Abstract
Background:
Loeys-Dietz Syndrome (LDS) is an inherited disorder predisposing individuals to thoracic aortic aneurysm and dissection (TAAD). Currently, there are no medical treatments except surgical resection. Although the genetic basis of LDS is well-understood, molecular mechanisms underlying the disease remain elusive impeding the development of a therapeutic strategy. In addition, aortic smooth muscle cells (SMC) have heterogenous embryonic origins depending on their spatial location, and lineage-specific effects of pathogenic variants on SMC function, likely causing regionally constrained LDS manifestations, have been unexplored.
Methods:
We identified an LDS family with a dominant pathogenic variant in TGFBR1 gene (TGFBR1A230T) causing aortic root aneurysm and dissection. To accurately model the molecular defects caused by this mutation, we used human-induced pluripotent stem cells (hiPSC) from a subject with normal aorta to generate hiPSC carrying TGFBR1A230T, and corrected the mutation in patient-derived hiPSC using CRISPR-Cas9 gene editing. Following their lineage-specific SMC differentiation through cardiovascular progenitor cell (CPC) and neural crest stem cell (NCSC) lineages, we employed conventional molecular techniques and single-cell RNA-sequencing (scRNA-seq) to characterize the molecular defects. The resulting data led to subsequent molecular and functional rescue experiments employing Activin A and rapamycin.
Results:
Our results indicate the TGFBR1A230T mutation impairs contractile transcript and protein levels, and function in CPC-SMC, but not in NCSC-SMC. ScRNA-seq results implicate defective differentiation even in TGFBR1A230T/+ CPC-SMC including disruption of SMC contraction, and extracellular matrix formation. Comparison of patient-derived and mutation-corrected cells supported the contractile phenotype observed in the mutant CPC-SMC. TGFBR1A230T selectively disrupted SMAD3 and AKT activation in CPC-SMC, and led to increased cell proliferation. Consistently, scRNA-seq revealed molecular similarities between a loss-of-function SMAD3 mutation (SMAD3c.652delA/+) and TGFBR1A230T/+. Lastly, combination treatment with Activin A and rapamycin during or after SMC differentiation significantly improved the mutant CPC-SMC contractile gene expression, and function; and rescued the mechanical properties of mutant CPC-SMC tissue constructs.
Conclusions:
This study reveals that a pathogenic TGFBR1 variant causes lineage-specific SMC defects informing the etiology of LDS-associated aortic root aneurysm. As a potential pharmacological strategy, our results highlight a combination treatment with Activin A and rapamycin that can rescue the SMC defects caused by the variant.
Keywords: Loeys-Dietz Syndrome, TGFBR1, hiPSC, Vascular biology, Smooth muscle cell differentiation, Single-cell RNA-Sequencing, Activin A, Rapamycin
Introduction
Thoracic aortic aneurysm (TAA) is a focal dilatation of the thoracic aorta that is highly hereditary. There are few pharmacological treatments to prevent or reverse TAA, and the only therapy to prevent subsequent aortic dissections is surgical repair.1 TAA formation is associated with dominant mutations in genes involved in canonical transforming growth factor-β (TGF-β) signaling, extracellular matrix (ECM) modeling, and vascular SMC contraction.2–6 Transforming Growth Factor Beta Receptor type I and II (TGFBR1 and TGFBR2) mutations predispose patients to thoracic aortic aneurysm and dissection (TAAD), and is known as Loeys-Dietz Syndrome (LDS).2 The mouse model of LDS carrying the p.M318R mutation in Tgfbr1 also recapitulated aortic root aneurysms observed in humans.7 Early reports about LDS-related disorders including Marfan Syndrome (MFS) led to the speculation that elevated TGFβ signaling is the major driver of TAAD.8–10 However, subsequent molecular studies revealed that pathogenic variants in TGFβ pathway genes (TGFBR1, TGFBR2, SMAD3, TGFB2, and TGFB3) are loss-of-function mutations.11–17 The paradoxical elevation in TGFβ signaling in end-stage aneurysmal tissue from patients with inactivating mutations suggests that end-stage tissue may not be ideal to discern the early molecular events underlying TAAD.
Vascular SMC of different aortic regions have distinct embryonic origins. Ascending aorta SMC are derived from both neural crest stem cells (NCSC) and cardiovascular progenitor cells (CPC) in the second heart field while the SMC in the aortic root are mainly derived from CPC lineage.18,19 Given the cellular and ECM abnormalities of aortic medial layer in TAA patients, vascular SMC have been the central focus in understanding the pathophysiology of aneurysm formation. Individuals with loss-of-function mutations in SMC contractile genes or early loss of myosin light chain phosphorylation are predisposed to TAAD.20,21 Notably, TGFβ signaling plays a critical role in vascular SMC differentiation,22 and regulates contractile gene expression.23 Deletion of a TGFBR downstream effector, SMAD2, causes defective differentiation of NCSC-derived SMC (NCSC-SMC)24, while a loss-of-function SMAD3 mutation disrupts CPC-derived SMC (CPC-SMC) contractile function in a human induced pluripotent stem cell (hiPSC) disease model.25 However, it remains unclear how pathogenic variants of TGFβ pathway genes alter downstream signaling events and SMC differentiation from CPC and NCSC lineages.
Here we report an LDS family with TAAD, carrying an autosomal dominant pathogenic variant (TGFBR1A230T) in the TGFBR1 gene. To investigate the molecular defects caused by this mutation, we generated mutant hiPSC using hiPSC from a subject with normal aorta, and corrected the mutation in patient derived hiPSC using CRISPR-Cas9 gene editing. We observed that TGFBR1A230T altered the differentiation dynamics and contractile function in CPC-SMC, but not in NCSC-SMC. Single-cell profiling revealed significant differentiation defects in mutant SMC with de-enrichment of ECM organization, and contractile function genes. Comparison of patient and patient-corrected CPC-SMC recapitulated the molecular and functional phenotype associated with the mutation. In downstream signaling, TGFBR1A230T impaired SMAD3 and AKT activation. Consistent with this finding, a loss-of-function mutation in SMAD3 exhibited a largely overlapping molecular phenotype with TGFBR1A230T. Motivated by the molecular characterization, we rescued the molecular and functional defects caused by the mutation using a combination treatment with Activin A and Rapamycin in vitro, offering a potential pharmacological strategy to prevent aortic root aneurysm in LDS patients with TGFBR1 mutations.
Methods
All experiments were performed according to the protocols approved by the Institutional Review Board at the University of Michigan. All materials can be made available from the corresponding authors upon reasonable request. The raw and processed scRNA-seq data have been deposited to the Gene Expression Omnibus (GEO) database under the accession number GSE175647. Detailed descriptions of additional experimental procedures are available in the Supplemental Material.
Lineage-specific induction of hiPSC
Lineage-specific induction of hiPSC to generate CPC-SMC, and NCSC-SMC were performed as described previously.25–29 For CPC-SMC differentiation, hiPSC were seeded on Matrigel-coated plates at a density of 3×104 cells/cm2 in TesRE8 medium with 10μM Y27632 (Stemgent), and then incubated in CPC differentiation medium (DMEM/F12 (Gibco), B27 (without vitamin A, Life Technologies), 25ng/mL BMP4 (PeproTech), 8μM CHIR99021 (Biogen), 50μg/mL ascorbic acid (Sigma), 400μM 1-thioglycerol (Sigma), and 1% Penicillin-Streptomycin (Gibco)) for 3 days. The resulting CPC were seeded at a density of 1.5×104 cells/cm2 in CPC-SMC medium (DMEM/F12, B27, 1% Penicillin-Streptomycin, 400 μM 1-thioglycerol, 2 ng/mL TGF-β1 (PeproTech) and 10 ng/mL PDGFBB (PDGFβ; PeproTech)), and incubated for 7 days. For NCSC-SMC differentiation, hiPSC were seeded on Matrigel-coated dishes at a density of 2×104 cells/cm2 in TesRE8 medium with 10μM Y27632. After reaching ~50% confluency, they were incubated in NCSC differentiation medium (DMEM/F12, 1× N2 supplement (Life Technologies), 0.1% BSA (Sigma), 1% penicillin-streptomycin plus 10μM SB4315421 (Stemgent) and 1 μM LDN193189 (Stemgent)) for 6 days. From day 2 to day 6, 3μM CHIR99021 was added to the medium. The resulting NCSC were seeded at a density of 8×104 cells/cm2 in NCSC medium with 10μM Y27632. One day later, cells were induced with NCSC-SMC medium (DMEM/F12, 20% knockout serum replacement (Thermo Fisher Scientific), 1% Penicillin-Streptomycin, 2 ng/mL TGF-β1) for 8 days.
