Abstract
Cardiolipins (CLs) are an important, regulated lipid class both in prokaryotic and eukaryotic cells, yet they remain largely unexplored by matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) in tissues. For example, no in-depth optimization studies of label-free visualization of CLs in complex biological samples have been reported. Here we report a streamlined modification to our previously reported MALDI-MSI method for detection of endogenous CLs in prokaryotic and eukaryotic cells based on preparation with norharmane (NRM) matrix. Notably, the use of an NRM matrix permitted sensitive detection (4.7 pg/mm2) of spotted CL synthetic standards. By contrast, four other MALDI matrices commonly used for lipid analysis failed to generate CL ions. Using this NRM-based method, endogenous CLs were detected from two types of complex biological samples: dried bacterial arrays and mouse tissue sections. In both cases, using NRM resulted in better signal/noise for CL ions than the other matrices. Furthermore, inclusion of a washing step improved CL detection from tissue and this combined tissue preparation method (washing and NRM matrix) was used to profile normal mouse lungs. Mouse lung yielded 28 unique CLs that were mapped and identified. Consistent with previous findings, CLs containing polyunsaturated fatty acids (PUFAs) were found in abundance in the airway and vascular features of the lung. This work represents a comprehensive investigation of detection conditions for CL using MALDI-MSI in complex biological samples that resulted in a streamlined method that enables future studies of the biological role(s) of CL in tissue.
Keywords: MALDI, mass spectrometry imaging, norharmane (NRM), cardiolipin, eukaryotic lipids, prokaryotic lipids
1. INTRODUCTION:
Matrix-assisted laser desorption/ionization (MALDI) is one of the most successful “soft” ionization methods in the field of mass spectrometry and enables the analysis of a broad range of molecules, including lipids.1–2 Although a precise mechanism explaining MALDI ionization remains elusive, the importance of selecting the optimal matrix to assist desorption of a given class of analyte is an important experimental consideration.3 Both the development of and the search for matrices was, and still is, an empirical process and requires properties like vacuum stability4 and high absorption at the laser wavelength5 to be evaluated. Matrix selection is especially important for MALDI mass spectrometry imaging (MALDI-MSI). MALDI-MSI has become a versatile tool for visualizing the distribution of biomolecules in tissue sections without radiogenic labeling of analytes6–7 and matrix optimization is required to provide maximum signal/noise of targeted analytes on tissues.8 Research efforts in the field of MALDI-MSI are ongoing to find the best matrix for specifically targeted analytes. The first report of cardiolipin (CL) detection in mammalian tissue sections by MALDI-MSI appeared in 2007 using the conventional matrix 2,6-dihydroxyacetophenone (DHA), with and without the addition of cesium (decreasing the relative abundance of sodiated and potassiated CLs).9 However, comprehensive investigations on the detectability of CL by MALDI-MSI, are not yet available, even though CL has been detected in tissue imaging studies for over a decade.3, 9–10
The canonical CL structure comprises two phosphatidylglycerol lipids, joined by a central glycerol backbone, to form a dimeric tetra-acylated phospholipid. Notably, CLs are ubiquitously observed in biological matrices.11,12 In prokaryotic cells, CLs are dominant lipids that reside at the poles of the inner and outer membranes.13 Gram-negative bacterial CL is synthesized at the inner membrane and actively transport to the outer membrane by the transporter PgbA.14 CL transport to the outer membrane is an important factor in Salmonella virulence and survival during infection15, further highlighting the importance of understanding the role of CL (both host- and pathogen-borne) within tissue systems. Eukaryotic CLs have multiple structural and functional roles in bioenergetics, mitochondrial signaling, and cellular fate pathways.16–18 CLs are predominant lipids in the mitochondrial inner membrane19–20 and tissues rich in mitochondria (such as cardiac muscle) show robust CL patterns in MALDI-MSI.21 While the aggregate abundance of CLs in cells and tissues is low relative in comparison to other phospholipids (2–3 mol %); approximately one quarter of the phospholipid molecules in the inner mitochondrial membrane (IMM) are CLs.22 In addition, CLs have been described as biomarkers for discrimination between oncocytic tumors and non-oncocytic tumors.23 Moreover, regio‐specific CL changes have been demonstrated in traumatic brain injury (TBI).24 Subsequently, translocation of CL between the inner and outer mitochondrial membranes was found to drive mitophagy in a TBI model.25 Thus, the ability to detect and spatially resolve CL in various pathologies could lead to a better understanding of the regio-specific roles for CL at the tissue level.
Several methodologies have been reported for the analysis of CL in tissue, each with advantages and disadvantages, yet there have been no comprehensive comparative analyses of the reported methods. While CL mapping has been shown by multiple mass spectrometric imaging modalities beyond MALDI, including Desorption Electrospray Ionization (DESI)23 and Secondary Ion Mass Spectrometry (SIMS)26–27, the present study focuses exclusively on advancing CL detection using MALDI. Specifically, the formation of alkali metal adducts are a concern in lipid imaging experiments by MALDI as they can complicate data interpretation. Thus, minimizing alkali metal adducts is important in lipid imaging experiments by MALDI. The first report of CL detection using MALDI-MSI established the complexity of CL analysis due to adduct formation and addressed it using cesium.9 Another approach, demonstrated by Angel, et. al., was depletion of alkali adducts by washing tissue sections with an ammonium formate solution, prior to matrix application. In that study, the detection of phospholipids as quasi-molecular ions was significantly enhanced by elimination of alkali adduct ions.28 Adding to the complexity of CL analysis is the similarity of authentic CL to salt-bridged (non-covalently bonded) dimeric phospholipid species, which makes true CL ions difficult to assign without high resolving power instruments. In order to investigate oxidized CLs, in disease states, using MALDI, Amoscato et. al. developed a paired derivatization and enzymatic digest process that reduced complexity and enhanced specificity of CL detection.22 This work demonstrated that DHB is a useful matrix to detect CLs in brain sections using MALDI-MSI; however, the method required a tandem, on-tissue derivatization process, beginning with 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide treatment for carboxyl group modification, followed by phospholipase C treatment for elimination of unwanted interference from other lipids.22 Further, various MALDI matrices have been used for MALDI-MSI reports including 2,5-dihyroxybenzoic acid (DHB)29, 9-aminoacridine (9-AA)30, IR-78031, 1,5-diaminonaphthalene (DAN or 1,5 DAN)32, and norharmane (NRM)33. However, they have not been systematically evaluated for CL detection by MALDI-MSI.
