex cis-acting element. Interestingly, the two other AU-rich regions in the 3′-UTR do not appear to be involved in the regulation of reporter activity. The location of the cis element within the 3′-UTR is typical of most instability sequences described to date (1, 2, 23). However, the EGF-R mRNA cis-acting element is distinctive in that it is devoid of a consensus nonamer (34, 68); instead it has two classical pentamers (AUUUA) (9, 23, 42, 43) and two extended pentamers (AUUUUUA), residing in separate discrete ∼75-nt regions, EGF-R1A and EGF-R2A, respectively. Each of these regions is capable of independently regulating basal and EGF-stimulated Luc mRNA turnover, suggesting that both components of the cis element (EGF-R1A and EGF-R2A) play an important role in the EGF-induced stabilization of EGF-R mRNA. Based on our observations, we presume that EGF-R1A and EGF-R2A function in a cooperative manner to regulate EGF-R mRNA turnover. It is important to note, however, that neither the EGF-R1A nor the EGF-R2A sequence reduced reporter activity to the level of the c-fos ARE sequence, suggesting that these EGF-R AU-rich sequences are less potent than the well-characterized destabilizing nonamers in the c-fos ARE (34, 68). The EGF-R mRNA cis element has intermediate capacity to confer instability to a heterologous reporter, consistent with the intermediate basal endogenous EGF-R mRNA half-life of ∼6.5 to 9 h documented for these two cell lines. Interestingly, we found the use of the LightCycler significantly facilitated analysis of Luc mRNA half-life for turnover studies and also verified that in these cells, the Luc reporter protein assay was an excellent surrogate for Luc mRNA, as the two measurements closely followed each other.
Analysis of our mutant constructs in transfections provided insight into the potential contribution of each AU-extended pentamer to the regulation of EGF-R mRNA turnover. Interestingly, mutation of the 5′ extended pentamer in EGF-R2A (EGF-R22G) did not alter reporter activity in the presence or absence of EGF. However, mutation of the 3′ extended pentamer (EGF-R23G) upregulated basal reporter activity to ∼50% of the wild-type level (EGF-R23) while having a similar effect on EGF-induced reporter activity (∼2-fold increase). These data suggest that the 5′ extended pentamer does not contribute to the regulation of EGF-R mRNA turnover, while the 3′ extended pentamer is likely to be involved in regulating basal, but not EGF-induced, changes in EGF-R mRNA turnover.
Our REMSA and UVXL studies identified novel cytoplasmic proteins from ER-negative breast cancer cells that bound specifically to the 260-nt cis-acting element of EGF-R mRNA. These are the first proteins from breast cancer cells to be described that target the EGF-R mRNA. Interestingly, these proteins appear to be widely distributed in a variety of human breast and prostate cancer, as well as fibrosarcoma, cell lines. Taken together with our detection of a similar complex in epidermoid KB cells (45), these data support a central role for these proteins in the regulation of EGF-R gene expression in cancer cells. Interestingly, binding was detected in breast cancer cell lines that were ER positive, such as MCF7 cells, and also in ER-negative cells, such as MDA-MB-231 and MDA-MB-468 cells. Furthermore, the intensity of binding to the cis-acting element appeared unrelated to the cellular level of EGF-R expression.
We found that PMA and EGF both rapidly downregulated binding activity of these proteins to the EGF-R2A probe. The decrease in binding was detectable within 5 min and may be due to a change in the phosphorylation state of one or more of these proteins that modifies binding. The binding activity of several other AUBPs has been shown to be regulated by PMA (4). However, these have been predominantly proteins that target classic AREs comprising nonamers, such as AU-rich binding factor (AUBF) (43). In addition, phosphorylation of the iron regulatory protein by PMA induces rapid changes in binding to the iron-responsive element (54). Given that PMA and EGF both activate the protein kinase C pathway in these cells, this finding provides a ready explanation for the similar effects observed in REMSA with the two ligands. Further studies will be required to determine which isotype of protein kinase C mediates these effects, whether other downstream effectors of EGF signaling regulate binding, whether these proteins are ubiquitously expressed, and when they appear during development.
