Abstract
The genetic basis for most inherited neurodegenerative diseases has been identified, yet there are limited disease modifying therapies for these patients. A new class of drugs - antisense oligonucleotides (ASOs) - are showing promise as a therapeutic platform for treating neurological diseases. ASOs are designed to bind to the RNAs encoded by a target gene, thereby suppressing expression by catalyzing degradation of those RNAs or to elevate expression by correcting faulty RNA splicing. Following delivery by intrathecal injection into the cerebral spinal fluid that surrounds the brain and spinal cord, antisense agents distribute broadly into nervous tissues where they produce long-term effects. The recent approval of nusinersen as a treatment for spinal muscular atrophy validated antisense technology as a platform for neurodegenerative and other neurological diseases. Nusinersen demonstrated that effective treatments for neurodegenerative disease can be identified and that treatment not only slows disease progression but improves some symptoms. Antisense drugs are currently in development for amyotrophic lateral sclerosis, Huntington’s disease, Alzheimer’s disease, Parkinson’s, disease and Angelman syndrome. Several additional drugs are in late stage research for the treatment of spinocerebellar ataxias, sporadic forms of amyotrophic lateral sclerosis, infantile seizure disorders, and neurodevelopmental disorders, as well as multiple programs in drug discovery for the treatment of additional neurological diseases. This review will highlight the advances in antisense technology as potential treatments for neurological diseases.
Keywords: Alzheimer’s disease, Amyotrophic lateral sclerosis, Antisense oligonucleotide (ASO), Huntington’s disease, Parkinson’s disease, RNA, siRNA, spinal muscular atrophy
INTRODUCTION
Advances in molecular genetics in the 1980s and further refinement in the 1990s allowed the identification of causative genes for most of the inherited neurological disorders. These were heady times for the field, as a steady flow of news on identification of genes that causes devasting neurological diseases occurred throughout the 1990s. One of the more notable discoveries was the causative gene for Huntington’s disease (HD), caused by a triplet repeat (CAG) expansion in exon 1 of the huntingtin gene (1). The list of diseases due to repeat expansion has expanded to include a number of spinocerebellar ataxias, spinal and bulbar muscle atrophy, type 1and 2 myotonic dystrophy, fragile X syndrome and most recently a inherited form of amyotrophic lateral sclerosis (2–4). Completion of the sequencing of the human genome, which provides a genetic reference, and advances in DNA sequencing have dramatically accelerated the pace of identification of genetic changes that cause disease including the private mutations harbored by a single family or in small populations. Today, it is common to identify a putative disease-causing gene within a few weeks of patient presentation.
If clinical geneticists can quickly identify the genetic cause of a disease, why do we not yet have effective therapies for these inherited neurological diseases? The answers to this question are complex and multifactorial. Perhaps a key historical limitation was the absence of interest by large pharmaceutical companies to apply their resources towards the discovery and development of drugs for what were considered rare patient populations. Fortunately, this is changing with commercial success of several rare disease drugs. Additional hurdles include: lack of knowledge of the normal function of the gene and how the genetic change affects the gene function, development of model systems to evaluate potential therapies, adequate knowledge of the natural history of the disease, development of both disease relevant and target engagement biomarkers that can be used to reduce project risk, and in many cases, the lack of targetability of the affected gene by traditional drug discovery strategies. Importantly, expansion of drug discovery platforms beyond traditional small molecule drugs has led to development of designer DNA drugs, frequently referred to as antisense oligonucleotides (ASOs), to directly target expression of the genes causative of neurological diseases (5). Targeting RNA rather than protein simplifies the drug discovery process and dramatically expands the types of therapeutic targets for drug discovery.
Antisense drugs have been used as research tools for neuroscientists for almost 30 years. Some of the early applications were to help determine functions of various proteins in the central nervous system (6–10). These early studies were limited by the adverse effects observed with first generation ASOs and their limited duration of effect. Advances in antisense chemistry and better understanding of antisense mechanisms and biodistribution helped advance the platform not only as a research tool but importantly as a therapeutic agent for severe neurological diseases. Today there are five licensed antisense drugs for neurological diseases (Table 1), nine antisense drugs in clinical trials and a larger number of targets being pursued with antisense technology for neurological diseases. This review will summarize the pharmacological properties of antisense drugs being used to treat neurological diseases.
Table 1.