Previously, we showed that NRM was an efficient matrix for bacterial lipid A, a similarly high mass lipid to CL, and expected it might also be adapted for CL detection by MALDI-MSI.33 In this study, we demonstrate that NRM is an effective matrix for detection of CLs in both prokaryotic and eukaryotic samples; with minimal, streamlined sample preparation. We show that our method can efficiently detect CLs without the additional labor-intensive the derivatization procedures. This work further enables the study of CLs in biologically-relevant model systems.
2. EXPERIMENTAL
2.1. Materials
Methanol (MeOH, 98%), 9-AA (99%), DAN (99%), DHB (99%), IR-780 (99%), NRM (98%), and chloroform (HPLC grade) were purchased from Sigma Aldrich (St. Louis, MO, USA). Synthetic CL standards, 1’,3’-bis[1-palmitoyl-2-oleoyl-sn-glycero-3-phospho]-glycerol (referred to here as CL (64:0)) and 1’,3’-bis[1,2-dipalmitoyl-sn-glycero-3-phospho]-glycerol (CL (68:2), structure given in Figure S1) was purchased from Avanti Polar Lipid (AL, USA). Phosphate-buffered saline (PBS) and certified endotoxin-free water were sourced from Gibco (Grand Island, NY, USA). Focus MALDI plates were obtained from Hudson (Suwon, South Korea). All other chemicals were obtained from Sigma Aldrich unless otherwise noted.
2.2. Sample preparation
In all cases, matrices were prepared by dissolving 10 mg powder in 1 mL of chloroform:methanol (2:1, vol:vol) for spot analysis or for use in a pneumatic matrix sprayer. In order to directly compare ionization efficiency of NRM with previous reports, IR-780 was used as a matrix at 2 mg/mL (in methanol).31 Slides were washed (or left unwashed for comparison) by dipping in 50 mM ammonium formate (pH 6.7) for 30 seconds, as previously described28, and dried under vacuum prior to matrix application.
2.3. Fabrication of bacteria arrays on ITO surface
Bacteria were grown in Lysogeny Broth (LB, Becton-Dickenson, Hunt Valley, MD, USA) in shaking liquid culture (200 RPM), to mid-log phase, at 37°C. Pseudomonas aeruginosa (Pa) strain ATCC 31482 and Escherichia coli (Ec) strain ATCC 25922 were used in these studies. The same LB formulation, with the addition of 1.5% (w/v) Bacto agar (Becton-Dickenson), was used for growth on solid media. One microliter of bacterial suspensions in PBS (Pa and Ec at OD600 : 1.7±0.2, 1.0±0.1, 0.1±0.01, and 0.01±0.003), spotted on the conductive side of an ITO slide and the droplet was left to dry under ambient laboratory conditions prior to matrix application.
2.4. Tissue preparation and imaging
Twelve micron thick sections were cut using a ThermoFisher cryostat (Waltham, MA, USA), mounted onto a cold glass slide, thaw-mounted and incubated at 37°C until visibly dry. Inflated lung sections were cut at a thickness of 13 μm according to the previously established protocol.21 Spot analysis was performed on tissue, using uninfected, unfixed, frozen mouse kidney, spleen, brain and gelatin inflated lung tissue (with intact heart tissue) prepared using 2% porcine gelatin as previously described and is given as an expanded method in the Supplemental Information.21 Serial sections were used whenever head-to-head matrix condition comparisons were made. For MSI of endogenous CLs, matrices were applied with a SunCollect MALDI Sprayer, an automated pneumatic sprayer device (SunChrom, Napa, CA), with the flow rate set to 20 μL/min with 12 passages. MSI data were collected on an Autoflex Speed (Bruker) and rapifleX (Bruker), calibrated with red phosphorous clusters in the negative ion mode; conditions described below. H&E staining image of tissues after MALDI-MSI experiment were obtained post-acquisition after elimination of matrix on the tissues.34
2.5. Instrument and conditions
MALDI-TOF analyses were performed on a Bruker Autoflex Speed mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with a 2 kHz Nd:YAG (355 nm) laser for method development experiments. The laser power energy was adjusted between 0% and 100% to provide the laser pulse energy between 93.8 and 121.8 μJ. A typical laser pulse energy of 60% was applied in the experiments, except where otherwise stated. A MTP 384 massive target (serial number 03798) from Bruker Daltonics was used for the sample plate. The instrument was operated in linear mode, with accelerating voltage, grid voltage and delayed extraction time set to 19 kV, 90%, and 120 ns, respectively. Unless otherwise stated, each mass spectrum was acquired as an average of 500 laser shots, at 10.0 Hz frequency. Fine-scale spatial resolution imaging experiments (10–50 μm) were performed using a rapifleX MALDI-TOF Tissuetyper (Bruker Daltonics). The instrument is equipped with a Smart beam 3D 10 kHz Nd:YAG (355 nm) laser. CL imaging data was collected in negative ion mode from m/z 1000–2500, with 200 laser shots averaged per pixel. Image acquisition, normalization, and visualization were carried out using flexImaging 4.1 software and spectral post-analysis with mMass. High resolution (HR) MSI was performed on a MALDI LTQ-XL-Orbitrap (Thermo Fisher, San Jose, CA). Xcaliber version 2.2 and ImageQuest version 1.0.1 software were used for MALDI-MSI data acquisition and processing.
3. RESULTS AND DISCUSSION
3.1. Effect of matrix for efficient detection of CL
Initially, we evaluated the potential effectiveness of the following matrices for the detection of CLs, using their physicochemical properties as a guide for selection. Supplemental Table 1 details these properties, such as experimental and theoretical values (XLOGP3) of n-octanol/water partition coefficient, pKa, chemical formula, and molar mass of the individual matrices evaluated. The n-octanol/water partition coefficient represents the hydrophilicity/lipophilicity of the molecule by measuring ratio of a chemical’s concentration in the octanol phase to its concentration in the aqueous phase. The XLOGP3 value showed a theoretical n-octanol/water partition coefficient via a computational chemistry method. Of all of the matrices examined, the properties of NRM suggest it to be optimal for CL ionization from tissue. One of the possible scenarios explaining why NRM would be an efficient matrix for imaging CLs in tissue is the high hydrophobicity (partition coefficient = 3.17, XLOGP3 = 3.2) and pKa (13.2) as shown in Supplemental Table 1. NRM has the highest values in both cases. This was interpreted to mean NRM has the highest hydrophobicity and lipophilicity. CL has two phosphatidic acid moieties, which act as anionic loci at physiological pH (~7.4). Therefore, it is expected that positively charged matrices, such as 9-AA, NRM, and DAN interact with CLs via electrostatic interaction and NRM serves as an efficient base compared to the other matrices.