Statistical Analyses for qRT-PCRs, Western Blots, and Functional Assays
All quantitative data was presented as mean± standard deviation (std) with at least three biological replicates. We conducted a Shapiro-Wilk normality test prior to all analyses. When analyzing more than two groups that are normally distributed, we tested for equal variance using Brown-Forsythe test. If the standard deviations are not significantly different, we performed one-way ANOVA analysis with Dunnett’s multiple comparison test to compare the mean of each column with the mean of a control column. As a result, each p value is adjusted to account for multiple comparisons. When analyzing only two datasets that are normally distributed, we used unpaired t-test. If the variances are significantly different, we conducted unpaired t-test with Welch’s correction. The statistical analyses were performed using the recommended tests by GraphPad Prism 9 Software.
Results
Identification of a novel TGFBR1 variant causing Loeys-Dietz Syndrome and Lineage-specific hiPSC disease modeling
We identified a family with hereditary cardiovascular phenotypes including aortic root aneurysm and dissection at Michigan Medicine. Based on the clinical inquiry, seven family members spanning three generations were diagnosed with LDS (Figure 1A). One relative died from aortic dissection prior to the identification of the family. Imaging studies indicated aortic root aneurysm as the most common phenotype in the family (Figure 1B). Notably, patients IV-1 and IV-2 developed aortic root dilatation with early onset at ages 14, and 16 respectively (IV-1= Size: 38 mm, Z-score: 3.86). Clinical genetic testing (Connective Tissue Gene Test) revealed that all family members diagnosed with LDS carry a dominant pathogenic variant (c.688G>A) in TGFBR1 gene resulting in Alanine 230 to Threonine substitution (p.Ala230Thr), denoted as TGFBR1A230T.
Figure 1. A novel LDS mutation and Lineage-Specific hiPSC Disease Modeling.
A) A four-generation pedigree highlighting the family members diagnosed with LDS (filled symbol) or aortic dissection (AD). All LDS patients (+ symbol) underwent targeted gene sequencing, revealing that they carry the autosomal dominant TGFBR1A230T(c.G688A) variant (TGFBR1A230T/+). Slash symbol: not alive. B) Three-dimensional reconstruction of aortic root from Patients III-1 and IV-1. C) PDB structure of TGFBR1 protein (colored in green) in complex with ATP (spheres). The mutated residue is shown in magenta. In the mutant structure, the docked ATP is far away from residue 230. D) The Sanger sequencing results showing the position of the knock-in mutation, and the corrected base in the patient-derived hiPSC (red rectangle). E) Diagram summarizing the lineage-specific differentiation of hiPSC to CPC-SMC and NCSC-SMC.
To predict how the TGFBR1A230T variant affects TGFBR1 function, we conducted structural modeling of the protein using the COACH program, which generates ligand binding site predictions.30 Alanine 230 residue along with the nearby residues (211– 213, 219, 232, 260, 280–284, 286–287, 337–338, 340, 350–351) is located inside the ATP binding pocket. When ATP was docked in the protein using PatchDock31, the mutated residue was located far away from ATP suggesting the larger Threonine could fill up the binding pocket making the binding site inaccessible to ATP (Figure 1C, Figure IA in the Supplement). Based on this prediction, we concluded the TGFBR1A230T mutation potentially inhibits TGFBR1 kinase activity by disrupting its ATP binding ability.
For in vitro disease modeling of LDS, we used healthy donor cells (TGFBR1+/+) from a subject with normal aorta, and generated integration-free hiPSC carrying c.G688A mutation by CRISPR-Cas9 gene editing (Figure 1D).32 We generated two heterozygote (TGFBR1A230T/+), and two homozygote (TGFBR1A230T/A230T) clones to compare with the isogenic donor cells (Figure 1D). Importantly, TGFBR1A230T mutation did not affect the hiPSC pluripotency (Figure IB in the Supplement). Furthermore, we generated hiPSC (Patient A230T/+) from Patient IV-1 peripheral blood mononuclear cells (Figure 1A, 1D), and corrected the mutation in the patient-derived hiPSC (iWT +/+). All CRISPR-Cas9 edited cell lines were verified by Sanger sequencing. To address the lineage-specific effects of TGFBR1A230T, hiPSC were differentiated into CPC-SMC, and NCSC-SMC using conventional in vitro differentiation protocols (Figure 1E).25–29 Although the resulting SMC express low levels of late-differentiation marker MYH11, they have robust expression of SMC contractile genes including ACTA2, SM22α (TAGLN), CNN1, as well as MYOCD, a master regulator of SMC contractile gene expression.33
TGFBR1A230T mutation impairs contractile gene expression and function in CPC-SMC, but not in NCSC-SMC
Following the lineage-specific differentiation of hiPSC, we compared the contractile transcript and protein levels, and function in the mutant (TGFBR1A230T/+, TGFBR1A230T/A230T) and isogenic control cells (TGFBR1+/+). Since LDS is inherited in an autosomal dominant manner (Figure 1A), TGFBR1A230T/+ SMC likely represent a better in vitro model to mimic the disease phenotype. Compared to the control CPC-SMC, mutant cells, including TGFBR1A230T/+ CPC-SMC, showed decreased expression of SMC markers (p < 0.01); ACTA2, CNN1, SM22α, and MYOCD (Figure 2A). Interestingly, we did not detect significant changes in contractile gene expression both in TGFBR1A230T/+, and TGFBR1A230T/A230T NCSC-SMC, indicating the lineage-specific transcriptional defects caused by the mutation (Figure 2A).
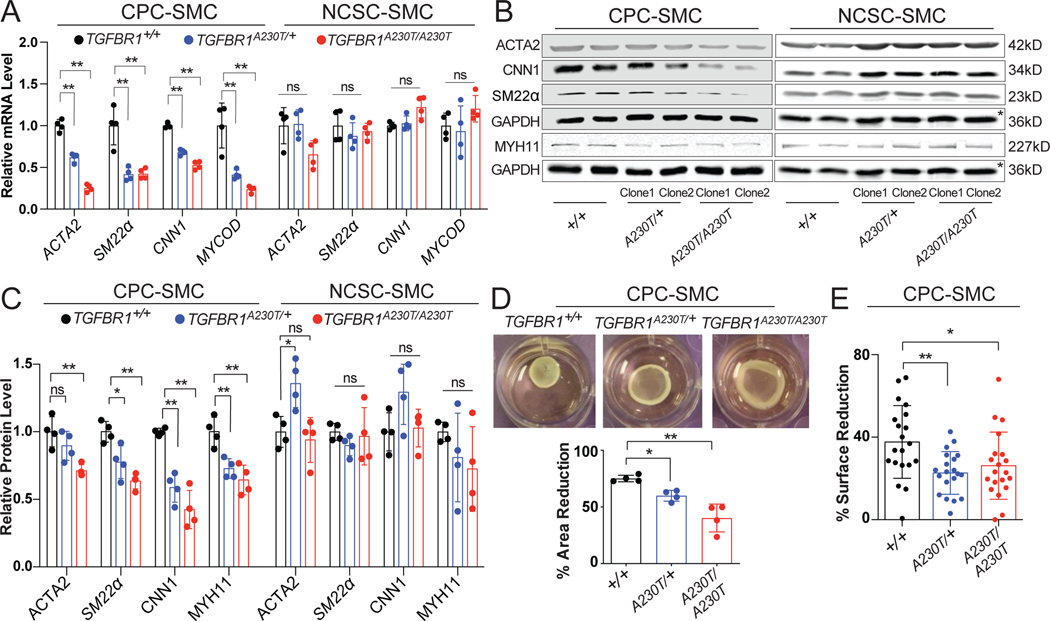
Figure 2. A230T substitution impairs the contractile phenotype only in CPC-SMC.
A) Relative mRNA levels of SMC markers in TGFBR1+/+ (black), TGFBR1A230T/+ (blue), and TGFBR1A230T/A230T (red) CPC- and NCSC-SMC. The average expression levels in the control samples were set to 1. n= 4 biological replicates, mean +/− standard deviation (std). *p < 0.05; **p < 0.01; NS, not significant; one-way ANOVA with Dunnett’s multiple comparison test. B) Western blots of protein extracts from the control and the mutant SMC. Two independent clones (Clone1 and Clone2) for each condition showed consistent patterns. Due to the low MYH11 levels in CPC-SMC, separate immunoblots with higher protein input were performed to measure MYH11 levels with respect to GAPDH. The asterisks highlight the same GAPDH control used for the quantification of SMC markers in NCSC-SMC. C) Quantification of western blot data showing relative protein levels of SMC markers in TGFBR1+/+ (black), TGFBR1A230T/+ (blue), and TGFBR1A230T/A230T (red) CPC- and NCSC-SMC. n= 4, mean +/− std, one-way ANOVA with multiple comparison test. D) Representative gel contraction images from the control and the mutant CPC-SMC, and quantification of the area reduction in percentages. n= 4, mean +/− std, one-way ANOVA with multiple comparison test. E) Quantification of surface reduction in CPC-SMC after carbachol treatment. n= 20 cells; mean +/− std; one-way ANOVA with multiple comparison test.