Multiple mRNA-binding proteins that target AREs in early response genes and cytokines have been described in the past few years. The vast majority of these proteins have molecular masses in the range of 30 to 50 kDa. The first AUBP described that bound an ARE was termed AUBF (∼44 kDa) (42). Binding of this protein (a three-subunit complex) was upregulated by PMA and calcium ionophores (43). Subsequently, other ARE-binding proteins have been characterized, and some have been cloned. AUF1, a 37-kDa protein that contains two RRMs, was recently cloned (66). Three other proteins, termed AU-A, AU-B, and AU-C, that bound to AREs were described 2 years earlier (5). Several other studies have identified proteins of 82, 71, 66, and 37 kDa (66), as well as 70, 45, 40, 38, and 32.5 kDa (50), that target AREs within the c-fos and GM-CSF mRNAs. Several of these proteins appear to be cross-reactive with AUF1 antibodies (45, 40, and 38 kDa) or hnRNP antibodies (40 and 38 kDa) (50). Most recently, HuR, a member of the elav family of RRM-containing RNA-binding proteins, was cloned (41). HuR is a ubiquitously expressed ∼32- to 38-kDa protein that binds with high affinity to AREs present in a number of early response genes. HuR is remarkable in that it shuttles between the nucleus and cytoplasm (18) and has been shown to be an important component of the mRNA destabilization machinery (48). Almost all of the studies involving the above proteins have utilized sequences based on nonamer-pentamer repeats, and none to our knowledge have focused on the characterization of proteins that bind to extended AU pentamers. In this context, several lines of evidence suggest that the proteins we identified are not classical AU-rich nonamer-binding proteins. First, the AU-rich sequences in EGF-R1A and EGF-R2A are pentamers and extended pentamers and do not contain any nonamers. Second, antibodies to two well characterized AUBFs, HuR and AUF1, did not supershift the complex with either probe. Third, competition studies with excess unlabeled nonamer binding probe (EGF-R9T) riboprobe had little effect on the EGF-R1A and EGF-R2A probes. Last, the sizes of the proteins detected (∼55 to 80 kDa) were in a range above that of most of the AUBFs described to date (∼30 to 50 kDa) (7, 49, 54, 60).
The EGF-R2A sequence was of great interest because of the complex set of proteins binding on UVXL, the presence of two extended pentamers, and its stable stem-loop structure. Interestingly, UVXL showed that the 5′ end of EGF-R2A mRNA, EGF-R22, appeared to bind only one of the proteins observed with the EGF-R2A. In contrast, the 3′ end of EGF-R2A, EGF-R23, appeared to contribute the majority of binding, as we detected all of the RNA-protein complexes observed with the EGF-R2A probe, as well as additional smaller complexes at ∼35 and 45 kDa. To further define the major binding motif in EGF-R2A, we used a combination of DNA oligomers and mutant probes. The oligomer data suggested, somewhat unexpectedly, that the 5′ AU-extended pentamer in EGF-R2A was unlikely to be involved in protein binding. Instead, the 12 nt 5′ of the extended pentamer was shown to be critical for binding a single protein of ∼55 kDa. These findings were corroborated when we showed that the triple-G mutant, designed to disrupt the 5′ AU-extended pentamer and significantly modify the predicted stem-loop structure (EGF-R22G), bound with increased affinity to the protein extract in both REMSA and, in particular, UVXL. Thus, introduction of the mutation in the 5′ extended pentamer of EGF-R2A induced changes that increased rather than abolished binding. Studies using the EGF-R23 probe were quite different. The DNA oligomer data with portions of the EGF-R23 sequence suggested that the 3′ end of the sequence contained the major protein binding site. When we examined the binding profile of EGF-R23G, which was designed to disrupt the 3′ extended pentamer, we found that two RNA-protein complexes (∼35 and 45 kDa) were abolished, and there was increased binding of the other major protein moieties at ∼55 and 80 kDa. This confirmed a direct role for the extended pentamer in binding of at least two of the several proteins binding to EGF-R23. However, it also indicated that a significant proportion of protein binding to the EGF-R23 sequence occurs outside the extended AU pentamer region. Thus, there is differential binding of proteins to the two extended pentamers within the EGF-R2A sequence, and the majority of binding to EGF-R2A is directed to non-AU-rich sequence. These data suggest a model in which the 5′ extended AU pentamer is not involved in the regulation of EGF-R mRNA stability or binding of trans-acting factors, while the 3′ extended pentamer plays an important role in regulating basal mRNA stability, in part, through binding of specific trans-acting factors.