Figure 1 demonstrates the effectiveness of NRM as a MALDI matrix to detect CLs in the negative ion mode. Using a commercial synthetic CL (68:2) as a standard, we determined the limit of detection (LOD) for common lipid matrices (9-AA, DAN, and DHB), compared to NRM. As shown in Figure 1a, NRM was the only matrix that showed CL detection at a level of 4.7 pg/mm2. Additionally, given that DAN generated interfering ions in the m/z region 1000 to 2000, which bracketed the expected CL distribution, it was excluded from further studies. The matrix IR-780 was recently reported as a matrix substance for the detection of high molecular weight lipids31 and was subsequently tested for detection of spotted CL standards. In the present study, IR-780 showed limited sensitivity compared to NRM (Figure S2) and was also excluded from further studies.
Figure 1.
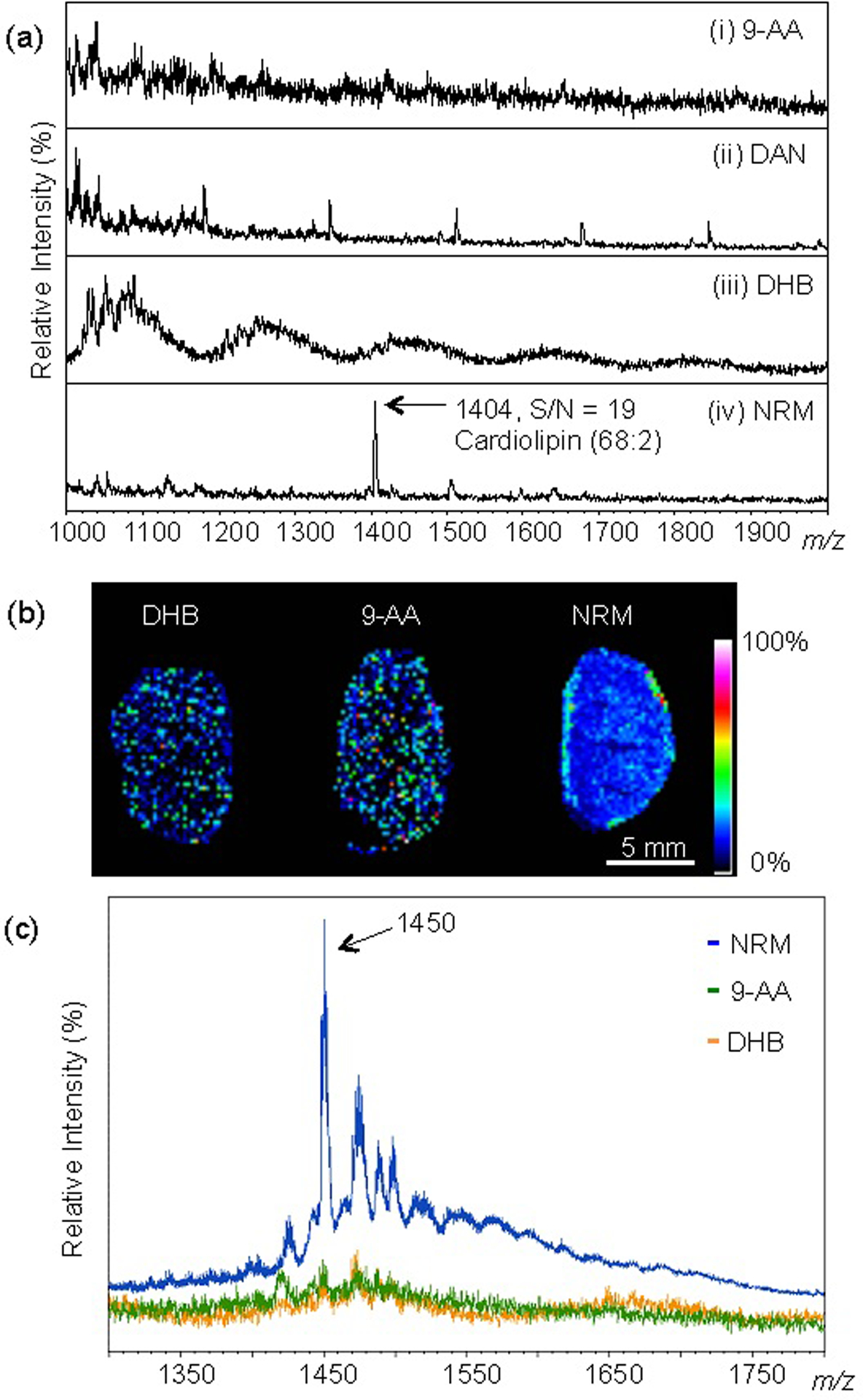
Norharmane (NRM) matrix facilitates detection of cardiolipin synthetic standard and endogenous CL from mouse tissue. (a) MALDI mass spectra of various matrices (i) 9-AA, (ii) DAN, (iii) DHB, and (iv) NRM with 4.7 pg/mm2 of synthetic CL(68:2) detected as the ion m/z 1404, [M-H]−. (b) MALDI-MSI, mouse kidney section prepared with given matrix, ion m/z 1450 shown, spatial resolution 200 μm, normalized by total ion current (TIC). (c) Averaged mass spectra of each tissue from (b) prepared with 9-AA, DHB, and NRM. Negative ion mode Autoflex MALDI-TOF.
