Abstract
Inflammatory bowel diseases [IBD] are a heterogeneous spectrum with two extreme phenotypes, Crohn’s disease [CD] and ulcerative colitis [UC], which both represent numerous phenotypical variations. Hence, we should no longer approach all IBD patients similarly, but rather aim to rethink clinical classifications and modify treatment algorithms to usher in a new era of precision medicine in IBD. This scientific ECCO workshop aims to provide a state-of-the-art overview on prognostic and predictive markers, shed light on key questions in biomarker development, propose best practices in IBD biomarker development [including trial design], and discuss the potential for multi-omic data integration to help drive further advances to make precision medicine a reality in IBD.
Keywords: Precision medicine, personalised medicine, inflammatory bowel diseases, disease prognosis, disease outcome, response to therapy
1. Introduction
Inflammatory bowel disease [IBD] consists of a spectrum of various phenotypes within the two broader classifications of Crohn’s disease [CD] and ulcerative colitis [UC].1,2 Even considering such broad diagnostic umbrella terms, is it important to note that there is not just one CD or one UC entity, but rather numerous variations that differ in clinical presentation and behaviour, which likely reflects differences in underlying pathogenic mechanisms. Hence, we can no longer approach all IBD patients similarly. With rapidly expanding technologies that allow detailed molecular profiling,3 we should rethink clinical classifications and modify treatment algorithms to usher in a new era of precision medicine in IBD [Table 1].
Table 1.
BEST biomarkers proposed by the FDA/NIH Working Group.a
| Biomarker type | Biomarker definition |
|---|---|
| Susceptibility/risk biomarker | A biomarker that indicates the potential for developing a disease or medical condition in an individual who does not currently have clinically apparent disease or the medical condition [see Torres et al.4] |
| Predictive biomarker | A biomarker used to identify individuals who are more likely than similar individuals without the biomarker to experience a favourable or unfavourable effect from exposure to a medical product or an environmental agent |
| Diagnostic biomarker | A biomarker used to detect or confirm presence of a disease or condition of interest or to identify individuals with a subtype of the disease |
| Monitoring biomarker | A biomarker measured serially for assessing status of a disease or medical condition or for evidence of exposure to [or effect of] a medical product or an environmental agent |
| Pharmacodynamic/response biomarker | A biomarker used to show that a biological response has occurred in an individual who has been exposed to a medical product or an environmental agent |
| Safety biomarker | A biomarker measured before or after an exposure to a medical product or an environmental agent to indicate the likelihood, presence, or extent of toxicity as an adverse effect |
| Prognostic biomarker | A biomarker used to identify likelihood of a clinical event, disease recurrence, or progression in patients who have the disease or medical condition of interest |
| Reasonably likely surrogate endpoint | An endpoint supported by strong mechanistic and/or epidemiological rationale such that an effect on the surrogate endpoint is expected to be correlated with an endpoint intended to assess clinical benefit in clinical trials, but without sufficient clinical data to show that it is a validated surrogate endpoint |
aFDA-NIH Biomarker Working Group. BEST [Biomarkers, EndpointS, and other Tools] Resource [Internet]. Silver Spring [MD]: Food and Drug Administration [US]; 2016. Bethesda MD: co-published by National Institutes of Health [US]; 2016.5
Until now, clinicians have mainly relied on clinical markers with poor accuracy and limited predictive value.6 This is due to the retrospective nature of most studies, loose definitions of predicted outcomes, and the heterogeneous medical approach to patients. Although a treat-to-target approach currently supports rigorous monitoring, with treatment changes if predefined goals are not met,7,8 the identification of prognostic and predictive markers provides significant promise to further improve clinical care. A long-held goal within the field has been to accurately predict disease course at diagnosis and respond accordingly by administering therapies that are optimal for the individual patient at the molecular level, and would thus optimise treatment response. Although much progress has been made, routine and accurate application of precision medicine biomarkers is not currently possible in IBD clinical practice.
Precision medicine in IBD might sound unrealistic, especially due to the heterogeneity and complex pathophysiology of the disease. However, achievements in other fields, such as oncology,9–11 suggest otherwise and support our belief that the IBD community should invest in prognostic and predictive biomarkers. Although progress has been made in recent years, translating findings into daily clinical practice is a lengthy process, as observed in other diseases such as cystic fibrosis.12 This is not simply due to waiting for clinical trials. Rather, the poor understanding of the molecular mechanisms that drive disease heterogeneity is the greatest bottleneck, which should be addressed to deliver biomarkers with robust clinical potential.
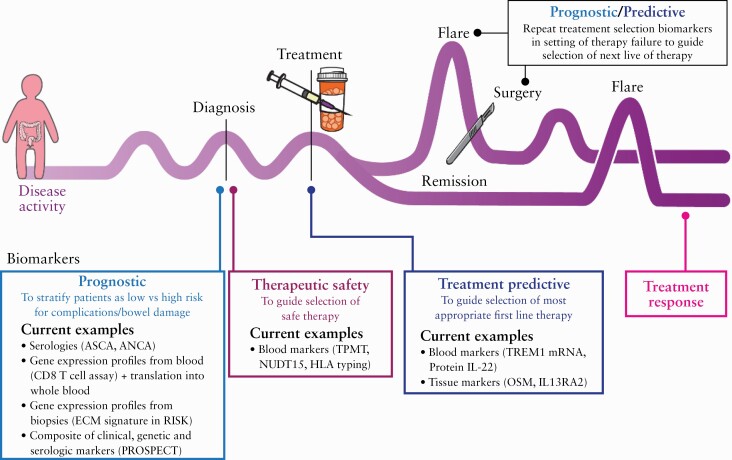
This ECCO scientific workshop on precision medicine aims to provide a state-of-the-art overview on prognostic and predictive markers [Figure 1], shed light on key questions to be answered in biomarker development, propose best practices in IBD biomarker development [including trial design], and discuss the potential for multi-omic data integration to help drive further advances to make precision medicine a reality in IBD.
Figure 1.
Utility of prognostic and predictive biomarkers along the inflammatory bowel disease [IBD] patient’s journey. Published with permission from ©Mount Sinai Health System.
2. Prognostic Biomarkers, a State-of-the-art Overview
It is widely recognised that addressing and understanding disease prognosis is one of the cornerstones for developing a precision-medicine approach in any condition,13 including IBD.14,15 While clinical features predictive of future IBD outcomes have been suggested,16 many of these have not been validated or are not sufficiently prognostic to guide therapeutic decision making.
A focus of biomarker development in IBD has been the study of intestinal biopsies, the site of active inflammation. The most important prognostic findings from biopsy studies were revealed in the RISK study; an extracellular matrix signature derived from intestinal biopsies of newly-diagnosed paediatric CD patients was predictive of developing stricturing disease when patients were followed up for 3 years.17 The RISK study group demonstrated that combining age, race, disease location, antimicrobial serologies, and ileal gene signatures provided a promising prediction model, which needs further validation in independent paediatric and adult cohorts. The RISK study not only underscored the value of large, well-characterised, prospective cohorts before initiation of treatment, but also highlighted important limitations in observational, multicentre cohorts; namely, treatments are often non-protocolised, resulting in highly varied management between sites, and the lack of specificity of phenotype classifications may hinder generalisability for ascertainment of outcomes.18 Additionally, the initial prognostic performance was assessed using a split-sample validation or leave-one-out cross-validation, which involves re-sampling of individuals from the same cohort as the patients in which the biomarker was discovered. This approach increases the risk of ‘overfitting’, which occurs when data have been so tightly modelled on that of the discovery cohort that it subsequently does not replicate in independent cohorts.19 This is likely to be a major reason why many described biomarkers fail to validate when tested in independent cohorts, thus highlighting the critical need for additional validation to ensure analytical validity.20
Given the invasiveness of endoscopic procedures, the development of accurate non-invasive biomarkers becomes a priority. Therefore focus has turned toward blood-based biomarkers, which would be simple and acceptable to most patients. Based on initial promising prognostic DNA methylation signatures from intestinal epithelial cells,21 DNA methylation signatures in peripheral blood leukocytes have been assessed, although no association with clinical outcome was demonstrated.22 Genome-wide association studies [GWAS] in IBD have also identified a genetic contribution to disease prognosis.23,24 However, given the low odds ratios [OR] observed for associated variants, these findings are not sufficient to guide IBD patient stratification. Consistent with this, several other blood-based tests, serological, genetic, or a combination of both, may have prognostic potential.25–27 However, none of these have been implemented in daily clinical practice for several reasons. These include the fact that these findings were mostly derived from associative studies, with limited effect sizes to impact on clinical decision making and without [prospective] replication. To date, there is only one validated prognostic blood test,28 which was originally derived from a gene-expression signature of CD8 + T cells in treatment-naïve, newly-diagnosed patients with IBD.29 Accordingly, this blood-based prognostic biomarker is being assessed in the PROFILE trial, where newly diagnosed patients with CD are stratified based on their biomarker status to assess if it reliably informs the selection of the most effective treatment strategy for each individual patient.30 This whole-blood prognostic signature illustrates the inherent challenges regarding the time required for discovery, validation, and translation for any successful biomarker.