In summary, these data provide compelling evidence for a central role of mRNA turnover in the regulation of EGF-R gene expression in ER-negative breast cancer cells. The control of EGF-R mRNA stability is a complex process involving regulated interactions between a novel 3′-UTR cis element and multiple trans-acting protein factors. The cis element contains AU pentamers and extended pentamers and is a target for EGF-regulated trans-acting proteins. Interestingly, only one of the extended pentamers in this region is likely to contribute, in part, to the regulation of EGF-R mRNA stability and binding of specific proteins. These EGF-R mRNA-binding proteins are widely distributed in multiple cancer cell types, and their size and binding characteristics suggest that they are not classical ARE-binding proteins. The identification of a novel cis element in EGF-R mRNA will allow detailed assessment of the mechanisms governing mRNA turnover in a variety of relevant cell types in which overexpression of EGF-R is linked with proliferation. Furthermore, the isolation and cloning of proteins that target the cis element should provide valuable insight into the regulation of EGF-R mRNA expression in breast and other cancer cells and may provide the basis for designing strategies to disrupt EGF-R expression and consequently alter rates of cellular proliferation and growth.
ACKNOWLEDGMENTS
We thank Ana Zubiaga for providing the c-fos ARE plasmid, Roger Davis for the pBluescript EGF-R plasmid, Henry Furneaux for the HuR antibody, Gary Brewer for the AUF1 antibody, Robert Medcalf for the 9SWT plasmid, John Daly and Janelle Staton for technical assistance, and the Medical Illustrations Department at Royal Perth Hospital for the figures.
This work was supported by grants from the National Health and Medical Research Council of Australia, Royal Australasian College of Physicians, Cancer Foundation of Western Australia, and Royal Perth Hospital Medical Research Foundation to P.J.L.
REFERENCES
- 1.Anant S, Davidson N O. An AU-rich sequence element (UUUN[A/U]U) downstream of the edited C in apolipoprotein B mRNA is a high-affinity binding site for apobec-1: binding of apobec-1 to this motif in the 3′ untranslated region of c-myc increases mRNA stability. Mol Cell Biol. 2000;20:1982–1992. doi: 10.1128/mcb.20.6.1982-1992.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belasco J. mRNA degradation in prokaryotic cells: an overview. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press; 1993. pp. 3–12. [Google Scholar]
- 3.Bjorge J D, Paterson A J, Kudlow J E. Phorbol ester or epidermal growth factor (EGF) stimulates the concurrent accumulation of mRNA for the EGF receptor and its ligand transforming growth factor-alpha in a breast cancer cell line. J Biol Chem. 1989;264:4021–4027. [PubMed] [Google Scholar]
- 4.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3′ untranslated region of lymphokine mRNA. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992;267:6302–6309. [PubMed] [Google Scholar]
- 6.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F Y, Amara F M, Wright J A. Defining a novel ribonucleotide reductase R1 mRNA cis element that binds to a unique cytoplasmic trans-acting protein. Nucleic Acids Res. 1994;22:4796–4797. doi: 10.1093/nar/22.22.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C Y, Chen T M, Shyu A B. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 10.Chen C Y, De Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Clark A J L, Ishii S, Richert N, Merlino G T, Pastan I. Epidermal growth factor regulates the expression of its own receptor. Proc Natl Acad Sci USA. 1985;82:8374–8378. doi: 10.1073/pnas.82.24.8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleveland D W, Yen T J. Multiple determinates of eukaryotic mRNA stability. New Biol. 1989;1:121–126. [PubMed] [Google Scholar]
- 14.Davis R J, Czech M P. Tumor-promoting phorbol diesters cause the phosphorylation of epidermal growth factor receptors in normal human fibroblasts at threonine-654. Proc Natl Acad Sci USA. 1985;82:1974–1978. doi: 10.1073/pnas.82.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 16.DeMaria C T, Sun Y, Long L, Wagner B J, Brewer G. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J Biol Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- 17.Downward J, Yarden Y, Mayes E, Scarce G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield M D. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 18.Fan X C, Steitz J A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Pol J A, Hamilton P D, Klos D J. Transcriptional regulation of proto-oncogene expression by epidermal growth factor, transforming growth factor beta 1, and triiodothyronine in MDA-MB-468 cells. J Biol Chem. 1989;264:4151–4166. [PubMed] [Google Scholar]