We next compared matrix efficiency to detect endogenous CL in mouse tissue, using MALDI-MSI. Mouse kidney sections were used as a benchmark tissue for method comparison by MALDI-MSI. Normal mouse kidneys were harvested and prepared on ITO slides, and coated with NRM matrix as described. As shown in Figure 1b, we observed a unique ion distribution (m/z 1450, subsequently identified as CL 72:7) in the kidney tissue prepared with NRM; this ion was localized in the outer cortex region, at the tissue boundary. In contrast, there was no discernable tissue pattern, for the same ion (m/z 1450) in serial sections prepared with DHB and 9-AA. The peak intensity of the ion m/z 1450 was evaluated as an average mass spectrum of each tissue prepared with 9-AA, DHB, or NRM (Figure 1c). Based on the ion intensity (m/z 1450), the average spectra showed an approximately five times stronger signal-to-noise (S/N) ratio with NRM versus 9-AA and DHB. Together, these results demonstrated that NRM is an efficient lipid matrix, with an improved detection threshold of spotted CL standards and enabled CL mapping by MALDI-MSI in a single step (i.e. NRM matrix application alone).
3.2. Direct profiling of CL from bacteria
An abundant biological source of CLs is the Gram-negative bacterial membrane. CLs are estimated to constitute 5–30% of the membrane lipid content, dependent upon growth phase and strain.35 To further extend the characterization of NRM as an efficient matrix for CL, bacteria (Ec and Pa) were directly deposited on a target plate and used as imaging samples. For all experiments, bacterial cultures grown to mid-log and adjusted to a uniform turbidity using optical density (OD600) = 0.5 (approximately 0.5×109 cfu/mL of bacteria) were used. For MALDI-MSI analysis, 1 μL of the bacterial culture was spotted, followed with 1 μL of the respective matrix solution (9-AA, DHB, or NRM). Figure 2a (panels i, iii, v) represents mass spectra of intact Ec bacteria spotted directly on target. The ion m/z 1376 in Figure 2a (panel v) corresponds to CL (66:2). Additional ions corresponding to CLs, such as CL (64:2) and (68:2) were observed in the mass spectrum. Figure 2a (panels ii, iv, vi) represent mass spectra of intact Pa, the ion m/z 1404 in Figure 2a (panel vi) corresponds to CL (68:2). Notably, NRM was the only matrix to generate CL ions directly from Ec and Pa.
Figure 2.
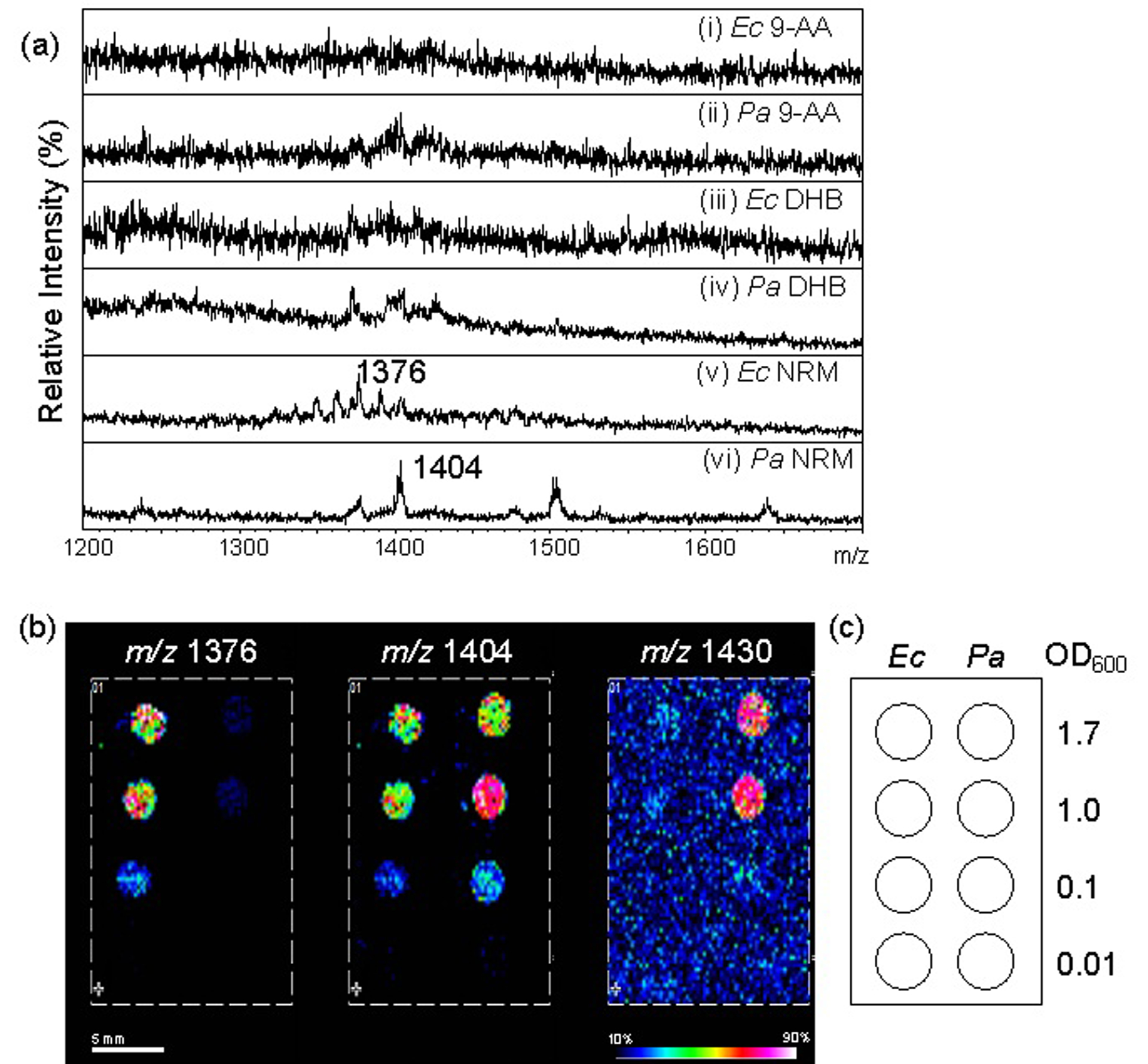
Cardiolipin detection from bacterial spots by MALDI-MSI using NRM matrix. (a) MALDI mass spectra resulting from various matrices (i) and (ii) 9-AA, (iii) and (iv) DHB, and (v) and (vi) NRM with OD 0.5 of Ec ((i), (iii), and (v)) and Pa ((ii), (iv), and (vi)). MALDI-TOF linear negative, Autoflex. (b) MALDI-MSI of CL ions on a bacterial array in the arrangement given in (c). (b) MALDI-TOF reflectron negative, rapifleX. Normalized, TIC.