Recent knowledge suggests that metabolomic or metagenomic profiles, or both, may be powerful biomarkers in the future. However, the methodology for such profiles is less established and findings appear to vary considerably across individuals and studies. Several studies report an enumeration of bacterial taxa that are associated with disease activity, but none of these taxa currently has proven clinical utility as a prognostic marker.31 This limitation may be due to the broad inter- and intra-individual variability in the human microbiome.32
Another emerging area covers radiomic or imaging-based biomarkers. Findings at an initial scan before initiation of therapy may prognosticate disease course by recognising underlying bowel damage from long-term inflammation. One potential radiomic tool is the Lémann index [LI], which is a scoring system that uses clinical, endoscopic, and magnetic resonance enterography [MRE] information to assess cumulative disease burden.33,34 A comparison of disease activity by the Magnetic Resonance Index of Activity [MaRIA] and bowel damage by LI revealed that bowel damage and the LI [but not MaRIA] are independent prognostic factors for intestinal surgery (hazard ratio [HR]: 3.2 and 1.1, respectively) and of CD-related hospitalisation [HR: 1.88 and 1.08, respectively].35 Additional MRE prognostic features include length of ileal disease and bowel wall thickness.36 Likewise, small intestine oral contrast ultrasound scores have been developed that assess bowel wall thickness, lumen diameter, lesion length, number of lesion sites, presence of fistula, mesenteric adipose tissue alteration, abscesses, and lymph nodes. The sonographic lesion index for CD can be prognostic for surgery within a 1-year follow-up.37 There are ongoing efforts to use artificial intelligence to capture key radiological features of predictive value.
3. Predictive Efficacy and Safety Biomarkers, a State-of-the-art Overview
Easily accessible biomarkers that can predict therapy [non-]response would greatly improve patient outcomes. Primary non-response to anti-tumour necrosis factor [TNF] agents has been linked to various clinical features, including age at treatment initiation and previous surgery,38 whereas associations with baseline inflammatory burden have been conflicting.39–41 Similar scoring tools have been developed to predict vedolizumab and ustekinumab response,42,43 including clinical features such as previous surgery, anti-TNF exposure, baseline serum albumin, and draining fistula. Given the large overlap in predictive features between individual therapies, treatment stratification based purely on clinical features has been regarded as ultimately having limited capacity for selecting molecular pathway-based specific treatments. Therefore, there is a high unmet need for identifying molecular predictive biomarkers.
One of the first applications of genetics in IBD therapeutics aimed to identify patients at higher risk of side effects. A historical example was successfully translated to routine clinical practice, specifically the association of polymorphisms in the enzyme thiopurine methyltransferase [TPMT] and thiopurine-induced myelosuppression.44 Testing for TPMT before commencement of thiopurines and appropriate dose reduction reduces the risk of myelosuppression by 10-fold in risk allele carriers.45 GWAS also identified an additional susceptibility locus in the nudix hydrolase 15 [NUDT15] gene region, which is associated with bone marrow suppression.46,47 Polymorphisms in these two genes explain almost 50% of thiopurine-induced myelotoxicity, providing a strong rationale for genotype screening before thiopurine initation.48 GWAS approaches have also identified genetic factors that predispose to aminosalicylate-associated nephrotoxicity. A strong association with the human leukocyte antigen [HLA] region has been demonstrated, but the rarity of this adverse event has precluded its adoption in clinical practice.49
Multiple genetic association studies have also explored the relationship of genetic variants for response to therapy in a retrospective manner, often limited by low sample size.50 In one of the largest observational studies addressing the topic of immunogenicity [a common cause for loss of response to anti-TNF agents51], the PANTS study identified through whole-exome sequencing an association between the HLA-DQA1*05 haplotype and the risk of immunogenicity to anti-TNF agents.52,53 Considering that HLA-DQA1*05 carriage almost doubles the risk of immunogenicity, pre-treatment screening may allow for a more informed decision on the benefit-risk balance of monotherapy versus combination therapy.
Transcriptional profiling has been at the forefront in predictive biomarker development, especially at the tissue level. Early microarray studies that profiled mucosal gene expression of anti-TNF naïve patients with colonic CD or UC identified four genes [IL13RA2, IL-11, IL-6, TNFAIP6] that accurately separated responders from non-responders to infliximab induction.54,55 This IL13RA2 signal was later replicated in independent cohorts.56,57 An attempt to refine the original signature generated by using samples from the ACT1 trial, and to validate it in a phase 2a trial of golimumab in UC, was unsuccessful.55,58,59 Despite the negative result, this pilot study highlighted key factors for successful biomarker discovery. These include the need for tight outcome definitions and for addressing other causes of non-response, including immunogenicity or drug underdosing. Due to the increasing number of publicly available datasets, a meta-analysis across studies may provide further insights into both disease pathogenesis and biomarker discovery. For example, Oncostatin M [OSM] is one of the most highly expressed cytokine genes in the mucosa of anti-TNF non-responders. Using an elegant experimental set-up, the authors not only validated the predictive value of OSM mRNA quantification in multiple cohorts, but also provided mechanistic proof that it may be an alternative, targetable, pathway.60
The combination of single-cell approaches with transcriptomic profiling has allowed for a much more granular overview of gene expression within cellular clusters. Using a combination of transcriptomic and proteomic single-cell platforms, Martin et al. identified a functionally interconnected group of activated cells [inflammatory macrophages, IgG+ plasma cells, activated T cells, dendritic cells, and stromal cells] enriched in the mucosa of anti-TNF CD non-responders.61 A significant challenge in translating these findings remains the transposition of a complex genetic signature to a test that can be easily used in routine clinical care. Whether these signatures of non-response are anti-TNF specific and not simply representing a generic treatment-refractory phenotype remains to be established, especially as this indeed seems to be the case with OSM.62 Reassuringly, studies exploring biomarkers to anti-trafficking agents have not identified a relevant overlap with transcriptional biomarkers of response to anti-TNF agents. In a biomarker discovery study performed in tandem with a phase 2 randomised controlled trial [RCT] testing the efficacy of etrolizumab in anti-TNF naïve UC patients, baseline levels of integrin alpha ε [ITGAE] and granzyme A [GZMA] in the intestinal mucosa predicted endoscopic response.63 Additionally, baseline expression levels of four genes [PIWIL1, MAATS1, RGS13, DCHS2] were identified to predict endoscopic response to vedolizumab.64 None of these genes predicted response to anti-TNF therapy. Serum IL-22 was predictive for anti-IL23p19 success [MEDI2070] in a recent phase 2 RCT trial in patients with CD.65
Transcriptional profiling of peripheral blood may also hold promise for biomarker discovery. Baseline expression of Triggering Receptor Expressed in Myeloid Cells-1 [TREM1] in both gut and blood accurately predicted poor response to anti-TNF therapy, although conflicting results have been published.66–68 Hence, larger validation studies are necessary.