To confirm the transcript changes at the protein level, we performed immunoblots for key SMC markers. We observed that ACTA2, CNN1, SM22α, and MYH11 levels in the mutant CPC-SMC were significantly reduced compared to the control (Figure 2B–C). Strikingly, the contractile protein levels, except ACTA2, were unaltered in TGFBR1A230T/+, and TGFBR1A230T/A230T NCSC-SMC (Figure 2B–C). Next, we measured the contractile function in mutant CPC-SMC by collagen gel contraction and carbachol assays. Both assays revealed decreased contractility in the mutant CPC-SMC compared to their isogenic control (Figure 2D–E). As expected, TGFBR1A230T mutation did not affect contractile function in NCSC-SMC (Figure IC in the Supplement). Collectively, these data suggest that TGFBR1A230T mutation impairs SMC contractile phenotype in a lineage-specific manner targeting CPC-SMC. Given that aortic root is mostly populated with CPC-SMC, our in vitro findings suggest a potential explanation for the aortic root aneurysm observed in TGFBR1A230T/+ LDS patients.
Single Cell Profiling of TGFBR1+ and TGFBR1A230T CPC-SMC
To elucidate the transcriptional changes caused by the mutation, we performed large-scale, single cell RNA-sequencing (scRNA-seq) using an automated microwell-based platform.34–36 We obtained scRNA-seq profiles from three CPC-SMC populations (Figure 3A, Figure IIA in the Supplement): TGFBR1+/+ (6,929 cells), TGFBR1A230T/+ (6,021 cells), and TGFBR1A230T/A230T (5,980 cells). To identify molecularly distinct cell clusters in an unsupervised manner, we used Louvain community detection as implemented by Phenograph37 and embedded the scRNA-seq profiles in two dimensions using Uniform Manifold Approximation and Projection (UMAP).38 Binomial specificity analyses revealed specific and pervasive markers in each cluster (Supplemental Excel File I).39
Figure 3. Single-cell Profiling reveals the molecular defects in TGFBR1A230T CPC-SMC.
A) UMAP projection of the combined dataset of 18,930 scRNA-seq profiles. Individual samples are highlighted in different colors. B) Left: UMAP projection colored based on cluster identity. Right: Fraction of each sample in different clusters (normalized to the total cell number in each sample); TGFBR1+/+: black, TGFBR1A230T/+: blue, TGFBR1A230T/A230T: red. Cell clusters were ranked based on their abundance with C8 being the least abundant cluster in the combined dataset. C) Scaled expression of contractile, and cell proliferation genes on UMAPs. D) Heatmap (scaled by row) showing average expression of binomially specific cluster markers (FDR < 0.01) as well as contractile genes across eight cell clusters. E) Log-transformed FC values for protein-coding genes that are differentially expressed in both TGFBR1A230T/A230T vs. TGFBR1+/+ and TGFBR1A230T/+ vs. TGFBR1+/+ comparisons (padj < 0.001). F) GSEA showing significantly regulated gene ontology gene sets across two comparisons. Circle sizes indicate the log-transformed p values. NES, normalized enrichment score. G) GSEA showing the cluster-level enrichment or depletion of canonical pathway gene sets from Reactome Pathway Database. The clusters are colored based on log-transformed p-values. H) Percent of EdU+ cells in the control and mutant CPC-SMC. n=5; mean +/− std; *p < 0.05; **p < 0.01; one-way ANOVA with multiple comparison test.
We detected eight cell clusters (denoted as C1-C8), many of which were differentially distributed among the control and mutant SMC (Figure 3B). C2, and C7 were enriched in TGFBR1+/+; C4-C6 were more abundant in TGFBR1A230T/+; and C3, and C8 were enriched in TGFBR1A230T/A230T (Figure 3B). We also identified a more evenly distributed and pervasive C1 cluster enriched in several fibroblast markers (Supplemental Excel File I). Interestingly, the identification of multiple unique clusters including fibroblast-like cells among TGFBR1+/+ cells indicate the molecular heterogeneity of CPC-SMC generated using the standard differentiation protocol.25–27,29 Less than 3% of TGFBR1+/+ cells clustered in TGFBR1A230T/A230T enriched C3, and C8 clusters, highlighting the large expression differences caused by the mutation. The distribution of TGFBR1A230T/+ cells were intermediate between the control and TGFBR1A230T/A230T cells. The SMC contractile markers validated in Figure 2 were downregulated in C3, and C8 clusters, while cell division markers were upregulated in mutant enriched clusters implying increased proliferation among the mutant cells (Figure 3C).
Next, we plotted the cluster-level average expression of top binomially-specific markers (Fold-change (FC) > 2, FDR < 0.01) and the contractile markers across different clusters (Figure 3D, Supplemental Excel File I). The SMC contractile markers were significantly enriched in C2, C6, and C7 (FDR < 0.01, Supplemental Excel File I); which we named as SMC1, SMC2 and SMC3 (Figure 3D). Nearly 70% TGFBR1+/+ cells were clustered in SMC1, SMC2 and SMC3 compared to only 23% of TGFBR1A230T/+, and 5% TGFBR1A230T/A230T cells indicating the decreased SMC identity in the mutant cells. C4, C5, and C8 clusters were enriched in cell cycle-related genes. Fibroblast marker DCN was significantly enriched in C1, C3, and C8 suggesting a fibroblast-like identity for the majority of TGFBR1A230T/A230T cells (FDR < 0.01, Figure 3D).40 SMC phenotypic modulation to fibroblast-like cells was previously reported in scRNA-seq studies of human atherosclerotic lesions, and murine MFS aortic aneurysm model.41,42 Modulated SMC markers such as SERPINE1, and TCF21 were significantly enriched in TGFBR1A230T/+ CPC-SMC enriched clusters C4-C6 (Supplemental Excel File I). SMC1 cluster was highly enriched in BGN (FDR < 0.01, Figure 3D, Supplemental Excel File I), loss-of-function mutation of which causes TAAD43, while SMC3 was enriched in an early SMC differentiation marker; ACTG2 (FDR < 0.01, Figure 3D, Supplemental Excel File I), implying that SMC1, and SMC3 clusters could be representing similar cell types at different stages of development. Furthermore, SMC1 and SMC3 were significantly enriched in TGFBI (Transforming Growth Factor Beta Induced), expression of which is induced by TGFβ signaling.
To further characterize the molecular differences, we performed single-cell differential expression (SCDE) analysis comparing the control and mutant CPC-SMC (Supplemental Excel File II).44 We identified thousands of differentially expressed genes in each comparison, highlighting the drastic differentiation defects. Many contractile and ECM genes including TAAD-associated MMP2, and COL1A145,46 were downregulated both in TGFBR1A230T/+, and TGFBR1A230T/A230T CPC-SMC (Figure 3E). Strikingly, a recent scRNA-seq comparison of control and aneurysmal human aortic tissue suggested ERG as a regulator of aortic wall function47, which was also significantly downregulated in the mutant CPC-SMC. Using GeneSet enrichment Analysis (GSEA), we identified Gene Ontology sets enriched in each condition.48 GSEA revealed that key biological processes for SMC physiology including ECM remodeling, cell adhesion, collagen fiber formation and SMC migration were downregulated, while cell cycle-related, and RNA processing gene sets were upregulated in the mutant CPC-SMC (Figure 3F, Supplemental Excel File III). TGFβR signaling gene set showed enrichment in the control consistent with the structural prediction about the inactivating nature of the mutation (Figure 3F). We also performed scRNA-seq of TGFBR1+/+ NCSC-SMC to infer lineage-specific differences based on the gene expression profiles (Figure IIB in the Supplement). We identified many transcripts differentially expressed in CPC-SMC vs. NCSC-SMC comparison (Supplemental Excel File II). MFS-associated FBN1 was significantly enriched in CPC-SMC (Figure IIC in the Supplement, Supplemental Excel File II), while MFAP5 and PTPN11, mutations of which cause ascending aneurysm and root dilation were enriched in NCSC-SMC.5 This implies potential lineage-specific effects of other aneurysm-associated mutations. Furthermore, NCSC-SMC were significantly enriched in JNK, MAPK and Ras Protein Signaling Cascades (Figure IID in the Supplement, Supplemental Excel File III), suggesting the involvement of additional signaling components in NCSC-SMC regulation.