To determine whether these findings could be extended to a two-dimensional imaging method, a bacterial array was fabricated on ITO glass surface and the array was analyzed by MALDI-MSI using NRM as the matrix. Four concentrations of bacteria were spotted (OD600 = 1.7, 1.0, 0.1, and 0.01) for each of the bacterial species and a dilution effect was observed for each of the three CLs (Figure 2b). Three major ions were detected in the bacterial array (m/z 1376, 1404, and 1430; Figure 2b) and were consistent with those observed in the spot analysis (Figure 2a); these ions correspond to CL (64:2), (68:2), and (70:3), respectively. A different CL (64:2) ion was detected in Ec, than the CL (70:3) ion mainly detected in Pa. However, one CL (68:2) ion was detected in both bacterial backgrounds. The LOD of endogenous CL (68:2) in Ec and Pa represents a cutoff of approximately 104 cfu/μL in this preparation method and volume. The results of this bacterial array suggested that CLs could be further developed as a potential biomarker for identification of individual bacteria species. In contrast to 9-AA and DHB, NRM yielded CL ions from direct analysis of dried bacteria without extraction, derivatization, or other preparation.
3.3. Aqueous washing improves detection of CL
Recently, it was demonstrated that washing tissue samples before analysis, using an aqueous solution of formate, enhanced detection of lipid species by MALDI-MSI, presumably due to desalting.28 To determine whether prior washing of murine tissue samples in a similar fashion before matrix deposition and analysis enhanced the detection of CLs, multiple tissue types (kidney, spleen, and inflated lung sections) were evaluated. Slides were washed with 50 mM ammonium formate (pH 6.7), prior to applying NRM matrix and evaluation by MALDI-MSI. Increased intensity of the quasi-molecular ion m/z 1448.0 (CL (72:8)) and the complementary reduction in the sodium adduct observed as m/z 1470.0 were observed after the washing step. Figure S3 demonstrates the washing results for lung and kidney samples. The trend was similar in all tissues we tested. Thus, we concluded that inclusion of a desalting wash step prior to tissue preparation with NRM for MALDI-MSI improved the detection of CL and all subsequent experiments were performed with the washing step added.
3.4. Evaluation of NRM for CL mapping at high spatial resolution by MALDI-MSI and CL identification
Using the methods established above, 50 μm spatial resolution images of CLs in multiple tissue types were obtained. The distributions of CLs were demonstrated on lung (Figure 3a) and horizontally sectioned brain tissue (Figure S4). In lung sections, the majority of CL is localized near the midline (cardiac area) and in endothelial-lined vascular structures of the lung. This result was consistent with our previous report of the location of CLs in inflated lung tissues.21 As in that study, the presence of polyunsaturated fatty acid (PUFA)-containing lipids tended to co-localize with airway and vascular structures in the lung, recapitulated here with PUFA-CLs including CL (72:8), (72:7), and (72:6) (m/z 1448, 1450, 1452, respectively). Interestingly, the extensively unsaturated CLs detected in the lung (74:10, 76:12, 76:11,76:10, and 76:9; m/z 1404, 1472, 1496, 1498, and 1500, respectively) showed a more diffuse pattern suggestive of a parenchymal association, which will be evaluated in future studies. The added sensitivity of desalting and preparation with NRM matrix enabled the detailed mapping of unique CL distributions in mouse lung tissues.
Figure 3.
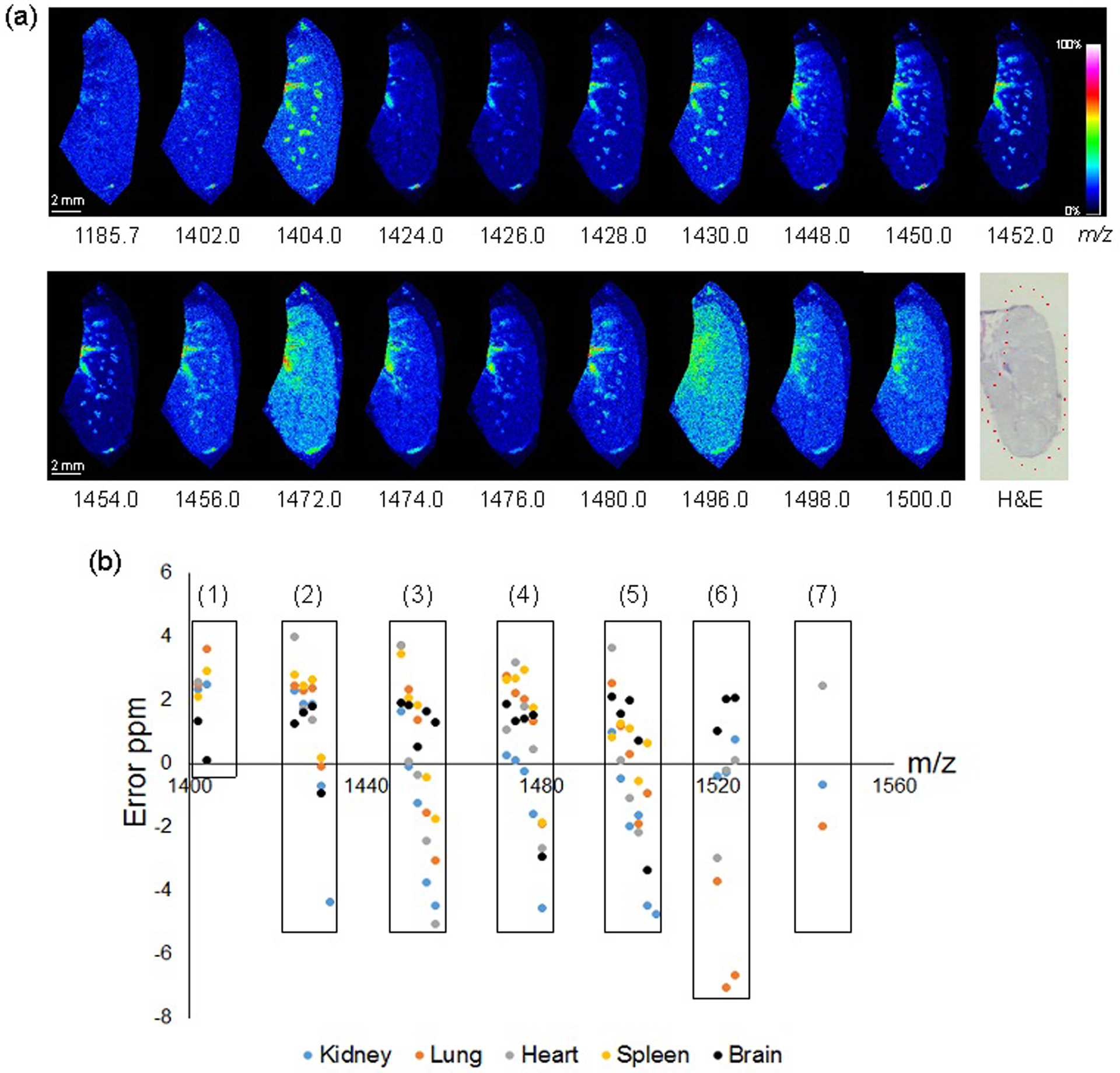
MALDI detection of CL in mouse tissues. (a) MALDI-MSI of CLs in inflated mouse lung, spatial resolution 50 μm, normalized (TIC), reflectron negative rapifleX MALDI-TOF. Post-acquisition H&E given with scan area outlined in red. (b) Identified CLs as ions plotted by mass error ppm. Numerals represent acyl carbon sum compositions: (1) 68, (2) 70, (3) 72 (4) 74, (5) 76, (6) 78, and (7) 80. MALDI-LTQ/Orbitrap MS, summarized from MSI with spatial resolution 200 μm, normalized (TIC).