Examples of Marketed Antisense Drugs or In Clinical Development for Neurological Diseases
| Drug | Target | Indication | Mechanism | Chemistry | Delivery Route | Status | Reference |
|---|---|---|---|---|---|---|---|
| Eteplirsen | Dystrophin | Duchene muscular dystrophy-exon 51 | Splicing modulation | PMO | Intravenous | Marketed | (153; 154) |
| Golodirsen | Dystrophin | Duchene muscular dystrophy-exon 53 | Splicing modulations | PMO | Intravenous | Marketed | (155) |
| Inotersen | transthyretin | Familial amyloid polyneuropathy & cardiomyopathy | RNase H | 2’-MOE/DNA chimera | Subcutaneous | Marketed | (15) |
| Patisiran | transthyretin | Familial amyloid polyneuropathy | siRNA | Liposomal formulation | Intravenous | Marketed | (17) |
| Nusinersen | Survival motor neuron 2 | Spinal muscular atrophy | Splicing modulation | Uniform 2-MOE | Intrathecal | Marketed | (22; 85; 86) |
| Tominersen | Huntingtin | Huntington’s disease | RNase H | 2’-MOE/DNA chimera | Intrathecal | Phase 3 | (147) |
| Tofersen | Superoxide dismutase 1 | Familial ALS due to mutations in SOD1 | RNase H | 2’-MOE/DNA chimera | Intrathecal | Phase 3 | (46; 101) |
| Wve-120101 | Huntingtin | Huntington’s disease | RNase H | Unknown | Intrathecal | Phase 1/2 | * |
| Wve-120102 | Huntingtin | Huntington’s disease | RNase H | Unknown | Intrathecal | Phase 1/2 | * |
| IONIS-MAPTRx/ BIIB080 | MAPT (Tau) | Alzheimer’s disease/ Primary Tauopathies | RNase H | 2’-MOE/DNA chimera | Intrathecal | Phase 1/2 | ** |
| IONIS-C9Rx/ BIIB078 | C9orf72 | Familial ALS due to mutations in C9orf72 | RNase H | 2’-MOE/DNA chimera | Intrathecal | Phase 1/2 | ** |
| ION859/ BIIB094 | LRRK2 | Parkinson’s disease | RNase H | 2’-MOE/DNA chimera | Intrathecal | Phase 1/2 | ** |
| GTX-101 | UBE3A-ATS | Angelman syndrome | RNase H | Unknown | Intrathecal | Phase 1/2 | *** |
| IONIS-DMN2–2.5Rx | Dynamin 2 | Centronuclear myopathy | RNase H | cET/DNA chimera | Intravenous | Phase 1/2 | ** |
Unable to find published literature on the drug. Information obtained from Wave Life Science corporate website.
Unable to find published literature on the drugs. Information obtained from Ionis Pharmaceuticals or Biogen corporate website
Unable to find published literature on the drug. Information obtained from GeneTx Biotherapeutics corporate website
ANTISENSE MECHANISMS OF ACTION
Antisense drugs bind to RNAs through normal Watson-Crick base pairing, although non-canonical base pairing (e.g. G to U) is also a possibility. Following binding to a target RNA, an antisense drug can modulate the RNA through a variety of different mechanisms, including degradation of the targeted RNA through endogenous nucleases; altering the splicing of the RNA; displacement of proteins bound to the RNA; and disruption of regulatory structures in the target RNA (Figures 1 and 2). It is expected that additional mechanisms for ASOs will be identified. Both the chemistry of the oligonucleotide and the positioning on the target RNA to which the ASO binds, are major design elements that dictate which antisense mechanism is utilized (5; 11). No single antisense mechanism meets all therapeutic needs and no mechanism is vastly superior to other mechanisms of action.