To identify molecular signatures of each CPC-SMC cluster, we conducted cluster-level GSEA using Reactome pathway database gene sets. SMC Contraction, Cell Surface Interactions at the Vascular Wall, and IGF Transport/Uptake gene sets were significantly enriched in SMC1 cluster, while TGFβR signaling activates SMADs gene set was highly enriched in SMC3 cluster (Figure 3G, Supplemental Excel File III). Furthermore, SMC1, SMC2, and SMC3 showed enrichment in Elastic Fiber Formation gene set. In contrast, mutant cell enriched clusters were upregulated for WNT signaling and Rho GTPase effectors gene sets, which are involved in vascular SMC proliferation (Figure 3G, Supplemental Excel File III).49,50 Based on these findings, we validated higher proliferative activity in the mutant cells by EdU stainings, which label mitotically active cells (Figure 3H). In sum, our cluster-level GSEA analysis confirmed the molecular deficits and reduced SMC identity in the mutant CPC-SMC, and revealed key biological processes such as Elastic Fiber Formation, Collagen Formation and IGF Transport/Uptake that are co-regulated with contractile gene expression.
TGFBR1A230T mutation impairs CPC-SMC contractile gene expression by disrupting SMAD3 and AKT signaling
To further validate the role of TGFBR1A230T mutation in CPC-SMC gene expression and function, we generated hiPSC (Patient A230T/+) from peripheral blood mononuclear cells harvested from female Patient IV-1 (IV-1, Figure 1A), and corrected the mutation (iWT+/+). iWT+/+ CPC-SMC had significantly higher expression of ACTA2, CNN1, SM22α, and MYOCD compared to Patient A230T/+ (Figure 4A). Consistently, the contractile function of iWT+/+ cells was significantly improved in gel contraction assay (Figure 4B). To uncover genome-wide transcriptional changes, we performed scRNA-seq on Patient A230T/+ (3,473 cells) and iWT+/+ CPC-SMC (1,483 cells), and compared their expression profiles with TGFBR1A230T/+ and TGFBR1+/+. To do this, we embedded the scRNA-seq profiles of TGFBR1+/+, TGFBR1A230T/+, TGFBR1A230T/A230T CPC-SMC in two dimensions using UMAP as in Figure 3A, and projected the single-cell profiles of Patient A230T/+ and iWT+/+ CPC-SMC onto this embedding (Figure 4C).51 Kernel density estimates for the projections of Patient A230T/+ and iWT+/+ cells were represented with contour lines. Patient A230T/+ cells projected onto mutant CPC-SMC enriched clusters (Figure 3B) with negligible density on SMC1, and SMC3 clusters. Interestingly, iWT+/+ cells had significant projection density on TGFBR1+/+ enriched SMC1, and SMC3 clusters indicating the potent effects of mutation-correction on global CPC-SMC transcription (Figure 4C). Overall, these results demonstrate that correction of TGFBR1A230T mutation in patient-derived cells significantly improves contractile gene expression and function in CPC-SMC.
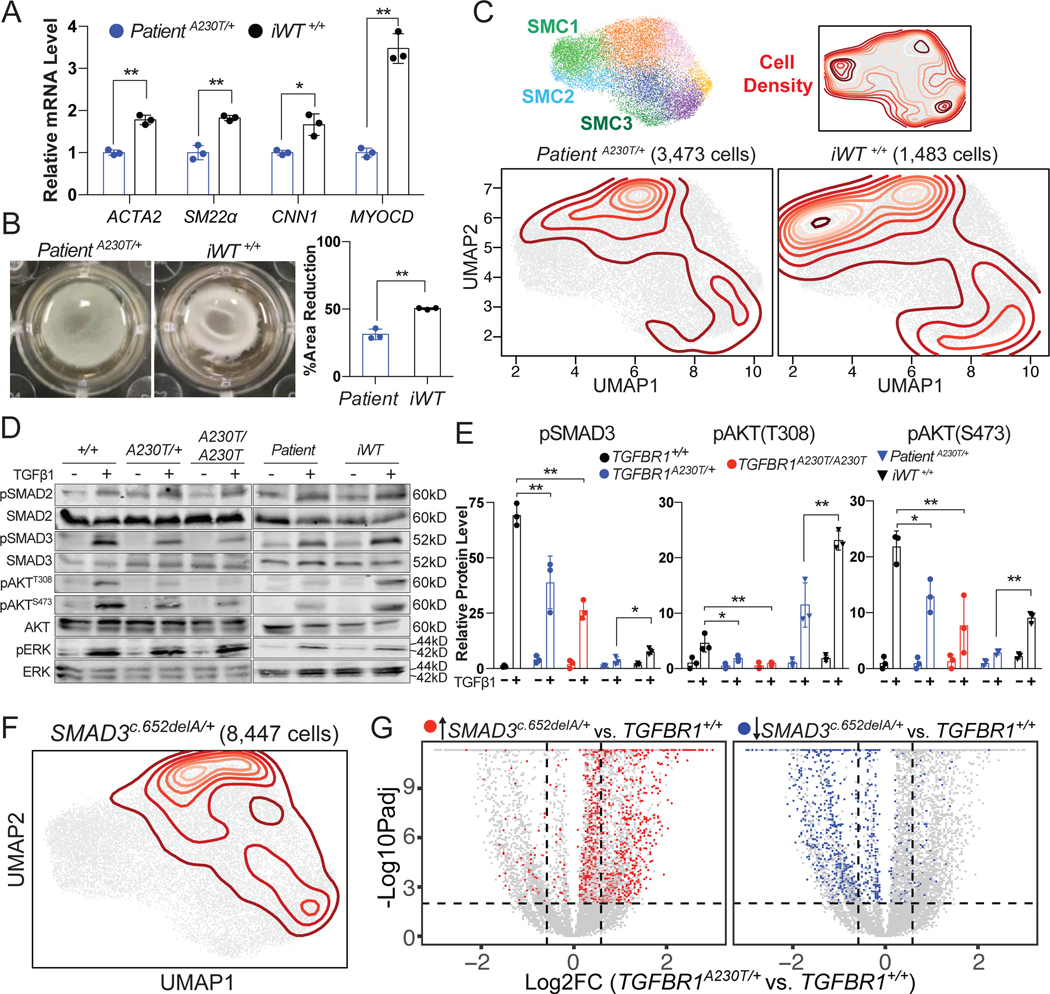
Figure 4. TGFBR1A230T mutation impairs CPC-SMC contractility by disrupting SMAD3 and AKT signaling.
A) Relative mRNA levels of SMC markers in Patient A230T/+ (blue) and iWT+/+ CPC-SMC (black). The average expression levels in the control samples were set to 1. n= 3, mean +/− standard deviation (std). *p < 0.05; **p < 0.01; unpaired t-test. B) Representative gel contraction images from the control and the mutant CPC-SMC, and the quantification of the area reduction. n= 3, mean +/− std, unpaired t-test. C) Top: Combined UMAP embedding of TGFBR1+/+, TGFBR1A230T/+ and TGFBR1A230T/A230T CPC-SMC, and the location of SMC1, SMC2, and SMC3 clusters as in Figure 3B. Bottom: Patient A230T/+ and iWT+/+ CPC-SMC are projected onto the combined UMAP embedding, and their density is represented with contours. D) Western blots of intracellular mediators from the control and the mutant SMC with or without TGFβ1 treatment. E) Quantification of western blot data for pSMAD3, pAKT(S473), and pAKT(T308). For quantifications, each phosphorylated protein was normalized to its respective non-phosphorylated form. n= 3, mean +/− std. *p < 0.05; **p < 0.01; NS, not significant; one-way ANOVA with multiple comparison test or unpaired t-test (Patient A230T/+ vs. iWT+/+ comparison). F) UMAP projection of SMAD3c.652delA/+ CPC-SMC onto TGFBR1+/+, TGFBR1A230T/+ and TGFBR1A230T/A230T CPC-SMC embedding. G) Scatterplot of fold change (FC) versus log-transformed FDR adjusted p-values (padj) for all transcripts in the TGFBR1A230T/+ vs. TGFBR1+/+ comparison (grey). Significantly enriched (red) and depleted (blue) genes (FC > 2, padj < 0.01) in SMAD3c.652delA/+ vs. TGFBR1+/+ comparison are highlighted on the scatterplot.