Distribution of CLs in mouse brain was also evaluated and clearly illustrated the region-specific nature of CL-species distribution (Figure S4). These results further highlight the need for more in-depth study of individual CLs in organization and maintenance of brain structure. Finally, to further test the spatial resolution limits of using NRM for endogenous CLs mapping by MALDI-MSI, we analyzed the fine structure of mouse liver at 10 μm spatial resolution and the expected lacey cellular structures, typical of liver tissue (illustrated by the ion m/z 1450 (CL (72:7)) were observed (Figure S5).
Table 1 details CLs detected from lung tissue using High-Resolution (HR)-MSI. Due to the addition of a formate washing step, the alkali cation adducted ion species were excluded from the candidates that only describe the quasi-molecular ion [M-H]−. We observed 26 species of CLs of various carbon chain lengths and double bond arrangements in lung tissue. HR-MSI generates high accuracy data which complemented by MS/MS performed on ions in the region, allowed us to putatively identify CL compositions at less than ± 3 ppm error. Several ions are tentatively identified as CLs at less than ± 7 ppm and are included in Table 1 and all Supplemental Tables 2–5 for the other tissues evaluated. The detailed information regarding isomeric acyl positioning or location of double bond(s) of the detected CLs is not considered in this work. Supplemental Tables 2–5 list the CLs identified in kidney, heart, spleen, and brain tissues, respectively, and sample collision induced dissociation spectra of various CLs are shown in Figure S6. Figure 3b illustrates the error in ppm between experimental and theoretical m/z among the identified CLs from the five tissue types we analyzed. The X-axis represents m/z value from 1400 to 1560. In most cases, the error was < ± 5 ppm, except for two CLs (78:13 and 78:12) from lung tissue (Table 1). The longest chain length of CL (80:16) was only detected in kidney, lung, and heart, whereas in spleen the longest was a slightly shorter CL (76:8) (Supplemental Table 3). The other long chain lengths, such as 78 or 80 were not detected in spleen. In the case of brain, the most intense CL peak in the averaged mass spectrum was CL (74:7) at m/z 1478.0142 (± 1.56 ppm). This result matched well with previous reports.36 We found various chain lengths and double bond compositions of CLs using HRMS, with acyl carbon sums grouped (boxes) in Figure 3b. As the number of double bonds in CLs decreased, the mass error (given as ppm) showed a negative trend for CLs with the same number of acyl carbons. This trend was particularly noticeable for CLs with 70, 72, 74, and 76 acyl carbons (Figure 3, classes 2–5) and does not appear to have impacted the confidence of assignments. The cause of the trend could not be ascertained and is under investigation.
Table 1.
List of identified CLs from mouse lung.
| No. | CL List | m/z (Observed) | m/z (Theoretical) | Molecular Formula | Error (ppm) |
|---|---|---|---|---|---|
| 1 | CL (54:6) | 1185.7395 | 1185.7353 | C63H112O16P2 | 3.54 |
| 2 | CL (68:3) | 1401.9841 | 1401.9806 | C77H144O17P2 | 2.50 |
| 3 | CL (68:2) | 1404.0014 | 1403.9963 | C77H146O17P2 | 3.63 |
| 4 | CL (70:6) | 1423.9684 | 1423.9649 | C79H142O17P2 | 2.46 |
| 5 | CL (70:5) | 1425.9839 | 1425.9806 | C79H144O17P2 | 2.31 |
| 6 | CL (70:4) | 1427.9996 | 1427.9962 | C79H146O17P2 | 2.38 |
| 7 | CL (70:3) | 1430.0118 | 1430.0119 | C79H148O17P2 | −0.07 |
| 8 | CL (70:2) | N.D. | 1432.0275 | C79H150O17P2 | N.A. |
| 9 | CL (72:8) | 1447.9704 | 1447.965 | C81H142O17P2 | 3.73 |
| 10 | CL (72:7) | 1449.984 | 1449.9806 | C81H144O17P2 | 2.34 |
| 11 | CL (72:6) | 1451.9982 | 1451.9962 | C81H146O17P2 | 1.38 |
| 12 | CL (72:5) | 1454.0097 | 1454.0119 | C81H148O17P2 | −1.51 |
| 13 | CL (72:4) | 1456.0231 | 1456.0275 | C81H150O17P2 | −3.02 |
| 14 | CL (74:10) | 1471.969 | 1471.9649 | C83H142O17P2 | 2.79 |
| 15 | CL (74:9) | 1473.9839 | 1473.9806 | C83H144O17P2 | 2.24 |
| 16 | CL (74:8) | 1475.9992 | 1475.9962 | C83H146O17P2 | 2.03 |
| 17 | CL (74:7) | 1478.0139 | 1478.0119 | C83H148O17P2 | 1.35 |
| 18 | CL (74:6) | 1480.0247 | 1480.0275 | C83H150O17P2 | −1.89 |
| 19 | CL (76:12) | 1495.9687 | 1495.9649 | C85H142O17P2 | 2.54 |
| 20 | CL (76:11) | 1497.9824 | 1497.9806 | C85H144O17P2 | 1.20 |
| 21 | CL (76:10) | 1499.9967 | 1499.9962 | C85H146O17P2 | 0.33 |
| 22 | CL (76:9) | 1502.0091 | 1502.0119 | C85H148O17P2 | −1.86 |
| 23 | CL (76:8) | 1504.0261 | 1504.0275 | C85H150O17P2 | −0.93 |
| 24 | CL (76:7) | N.D. | 1506.0432 | C85H152O17P2 | N.A. |
| 25 | CL (78:14) | 1519.9593 | 1519.9649 | C87H142O17P2 | −3.68 |
| 26 | CL (78:13) | 1521.9699 | 1521.9806 | C87H144O17P2 | −7.03 |
| 27 | CL (78:12) | 1523.9861 | 1523.9962 | C87H146O17P2 | −6.63 |
| 28 | CL (80:16) | 1543.9619 | 1543.9649 | C89H142O17P2 | −1.94 |
N.D., Not Detected. N.A., Not Applicable. MALDI Ion Trap/Orbitrap MSI.