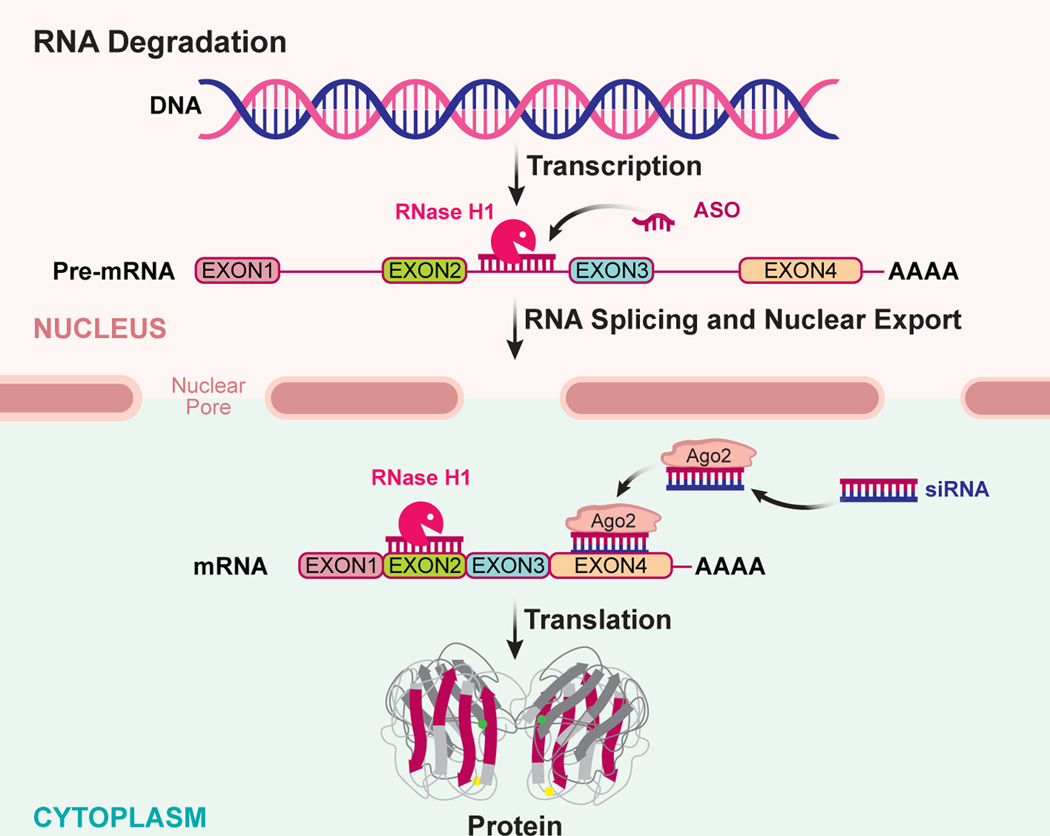
Figure 1:
RNA degradation antisense mechanisms. RNA is transcribed from DNA into a precursor form (pre-mRNA) which undergoes several post-transcriptional processing events, such as splicing to remove intronic sequence forming the mature RNA (mRNA). The mRNA is exported out of the nucleus to the cytoplasm where it is translated into its protein product. Two broadly used antisense mechanisms result in selective degradation of the targeted RNA. Single stranded ASOs that work through the RNase H1 mechanism bind to the targeted RNA and recruit RNase H1 to the ASO-RNA heteroduplex in either the nucleus or cytoplasm. RNase H1 catalyzes the degradation of the RNA strand releasing the ASO to bind to another target RNA. A second common antisense mechanism, siRNA, utilizes double stranded RNA or RNA analogs which are dissociated within the cell and the antisense strand (also commonly referred to as the guide strand) binds to argonaute 2 (Ago2) protein in a facilitated manner. The antisense strand (guide RNA) bound to the Ago2 protein directs the complex to the targeted RNA through Watson-Crick base pairing to a complementary sequence in the targeted RNA. Ago2 cleaves the targeted RNA and after cleavage the Ago-2/RNA complex is released allowing it to bind and cleave additional target RNAs.