The combination of metabolomic, metataxonomic, or metagenomic profiling has been explored for biomarker discovery.69,70 Overall, patients with a more diverse baseline microbiome and higher microbial diversity show better response to anti-TNF agents, vedolizumab, and ustekinumab. Fewer mucus-colonising bacteria, a higher abundance of short-chain fatty acid-producing bacteria, and lower abundance of pro-inflammatory bacteria are also associated with a favourable outcome.31,71 Models based on a combination of clinical data and microbiota have good predictive accuracy [vedoNet being an excellent example] and certainly merit further investigation as a potential predictive tool of the future.72
Integration of profiles targeting different omics layers may provide an advantage in biomarker discovery. In an inception cohort study of 428 paediatric UC patients receiving standardised mesalamine or corticosteroids and with a pre-defined protocol for therapy escalation, the combination of mucosal gene-expression signatures and abundance of bacterial species with clinical indices could predict upfront who would require escalation to anti-TNF therapy.73 Due to the increasing complexity of such multi-omic data, machine-learning approaches will be increasingly essential to use the full power of these analyses.3
4. Key Questions in Biomarker Development
Effective integration of biomarkers into IBD clinical practice will ultimately rely on well-designed and coordinated efforts in biomarker research. To advance biomarker development, careful consideration must be given to the specific questions and criteria used in study designs and evaluation of potential markers. We recommend the following key areas as important considerations and priorities when developing biomarkers of prognosis and response to therapy in IBD in Box 1.
Box 1. Priorities for research in predicting disease outcome and response to therapy in inflammatory bowel disease [IBD].
| – Defining clear, objective, and clinically relevant outcomes |
| ◦ Prognostic: IBD–related surgery versus IBD-related hospitalisation versus new disease-related complications versus need for biologic/small-molecule agents versus IBD-DI [Disability Index] versus cumulative bowel damage scores [such as Lémann index] versus … |
| ◦ Predictive: mucosal versus transmural healing, and definitions for these |
| – Incorporating adequate drug concentration for biomarkers of response to therapy |
| – Assessing biomarkers in specific patient populations and/or timepoints in the disease course |
| ◦ Prognostic: at diagnosis and in postoperative setting |
| ◦ Predictive: biologic-naïve patients versus at the time of biologic failure [considering prior/current drug exposures] |
| – Defining criteria for evaluating performance and impact of new biomarkers |
| – Standardization of baseline clinical data and phenotyping, including response, across independent studies, allowing pooled data analysis and direct comparisons |
| – Biomarker incorporation into clinical trials: early-phase interventional trials in IBD should have exploratory biomarker development projects built into funding, design, and analysis. Results should be published, even if negative. Appreciation for the length of time and preparation required prior to initiation of such designs, and the general difficulties of obtaining funding for pre-trial methodology and planning activities |
| – Increased academia-industry collaborations to combine intellectual expertise and experience and to mine randomized controlled trial datasets and samples |
| – Well-powered ambitious prospective studies that are specifically designed towards accumulation of data for several molecular layers |
4.1. Defining clear, objective, and relevant clinical outcomes
Clear and objective outcomes are essential for robust endpoints in biomarker research, where reproducibility is critical. We first must define what it is we want to predict, as endpoints will directly inform the performance of any biomarker. For prognosis, outcomes should reflect adverse disease-related events from both patient and health system perspectives. We propose that outcomes for prognostic biomarker performance should include IBD-related surgery, IBD-related hospitalisation, need for biological or small-molecule therapy, and new disease-related complications by Montreal classification [new stricture, abdominal fistula, or abscess, or perianal fistula or abscess for CD, and disease extension for UC]. Each of these outcomes has been used in previous studies investigating impact of therapy and early endoscopic healing on the prognosis of IBD.8,74,75 These outcomes could be used individually, or ideally as a composite, to capture a range of events that would be considered as comprising poor prognosis. In addition, newer outcomes of interest may be used in future prognostic biomarker research, including the IBD Disability Index [IBD-DI] and the LI.34,76 The IBD-DI and LI may become more widely used outcomes as local expertise and familiarity become more common.
Outcomes to assess biomarkers of response can incorporate shorter- or longer-term endpoints, or both. Given the increased importance of objective metrics of response, outcomes for therapeutic response should ideally be based on endoscopic, histological, or radiological assessments [or combinations thereof] of disease activity within 6–12 months after initiating a new therapy or at time of loss of response.77,78 Various definitions of endoscopic remission have been proposed, including specific endoscopic score cut-offs, percentage decrease from baseline score, or absence of ulcerations.79 Although these are reasonable endpoints, the ideal endoscopic target is still a subject of active investigation, as is the radiological target. Whether we should aim for mucosal or transmural healing is still debated, although this will undoubtedly affect biomarker development. To align RCT data as much as possible with real-world observational cohorts, time points for response assessment in biomarker studies should parallel RCT endpoints that are commonly at the end of induction [primary non-response] and the end of the first year of maintenance [secondary non-response]. Alternative outcomes may include longer-term durable steroid-free response, with incorporation of objective resolution of inflammation [non-invasive blood or stool markers, endoscopy, or combinations thereof], fibrosis, or both.
4.2. Incorporating adequate drug exposure in assessment of response to therapy
Although therapeutic drug monitoring has been increasingly used in clinical practice, especially for anti-TNF agents, to date it has rarely been considered in IBD biomarker research. Incorporation of drug-concentration data into biomarker study designs will be key to developing more robust predictors of response to therapy. Outcomes in these studies should be conditioned on having adequate drug concentrations present at the time of response assessment, minimising the risk that mechanistic or molecular responders are considered non-responders because of inadequate exposure. Whereas the ‘optimal’ drug concentrations are not firmly established and may differ from patient to patient, biomarker studies should at least require the presence of drug in the blood at the time of outcome assessment or a minimum concentration as currently accepted.80,81
4.3. Assessing biomarkers in specific patient populations and/or timepoints in disease course
To date, many biomarker results have not been consistently replicated in other cohorts or have been conflicting, as recently highlighted with whole-blood TREM1.66–68 This is likely due to the heterogeneity of studies, including differences in patient populations and outcome definitions. Biomarkers may be specific to certain disease states and may depend on the specific IBD patient clinical scenario. Prognostic biomarkers will be of greatest utility at specific time points in the disease course, especially at diagnosis [Figure 1], to inform treatment and disease monitoring strategies. Another key juncture where prognostic biomarkers can have significant value is in the postoperative setting, to help determine which patients need immediate post operative prophylaxis versus those who may only require therapy monitoring. It is possible that, following therapy introduction, biomarkers change over time and provide a more dynamic and accurate prognostication of disease course. This is another area that warrants further research. However, such biomarker changes might also complicate their use and their potential for generalisation in daily clinical practice.
Biomarkers for therapy response should also consider disease state, in particular current and previous treatment exposures. For example, RCTs have consistently revealed lower response rates in patients with previous anti-TNF exposure, so this group of patients may require a different set of biomarkers to select subsequent treatment, which are distinct from patients who are biologic naïve. Biomarker study designs should focus on specific common clinical scenarios to inform patient selection. For example, such studies should focus on biologic-naïve patients to inform selection of first-line biologic therapy and on the selection of a subsequent therapy at the time of biologic failure. As many health systems and insurers may promote anti-TNF as first-line therapy, particularly with the cost savings of biosimilars, predictive biomarkers for therapy selection following anti-TNF failure is a significant unmet need. Factors such as disease duration or disease phenotype [fistulising versus stricturing disease], and potentially even gender,82 are likely also important stratifying variables when analysing predictive biomarkers for therapeutic response. Last, longitudinal assessment of biomarkers may provide additional predictive value by informing decisions on when to terminate therapy early.