4. CONCLUSIONS
Here, we studied the effect of a range of different MALDI matrices to determine the most efficient means to analyze CLs in various biological samples, including Gram-negative bacteria and mouse tissues. We identified that NRM was the most effective matrix among the five matrices analyzed, which included 9-AA, DHB, DAN, IR-780, and NRM. The LOD for NRM was found to be 4.7 pg/mm2 for commercial synthetic CL standards, which translated to the ability to readily detect CLs in relevant biological samples. We showed that CLs may be further developed as a tool for bacterial identification. Furthermore, washing tissue sections prior to NRM matrix coating increased detection of CLs as deprotonated ions. This simple washing step incorporated before matrix application represents a rapid tissue preparation for MALDI-MSI analysis of CL in tissue that does not involve additions of metal ions or enzymatic digestions steps. Using this method, we identified similar CL populations in each of the tissues we profiled, including mouse lung, heart, spleen, kidney, and brain. This streamlined method of desalting and subsequent NRM matrix application can be easily applied for profiling CLs in diverse types of biological samples when analyzing by MALDI-coupled techniques and will lead to further expansion of our understanding of the role of CL in health and disease.
Supplementary Material
ACKNOWLEDGMENT
The instruments of the NIDA IRP structural biology unit were critical in advancing this work as were the resources of the Johns Hopkins Applied Imaging Mass Spectrometry (AIMS) Core at the Johns Hopkins Medical Institutes. The authors thank David J. Varisco for reading the manuscript and providing insightful comments. We acknowledge the following funding sources supporting this work: Cystic Fibrosis Foundation Research Grant to R.K.E. (ERNST18GO), National Institutes of Health Grants to R.K.E. and D.R.G. (R01GM111066 and 1R01 AI147314-01A1), D.R.G. thanks the International Centre for Cancer Vaccine Science project carried out within the International Research Agendas program of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund (MAB/2017/03) for support.
Footnotes
Supporting Information
Supplemental Information File containing expanded methods and additional data is available as a PDF.
REFERENCES
- (1).Kaufmann R Matrix-Assisted Laser Desorption Ionization (Maldi) Mass Spectrometry: A Novel Analytical Tool in Molecular Biology and Biotechnology; J Biotechnol,1995, 41, 155–175. [DOI] [PubMed] [Google Scholar]
- (2).Woods AS; Jackson SN Brain Tissue Lipidomics: Direct Probing Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry; AAPS J,2006, 8, E391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Leopold J; Popkova Y; Engel KM; Schiller J Recent Developments of Useful Maldi Matrices for the Mass Spectrometric Characterization of Lipids; Biomolecules,2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yang J; Norris JL; Caprioli R Novel Vacuum Stable Ketone-Based Matrices for High Spatial Resolution Maldi Imaging Mass Spectrometry; J Mass Spectrom,2018, 53, 1005–1012. [DOI] [PubMed] [Google Scholar]
- (5).Robinson KN; Steven RT; Bunch J Matrix Optical Absorption in Uv-Maldi Ms; J Am Soc Mass Spectrom,2018, 29, 501–511. [DOI] [PubMed] [Google Scholar]
- (6).Cornett DS; Reyzer ML; Chaurand P; Caprioli RM Maldi Imaging Mass Spectrometry: Molecular Snapshots of Biochemical Systems; Nat Methods,2007, 4, 828–833. [DOI] [PubMed] [Google Scholar]
- (7).Franck J; Arafah K; Elayed M; Bonnel D; Vergara D; Jacquet A; Vinatier D; Wisztorski M; Day R; Fournier I; Salzet M Maldi Imaging Mass Spectrometry: State of the Art Technology in Clinical Proteomics; Mol Cell Proteomics,2009, 8, 2023–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Norris JL; Caprioli RM Analysis of Tissue Specimens by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry in Biological and Clinical Research; Chem Rev,2013, 113, 2309–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang HY; Jackson SN; Woods AS Direct Maldi-Ms Analysis of Cardiolipin from Rat Organs Sections; J Am Soc Mass Spectrom,2007, 18, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sparvero LJ; Amoscato AA; Dixon CE; Long JB; Kochanek PM; Pitt BR; Bayir H; Kagan VE Mapping of Phospholipids by Maldi Imaging (Maldi-Msi): Realities and Expectations; Chem Phys Lipids,2012, 165, 545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kagan VE; Tyurina YY; Tyurin VA; Mohammadyani D; Angeli JP; Baranov SV; Klein-Seetharaman J; Friedlander RM; Mallampalli RK; Conrad M; Bayir H Cardiolipin Signaling Mechanisms: Collapse of Asymmetry and Oxidation; Antioxid Redox Signal,2015, 22, 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).De Bruijn JH Chemical Structure and Serological Activity of Natural and Synthetic Cardiolipin and Related Compounds; Br J Vener Dis,1966, 42, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mileykovskaya E; Dowhan W Cardiolipin Membrane Domains in Prokaryotes and Eukaryotes; Biochim Biophys Acta,2009, 1788, 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dong H; Zhang Z; Tang X; Huang S; Li H; Peng B; Dong C Structural Insights into Cardiolipin Transfer from the Inner Membrane to the Outer Membrane by Pbga in Gram-Negative Bacteria; Sci Rep,2016, 6, 30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dalebroux ZD; Edrozo MB; Pfuetzner RA; Ressl S; Kulasekara BR; Blanc MP; Miller SI Delivery of Cardiolipins to the Salmonella Outer Membrane Is Necessary for Survival within Host Tissues and Virulence; Cell Host Microbe,2015, 17, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Paradies G; Paradies V; De Benedictis V; Ruggiero FM; Petrosillo G Functional Role of Cardiolipin in Mitochondrial Bioenergetics; Biochim Biophys Acta,2014, 1837, 408–417. [DOI] [PubMed] [Google Scholar]
