Abstract
A single vanD-containing Enterococcus faecium strain (N97-330) was isolated in Canada. The vanD-containing region was cloned and sequenced. Although the proteins have more than 96% identity to a previously described vanD region in BM4339, the vanSD gene contains a frameshift mutation that leads to a predicted truncated protein. Furthermore, sequence analysis of the ddl gene revealed the presence of an IS982-like element (ISEfm1) which interrupted the d-Ala–d-Ala ligase. This suggested the constitutive expression of the vanD operon, which was confirmed. Pulsed-field gel electrophoresis fingerprinting demonstrated that BM4339 was not related to N97-330 (>15 band differences). Both strains contained multiple copies of the IS982-like element.
Vancomycin-resistant enterococci produce modified precursors that terminate in either d-alanyl-d-lactate (d-Ala-d-Lac) or d-alanyl-d-serine (d-Ala-d-Ser), which have a much lower affinity for glycopeptides than do unmodified precursors (2, 5). The genetic basis for resistance lies in genes whose products have homology to the bacterial d-Ala-d-Ala ligases, encoded by ddl genes, which produce the dipeptide target for glycopeptide antibiotics. High-level vancomycin resistance is conferred either by the transferable, inducible VanA or VanB d-Ala-d-Lac ligases (4, 11, 18) or by the nontransferable, constitutive VanD d-Ala-d-Lac ligase, which thus far has been described only for Enterococcus faecium BM4339 (7, 17) and E. faecium A902 (15). This report describes the genetic characterization of vancomycin resistance in a strain of E. faecium which is the first VanD-type strain isolated in Canada.
A vancomycin-resistant enterococcus was isolated from a stool specimen from a 59-year-old Ontario man who had had an orthoptic liver transplant 46 days before and had received multiple courses of antibiotics. Multiple attempts to amplify vanA, vanB, and vanC1 using PCR with the previously reported primer sets (8) were negative. Initial confirmation of the isolate as a possible VanD-type strain was done courtesy of P. Courvalin (Institut Pasteur) using primers which amplify a 0.46-kb vanD fragment (17). The strain (N97-330) was identified as an E. faecium strain by using standard biochemical tests for strain identification (9). The MICs (micrograms per milliliter) of a number of antibiotics as determined by agar dilution (14) are shown in Table 1 and compared to those for the vanD strains BM4339 and A902 (15, 17). Expression of the vancomycin-resistance phenotype was not inducible in N97-330 (data not shown), suggesting constitutive expression of resistance genes similar to that in BM4339 (17) but unlike that in A902 (15). Macrorestriction analysis was carried out by separating ApaI- or SmaI-digested genomic DNA by pulsed-field gel electrophoresis using 1.1% agarose gels and a CHEF-DRIII (Bio-Rad, Hercules, Calif.) with the following pulse times: 1 to 10 s for 12 h followed by 1 to 35 s for 31 h at 200 V in 0.5× Tris-borate-EDTA at 14°C. A comparison of the DNA fingerprints of N97-330 and BM4339 (courtesy of F. Tenover, Centers for Disease Control and Prevention) did not reveal any genetic relationship between the two strains (13, 23). E. faecium A902, which differed from BM4339 (≥6 band differences) (15), also appears not to be related to N97-330 (data not shown).
TABLE 1.
Antibiograms of vanD-containing E. faecium strains N97-330, BM4339 (17), and A902 (15)
| Antibiotic | MIC (μg/ml)a
|
||
|---|---|---|---|
| N97-330 | BM4339 | A902 | |
| Ampicillin | 128 | NA | 64–128 |
| Penicillin G | 256 | 256 | 256 |
| Chloramphenicol | 8 | NA | S |
| Doxycycline | 0.12 | NA | NA |
| Gentamicin | <500 | >2,000 | R |
| Streptomycin | >2,000 | >2,000 | R |
| Tetracycline | 0.25 | 16 | S |
| Vancomycin | >256 | 64 | 128 |
| Teicoplanin | 64 | 4 | 4b |
MICs for N97-330 and A902 were determined by agar dilution; the method of determining the MICs for BM4339 was not given. NA, MIC not available in the publication; S, susceptible to the antibiotic; R, resistant to high levels of the antibiotic.