4.4. Defining criteria for evaluating performance and impact of new biomarkers
To incorporate prospective biomarkers into clinical practice, they should show incremental value in addition to current common clinical and commercially available laboratory parameters. Although evaluating the prognostic or predictive performance of a biomarker alone is an important step, such performance must ultimately be compared with other clinical variables. It is possible that biomarkers, particularly if mostly indicative of inflammatory burden, may perform similarly to C-reactive protein or faecal calprotectin. We recommend that IBD biomarker studies compare the predictive performance of selected markers alone and with clinical parameters [such as age, disease phenotype, clinical laboratory values] and then verify whether the composite provides a significant improvement in prediction for biomarker alone or composite models. In IBD prognosis biomarker studies, comparisons should be made with clinical risk factors for more aggressive disease and with consideration of transcriptional tests or serologies as comparison biomarkers that have demonstrated some prognostic capability.28,83,84
5. Biomarker Development in Trial Design
Many of the major advances in IBD from recent decades have been facilitated by carefully performed RCTs, which have rightly been considered the gold standard for investigation of new interventions.85 However, despite the increasing number of therapeutic options for IBD, remission rates across all mechanisms of action seem to plateau at approximately 30% for all drugs. Furthermore, there has been a worrying decline in recruitment to IBD trials globally.86 This has been compounded by the detrimental effect of coronavirus disease 2019 [COVID-19] on trial recruitment and a need to consider alternative methods for future IBD trial conduct.87 Accordingly, there have been growing concerns in the field that such a trajectory is not sustainable and that there is a pressing need to address some of the inefficiencies of clinical trials in IBD.88
5.1. General trial aspects in biomarker development
There are multiple focus areas that can address clinical trial efficiency. These include greater use of adaptive platform designs and appropriate selection of outcome measures to reduce the [historically high] placebo response rates in IBD.89,90 Whereas these topics are beyond the scope of this manuscript, biomarker incorporation into RCTs is another way to improve efficiency of trial design and eventually clinical practice. Many of the developments in clinical trials will be driven and complemented by studies that provide a greater overall understanding of disease pathogenesis. This will ultimately result in better target selection,91 more accurate biomarkers, and key efficiency savings for pharmaceutical companies in drug development programmes.
All the aforementioned biomarkers in IBD perfectly illustrate that there is no single correct approach to biomarker development; biomarkers with strong credentials can be, and have been, developed from both prospective observational cohorts and prospective clinical trials. The development and validation of ITGAE and OSM gene expression levels using datasets from RCTs60,63 highlights the extremely valuable information that can be gained from post-hoc analysis of RCT data. Clinical trials in IBD, particularly industry-sponsored trials, typically collect numerous biological specimens at fixed and regular time points and have highly curated and detailed clinical phenotyping databases from prospective follow-up of patients during the trial. In addition, there is often longer-term follow-up through open-label extension programmes.
We note the recent and commendable efforts by the pharmaceutical industry to build molecular and translational analyses into trials, to allow for identification and validation of drug-specific biomarkers for response.65 However, as not all of these analyses have been reported, we would strongly recommend publication of such biomarker studies, regardless of the results obtained, to help advance knowledge and understanding in the field. Given the considerable inefficiency and the time required to recruit and follow up large cohorts of patients with IBD, the availability of already collected, archived samples and detailed datasets from clinical trials provides a rich opportunity for exploration and collaboration.92 Accordingly, we strongly encourage increased academia-industry collaboration in the near future to combine intellectual expertise and experience and to mine RCT datasets and samples to ultimately drive biomarker development, validation, and further progress in the field.
With regard to trial design, it is important to carefully plan and account for which samples could be collected for development of future biomarkers. The number and nature of both procedures and samples will be dictated by multiple factors, such as funding, ethical approval, trial duration, and availability of staff and facilities, among other considerations. However, sponsors should consider collection [and appropriate storage] of whole blood, serum, stool, peripheral blood mononuclear cells, biopsy samples, and endoscopic and imaging results whenever possible.
5.2. Biomarker development embedded in early drug development
It is worth noting that in analogous clinical trials in oncology, prospective therapeutics typically do not proceed beyond early-phase trial investigation if there is no accompanying predictive biomarker identified, regardless of possible efficacy signals for the therapeutic agent.93 Although this may not be the case yet for IBD, we urge all early-phase interventional trials in IBD to have exploratory biomarker development projects built into trial funding, design, and analysis. Reassuringly, we note that this is already consistent with practices for many commercial companies and ongoing trials in IBD.94
Coupled with furthering progress in the field and the possibility for improved clinical outcomes for patients, there is also significant benefit to trial sponsors from focusing on biomarker development in the early-phase setting. If companion biomarkers to predict response or non-response to therapies can be identified, this would allow for more efficient designs and smaller sample sizes to be used in late-phase trials to affirm validity/clinical utility. This would ultimately reduce costs for each individual trial and potentially expedite the approval of regulatory submission packages following trial completion.
5.3. A need to determine clinical utility
For biomarkers that have demonstrated analytical validity, the most important next step is determining whether there is clinical utility, which would provide definitive evidence to guide therapeutic decision making. Whether this clinical utility requires assessment in a biomarker-guided RCT or in a real-world setting is still a matter of debate and largely depends on the type of biomarker and the credentials of the marker itself. An important point is that TPMT testing, assessed before initiation of thiopurine treatment, has never undergone assessment for clinical utility in an RCT setting. However, TPMT testing is widely used in most health care settings around the world.
For prognostic biomarkers, it is possible to have a biomarker with strong analytical validity that may not necessarily equate to clinical utility. In such instances, assessment in a biomarker-guided RCT is generally advised.95 For predictive biomarkers, such as those for therapeutic response and non-response, if the biomarker robustness is considered strong, the biomarker should be considered for use in clinical practice and assessed for clinical utility in a real-world setting, ideally through well-designed prospective observational studies. It is interesting to note that there are already anecdotal reports from centres adopting some of the novel predictive biomarkers into their standard of care, such as NUDT15 testing before initiation of thiopurines and HLA-DQA1*05 testing before initiation of anti-TNF agents. However, there are other considerations for biomarkers, including cost, reimbursement considerations, approval from health care technology agencies [HTAs], and regulatory approval. Accordingly, there may be reluctance by many to use biomarkers in clinical practice without assessment in an RCT.
Biomarker-guided RCTs to assess clinical utility can be broadly divided into the following two categories: biomarker-guided trials using an adaptive design and biomarker-guided trials using more traditional, non-adaptive designs.96,97 Adaptive clinical trials allow modification of design features based on accruing information during the course of a trial; these designs are increasingly used in drug development programmes.98 Importantly, there are many individual biomarker-guided designs, with similar terminology to describe them and subtle variations between each individual approach.97 Given the number of possible biomarker-guided trial designs, choosing an appropriate design is challenging and requires careful consideration and close discussions with statisticians and trial methodologists with expertise in the area. The critical points to consider are the credentials of the biomarker, including the prevalence of the biomarker in the population of interest, and the number and type of therapies available.
A further important consideration is the role of regulators in helping to support and drive innovation in RCTs to enable discovery, validation, and assessment for clinical utility across different phases of clinical trials. Whereas the Food and Drug Administration of the USA and the European Medicines Agency both encourage the collection of data to develop or validate biomarkers, there has been a lack of clear guidance regarding the use of specific trial designs and what would be considered acceptable coefficients or metrics for use in RCTs.99 Moreover, it is currently unclear whether regulators or HTAs would accept findings from either RCTs or prospective validation cohorts in external territories, before confirmation for either regulatory approval or reimbursement, respectively. Likewise, the International Conference on Harmonisation [ICH] guidance [E16] on biomarkers was developed in 2011 and is in need of an update to reflect progress over the past decade.100 We believe that future regulatory advice and indeed greater uptake of biomarker-guided trials will likely be driven by case studies demonstrating successful implementation across the field.
5.4. Various trial designs, depending on the biomarker context
A biomarker-stratified approach is a potentially suitable option for investigation of single biomarkers with strong credentials.95 Indeed, this approach was taken in the PROFILE trial. PROFILE is the first biomarker-guided trial in IBD where investigators used a biomarker-stratified design to assess a prognostic biomarker for clinical utility to guide choice of therapeutic strategy following diagnosis.30 Accordingly, several other groups around the world investigating single biomarkers are embarking on similar approaches to evaluate clinical utility, including the INHERIT trial [NCT04109300] to assess clinical utility of HLA-DQA1*05 before initiation of infliximab in a Canadian IBD cohort.