To reveal the downstream signaling defects caused by TGFBR1A230T mutation, we assayed the phosphorylation status of key TGFBR downstream effectors in CPC-SMC and NCSC-SMC by immunoblots. TGFBR can activate various downstream signaling mechanisms via SMAD-dependent and independent pathways including the MAPK and AKT signaling cascades.52 To induce phosphorylation of TGFBR1 downstream effectors, we treated differentiated SMC with 2 ng/ml TGFβ1 for 1 hour prior to protein extraction. We observed that p-SMAD3, p-AKT-T308, and pAKT-S473 levels were significantly reduced both in TGFBR1A230T/+ and TGFBR1A230T/A230T CPC-SMC compared to TGFBR1+/+ CPC-SMC (Figure 4D–E). Consistent with the contractile function recovery in iWT+/+ CPC-SMC, the phosphorylation levels of these proteins were elevated in iWT+/+ compared to Patient A230T/+ cells (Figure 4D–E). Strikingly, pERK, pSMAD2, and pMTOR levels were not different between the control and mutant CPC-SMC suggesting unaltered SMAD2, MAPK and MTOR regulation in the mutant cells (Figure 4D, Figure IIIA in the Supplement). The phosphorylation level of WNT signaling intracellular signal transducer, β-Catenin (CTNNB), was also unaltered between the control and mutant CPC-SMC after TGFβ1 stimulation (Figure IIIA in the Supplement). In agreement with the lack of contractile phenotype in TGFBR1A230T NCSC-SMC, p-SMAD3, p-AKT-T308, and pAKT-S473 levels did not change between the control and mutant NCSC-SMC (Figure IIE in the Supplement). These results suggest that TGFBR1-SMAD3, and TGFBR1-AKT cascades play key roles in mediating the cellular response to TGFBR1 activation.
Based on these results, we hypothesized that transcriptional changes caused by TGFBR1A230T and loss-of-function SMAD3 mutations in CPC-SMC should resemble each other. To compare the molecular deficits caused by TGFBR1A230T mutation and SMAD3 deficiency in the same isogenic background, we utilized hiPSC carrying heterozygous SMAD3c.652delA frameshift mutation, which is associated with TAAD.25 This mutation causes non-sense mediated decay of SMAD3 protein as confirmed by immunoblots (Figure IIIB in the Supplement). We performed large-scale scRNA-seq on SMAD3c.652delA/+ CPC-SMC (8,447 cells), and projected their single cell profiles onto the expression profiles of TGFBR1+/+, TGFBR1A230T/+, TGFBR1A230T/A230T CPC-SMC (Figure 4F). We observed significant projection density of SMAD3c.652delA/+ CPC-SMC on mutant CPC-SMC, with negligible projection density on SMC1, SMC2, and SMC3 clusters (Figure 4F). To compare global transcriptional defects caused by SMAD3c.652delA/+, and TGFBR1A230T/+ mutations, we performed SCDE analysis comparing isogenic TGFBR1+/+, and SMAD3c.652delA/+ CPC-SMC. Many significantly enriched transcripts in SMAD3c.652delA/+ versus TGFBR1+/+ comparison (FC > 2, FDR adjusted p values (padj) < 0.01) were also enriched in TGFBR1A230T/+ versus TGFBR1+/+ comparison (Figure 4G, Supplemental Excel File II). Similarly, significantly downregulated genes largely overlapped between both comparisons (Figure 4G, Supplemental Excel File II) suggesting that SMAD3c.652delA/+ mutation largely mimics the global transcriptional defects caused by TGFBR1A230T/+ mutation in CPC-SMC. However, there were additional ECM genes such as Col8a2 and Col12a1 downregulated in TGFBR1A230T/+ CPC-SMC (Supplemental Excel File II) indicating the contribution of SMAD3-independent signaling deficits in TGFBR1A230T phenotype. To confirm the effects of loss-of-function SMAD3 mutations on contractile protein levels, we also performed immunoblots using a SMAD3 mutant human aortic root aneurysmal sample, which showed decreased contractile protein levels consistent with our findings (Figure IIIC in the Supplement).
Combination treatment with Activin A and Rapamycin increases contractile mRNA and protein levels in TGFBR1A230T CPC-SMC
Our molecular characterization revealed SMAD3 and AKT as potential mediators of TGFBR1 signaling in CPC-SMC. Furthermore, mutant CPC-SMC are more proliferative compared to control SMC which can influence contractile gene expression. Based on these findings, we reasoned that mutant SMC contractile gene expression and function can be improved by activating SMAD3 and AKT cascades, while inhibiting SMC proliferation in a TGFBR1-independent manner. Similar to TGFβ1, Activin A (ACA), a member of TGFβ superfamily, induces intracellular signaling via SMAD2/3 signaling cascade.53,54 Importantly, ACA signals through Activin receptors, providing an alternative strategy to activate SMAD3 bypassing TGFBR1. Activin receptors are abundantly expressed in both control and mutant CPC-SMC according to our scRNA-seq data. Rapamycin is a widely used AKT activator55,56 that can be exploited to activate AKT in a TGFBR1 independent manner (Figure 5A). Rapamycin also inhibits aortic SMC proliferation57 allowing us to reduce SMC proliferation while activating AKT cascade.
Figure 5. Combination treatment with Activin A and Rapamycin increases contractile mRNA and protein levels in TGFBR1A230T CPC-SMC.
A) Diagram summarizing the strategy to activate SMAD3 and AKT while inhibiting cell proliferation in TGFBR1A230T CPC-SMC. B) Relative mRNA levels of SMC markers in TGFBR1+/+ (black), TGFBR1A230T/+ (blue), and TGFBR1A230T/A230T (red) CPC-SMC treated with vehicle (DMSO, denoted as -), Activin A only (ACA), Rapamycin only (Rapa.), and Activin A and Rapamycin combination (ACA+Rapa.) during SMC differentiation (early treatment). n= 3, mean +/− std. *p < 0.05; **p < 0.01; one-way ANOVA with Dunnett’s multiple comparison test. C) Representative western blot images of contractile proteins in TGFBR1A230T/+, and TGFBR1A230T/A230T CPC-SMC treated with vehicle (−) or Activin A and Rapamycin combination (ACA+Rapa.) during SMC differentiation. D) Relative protein levels of SMC markers in TGFBR1+/+ (black), TGFBR1A230T/+ (blue), and TGFBR1A230T/A230T (red) CPC-SMC under different conditions. n= 4, mean +/− std, unpaired t-test. E) Top: Diagram showing the late treatment paradigm. Bottom: Relative mRNA levels of SMC markers in different CPC-SMC populations treated with vehicle (−), or ACA+Rapamycin for two days after SMC differentiation. n= 3, mean +/− std. *p < 0.05; **p < 0.01; unpaired t-test. F) Representative western blot images of contractile proteins in TGFBR1A230T/+, and TGFBR1A230T/A230T CPC-SMC treated with vehicle (−) or ACA+Rapamycin for two days after SMC differentiation. G) Relative protein levels of SMC markers in in different CPC-SMC populations. n= 3, mean +/− std, unpaired t-test.
To induce mutant CPC-SMC contractile gene expression, we first treated them with vehicle (DMSO), 2ng/ml ACA, 20nM rapamycin, or a combination of both agents (ACA+Rapamycin) during the conversion of CPC to CPC-SMC (early treatment). We confirmed that TGFBR1A230T CPC-SMC treated with both agents have higher SMAD3, AKT as well as SMAD2 phosphorylation (Figure IIID in the Supplement). Rapamycin treatment also inhibited CPC-SMC proliferation as confirmed by EdU stainings (Figure IVA in the Supplement). ACA treatment increased ACTA2, SM22α, CNN1, and MYOCD expression in mutant CPC-SMC, while rapamycin’s effect on contractile gene expression appeared more targeted on SM22α (Figure 5B). Strikingly, treatment of mutant cells with both agents elevated contractile gene expression considerably more than individual treatments suggesting their combinatorial activity (Figure 5B). ACA+Rapamycin treated TGFBR1+/+ and TGFBR1A230T/+ cells had higher contractile mRNA levels than untreated TGFBR1+/+ indicating that saturating TGFBR1-SMAD3 and TGFBR1-AKT cascades promote contractile gene expression even in the control CPC-SMC (Figure IVB in the Supplement). To validate the robust effects of the combination treatment on TGFBR1A230T CPC-SMC at the protein level, we measured ACTA2, SM22α, CNN1 and MYH11 levels by immunoblots (Figure 5C, Figure IVC in the Supplement). Consistent with the transcript levels, all four contractile proteins were elevated in the treated mutant CPC-SMC (p < 0.01, Figure 5D).
Next, we measured the effectiveness of combination treatment on differentiated CPC-SMC. To do this, CPC-SMC were reseeded at the end of the differentiation protocol (Figure 1E), and treated with vehicle (DMSO) or ACA+Rapamycin for two additional days (late treatment). ACA+Rapamycin treatment increased the contractile mRNA levels both in TGFBR1A230T/+ and TGFBR1A230T/A230T CPC-SMC (Figure 5E, Figure IVB in the Supplement). Immunoblots for ACTA2, SM22α, CNN1 and MYH11 were also consistent with qRT-PCR results (Figure 5F–G, Figure IVC in the Supplement). Overall, these results indicate that the combination treatment is effective in improving contractile mRNA and protein levels in mutant CPC-SMC, when administered during or after CPC-SMC differentiation. Furthermore, individual and combination treatment data support TGFBR1-SMAD3 cascade as the principal target of TGFBR1 signaling. Although not effective independently, TGFBR1-AKT signaling and inhibition of cell proliferation appear to be crucial in reinforcing SMAD3-dependent contractile gene expression.