16 to 32 μg/ml by broth microdilution.
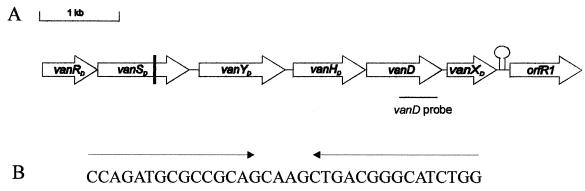
By using standard methods (20) and the 0.46-kb vanD PCR product as a probe, two overlapping clones were isolated from an N97-330 genomic library constructed in λEMBL3 (Promega, Madison, Wis.). These clones were subjected to partial sequence analysis (Fig. 1). The sequence of the 5′ end of the vanRD gene was obtained by sequencing a PCR product amplified from N97-330 genomic DNA using a primer designed based on a sequence upstream of BM4339 vanRD (7) and a primer from a previously sequenced region of the N97-330 vanRD gene. The complete nucleotide sequence of the 6,793-bp vanD region is shown in Fig. 1A. The BLAST programs (1) (http://www.ncbi.nlm.nih/BLAST) were used to identify putative products of detected open reading frames (ORFs). Six genes were found to have the same genetic organization as that of the vanD operon of E. faecium BM4339 (Fig. 1) (7). Comparisons of the amino acid products from the two strains revealed that pairs of homologous genes had at least 96% identity (data not shown). Further comparisons of the vanD sequence from N97-330 revealed 2.1 and 3.1% base pair differences with the genes from BM4339 and E. faecium A902 (15), respectively. These differences are less than those exhibited by the three designated vanB subtypes (3.6 to 5%) (10, 16) but the genes may still be considered vanD variants.
FIG. 1.
Schematic representation of the vanD gene cluster and sequence of a putative stem-loop structure. (A) Predicted ORFs from a 6,793-bp region sequenced from two λ clones and a PCR product. Open arrows represent the coding sequences for the vanRD, vanSD, vanYD, vanHD, vanD, vanXD, and orfR1 genes. The vertical line in the vanSD ORF represents the position of the frameshift mutation leading to a predicted truncated protein. The vanD probe used in hybridization experiments to isolate the λ clones is shown below the corresponding region. (B) Sequence of the putative stem-loop structure downstream of vanXD.
Beginning 161 bp downstream of the vanXD stop codon is an ORF of 909 bp (orfR1) coding for a putative product of 302 amino acids that shows low but significant homology to several regulatory proteins. The highest homology was with the YobV protein (313 residues) from Bacillus subtilis (accession no. AF027868), with which it has 30% identity. We have detected a region of dyad symmetry beginning 10 bp downstream of the vanXD stop codon that could form a putative hairpin structure with a −ΔG of 96 kJ/mol that may play a regulatory role (Fig. 1B). Transcription studies are needed to determine if orfR1 is part of the vanD operon. Fifty base pairs of sequence downstream of the BM4339 vanXD is available (7), and an alignment with the N97-330 sequence (data not shown) reveals significant divergence, with the regions sharing only 40% identity. This may reflect a different genetic location of the vanD region in BM4339 compared to that in N97-330 or may be due to interstrain sequence divergence.
Comparison of the vanSD genes from BM4339 and N97-330 revealed that the latter has a 1-bp deletion at nucleotide position 670 of the BM4339 gene sequence, which results in a frameshift leading to, presumably, a truncated nonfunctional protein of 233 amino acids. The vanA and vanB operons are activated in the presence of an inducer when the VanR protein becomes phosphorylated either by the cognate autophosphorylated VanS (VanS ∼ P) or by an unknown kinase (3, 22). In N97-330, the absence of a functional VanSD protein may lead to a high steady-state level of VanR ∼ P and thus to constitutive expression of the vancomycin resistance operon, as growth studies have shown. Vancomycin resistance is constitutive in BM4339, although it carries an intact vanSD gene (7, 17).