- 20.Fernandez-Pol J A. Modulation of EGF receptor protooncogene expression by growth factors and hormones in human breast carcinoma cells. Crit Rev Oncog. 1991;2:173–185. [PubMed] [Google Scholar]
- 21.Filmus J, Pollak M N, Cailleau R, Buick R N. MDA-MB-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochem Biophys Res Commun. 1985;128:898–905. doi: 10.1016/0006-291x(85)90131-7. [DOI] [PubMed] [Google Scholar]
- 22.Ford L P, Watson J, Keene J D, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillis P, Malter J S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991;266:3172–3177. [PubMed] [Google Scholar]
- 24.Gorospe M, Baglioni C. Degradation of unstable interleukin-1 alpha mRNA in a rabbit reticulocyte cell-free system. Localization of an instability determinant to a cluster of AUUUA motifs. J Biol Chem. 1994;269:11845–11851. [PubMed] [Google Scholar]
- 25.Iwai Y, Bickel M, Pluznik D H, Cohen R B. Identification of sequences within the murine granulocyte-macrophage colony-stimulating factor mRNA 3′-untranslated region that mediate stabilization induced by mitogen treatment of EL-4 thymoma cells. J Biol Chem. 1991;266:17959–17965. [PubMed] [Google Scholar]
- 26.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3–L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joseph B, Orlian M, Furneaux H. P21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilization proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 28.Kesavan P, Mukhopadhayay S, Murphy S, Rengaraju M, Lazer M A, Das M. Thyroid hormone decreases the expression of epidermal growth factor receptor. J Biol Chem. 1991;266:10282–10286. [PubMed] [Google Scholar]
- 29.King C R, Kraus M H, Williams L T, Merlino G T, Pastan I H, Aaronson S A. Human tumor cell lines with EGF receptor gene amplification in the absence of aberrant sized mRNAs. Nucleic Acids Res. 1985;13:8477–8486. doi: 10.1093/nar/13.23.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klijn J G, Berns P M, Schmitz P I, Foekens J A. The clinical significance of epidermal growth factor receptor (EGF-R) in human breast cancer: a review on 5232 patients. Endocrine Rev. 1992;13:3–17. doi: 10.1210/edrv-13-1-3. [DOI] [PubMed] [Google Scholar]
- 31.Kontoyiannis D, Pasparakis M, Pizarro T T, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 32.Kudlow J E, Cheung C Y, Bjorge J D. Epidermal growth factor stimulates the synthesis of its own receptor in a human breast cancer cell line. J Biol Chem. 1986;26:4134–4138. [PubMed] [Google Scholar]
- 33.Lafon I, Carballes F, Brewer G, Poiret M, Morello D. Developmental expression of AUF1 and HUR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- 34.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leedman P J, Stein A R, Chin W W. Regulated specific protein binding to a conserved region of the 3′-untranslated region of thyrotropin beta-subunit mRNA. Mol Endocrinol. 1995;9:375–387. doi: 10.1210/mend.9.3.7776983. [DOI] [PubMed] [Google Scholar]
- 36.Leedman P J, Stein A R, Chen W W, Rogers J T. Thyroid hormone modulates the interaction between iron regulatory proteins and the ferritin mRNA iron-responsive element. J Biol Chem. 1996;271:12017–12023. doi: 10.1074/jbc.271.20.12017. [DOI] [PubMed] [Google Scholar]
- 37.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 39.Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumors of glial organ. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 40.Lin C R, Chen W S, Kruiger W, Stolarsky L S, Weber W, Evans R M, Verma I M, Gill G N, Rosenfeld M G. Expression cloning of human EGF receptor complementary DNA: gene amplification and three related messenger RNA products in A431 cells. Science. 1984;224:843–848. doi: 10.1126/science.6326261. [DOI] [PubMed] [Google Scholar]
- 41.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 42.Malter J S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989;246:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 43.Malter J S, Hong Y. A redox switch and phosphorylation are involved in the post-translational up-regulation of the adenosine-uridine binding factor by phorbol ester and ionophore. J Biol Chem. 1991;266:3167–3171. [PubMed] [Google Scholar]
- 44.Maurer F, Tierney M, Medcalf R L. An AU-rich sequence in the 3′-UTR of plasminogen activator inhibitor type 2 (PAI-2) mRNA promotes PAI-2 mRNA decay and provides a binding site for nuclear HuR. Nucleic Acids Res. 1999;27:1664–1673. doi: 10.1093/nar/27.7.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCulloch R K, Walker C E, Chakera A, Jazayeri J, Leedman P J. Regulation of EGF-receptor expression by EGF and TGF alpha in epidermoid cancer cells is cell type-specific. Int J Biochem Cell Biol. 1998;30:1265–1278. doi: 10.1016/s1357-2725(98)00056-9. [DOI] [PubMed] [Google Scholar]