When evaluating multiple therapies and multiple biomarkers, a biomarker-stratified design is typically inefficient, given that the trial size should be based on the effect observed in all patients, which is likely to be [at best] modest.101 In this respect, much can be learned from the oncology field.102 Presented with a similar challenge in the setting of metastatic colorectal cancer, the FOCUS4 trial [ISRCTN90061546] was set up to investigate multiple molecular biomarkers and multiple therapies and did so using an umbrella, platform protocol with a multi-arm, multi-stage [MAMS] approach.103 One of the advantages of a seamless phase 2/3 approach in the FOCUS4 platform was that biomarkers or treatments demonstrating preliminary lack of efficacy were dropped early from the platform, and promising new biomarkers or treatments were added as the trial progressed.104
The benefits and efficiencies of biomarker-guided trials have been described, but it is important to note the limitations and challenges experienced from using such novel approaches and designs.105 In particular, the length of time and preparation required before initiation of such designs, and the general difficulties of obtaining funding for pre-trial methodology and planning activities, should be considered. This may be an area of focus for sponsors and IBD societies to consider in the future. To allow greater adoption across the IBD field, there is also a requirement for increased training and development to enable both familiarity and understanding of complex innovative trial designs.106
There are also many unknown aspects of conduct for biomarker-guided trials. These include effects on recruitment and retention from use of more innovative [and potentially more complicated] trial designs, whether biomarker results should be blinded to patients or clinicians during trial conduct, the best comparison groups [ie, which standard-of-care treatments to incorporate], how best to communicate these results to both clinicians and patients and, importantly, how the results affect clinician decision making and patient behaviour. These questions, and many more, could and should be addressed by ongoing and future biomarker-guided trials in the field.
6. Multi-omics Paving the Way to Incorporate the IBD Interactome in Biomarker Development
Learning from ongoing and future clinical trials in IBD and from related medical fields should enable greater progress toward delivering the goal of precision medicine. How we subsequently analyse and integrate generated data is rapidly changing with the increasing availability of high-throughput profiling. Personalised medicine is no longer a theoretical concept but is already happening3; the use of intestinal organoids as an in-vitro model for cystic fibrosis is a key example.107
Multi-omic profiling has generated the IBD interactome, which has provided insights on the mechanisms involved in IBD.3,108 Tentatively, a strategy towards biomarker discovery that summarises specific physiological phenomena associated with prognosis and response to therapy might serve to achieve predictors with clinical utility. Availability of such multi-omic based tools, especially in the early stages of the disease, would maximise the window of opportunity to reduce time to remission and the lower quality of life associated with the current approach in treating IBD patients.6
However, several challenges preclude the discovery and implementation of multi-omic solutions in standard management of patients [Box 2]. First, the deliverables that can be obtained from multi-omic biomarkers is uncertain. For example, predictions based on genetic biomarkers necessarily present an upper limit related to heritability of the investigated trait.109 Although profiles based on more dynamic layers, such as transcriptomic or proteomic data, are certainly closer to the pathogenic events at the molecular level, it is often not appreciated that univocal positive predictions might not be achievable. Rather than striving for individualisation, envisioning ways to incorporate molecular-based stratification [eg, good versus poor response] might be the way for successful integration into the clinical setting. Second, basic studies for molecular biomarkers are often affected by biases that impede distinguishing between cause and consequence.84 Although this problem also affects observational studies for clinical risk factors, prospective and well-powered studies for multi-omic biomarkers can be very costly. This relates to a third aspect associated with the inherent complexity of multi-omic profiling. For instance, even if profiling of many technologies in several tissues allows for refinement of biomarkers with large predictive power, collecting many different biosamples might be impractical in the clinical setting.110 Fourth, the integration of multi-omic biomarkers into clinical decision making can be difficult. Measurements from molecular-based biomarkers can often be noisy due to inherent real within-patient variability. For instance, protein levels might make for an excellent biomarker for longitudinal purposes, but genetic and lifestyle factors unrelated to the disease may complicate obtaining reference intervals.111 Hence, even if molecular biomarkers can provide orthogonal information that complements clinical factors easily obtained in routine patient care,17 those leading to well-defined thresholds and outcomes must be prioritised for successful integrative risk modelling in the context of clinical trials. Finally, datasets should be made publicly available upon publication of the results, allowing re-analysis for different research questions and enabling pooled analyses and thus increasing power. Publishers and funders should not simply encourage, or even better require, raw data deposition in the public domain, but should also insist on the availability of relevant metadata [which should ideally be standardised across the field].
Box 2. Best practices and main challenges for biomarker development.
| Best practices |
| – Well-defined ‘gold standard’ outcomes that are easily assessable/reproducible and clinically relevant [see Box 1] |
| – Compare the prognostic/predictive performance of identified biomarkers with clinically established prognostic/predictive parameters |
| – Development in large, well-characterised prospective cohorts |
| – Reproducibility: independent validation to confirm the absence of overfitting |
| – Confirm clinical utility in a [randomised] trial format |
| – Deposit raw data and detailed metadata in the public domain |
| Main challenges |
| – Highly varied management between patients due to lack of standardised treatment protocols and phenotyping in observational studies |
| – Variable, non-standardised endpoints in observational studies |
| – Biomarker applicability may differ in diverse populations with different genetic backgrounds, environmental exposures … |
| – Translating a complex signature to a test that can be easily used in routine clinical care, especially in the context of a multi-omic biomarker |
| – Distinguishing cause and consequence, advancing beyond pure associative studies |
| – Prospective and well-powered multi-omic studies are cost-prohibitive |
| – Lack of data dissemination and access to relevant data warehouses as available in other fields [ie, TCGA for cancer] |
| – Making relevant multi-omics datasets publicly accessible, including standardised phenotypic data and metadata |
The scope for multi-omic biomarkers for prognosis and response to therapy depends on finding the right balance between cutting-edge technology and easy-to-implement clinical tools. We acknowledge the current problems clinicians face in implementing biomarkers, including lack of robustness, invasiveness, and difficulties in measurements. However, clinical trials such as PROFILE demonstrate that omic-based biomarkers can have actionable potential and can be incorporated into the algorithms and clinical toolkit used for longitudinal management of patients.28,112 Instead of promoting a patchy strategy that targets specific omic technologies in unpowered studies, a renewed focus on well-powered ambitious prospective studies that are specifically designed towards accumulation of data for several molecular layers may be the key to discovering unexpected yet reliable biomarkers associated with specific responses of interest.113 This is the objective of several Horizon2020 Innovative Medicine Initiatives [IMI] projects [ImmUniverse, 3TR], IBD plexus, and many other collaborative initiatives.114 Besides allowing for integrative studies that can better refine our knowledge about the biological mechanisms associated with prognosis and drug response, this strategy could ultimately yield a panel of multiple markers that together provide likelihood of response to each mechanism and subgroup associated with these events.6,115
7. Conclusion
Over the past decade there have been major advances in understanding the prognosis of IBD and prediction of treatment response or non-response, with several promising biomarkers either undergoing validation in large prospective cohorts or currently being assessed for clinical utility. Ultimately, the best biomarkers will be those that can effectively guide treatment decisions and subsequently change disease course in the long term, which will likely involve the use of multiple complementary approaches [such as the IBD interactome].3 We have highlighted several best-practice examples, including the main challenges encountered in biomarker development to date, and have outlined key research priorities for consideration by funders and research groups to help further advance progress towards precision medicine in IBD.
Conflict of Interest
ECCO has diligently maintained a disclosure policy of potential conflicts of interests [CoI]. The conflict of interest declaration is based on a form used by the International Committee of Medical Journal Editors [ICMJE]. The CoI disclosures are not only stored at the ECCO Office and the editorial office of JCC, but are also open to public scrutiny on the ECCO website [https://www.ecco-ibd.eu/about-ecco/ecco-disclosures.html], providing a comprehensive overview of potential conflicts of interest of the authors.
Acknowledgements
We would like to acknowledge academic medical illustrator Jill Gregory, CMI, FAMI, for assistance with figure design.
Contributor Information
Scientific Workshop Steering Committee:
Scientific Workshop Steering Committee
Bram Verstockta,b, Claudio Fiocchil, Joana Torresk, Michael Scharlm
aUniversity Hospitals Leuven Department of Gastroenterology and Hepatology, KU Leuven, Leuven, Belgium
bKU Leuven Department of Chronic Diseases, Metabolism and Ageing, Translational Research Center for Gastrointestinal Disorders [TARGID], Leuven, Belgium
kDivision of Gastroenterology, Hospital Beatriz Ângelo, Loures, Portugal
lDepartment of Inflammation & Immunity, Lerner Research Institute, and Department of Gastroenterology, Hepatology & Nutrition, Digestive Disease Institute, Cleveland Clinic, Cleveland, OH, USA
mDepartment of Gastroenterology and Hepatology, University Hospital Zürich, Switzerland.