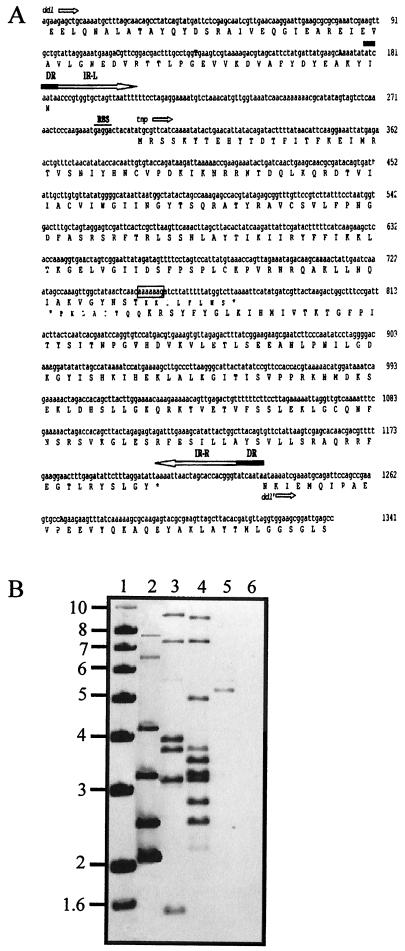
During initial characterization of N97-330, PCR was carried out with primers specific for the ddl gene of E. faecium (8). A product approximately 1 kb larger than the expected size was obtained (data not shown). Sequence analysis of 1,341 bp from this region revealed that a 1,041-bp insertion sequence (IS), defined by 22-bp perfect terminal inverted repeats, had been inserted in the ddl gene (Fig. 2A). Two overlapping ORFs were identified in the IS, and putative translation products showed between 38 and 43% identity with parts of the putative transposase protein (Tnp) from the IS982 family of insertion elements (12). Further analysis of the two ORFs revealed that a translational frameshift in the region from bases 748 to 754 (Fig. 2A) could lead to the translation of a fusion protein of 302 residues which can be aligned to the Tnps of IS982 family members. Control of transposition by programmed translational frameshifting is common in a number of bacterial tnp genes, though it has not been found in the IS982 family (12). Alternatively, a frameshift mutation may have occurred in the tnp gene after the IS was acquired by the genome, resulting in a truncated inactive protein. We propose naming this element ISEfm1 (12). In order to determine the number of copies of ISEfm1 in various strains of enterococci, we probed ClaI digests with a fragment of the tnp gene generated by PCR (Fig. 2B). Multiple copies exist in the E. faecium vanA and vanD strains, whereas a vancomycin-sensitive strain (ATCC 19434) appears to have a single copy. The probe did not hybridize to the type strain E. faecalis ATCC 29212. Several faint bands visible on the autoradiograph in the E. faecium lanes may be due to a low level of homology between the probe and the tnp genes of other IS elements found in E. faecium. It will be interesting to expand this work to include additional species of Enterococcus to determine if ISEfm1 is specific to E. faecium.
FIG. 2.
Sequence of the ISEfm1 element found in the ddl gene and prevalence of the insertion element in selected strains. (A) Complete nucleotide sequence and predicted amino acid sequence of the ISEfm1 element and partial ddl gene. Capital letters indicate nucleotide differences in ddl compared to the published sequence of E. faecium BM4147-1. Open arrows indicate the coding sequence for the ddl and tnp genes. ddl′ denotes the sequence of the ddl gene downstream of the ISEfm1 element. DR, IR-L, and IR-R indicate direct repeats, the left inverted repeat, and the right inverted repeat, respectively. The open box indicates the translational frameshift region. (B) Southern blot of ClaI-digested DNA probed with the ISEfm1 PCR product. Lane 1, 1-kb extension ladder (Life Technologies; sizes, in kilobases, of the fragments are indicated at the left); lane 2, E. faecium N97-330 (vanD); lane 3, E. faecium BM4339 (vanD); lane 4, E. faecium N98-638 (vanA); lane 5, E. faecium ATCC 19434; lane 6, E. faecalis ATCC 29212.