Combination treatment rescues contractile function and mechanical properties of TGFBR1A230T CPC-SMC
Given the effectiveness of combination treatment at the molecular level, we asked how increased contractile protein levels in the mutant CPC-SMC translate into contractile function. To evaluate functional recovery, we treated control and mutant CPC-SMC with both agents during or after SMC differentiation (early and late treatment), and examined them by gel contraction assay. Contractile function in treated mutant CPC-SMC significantly improved with TGFBR1A230T/+ CPC-SMC reaching control CPC-SMC levels (Figure 6A–B). Mutant CPC-SMC contractile function significantly increased even after late treatment indicating its efficiency on differentiated cells (Figure 6A–B).
Figure 6. Combination treatment rescues contractile phenotype in TGFBR1A230T CPC-SMC.
A) Representative gel contraction images from TGFBR1A230T/+, and TGFBR1A230T/A230T CPC-SMC treated with vehicle (−), or ACA+Rapamycin during or after SMC differentiation (early and late treatment). B) Quantification of the relative area reduction in ACA+Rapamycin treated CPC-SMC during or after SMC differentiation. n= 4, mean +/− std. *p < 0.05; **p < 0.01; unpaired t-test. C) Representative stress-strain curves from different samples. D) Mechanical parameter of maximum tangent modulus compared between different samples. n= 5, mean +/− std, **p < 0.01, one-way ANOVA with multiple comparison test. E) Immunostainings of CPC-SMC tissue rings for CNN1 and MYH11. Scale bars: 50 μm.
To investigate the functional properties of treated CPC-SMC, we measured their mechanical properties using ring-shaped tissue constructs (tissue rings).58–60 To accomplish this, TGFBR1+/+ and TGFBR1A230T/+ CPC-SMC were seeded around 2 mm agarose molds to allow them to form tissue rings for seven days. We then treated them with both agents for seven additional days. We measured their tensile strength (maximum stress) and stiffness (maximum tangent modulus) by performing uniaxial tensile test.59 Both tensile strength and stiffness were reduced in mutant CPC-SMC tissue rings, but showed significant recovery after the treatment (Figure 6C–D, Figure IVD in the Supplement). Strikingly, the mechanical properties of treated mutant CPC-SMC rings were comparable to control CPC-SMC rings demonstrating the potency of combination treatment (Figure 6C–D, Figure IVD in the Supplement). To assess SMC organization and contractile protein levels in tissue rings, we performed MYH11 and CNN1 immunostainings. Mutant CPC-SMC rings were disorganized with reduced MYH11 and CNN1 levels compared to the controls (Figure 6E). Following the treatment, mutant CPC-SMC had elevated levels of contractile proteins, and elongated morphology along the edges, forming a healthy contiguous cell layer (Figure 6E, Figure IVE in the Supplement). In sum, the combination treatment significantly improved the mechanical properties and contractility of mutant CPC-SMC, providing a pharmacological strategy to improve mutant SMC function.
Discussion
Although the LDS mutations clearly indicate that loss of TGFβ signaling is responsible for the aortic disease, the role of secondary increases in TGFβ signaling is still debated. Previously, we reported that the pathogenic SMAD3c.652delA variant causes defective differentiation of CPC-SMC sparing NCSC-SMC.25 In this study, we identified the pathogenic TGFBR1A230T variant in an LDS family with aortic root aneurysm. Our results demonstrate that TGFBR1A230T mutation also preferentially affects CPC-SMC. Strikingly, TGFBR1A230T mutation significantly impaired SMAD3 activation, and SMAD3c.652delA/+ mutation largely recapitulated genome-wide transcriptional changes in TGFBR1A230T/+ CPC-SMC, indicating the importance of SMAD3 in mediating TGFBR1 downstream signaling. Given that the aortic root is mostly populated with CPC-SMC, lineage specific effects of TGFBR1A230T implicate an etiology for LDS-associated aortic root aneurysm. This is consistent with the clinical presentations in patients with TGFBR1 and SMAD3 mutations who are prone to developing aortic root aneurysms sparing the ascending aorta. Our findings are also consistent with recent observations in the Tgfbr1M318R/+ LDS mouse model, which reported dysregulation of TGFβ signaling in CPC-derived SMC contributes to aortic root aneurysm formation.7
To characterize the molecular changes, we employed both conventional molecular methods and scRNA-seq, and compared knock-in TGFBR1A230T mutant cells, and patient hiPSC-derived SMC with their respective isogenic controls. This allowed us to analyze not only the standard SMC markers but also global gene expression changes caused by the mutation. Significant global transcriptional defects were noted in mutant CPC-SMC including TGFBR1A230T/+ CPC-SMC demonstrating the potent effects of this pathogenic mutation mimicking the autosomal dominant genotype in the LDS family. Several biological processes critical for SMC function, including SMC contraction and TGFBR signaling, were de-enriched in mutant CPC-SMC confirming the impaired contractile phenotype, and the inactivating nature of the mutation, while cell proliferation has increased as confirmed by EdU stainings. Furthermore, ECM organization and fiber formation gene sets were significantly downregulated in the mutant CPC-SMC, likely contributing to aneurysm development and other connective tissue abnormalities observed in LDS patients such as translucent skin, easy bruising and arterial tortuosity.
Our work also highlights the advantages of hiPSC disease modeling over the study of primary human aortic SMC and end-stage aneurysmal tissue in investigating the molecular defects caused by TAAD-associated pathogenic variants. Primary aortic SMC exhibit a significantly altered phenotype after a few passages, and surgical operation on patients with TGFBR1 mutations is rare, making it difficult to obtain enough SMC for molecular studies compared to the unlimited source of cells in hiPSC model. Furthermore, primary SMC are usually a mixture of NCSC- and CPC-derived SMC preventing the study of lineage-specific pathogenicity. Meanwhile, dissected human tissue has epigenetic and compensatory changes that accumulate over many years61,62, obscuring the early events in disease development. Increased SMAD2/3 phosphorylation is observed in end-stage aneurysmal samples from both LDS patients and animal models11,14,15, while our data shows the defective TGFBR1-SMAD3 cascade as the key driver of the mutant phenotype. This discrepancy suggests compensatory mechanisms being activated overtime in response to SMC abnormalities caused by the mutations.
Currently, there is no pharmacological treatment that could prevent TAAD formation. Although losartan, an Angiotensin II receptor antagonist and anti-hypertensive agent, reversed aortic dilatation in mouse models, a clinical trial revealed it is not more effective than β-blocker.9,63–65 Importantly, it is unclear whether any physiological effect of losartan is mediated by lowering systemic blood pressure rather than targeting the molecular mechanisms underlying aneurysm development. By detailed molecular characterization, we identified SMAD3 and AKT cascades as the TGFBR1 targets in CPC-SMC, guiding our pharmacological approach. Our hiPSC model allowed us to examine the early defects caused by the TGFBR1A230T variant, and we speculate that these early events may activate compensatory feedback loops and indirect stress-related changes that lead to the activation of other signaling components such as ERK overtime. Furthermore, NCSC-SMC are more dependent on SMAD2 activity compared to CPC-SMC24, which could explain the lack of SMAD2 regulation in CPC-SMC in our model. We also recently reported that SMAD3 deficiency significantly impairs in vitro differentiation of CPC-SMC but not NCSC-SMC further supporting the lineage-specific roles of SMADs.25
Our data indicates impaired TGFBR1-SMAD3 cascade is the main culprit for the mutant phenotype. AKT is a negative regulator of FOXO transcription factors, which inhibit MYOCD transcriptional activity. TGFBR1-SMAD3, and TGFBR1-AKT cascades likely converge on SMAD3 given that MYOCD and SMAD3 directly interact with each other to promote TGFβ induced downstream signaling.66,67 FOXO4, an inhibitor of MYOCD activity in SMC,68 could be at the intersection of TGFBR1-SMAD3 and TGFBR1-AKT cascades, as FOXO4 phosphorylation is induced by the combination treatment (Figure IIIE in the Supplement). By targeting TGFBR1-SMAD3 and TGFBR1-AKT cascades and inhibiting cell proliferation using ACA and Rapamycin, we significantly improved contractile mRNA and protein levels, and function in mutant CPC-SMC in vitro. The combination treatment also effectively rescued the mechanical properties of the mutant CPC-SMC tissue constructs. Although in vivo efficiency of this treatment is currently unknown, our hiPSC model provides key molecular insights as to how to treat patients carrying pathogenic variants, and a robust platform to screen drugs against other pathogenic variants associated with syndromic aortic aneurysm.