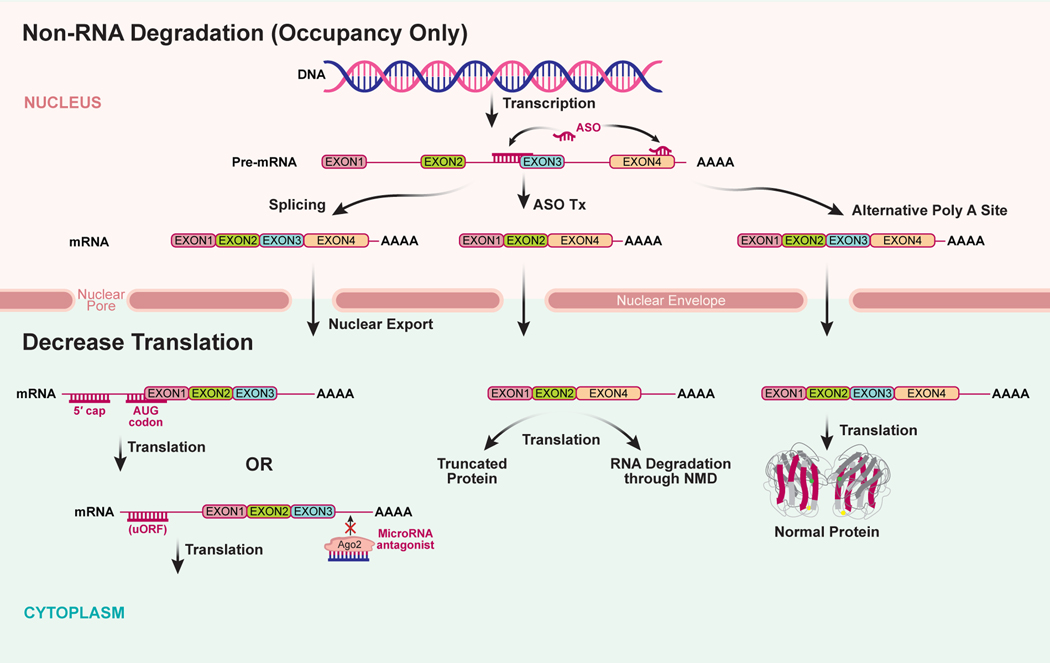
Figure 2:
Non-RNA degradation antisense mechanisms (occupancy only). RNA is transcribed from DNA into a precursor form (pre-mRNA) which is undergoes several post-transcriptional processing events, such as splicing to remove intronic sequences and polyadenylation to form the mature RNA (mRNA). The mRNA is exported out of the nucleus to the cytoplasm where it is translated into its protein product. ASOs can be designed to modify RNA processing events in the nucleus such as modulate RNA splicing to exclude or include a protein coding exon. In some cases, excluding specific exons could result in a truncated protein product or alternatively, the RNA missing a specific exon is recognized by the cell as mis-spliced and is degraded by the non-sense mediated decay (NMD) pathway. ASOs can also be designed to alter polyadenylation site selection resulting in loss of RNA regulatory sequences in 3’-untranslated regions of the RNA. In the cytoplasm, ASOs can be designed to decrease translation of the RNA into proteins by binding to sequences in the 5’-untranslated region of the mRNA or by blocking microRNA binding to the RNA. ASOs can also be designed to block translation starting at an upstream open reading frame (uORF) or disrupt regulatory RNA structures resulting in an increase in protein translation.
Degradation of the targeted RNA by either recruiting RNase H1 or binding to and activating argonaute 2 (Ago2) (Figure 1) are commonly used to promote degradation of the targeted RNA (5; 11). RNase H1 is a ubiquitously expressed enzyme involved in DNA replication and DNA transcription which cleaves the RNA strand of a DNA-RNA heteroduplex (12). Antisense drugs which have at least 5 to 7 consecutive DNA nucleotides are capable of supporting the RNase H mechanism (13). There are several approved antisense drugs that work through the RNase H1 mechanism and multiple drugs in clinical development (5; 14). One approved RNase H1 drug, inotersen, is for the treatment of the neurological disease hereditary transthyretin polyneuropathy (15) and nine RNase H ASOs are in clinical development (Table 1). ASOs that work through the RNA interference pathway (e.g. siRNAs) are generally delivered to cells as an RNA duplex, or modified RNA, where the two strands dissociate within the cell and the antisense strand (also referred to as the guide RNA) binds to Ago2. Ago 2 is a member of the argonaute family of proteins, which contains an RNase H like domain. The bound antisense strand directs the RNA-Ago2 complex to the targeted RNA where it cleaves the RNA, releasing the complex to bind to another RNA. ASOs that utilize the RNA interference mechanisms are also commonly used as research tools and are gaining momentum as therapeutic agents with the recent approval of siRNA drugs for the treatment of acute hepatic porphyria (5; 16) and a second, patisiran, recently approved for hereditary transthyretin polyneuropathy (17). To date, we know of no siRNA drugs in clinical development for central nervous system diseases, although progress is being made optimizing design and delivery of siRNAs for CNS diseases (18).