Enterococci with impaired Ddl activity that require the presence of a glycopeptide for growth have been reported (6, 19, 21, 24). Although N97-330 appears to have a nonfunctional Ddl due to insertion of an IS element, a glycopeptide is not required for growth due to a frameshift mutation in vanSD, which most likely leads to the constitutive expression of the van operon.
Nucleotide sequence accession numbers.
The complete nucleotide sequence of the 6,793-bp vanD region shown in Fig. 1A has been deposited in the GenBank database under accession number AF175293. The sequence of the ddl region containing ISEfm1 (Fig. 2A) has been deposited in the GenBank database under the accession number AF138282.
Acknowledgments
We gratefully acknowledge P. Courvalin for initial identification of a vanD amplicon from E. faecium N97-330 and R. Easy, R. Bosey, C. Murphy, and R. Hizon for valuable technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 6.Baptista M, Depardieu F, Reynolds P, Courvalin P, Arthur M. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol Microbiol. 1997;25:93–105. doi: 10.1046/j.1365-2958.1997.4401812.x. [DOI] [PubMed] [Google Scholar]
- 7.Casadewall B, Courvalin P. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol. 1999;181:3644–3648. doi: 10.1128/jb.181.12.3644-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facklam R R, Sahm D F. Enterococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 308–314. [Google Scholar]
- 10.Gold H S, Ünal S, Cerenado E, Thauvin-Eliopoulos C, Eliopoulos G M, Wennersten C B, Moellering R C., Jr A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39:2446–2453. doi: 10.1128/aac.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison D, Woodford N, Barrett S P, Sisson P, Cookson B D. DNA banding pattern polymorphism in vancomycin-resistant Enterococcus faecium and criteria for defining strains. J Clin Microbiol. 1999;37:1084–1091. doi: 10.1128/jcm.37.4.1084-1091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 15.Ostrowsky B E, Clark N C, Thauvin-Eliopoulos C, Venkataraman L, Samore M H, Tenover F C, Eliopoulos G M, Moellering R C, Jr, Gold H S. A cluster of VanD vancomycin-resistant Enterococcus faecium: molecular characterization and clinical epidemiology. J Infect Dis. 1999;180:1177–1185. doi: 10.1086/315030. [DOI] [PubMed] [Google Scholar]
- 16.Patel R, Uhl J R, Kohner P, Hopkins M K, Steckelberg J M, Kline B, Cockerill F R., III DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob Agents Chemother. 1998;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintiliani R, Courvalin P. Characterization of Tn1547, a composite transposon flanked by IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 19.Rosato A, Pierre J, Billot-Klein D, Buu-Hoi A, Gutmann L. Inducible and constitutive expression of resistance to glycopeptides and vancomycin dependence in glycopeptide-resistant Enterococcus avium. Antimicrob Agents Chemother. 1995;39:830–833. doi: 10.1128/aac.39.4.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Sifaoui F, Gutmann L. Vancomycin dependence in a VanA-producing Enterococcus avium strain with a nonsense mutation in the natural d-Ala-d-Ala ligase gene. Antimicrob Agents Chemother. 1997;41:1409. doi: 10.1128/aac.41.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva J C, Haldimann A, Prahalad M K, Walsh C T, Wanner B L. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:11951–11956. doi: 10.1073/pnas.95.20.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Bambeke F, Chauvel M, Reynolds P E, Fraimow H S, Courvalin P. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob Agents Chemother. 1999;43:41–47. doi: 10.1128/aac.43.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]