Supplementary Material
Clinical Perspective.
What is new?
We characterize lineage-specific cell signaling defects associated with a pathogenic TGFBR1 variant.
We provide a molecular-phenotype tailored treatment to prevent the SMC contractile defects caused by an LDS variant.
We highlight an hiPSC-based pipeline to gain mechanistic insight into pathogenic variants associated with syndromic TAAD.
What are the clinical implications?
Our results indicate that selective defects on CPC lineage SMC trigger aortic root aneurysm in LDS patients.
Using hiPSC disease modeling, we identify potential therapeutic agents for the treatment of root aneurysm in LDS.
Acknowledgments
Sources of Funding
This study was partially supported by National Institutes of Health grants HL130614, HL141891, and HL151776 (B.Y.), HL136231, HL109946, HL134569 (Y.E.C.), HL138139(J.Z.), Aikens Aortic Discovery Research Program (B.Y, J.Z., and Y.E.C.), Phil Jenkins and Darlene & Stephen J. Szatmari Funds (B.Y. and Y.E.C.).
Non-standard Abbreviations and Acronyms
- TAAD
Thoracic aortic aneurysm and dissection
- LDS
Loeys-Dietz Syndrome
- MFS
Marfan Syndrome
- SMC
Smooth muscle cells
- hiPSC
Human-induced pluripotent stem cells
- CPC
Cardiovascular progenitor cells
- NCSC
Neural crest stem cells
- TGFBR1
Transforming Growth Factor Beta Receptor 1
- A230T
Alanine 230 to Threonine substitution in TGFBR1
- iWT
Patient-corrected cells
- scRNA-Seq
Single cell RNA-Sequencing
- SCDE
Single cell differential expression analysis
- GSEA
Gene Set Enrichment Analysis
- UMAP
Uniform Manifold Approximation and Projection
- ACA
Activin A
Footnotes
Disclosures
Dogukan Mizrak is a co-inventor on a patent application related to microwell technology for single-cell sequencing that was filed by Columbia University.
References
- 1.Albornoz G, Coady MA, Roberts M, Davies RR, Tranquilli M, Rizzo JA and Elefteriades JA. Familial thoracic aortic aneurysms and dissections--incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg. 2006;82:1400–1405. [DOI] [PubMed] [Google Scholar]
- 2.Gillis E, Van Laer L and Loeys BL. Genetics of thoracic aortic aneurysm: at the crossroad of transforming growth factor-beta signaling and vascular smooth muscle cell contractility. Circ Res. 2013;113:327–340. [DOI] [PubMed] [Google Scholar]
- 3.Isselbacher EM, Lino Cardenas CL and Lindsay ME. Hereditary Influence in Thoracic Aortic Aneurysm and Dissection. Circulation. 2016;133:2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsay ME and Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinard A, Jones GT and Milewicz DM. Genetics of Thoracic and Abdominal Aortic Diseases. Circ Res. 2019;124:588–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milewicz DM, Guo DC, Tran-Fadulu V, Lafont AL, Papke CL, Inamoto S, Kwartler CS and Pannu H. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu Rev Genomics Hum Genet. 2008;9:283–302. [DOI] [PubMed] [Google Scholar]
- 7.MacFarlane EG, Parker SJ, Shin JY, Kang BE, Ziegler SG, Creamer TJ, Bagirzadeh R, Bedja D, Chen Y, Calderon JF, et al. Lineage-specific events underlie aortic root aneurysm pathogenesis in Loeys-Dietz syndrome. J Clin Invest. 2019;129:659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeys BL, Schwarze U, Holm T, Callewaert BL, Thomas GH, Pannu H, De Backer JF, Oswald GL, Symoens S, Manouvrier S, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. [DOI] [PubMed] [Google Scholar]
- 9.Holm TM, Habashi JP, Doyle JJ, Bedja D, Chen Y, van Erp C, Lindsay ME, Kim D, Schoenhoff F, Cohn RD, et al. Noncanonical TGFbeta signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science. 2011;332:358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY and Dietz HC. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. [DOI] [PubMed] [Google Scholar]
- 11.Bertoli-Avella AM, Gillis E, Morisaki H, Verhagen JMA, de Graaf BM, van de Beek G, Gallo E, Kruithof BPT, Venselaar H, Myers LA, et al. Mutations in a TGF-beta ligand, TGFB3, cause syndromic aortic aneurysms and dissections. J Am Coll Cardiol. 2015;65:1324–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardoso S, Robertson SP and Daniel PB. TGFBR1 mutations associated with Loeys-Dietz syndrome are inactivating. J Recept Signal Transduct Res. 2012;32:150–155. [DOI] [PubMed] [Google Scholar]
- 13.Inamoto S, Kwartler CS, Lafont AL, Liang YY, Fadulu VT, Duraisamy S, Willing M, Estrera A, Safi H, Hannibal MC, et al. TGFBR2 mutations alter smooth muscle cell phenotype and predispose to thoracic aortic aneurysms and dissections. Cardiovasc Res. 2010;88:520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsay ME, Schepers D, Bolar NA, Doyle JJ, Gallo E, Fert-Bober J, Kempers MJ, Fishman EK, Chen Y, Myers L, et al. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat Genet. 2012;44:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Laar IM, Oldenburg RA, Pals G, Roos-Hesselink JW, de Graaf BM, Verhagen JM, Hoedemaekers YM, Willemsen R, Severijnen LA, Venselaar H, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43:121–126. [DOI] [PubMed] [Google Scholar]
- 16.Micha D, Guo DC, Hilhorst-Hofstee Y, van Kooten F, Atmaja D, Overwater E, Cayami FK, Regalado ES, van Uffelen R, Venselaar H, et al. SMAD2 Mutations Are Associated with Arterial Aneurysms and Dissections. Hum Mutat. 2015;36:1145–1149. [DOI] [PubMed] [Google Scholar]
- 17.van de Laar IM, van der Linde D, Oei EH, Bos PK, Bessems JH, Bierma-Zeinstra SM, van Meer BL, Pals G, Oldenburg RA, Bekkers JA, et al. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J Med Genet. 2012;49:47–57. [DOI] [PubMed] [Google Scholar]
- 18.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. [DOI] [PubMed] [Google Scholar]
- 19.Sawada H, Rateri DL, Moorleghen JJ, Majesky MW and Daugherty A. Smooth Muscle Cells Derived From Second Heart Field and Cardiac Neural Crest Reside in Spatially Distinct Domains in the Media of the Ascending Aorta-Brief Report. Arterioscler Thromb Vasc Biol. 2017;37:1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo DC, Pannu H, Tran-Fadulu V, Papke CL, Yu RK, Avidan N, Bourgeois S, Estrera AL, Safi HJ, Sparks E, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. [DOI] [PubMed] [Google Scholar]
- 21.Zhu L, Vranckx R, Khau Van Kien P, Lalande A, Boisset N, Mathieu F, Wegman M, Glancy L, Gasc JM, Brunotte F, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38:343–349. [DOI] [PubMed] [Google Scholar]
- 22.Serralheiro P, Soares A, Costa Almeida CM and Verde I. TGF-beta1 in Vascular Wall Pathology: Unraveling Chronic Venous Insufficiency Pathophysiology. Int J Mol Sci. 2017;18:e2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Li Q, Jiao Y, Qin L, Ali R, Zhou J, Ferruzzi J, Kim RW, Geirsson A, Dietz HC, et al. Tgfbr2 disruption in postnatal smooth muscle impairs aortic wall homeostasis. J Clin Invest. 2014;124:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie WB, Li Z, Shi N, Guo X, Tang J, Ju W, Han J, Liu T, Bottinger EP, Chai Y, et al. Smad2 and myocardin-related transcription factor B cooperatively regulate vascular smooth muscle differentiation from neural crest cells. Circ Res. 2013;113:e76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong J, Zhou D, Jiang L, Qiu P, Milewicz DM, Chen YE and Yang B. In Vitro Lineage-Specific Differentiation of Vascular Smooth Muscle Cells in Response to SMAD3 Deficiency: Implications for SMAD3-Related Thoracic Aortic Aneurysm. Arterioscler Thromb Vasc Biol. 2020;40:1651–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Wang Y, Jiao J, Liu Z, Zhao C, Zhou Z, Zhang Z, Forde K, Wang L, Wang J, et al. Patient-specific cardiovascular progenitor cells derived from integration-free induced pluripotent stem cells for vascular tissue regeneration. Biomaterials. 2015;73:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z and Yang HT. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 2013;23:1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang A, Tang Z, Li X, Jiang Y, Tsou DA and Li S. Derivation of smooth muscle cells with neural crest origin from human induced pluripotent stem cells. Cells Tissues Organs. 2012;195:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O’Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. 2015;17:994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Roy A and Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneidman-Duhovny D, Inbar Y, Nussinov R and Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA and Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Wang DZ, Pipes GC and Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan J and Sims PA. An Automated Microwell Platform for Large-Scale Single Cell RNA-Seq. Sci Rep. 2016;6:33883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan J, Levitin HM, Frattini V, Bush EC, Boyett DM, Samanamud J, Ceccarelli M, Dovas A, Zanazzi G, Canoll P, et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018;10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizrak D, Levitin HM, Delgado AC, Crotet V, Yuan J, Chaker Z, Silva-Vargas V, Sims PA and Doetsch F. Single-Cell Analysis of Regional Differences in Adult V-SVZ Neural Stem Cell Lineages. Cell Rep. 2019;26:394–406 e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F and Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. [DOI] [PubMed] [Google Scholar]