- 46.Merlino G T, Xu Y H, Ishii S, Clark A J, Semba K, Toyoshima K, Yamamoto T, Pastan I. Amplification and enhanced expression of the epidermal growth factor receptor gene in A431 human carcinoma cells. Science. 1984;224:417–419. doi: 10.1126/science.6200934. [DOI] [PubMed] [Google Scholar]
- 47.Morris G L, Dodd J D. Epidermal growth factor receptor mRNA levels in human prostatic tumors and cell lines. J Urol. 1990;143:1272–1274. doi: 10.1016/s0022-5347(17)40253-9. [DOI] [PubMed] [Google Scholar]
- 48.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci USA. 1994;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamaki T, Imamura J, Brewer G, Tsuruoka N, Koeffler H P. Characterization of adenosine-uridine-rich RNA binding factors. J Cell Physiol. 1995;165:484–492. doi: 10.1002/jcp.1041650306. [DOI] [PubMed] [Google Scholar]
- 51.Peltz S W, Brewer G, Bernstein P, Hart P A, Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1:99–126. [PubMed] [Google Scholar]
- 52.Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- 53.Sainsbury J R, Farndon J R, Sherbet G V, Harris A L. Epidermal-growth-factor receptors and oestrogen receptors in human breast cancer. Lancet. 1985;i:364–366. doi: 10.1016/s0140-6736(85)91385-6. [DOI] [PubMed] [Google Scholar]
- 54.Schalinske K L, Eisenstein R S. Phosphorylation and activation of both iron regulatory proteins 1 and 2 in HL-60 cells. J Biol Chem. 1996;271:7168–7176. doi: 10.1074/jbc.271.12.7168. [DOI] [PubMed] [Google Scholar]
- 55.Seth D, Shaw K, Jazayeri J, Leedman P J. Complex post-transcriptional regulation of EGF-receptor expression by EGF and TGF-alpha in human prostate cancer cells. Br J Cancer. 1999;80:657–669. doi: 10.1038/sj.bjc.6690407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 57.Sheikh M S, Shao Z M, Chen J C, Li X S, Hussain A, Fontana J A. Expression of estrogen receptors in estrogen receptor-negative human breast carcinoma cells: modulation of epidermal growth factor-receptor (EGF-R) and transforming growth factor alpha (TGF alpha) gene expression. J Cell Biochem. 1994;54:289–298. doi: 10.1002/jcb.240540305. [DOI] [PubMed] [Google Scholar]
- 58.Thomson A M, Rogers J T, Walker C E, Staton J M, Leedman P J. Optimized RNA gel-shift and UV cross-linking assays for characterization of cytoplasmic RNA-protein interactions. BioTechniques. 1999;27:1032–1042. doi: 10.2144/99275rr03. [DOI] [PubMed] [Google Scholar]
- 59.Toi M, Tominaga T, Osaki A, Toge T. Role of epidermal growth factor receptor expression in primary breast cancer: results of a biochemical study and an immunocytochemical study. Breast Cancer Res Treat. 1994;29:51–59. doi: 10.1007/BF00666181. [DOI] [PubMed] [Google Scholar]
- 60.Vakalopoulou E, Schaack J, Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3′ untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991;11:3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson G M, Brewer G. Identification and characterization of proteins binding A+U-rich elements. Methods. 1999;17:74–83. doi: 10.1006/meth.1998.0709. [DOI] [PubMed] [Google Scholar]
- 62.Wittor H, Leying H, Hochhaus A, Miltenburg R V. Real-time quantitation of BCR-ABL mRNA transcripts using the LightCycler -t(9;22) Quantification Kit. Biochemica, no. 2. Indianapolis, Ind: Roche Diagnostics Corp.; 2000. [Google Scholar]
- 63.Xu Y H, Ishii S, Clark A J, Sullivan M, Wilson R K, Ma D P, Roe B A, Merlino G T, Pastan I. Human epidermal growth factor receptor cDNA is homologous to a variety of RNAs overproduced in A431 carcinoma cells. Nature. 1984;309:806–810. doi: 10.1038/309806a0. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto T, Nishida T, Miyajima N, Kawai S, Ooi T, Toyoshima K. The erbB gene of avian erythroblastosis virus is a member of the src gene family. Cell. 1983;35:71–78. doi: 10.1016/0092-8674(83)90209-x. [DOI] [PubMed] [Google Scholar]
- 65.Yeap B B, Krueger R G, Leedman P J. Differential posttranscriptional regulation of androgen receptor gene expression by androgen in prostate and breast cancer cells. Endocrinology. 1999;140:3282–3291. doi: 10.1210/endo.140.7.6769. [DOI] [PubMed] [Google Scholar]
- 66.You Y, Chen C Y, Shyu A B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3′ untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992;12:2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, De-Maria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;12:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zubiaga A M, Belasco J G, Greenberg M E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]