Disclaimer
ECCO Scientific Workshop Papers are targeted at health care professionals only and are based on a standardised drafting procedure. Any treatment decisions are a matter for the individual clinician and may not be based exclusively on the content of the ECCO Scientific Workshop Papers. The European Crohn’s and Colitis Organisation and/or any of its staff members and/or any paper contributor may not be held liable for any information published in good faith in the ECCO Scientific Workshop Papers.
References
- 1. Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet 2017;389:1741–55. [DOI] [PubMed] [Google Scholar]
- 2. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet 2017;389:1756–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fiocchi C, Dragoni G, Iliopoulos D, et al. Results of the seventh scientific workshop of the ECCO: precision medicine in IBD— what, why and how. J Crohns Colitis 2021. PMID: 33733656. [Google Scholar]
- 4. Torres J, Halfvarson J, Rodriguez-Lago I, et al. Results of the seventh scientific workshop of ECCO: prediction and prevention of inflammatory bowel diseases. J Crohns Colitis 2021. PMID: 33730755. [DOI] [PubMed] [Google Scholar]
- 5. FDA-NIH Biomarker Working Group. Best [biomarkers, endpoints, and other tools] resource.2016. https://www.ncbi.nlm.nih.gov/books/NBK326791/. Accessed 15 November 2020.
- 6. Noor NM, Verstockt B, Parkes M, Lee JC. Personalised medicine in Crohn’s disease. Lancet Gastroenterol Hepatol 2020;5:80–92. [DOI] [PubMed] [Google Scholar]
- 7. Colombel JF, Panaccione R, Bossuyt P, et al. Effect of tight control management on Crohn’s disease [CALM]: a multicentre, randomised, controlled phase 3 trial. Lancet 2017;390:2779–89. [DOI] [PubMed] [Google Scholar]
- 8. Ungaro RC, Yzet C, Bossuyt P, et al. Deep remission at 1 year prevents progression of early Crohn’s disease. Gastroenterology 2020;159:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cameron D, Piccart-Gebhart MJ, Gelber RD, et al. ; Herceptin Adjuvant [HERA] Trial Study Team . 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: final analysis of the HERceptin Adjuvant [HERA] trial. Lancet 2017;389:1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sparano JA, Gray RJ, Ravdin PM, et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. N Engl J Med 2019;380:2395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elborn JS, Ramsey BW, Boyle MP, et al. ; VX-809 TRAFFIC and TRANSPORT study groups . Efficacy and safety of lumacaftor/ivacaftor combination therapy in patients with cystic fibrosis homozygous for Phe508del CFTR by pulmonary function subgroup: a pooled analysis. Lancet Respir Med 2016;4:617–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hingorani AD, Windt DA, Riley RD, et al. ; PROGRESS Group . Prognosis research strategy [PROGRESS] 4: stratified medicine research. BMJ 2013;346:e5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solberg IC, Vatn MH, Høie O, et al. ; IBSEN Study Group . Clinical course in Crohn’s disease: results of a Norwegian population-based ten-year follow-up study. Clin Gastroenterol Hepatol 2007;5:1430–8. [DOI] [PubMed] [Google Scholar]
- 15. Wintjens D, Bergey F, Saccenti E, et al. Disease activity patterns of Crohn’s disease in the first 10 years after diagnosis in the population-based IBD South Limburg cohort. J Crohns Colitis 2021;15:391‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beaugerie L, Seksik P, Nion-Larmurier I, Gendre JP, Cosnes J. Predictors of Crohn’s disease. Gastroenterology 2006;130:650–6. [DOI] [PubMed] [Google Scholar]
- 17. Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389:1710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krishnakumar C, Ballengee CR, Liu C, et al. Variation in care in the management of children with Crohn’s disease: data from a multicenter inception cohort study. Inflamm Bowel Dis 2019;25:1208–17. [DOI] [PubMed] [Google Scholar]
- 19. Subramanian J, Simon R. Overfitting in prediction models ‐ is it a problem only in high dimensions? Contemp Clin Trials 2013;36:636–41. [DOI] [PubMed] [Google Scholar]
- 20. Korn EL, Freidlin B. Quantitative assessment of a prognostic or predictive biomarker panel. J Biopharm Stat 2018;28:264–81. [DOI] [PubMed] [Google Scholar]
- 21. Howell KJ, Kraiczy J, Nayak KM, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 2018;154:585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Somineni HK, Venkateswaran S, Kilaru V, et al. Blood-derived DNA methylation signatures of Crohn’s disease and severity of intestinal inflammation. Gastroenterology 2019;156:2254–65.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haritunians T, Taylor KD, Targan SR, et al. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis 2010;16:1830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee JC, Biasci D, Roberts R, et al. ; UK IBD Genetics Consortium . Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet 2017;49:262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dubinsky MC, Kugathasan S, Mei L, et al. ; Western Regional Pediatric IBD Research Alliance; Pediatric IBD Collaborative Research Group; Wisconsin Pediatric IBD Alliance . Increased immune reactivity predicts aggressive complicating Crohn’s disease in children. Clin Gastroenterol Hepatol 2008;6:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siegel CA, Horton H, Siegel LS, et al. A validated web-based tool to display individualised Crohn’s disease predicted outcomes based on clinical, serologic and genetic variables. Aliment Pharmacol Ther 2016;43:262–71. [DOI] [PubMed] [Google Scholar]
- 27. Wu J, Lubman DM, Kugathasan S, et al. Serum protein biomarkers of fibrosis aid in risk stratification of future stricturing complications in pediatric Crohn’s disease. Am J Gastroenterol 2019;114:777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biasci D, Lee JC, Noor NM, et al. A blood-based prognostic biomarker in IBD. Gut 2019;68:1386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee JC, Lyons PA, McKinney EF, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parkes M, Noor NM, Dowling F, et al. PRedicting outcomes for Crohn’s dIsease using a moLecular biomarkEr [PROFILE]: protocol for a multicentre, randomised, biomarker-stratified trial. BMJ Open 2018;8:e026767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caenepeel C, Sadat Seyed Tabib N, Vieira-Silva S, Vermeire S. Review article: how the intestinal microbiota may reflect disease activity and influence therapeutic outcome in inflammatory bowel disease. Aliment Pharmacol Ther 2020;52:1453–68. [DOI] [PubMed] [Google Scholar]
- 32. Integrative HMPRNC. The integrative human microbiome project. Nature 2019;569:641–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pariente B, Cosnes J, Danese S, et al. Development of the Crohn’s disease digestive damage score, the Lémann score. Inflamm Bowel Dis 2011;17:1415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pariente B, Mary JY, Danese S, et al. Development of the Lémann index to assess digestive tract damage in patients with Crohn’s disease. Gastroenterology 2015;148:52–63.e3. [DOI] [PubMed] [Google Scholar]
- 35. Fiorino G, Morin M, Bonovas S, et al. Prevalence of bowel damage assessed by cross-sectional imaging in early Crohn’s disease and its impact on disease outcome. J Crohns Colitis 2017;11:274–80. [DOI] [PubMed] [Google Scholar]
- 36. Jones GR, Fascì-Spurio F, Kennedy NA, et al. Faecal calprotectin and magnetic resonance enterography in ileal Crohn’s disease: correlations between disease activity and long-term follow-up. J Crohns Colitis 2019;13:442–50. [DOI] [PubMed] [Google Scholar]
- 37. Calabrese E, Zorzi F, Zuzzi S, et al. Development of a numerical index quantitating small bowel damage as detected by ultrasonography in Crohn’s disease. J Crohns Colitis 2012;6:852–60. [DOI] [PubMed] [Google Scholar]
- 38. Billiet T, Papamichael K, de Bruyn M, et al. A matrix-based model predicts primary response to infliximab in Crohn’s Disease. J Crohns Colitis 2015;9:1120–6. [DOI] [PubMed] [Google Scholar]