- (17).Mejia EM; Hatch GM Mitochondrial Phospholipids: Role in Mitochondrial Function; J Bioenerg Biomembr,2016, 48, 99–112. [DOI] [PubMed] [Google Scholar]
- (18).Dudek J Role of Cardiolipin in Mitochondrial Signaling Pathways; Front Cell Dev Biol,2017, 5, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Osman C; Voelker DR; Langer T Making Heads or Tails of Phospholipids in Mitochondria; J Cell Biol,2011, 192, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).van Meer G; Voelker DR; Feigenson GW Membrane Lipids: Where They Are and How They Behave; Nat Rev Mol Cell Biol,2008, 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Scott AJ; Chandler CE; Ellis SR; Heeren RMA; Ernst RK Maintenance of Deep Lung Architecture and Automated Airway Segmentation for 3d Mass Spectrometry Imaging; Sci Rep,2019, 9, 20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Amoscato AA; Sparvero LJ; He RR; Watkins S; Bayir H; Kagan VE Imaging Mass Spectrometry of Diversified Cardiolipin Molecular Species in the Brain; Anal Chem,2014, 86, 6587–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang J; Yu W; Ryu SW; Lin J; Buentello G; Tibshirani R; Suliburk J; Eberlin LS Cardiolipins Are Biomarkers of Mitochondria-Rich Thyroid Oncocytic Tumors; Cancer Res,2016, 76, 6588–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sparvero LJ; Amoscato AA; Fink AB; Anthonymuthu T; New LA; Kochanek PM; Watkins S; Kagan VE; Bayir H Imaging Mass Spectrometry Reveals Loss of Polyunsaturated Cardiolipins in the Cortical Contusion, Hippocampus, and Thalamus after Traumatic Brain Injury; J Neurochem,2016, 139, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chao H; Lin C; Zuo Q; Liu Y; Xiao M; Xu X; Li Z; Bao Z; Chen H; You Y; Kochanek PM; Yin H; Liu N; Kagan VE; Bayir H; Ji J Cardiolipin-Dependent Mitophagy Guides Outcome after Traumatic Brain Injury; J Neurosci,2019, 39, 1930–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tian H; Sparvero LJ; Amoscato AA; Bloom A; Bayir H; Kagan VE; Winograd N Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry High-Resolution Imaging of Cardiolipin Speciation in the Brain: Identification of Molecular Losses after Traumatic Injury; Anal Chem,2017, 89, 4611–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tian H; Sparvero LJ; Blenkinsopp P; Amoscato AA; Watkins SC; Bayir H; Kagan VE; Winograd N Secondary-Ion Mass Spectrometry Images Cardiolipins and Phosphatidylethanolamines at the Subcellular Level; Angew Chem Int Ed Engl,2019, 58, 3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Angel PM; Spraggins JM; Baldwin HS; Caprioli R Enhanced Sensitivity for High Spatial Resolution Lipid Analysis by Negative Ion Mode Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry; Anal Chem,2012, 84, 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Teearu A; Vahur S; Haljasorg U; Leito I; Haljasorg T; Toom L 2,5-Dihydroxybenzoic Acid Solution in Maldi-MS: Ageing and Use for Mass Calibration; J Mass Spectrom,2014, 49, 970–979. [DOI] [PubMed] [Google Scholar]
- (30).Cheng H; Sun G; Yang K; Gross RW; Han X Selective Desorption/Ionization of Sulfatides by Maldi-MS Facilitated Using 9-Aminoacridine as Matrix; J Lipid Res,2010, 51, 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Li N; Wang P; Liu X; Han C; Ren W; Li T; Li X; Tao F; Zhao Z Developing Ir-780 as a Novel Matrix for Enhanced Maldi MS Imaging of Endogenous High-Molecular-Weight Lipids in Brain Tissues; Anal Chem,2019, 91, 15873–15882. [DOI] [PubMed] [Google Scholar]
- (32).Caughlin S; Park DH; Yeung KK; Cechetto DF; Whitehead SN Sublimation of Dan Matrix for the Detection and Visualization of Gangliosides in Rat Brain Tissue for Maldi Imaging Mass Spectrometry; J Vis Exp,2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Scott AJ; Flinders B; Cappell J; Liang T; Pelc RS; Tran B; Kilgour DP; Heeren RM; Goodlett DR; Ernst RK Norharmane Matrix Enhances Detection of Endotoxin by Maldi-MS for Simultaneous Profiling of Pathogen, Host, and Vector Systems; Pathog Dis,2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Deutskens F; Yang J; Caprioli RM High Spatial Resolution Imaging Mass Spectrometry and Classical Histology on a Single Tissue Section; J Mass Spectrom,2011, 46, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Swain J; El Khoury M; Kempf J; Briee F; Van Der Smissen P; Decout JL; Mingeot-Leclercq MP Effect of Cardiolipin on the Antimicrobial Activity of a New Amphiphilic Aminoglycoside Derivative on Pseudomonas aeruginosa; PLoS One,2018, 13, e0201752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gao F; McDaniel J; Chen EY; Rockwell HE; Nguyen C; Lynes MD; Tseng YH; Sarangarajan R; Narain NR; Kiebish MA Adapted Ms/Ms(All) Shotgun Lipidomics Approach for Analysis of Cardiolipin Molecular Species; Lipids,2018, 53, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