ASO drugs can also bind to a target pre-RNA and interfere with its maturation or interfere with the function of the mature RNA through non-degradative mechanisms (Figure 2) (5; 11). Blocking protein translation by ASOs is an example of a mechanism that does not result in degradation of the RNA transcript, but decreases protein production (19). The best characterized examples of non-degradative mechanisms are ASOs that modulate pre-mRNA splicing to promote exclusion or inclusion of exons (20). For Duchenne muscular dystrophy, a disease caused by point mutations or deletions of one or more exons in the dystrophin gene, two antisense drugs (Table 1) have been approved that promote exon skipping to restore the correct reading frame and synthesis of a nearly full length dystrophin that retains partial dystrophin function (21). A second strategy is to promote exon inclusion, which was the strategy used for the treatment of spinal muscular atrophy (SMA) with nusinersen (22; 23). A third outcome of splicing modulation is to promote degradation of the RNA transcript through nonsense mediated decay, which eliminates mRNAs that harbor a premature termination codon (24). ASOs can also be used to block or displace access to the target RNA by proteins and other RNA. An example blockage of binding of exon junction complex proteins to the RNA downstream of a premature termination codon, thereby blocking degradation of the RNA by nonsense mediated decay (25). Another example is use of antisense drugs to modulate polyadenylation site selection (26). Finally, it should be noted that ASOs can be used to increase protein translation by blocking translation of upstream open reading frames, disrupting regulatory RNA structures, and blocking microRNA access to the 3’-UTR of the transcript (27–30).
OLIGONUCLEOTIDE CHEMICAL MODIFICATIONS
Unmodified DNA and RNA are unstable in biological systems and are not suitable as drugs. In developing nucleic acid therapeutics, two different strategies - formulations and chemical modifications - have been used to protect the nucleic acids from degradation. Chemical modification of ASOs have been used to enhance stability against endogenous nucleases, enhance binding affinity to RNA and decrease unwanted toxicities. Modifications to the phosphate backbone, ribose sugar and bases have all been examined for utility as potential antisense drugs (11; 31).
Nucleic acids are degraded by both endo- and exo- nucleases which cleave the phosphodiester bonds linking the nucleotides together. First-generation antisense chemistry focused on stabilizing the phosphodiester bond through chemical modifications including phosphorothioate, methyl phosphonate and phosphorodiamidate morpholinos. Of these, the phosphorothioate modification, in which one of the non-bridging oxygen atoms is replaced with sulfur, is the most widely utilized (32). Most of the approved antisense drugs as well as drugs currently in development have some phosphorothioate modifications. In addition to stabilizing the ASO against degradation by nucleases, the phosphorothioate modification enhances protein binding which can result in better tissue distribution and cell uptake, but protein binding also contributes to some unwanted side effects (11). Phosphorodiamidate morpholino oligonucleotides (PMO) are sugar-phosphate substitutions in which the sugar is replaced with a morpholine ring and the phosphate with a phosphorodiamidate linkage. Because of the neutral phosphorodiamidate linkage, morpholino oligonucleotides exhibit minimal protein binding and are rapidly excreted (33). The PMO modification does not support the RNase H1 or RNA interference mechanisms of action. PMOs are primarily used for modulating splicing, in which high doses are administered to compensate for the poor tissue distribution. There are two approved PMO drugs for the treatment of Duchenne muscular dystrophy (Table 1).
The ribose sugar has proven to be an important nucleic acid constituent in which chemical modifications have enhanced pharmacological effects. Sugar modifications are used to support splicing modulation, translation suppression, enhancement of translation, RISC-based mechanisms, and RNase H1 mechanisms (Figures 1 and 2). Most sugar modifications do not support RNase H1 activity, therefore they are generally incorporated at the ends of the oligonucleotide, leaving a DNA gap in the middle and are often referred to as “gapmers” or “second-generation” ASOs. The 2’-O-methyl modification is a naturally occurring ribose modification. The other commonly used sugar modifications are not naturally found in RNAs. Examples include 2’-O-methoxyethyl (MOE), 2’-fluoro and bridged 2’−4’ modifications such as locked nucleic acid (LNA) and constrained ethyl (cEt). Most sugar modifications show enhanced nuclease stability (the 2’-fluoro being an exception), and increased binding affinity to RNA. LNA and cEt modifications, which have a bridged 2’−4’ linkage, demonstrate the greatest binding affinity for their target RNAs (34–36). It should be noted that although the general trends are similar for the different sugar modifications, they each have unique attributes. Using affinity for RNA as an example, the ranking of the different sugar modifications is roughly DNA< RNA< 2’-O-methyl ≤ MOE <2’-fluoro < <cEt ~ LNA (36; 37). For resistance to nuclease degradation, the general ranking is RNA < DNA < 2’-fluoro < 2’-O-methyl < LNA < MOE < cEt. In addition, each modification has unique protein binding properties (38–40). Currently, the most effective drugs use a mixture of different chemical modifications which have been optimized for the specific indications.