- 39. Louis E, Vermeire S, Rutgeerts P, et al. A positive response to infliximab in Crohn disease: association with a higher systemic inflammation before treatment but not with -308 TNF gene polymorphism. Scand J Gastroenterol 2002;37:818–24. [PubMed] [Google Scholar]
- 40. Sandborn WJ, Colombel JF, D’Haens G, et al. Association of baseline C-reactive protein and prior anti-tumor necrosis factor therapy with need for weekly dosing during maintenance therapy with adalimumab in patients with moderate to severe Crohn’s disease. Curr Med Res Opin 2013;29:483–93. [DOI] [PubMed] [Google Scholar]
- 41. Verstockt B, Stylli J, Rahimian P, et al. An increased baseline mucosal TNF burden linked to adalimumab non-response: opportunities for therapeutic drug monitoring. In: 15th Conference of ECCO; Feb 12‐15, 2020; Vienna. [Google Scholar]
- 42. Dulai PS, Boland BS, Singh S, et al. Development and validation of a scoring system to predict outcomes of vedolizumab treatment in patients with Crohn’s disease. Gastroenterology 2018;155:687–95 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dulai PS, Guizzetti L, Ma T, et al. Clinical prediction model and decision support tool for ustekinumab in Crohn’s disease. Am J Gastroenterol 2019;114[Suppl];S373. [Google Scholar]
- 44. Relling MV, Schwab M, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther 2019;105:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Coenen MJ, de Jong DJ, van Marrewijk CJ, et al. ; TOPIC Recruitment Team . Identification of patients with variants in TPMT and dose reduction reduces hematologic events during thiopurine treatment of inflammatory bowel disease. Gastroenterology 2015;149:907–17.e7. [DOI] [PubMed] [Google Scholar]
- 46. Nishii R, Moriyama T, Janke LJ, et al. Preclinical evaluation of NUDT15-guided thiopurine therapy and its effects on toxicity and antileukemic efficacy. Blood 2018;131:2466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moriyama T, Nishii R, Perez-Andreu V, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet 2016;48:367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schaeffeler E, Jaeger SU, Klumpp V, et al. Impact of NUDT15 genetics on severe thiopurine-related hematotoxicity in patients with European ancestry. Genet Med 2019;21:2145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heap GA, So K, Weedon M, et al. Clinical features and HLA association of 5-aminosalicylate [5-ASA]-induced nephrotoxicity in inflammatory bowel disease. J Crohns Colitis 2016;10:149–58. [DOI] [PubMed] [Google Scholar]
- 50. Barber GE, Yajnik V, Khalili H, et al. Genetic markers predict primary non-response and durable response to anti-TNF biologic therapies in Crohn’s disease. Am J Gastroenterol 2016;111:1816–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kennedy NA, Heap GA, Green HD, et al. ; UK Inflammatory Bowel Disease Pharmacogenetics Study Group . Predictors of anti-TNF treatment failure in anti-TNF-naive patients with active luminal Crohn’s disease: a prospective, multicentre, cohort study. Lancet Gastroenterol Hepatol 2019;4:341–53. [DOI] [PubMed] [Google Scholar]
- 52. Sazonovs A, Kennedy NA, Moutsianas L, et al. ; PANTS Consortium . HLA-DQA1*05 carriage associated with development of anti-drug antibodies to infliximab and adalimumab in patients with Crohn’s disease. Gastroenterology 2020;158:189–99. [DOI] [PubMed] [Google Scholar]
- 53. Sazonovs A, Ahmad T, Anderson CA. Underpowered PANTS: a response to the conclusions of “Extended analysis identifies drug-specific association of two distinct HLA class II haplotypes for development of immunogenicity to adalimumab and infliximab. Gastroenterology 2021;160:470–1. [DOI] [PubMed] [Google Scholar]
- 54. Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis 2010;16:2090–8. [DOI] [PubMed] [Google Scholar]
- 55. Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009;58:1612–9. [DOI] [PubMed] [Google Scholar]
- 56. Verstockt B, Verstockt S, Creyns B, et al. Mucosal IL13RA2 expression predicts nonresponse to anti-TNF therapy in Crohn’s disease. Aliment Pharmacol Ther 2019;49:572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Toedter G, Li K, Marano C, et al. Gene expression profiling and response signatures associated with differential responses to infliximab treatment in ulcerative colitis. Am J Gastroenterol 2011;106:1272–80. [DOI] [PubMed] [Google Scholar]
- 58. Toedter G, Li K, Marano C, et al. Gene expression profiling and response signatures associated with differential responses to infliximab treatment in ulcerative colitis. Am J Gastroenterol 2011;106:1272–80. [DOI] [PubMed] [Google Scholar]
- 59. Telesco SE, Brodmerkel C, Zhang H, et al. Gene expression signature for prediction of golimumab response in a phase 2a open-label trial of patients with ulcerative colitis. Gastroenterology 2018;155:1008–11.e8. [DOI] [PubMed] [Google Scholar]
- 60. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators . Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019;178:1493–508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Verstockt S, Verstockt B, Vermeire S. Oncostatin M as a new diagnostic, prognostic and therapeutic target in inflammatory bowel disease [IBD]. Expert Opin Ther Targets 2019;23:943–54. [DOI] [PubMed] [Google Scholar]
- 63. Tew GW, Hackney JA, Gibbons D, et al. Association between response to etrolizumab and expression of integrin αE and granzyme A in colon biopsies of patients with ulcerative colitis. Gastroenterology 2016;150:477–87.e9. [DOI] [PubMed] [Google Scholar]
- 64. Verstockt B, Verstockt S, Veny M, et al. Expression levels of 4 genes in colon tissue might be used to predict which patients will enter endoscopic remission after vedolizumab therapy for inflammatory bowel diseases. Clin Gastroenterol Hepatol 2020;18:1142–51.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sands BE, Chen J, Feagan BG, et al. Efficacy and safety of MEDI2070, an antibody against interleukin 23, in patients with moderate to severe Crohn’s disease: a phase 2a study. Gastroenterology 2017;153:77–86.e6. [DOI] [PubMed] [Google Scholar]
- 66. Gaujoux R, Starosvetsky E, Maimon N, et al. ; Israeli IBD research Network [IIRN] . Cell-centred meta-analysis reveals baseline predictors of anti-TNFα non-response in biopsy and blood of patients with IBD. Gut 2019;68:604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Verstockt B, Verstockt S, Dehairs J, et al. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine 2019;40:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Verstockt B, Verstockt S, Blevi H, et al. TREM-1, the ideal predictive biomarker for endoscopic healing in anti-TNF-treated Crohn’s disease patients? Gut 2019;68:1531–3. [DOI] [PubMed] [Google Scholar]
- 69. Ding NS, McDonald JAK, Perdones-Montero A, et al. Metabonomics and the gut microbiome associated with primary response to anti-TNF therapy in Crohn’s disease. J Crohns Colitis 2020;14:1090–102. [DOI] [PubMed] [Google Scholar]
- 70. Aden K, Rehman A, Waschina S, et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel diseases. Gastroenterology 2019;157:1279–92.e11. [DOI] [PubMed] [Google Scholar]
- 71. Yilmaz B, Juillerat P, Øyås O, et al. ; Swiss IBD Cohort Investigators . Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med 2019;25:323–36. [DOI] [PubMed] [Google Scholar]
- 72. Ananthakrishnan AN, Luo C, Yajnik V, et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 2017;21:603–10.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hyams JS, Davis Thomas S, Gotman N, et al. Clinical and biological predictors of response to standardised paediatric colitis therapy [PROTECT]: a multicentre inception cohort study. Lancet 2019;393:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khanna R, Bressler B, Levesque BG, et al. ; REACT Study Investigators . Early combined immunosuppression for the management of Crohn’s disease [REACT]: a cluster randomised controlled trial. Lancet 2015;386:1825–34. [DOI] [PubMed] [Google Scholar]
- 75. Frøslie KF, Jahnsen J, Moum BA, Vatn MH; IBSEN Group . Mucosal healing in inflammatory bowel disease: results from a Norwegian population-based cohort. Gastroenterology 2007;133:412–22. [DOI] [PubMed] [Google Scholar]