- 39.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhl L, Genove G, Leptidis S, Liu J, He L, Mocci G, Sun Y, Gustafsson S, Buyandelger B, Chivukula IV, et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun. 2020;11:3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedroza AJ, Tashima Y, Shad R, Cheng P, Wirka R, Churovich S, Nakamura K, Yokoyama N, Cui JZ, Iosef C, et al. Single-Cell Transcriptomic Profiling of Vascular Smooth Muscle Cell Phenotype Modulation in Marfan Syndrome Aortic Aneurysm. Arterioscler Thromb Vasc Biol. 2020;40:2195–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meester JA, Vandeweyer G, Pintelon I, Lammens M, Van Hoorick L, De Belder S, Waitzman K, Young L, Markham LW, Vogt J, et al. Loss-of-function mutations in the X-linked biglycan gene cause a severe syndromic form of thoracic aortic aneurysms and dissections. Genet Med. 2017;19:386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kharchenko PV, Silberstein L and Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen M, Lee J, Basu R, Sakamuri SS, Wang X, Fan D and Kassiri Z. Divergent roles of matrix metalloproteinase 2 in pathogenesis of thoracic aortic aneurysm. Arterioscler Thromb Vasc Biol. 2015;35:888–898. [DOI] [PubMed] [Google Scholar]
- 46.Rahkonen O, Su M, Hakovirta H, Koskivirta I, Hormuzdi SG, Vuorio E, Bornstein P and Penttinen R. Mice with a deletion in the first intron of the Col1a1 gene develop age-dependent aortic dissection and rupture. Circ Res. 2004;94:83–90. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Ren P, Dawson A, Vasquez HG, Ageedi W, Zhang C, Luo W, Chen R, Li Y, Kim S, et al. Single-Cell Transcriptome Analysis Reveals Dynamic Cell Populations and Differential Gene Expression Patterns in Control and Aneurysmal Human Aortic Tissue. Circulation. 2020;142:1374–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bond M, Wu YJ, Sala-Newby GB and Newby AC. Rho GTPase, Rac1, regulates Skp2 levels, vascular smooth muscle cell proliferation, and intima formation in vitro and in vivo. Cardiovasc Res. 2008;80:290–298. [DOI] [PubMed] [Google Scholar]
- 50.Mill C and George SJ. Wnt signalling in smooth muscle cells and its role in cardiovascular disorders. Cardiovasc Res. 2012;95:233–240. [DOI] [PubMed] [Google Scholar]
- 51.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. 2019;10:4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, Liu X, Ren X, Tian Y, Chen Z, Xu X, Du Y, Jiang C, Fang Y, Liu Z, et al. Smad2 and Smad3 have differential sensitivity in relaying TGFbeta signaling and inversely regulate early lineage specification. Sci Rep. 2016;6:21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lebrun JJ, Takabe K, Chen Y and Vale W. Roles of pathway-specific and inhibitory Smads in activin receptor signaling. Mol Endocrinol. 1999;13:15–23. [DOI] [PubMed] [Google Scholar]
- 55.Martin KA, Merenick BL, Ding M, Fetalvero KM, Rzucidlo EM, Kozul CD, Brown DJ, Chiu HY, Shyu M, Drapeau BL, et al. Rapamycin promotes vascular smooth muscle cell differentiation through insulin receptor substrate-1/phosphatidylinositol 3-kinase/Akt2 feedback signaling. J Biol Chem. 2007;282:36112–36120. [DOI] [PubMed] [Google Scholar]
- 56.Jin Y, Xie Y, Ostriker AC, Zhang X, Liu R, Lee MY, Leslie KL, Tang W, Du J, Lee SH, et al. Opposing Actions of AKT (Protein Kinase B) Isoforms in Vascular Smooth Muscle Injury and Therapeutic Response. Arterioscler Thromb Vasc Biol. 2017;37:2311–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W, Li Q, Qin L, Ali R, Qyang Y, Tassabehji M, Pober BR, Sessa WC, Giordano FJ and Tellides G. Rapamycin inhibits smooth muscle cell proliferation and obstructive arteriopathy attributable to elastin deficiency. Arterioscler Thromb Vasc Biol. 2013;33:1028–1035. [DOI] [PubMed] [Google Scholar]
- 58.Gwyther TA, Hu JZ, Billiar KL and Rolle MW. Directed cellular self-assembly to fabricate cell-derived tissue rings for biomechanical analysis and tissue engineering. J Vis Exp. 2011;e3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gwyther TA, Hu JZ, Christakis AG, Skorinko JK, Shaw SM, Billiar KL and Rolle MW. Engineered vascular tissue fabricated from aggregated smooth muscle cells. Cells Tissues Organs. 2011;194:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dikina AD, Strobel HA, Lai BP, Rolle MW and Alsberg E. Engineered cartilaginous tubes for tracheal tissue replacement via self-assembly and fusion of human mesenchymal stem cell constructs. Biomaterials. 2015;52:452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomez D, Coyet A, Ollivier V, Jeunemaitre X, Jondeau G, Michel JB and Vranckx R. Epigenetic control of vascular smooth muscle cells in Marfan and non-Marfan thoracic aortic aneurysms. Cardiovasc Res. 2011;89:446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomez D, Kessler K, Michel JB and Vranckx R. Modifications of chromatin dynamics control Smad2 pathway activation in aneurysmal smooth muscle cells. Circ Res. 2013;113:881–890. [DOI] [PubMed] [Google Scholar]
- 63.Gallo EM, Loch DC, Habashi JP, Calderon JF, Chen Y, Bedja D, van Erp C, Gerber EE, Parker SJ, Sauls K, et al. Angiotensin II-dependent TGF-beta signaling contributes to Loeys-Dietz syndrome vascular pathogenesis. J Clin Invest. 2014;124:448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Habashi JP, Doyle JJ, Holm TM, Aziz H, Schoenhoff F, Bedja D, Chen Y, Modiri AN, Judge DP and Dietz HC. Angiotensin II type 2 receptor signaling attenuates aortic aneurysm in mice through ERK antagonism. Science. 2011;332:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lacro RV, Dietz HC, Sleeper LA, Yetman AT, Bradley TJ, Colan SD, Pearson GD, Selamet Tierney ES, Levine JC, Atz AM, et al. Atenolol versus losartan in children and young adults with Marfan’s syndrome. N Engl J Med. 2014;371:2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tong X, Wang S, Lei Z, Li C, Zhang C, Su Z, Liu X, Zhao J and Zhang HT. MYOCD and SMAD3/SMAD4 form a positive feedback loop and drive TGF-beta-induced epithelial-mesenchymal transition in non-small cell lung cancer. Oncogene. 2020;39:2890–2904. [DOI] [PubMed] [Google Scholar]
- 67.Qiu P, Ritchie RP, Fu Z, Cao D, Cumming J, Miano JM, Wang DZ, Li HJ and Li L. Myocardin enhances Smad3-mediated transforming growth factor-beta1 signaling in a CArG box-independent manner: Smad-binding element is an important cis element for SM22alpha transcription in vivo. Circ Res. 2005;97:983–991. [DOI] [PubMed] [Google Scholar]
- 68.Liu ZP, Wang Z, Yanagisawa H and Olson EN. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell. 2005;9:261–270. [DOI] [PubMed] [Google Scholar]
- 69.Mizrak D, Bayin NS, Yuan J, Liu Z, Suciu RM, Niphakis MJ, Ngo N, Lum KM, Cravatt BF, Joyner AL, et al. Single-Cell Profiling and SCOPE-Seq Reveal Lineage Dynamics of Adult Ventricular-Subventricular Zone Neurogenesis and NOTUM as a Key Regulator. Cell Rep. 2020;31:107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levitin HM, Yuan J, Cheng YL, Ruiz FJ, Bush EC, Bruce JN, Canoll P, Iavarone A, Lasorella A, Blei DM, et al. De novo gene signature identification from single-cell RNA-seq with hierarchical Poisson factorization. Mol Syst Biol. 2019;15:e8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.