- 76. Gower-Rousseau C, Sarter H, Savoye G, et al. ; International Programme to Develop New Indexes for Crohn’s Disease [IPNIC] group. Validation of the Inflammatory Bowel Disease Disability Index in a population-based cohort. Gut 2017;66:588–96. [DOI] [PubMed] [Google Scholar]
- 77. Colombel JF, D’haens G, Lee WJ, Petersson J, Panaccione R. Outcomes and strategies to support a treat-to-target approach in inflammatory bowel disease: a systematic review. J Crohns Colitis 2020;14:254–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ungaro R, Colombel JF, Lissoos T, Peyrin-Biroulet L. A treat-to-target update in ulcerative colitis: a systematic review. Am J Gastroenterol 2019;114:874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dulai PS, Levesque BG, Feagan BG, D’Haens G, Sandborn WJ. Assessment of mucosal healing in inflammatory bowel disease: review. Gastrointest Endosc 2015;82:246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Feuerstein JD, Nguyen GC, Kupfer SS, Falck-Ytter Y, Singh S; American Gastroenterological Association Institute Clinical Guidelines Committee . American Gastroenterological Association Institute guideline on therapeutic drug monitoring in inflammatory bowel disease. Gastroenterology 2017;153:827–34. [DOI] [PubMed] [Google Scholar]
- 81. Alsoud D, Vermeire S, Verstockt B. Monitoring vedolizumab and ustekinumab drug levels in patients with inflammatory bowel disease: hype or hope? Curr Opin Pharmacol 2020;55:17–30. [DOI] [PubMed] [Google Scholar]
- 82. Cutolo M, Straub RH. Sex steroids and autoimmune rheumatic diseases: state of the art. Nat Rev Rheumatol 2020;16:628–44. [DOI] [PubMed] [Google Scholar]
- 83. Dubinsky MC. Serologic and laboratory markers in prediction of the disease course in inflammatory bowel disease. World J Gastroenterol 2010;16:2604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Somineni HK, Venkateswaran S, Kilaru V, et al. Blood-derived DNA methylation signatures of Crohn’s disease and severity of intestinal inflammation. Gastroenterology 2019;156:2254–65.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Collins R, Bowman L, Landray M, Peto R. The magic of randomization versus the myth of real-world evidence. N Engl J Med 2020;382:674–8. [DOI] [PubMed] [Google Scholar]
- 86. Harris MS, Wichary J, Zadnik M, Reinisch W. Competition for clinical trials in inflammatory bowel diseases. Gastroenterology 2019;157:1457–61.e2. [DOI] [PubMed] [Google Scholar]
- 87. Noor NM, Hart AL, Irving PM, et al. Clinical trials [and tribulations]: the immediate effects of covid-19 on IBD clinical research activity in the United Kingdom. J Crohns Colitis 2020;14:1769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ma C, Guizzetti L, Jairath V. Improving clinical trial efficiency in gastroenterology. Gastroenterology 2019;157:892–3. [DOI] [PubMed] [Google Scholar]
- 89. Coalition APT. Adaptive platform trials: definition, design, conduct and reporting considerations. Nat Rev Drug Discov 2019;18:797–807. [DOI] [PubMed] [Google Scholar]
- 90. Jairath V, Jeyarajah J, Zou G, et al. A composite disease activity index for early drug development in ulcerative colitis: development and validation of the UC-100 score. Lancet Gastroenterol Hepatol 2019;4:63–70. [DOI] [PubMed] [Google Scholar]
- 91. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015;47: 856–60. [DOI] [PubMed] [Google Scholar]
- 92. Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nicolaides NC, O’Shannessy DJ, Albone E, Grasso L. Co-development of diagnostic vectors to support targeted therapies and theranostics: essential tools in personalized cancer therapy. Front Oncol 2014;4:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sandborn WJ, Vermeire S, Tyrrell H, et al. ; Etrolizumab Global Steering Committee . Etrolizumab for the treatment of ulcerative colitis and Crohn’s disease: an overview of the phase 3 clinical program. Adv Ther 2020;37:3417–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Freidlin B, Korn EL. Biomarker enrichment strategies: matching trial design to biomarker credentials. Nat Rev Clin Oncol 2014;11:81–90. [DOI] [PubMed] [Google Scholar]
- 96. Antoniou M, Jorgensen AL, Kolamunnage-Dona R. Biomarker-guided adaptive trial designs in phase II and phase III: a methodological review. PLoS One 2016;11:e0149803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Antoniou M, Jorgensen A, Kolamunnage-Dona R. Biomarker-guided Trial Designs [bigted]: an Online Tool to Help Develop Personalised Medicine. 2017. http://www.bigted.org Accessed Sept 15, 2020.
- 98. Bothwell LE, Avorn J, Khan NF, Kesselheim AS. Adaptive design clinical trials: a review of the literature and ClinicalTrials.gov. BMJ Open 2018;8:e018320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dulai PS, Peyrin-Biroulet L, Danese S, et al. Approaches to integrating biomarkers into clinical trials and care pathways as targets for the treatment of inflammatory bowel diseases. Gastroenterology 2019;157:1032–43.e1. [DOI] [PubMed] [Google Scholar]
- 100.US Food and Drug Administration. International conference on harmonisation; guidance on e16 biomarkers related to drug or biotechnology product development: context, structure, and format of qualification submissions; availability. Notice. Fed Regist 2011;76:49773–4. [PubMed] [Google Scholar]
- 101. Kaplan R, Maughan T, Crook A, et al. Evaluating many treatments and biomarkers in oncology: a new design. J Clin Oncol 2013;31:4562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Freidlin B, Allegra CJ, Korn EL. Moving molecular profiling to routine clinical practice: a way forward? J Natl Cancer Inst 2020;112:773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Adams R, Brown E, Brown L, et al. ; FOCUS4 Trial Investigators . Inhibition of EGFR, HER2, and HER3 signalling in patients with colorectal cancer wild-type for BRAF, PIK3CA, KRAS, and NRAS [FOCUS4-D]: a phase 2-3 randomised trial. Lancet Gastroenterol Hepatol 2018;3:162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Parmar MKB, Sydes MR, Cafferty FH, et al. Testing many treatments within a single protocol over 10 years at MRC CTU at UCL: multi-arm, multi stage platform, umbrella and basket protocols. Clin Trials 2017;14:451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Antoniou M, Kolamunnage-Dona R, Wason J, et al. Biomarker-guided trials: challenges in practice. Contemp Clin Trials Commun 2019;16:100493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Blagden SP, Billingham L, Brown LC, et al. ; Experimental Cancer Medicine Centres [ECMC] CID trials working group . Effective delivery of Complex Innovative Design [CID] cancer trials. A consensus statement. Br J Cancer 2020;122:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. de Poel E, Lefferts JW, Beekman JM. Intestinal organoids for cystic fibrosis research. J Cyst Fibros 2020;19[Suppl 1]:60–4. [DOI] [PubMed] [Google Scholar]
- 108. Sudhakar P, Verstockt B, Cremer J, et al. Understanding the molecular drivers of disease heterogeneity in crohn’s disease using multi-omic data integration and network analysis. Inflamm Bowel Dis 2020, Dec 14.. doi: 10.1093/ibd/izaa281. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Marigorta UM, Rodríguez JA, Gibson G, Navarro A. Replicability and prediction: lessons and challenges from GWAS. Trends Genet 2018;34:504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Boyapati RK, Kalla R, Satsangi J, Ho GT. Biomarkers in search of precision medicine in IBD. Am J Gastroenterol 2016;111:1682–90. [DOI] [PubMed] [Google Scholar]
- 111. Enroth S, Johansson A, Enroth SB, Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat Commun 2014;5:4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Whitcomb DC. What is personalized medicine and what should it replace? Nat Rev Gastroenterol Hepatol 2012;9:418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat Rev Genet 2018;19:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Seyed Tabib NS, Madgwick M, Sudhakar P, Verstockt B, Korcsmaros T, Vermeire S. Big data in IBD: big progress for clinical practice. Gut 2020;69:1520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gerich ME, McGovern DP. Towards personalized care in IBD. Nat Rev Gastroenterol Hepatol 2014;11:287–99. [DOI] [PubMed] [Google Scholar]