DISTRIBUTION OF ANTISENSE DRUGS IN THE CENTRAL NERVOUS SYSTEM
Most centrally delivered antisense oligonucleotides (ASOs) for neurological indications today are 18–20 bases in length and thus 6–8 KDa. These negatively charged macromolecules do not cross the blood brain barrier. To access the CNS in preclinical studies, ASOs are delivered directly into the CSF by injection into the lateral ventricles (ICV) or the lumbar space. Clinically, ASOs are delivered into the CSF by intrathecal injection into the lumbar space. Once introduced into the CSF, small water soluble molecules like ASOs distribute broadly throughout the CNS (22; 41–44). An ASO delivered into the non-human primate (NHP) CSF via intrathecal bolus injection can be found throughout the spinal cord and cortex, often with higher ASO concentrations in the cortex than in the spinal cord (45). ASOs are delivered to and are effective in deeper brain regions, including the hippocampus, pons and amygdala, though to a lesser extent than cortex or spinal cord (41; 45; 46).
The broad distribution of ASOs in the CNS is likely facilitated by intrinsic CSF dynamics. CSF surrounds the brain and spinal cord, occupying the open spaces of the ventricles, subarachnoid space, cisterns, sulci of the brain, and central canal of the spinal cord. In humans and NHPs, CSF turns over about 3–4 times per day and accounts for ~10% of the total fluid volume in the intracranial space (47). CSF is in constant movement, driven largely by CSF production, cardiac cycle and respiration (see reviews (48; 49)). In addition to bulk movement, CSF and interstitial fluid are continuously exchanged, with CSF moving through the ventricular ependymal layer, interstitial and perivascular space and perineural lymphatic channels (see reviews (50; 51)). The exchange of CSF and interstitial fluid is facilitated by convective influx of CSF along the periarterial space into the brain parenchyma through the glymphatic system, with these convective forces driving movement of macromolecules through the parenchyma (52). This constant mixing and movement of CSF is likely a key factor in the broad distribution of ASOs within the CNS.
A combination of immunohistochemistry and live imaging of labeled ASOs have allowed for detailed kinetics of early ASO distribution (53). These data support a model for ASO distribution where ASOs first associate with the pial membrane and the major cerebral surface arteries, suggesting that like other macromolecules, ASOs may access deeper levels of the parenchyma by traveling through intramural perivascular spaces. ASOs then progress into the parenchyma likely by direct migration through the glial limitans via gap-junctions or transcellular exchange and are detectable in the extracellular space. It is still not clear if ASO distribution into the parenchyma is primarily via passive diffusion or convective forces, or a combination of both. By 24 hours after dosing, an ASO is present in the intracellular space and pharmacologically active. Maximal onset of pharmacological action is typically between 1–3 days after injection and is then maintained for weeks after dosing.
Once in the parenchyma, ASOs are able to target the major cell types including neurons, astrocytes, microglia and oligodendrocytes (43; 54; 55). One cell population that appears partially resistant to the actions of ASOs are cerebellar granule cells, despite robust activity in neighboring Purkinje cells (56). The mechanism for this resistance is not yet known.
Distribution into the parenchyma is dose dependent, with an increase in neuronal ASO levels with increasing dose (53). Conversely, ASO levels in pia and perivascular spaces tend to be more constant across pharmacologically active dose levels. This is likely due to rapid absorption and saturation of vascular intramural basement membranes with ASOs, allowing for increased penetration across these membranes with increasing doses.
Insight into the pharmacology of ASOs can be gleaned from quantification of ASO activity in individual neurons in the dorsal root ganglia (DRG). Due to the unique architecture and stereotypical neuronal sizes of the DRG, it is possible to quantify pharmacology in 3 neuronal subtypes exposed to similar amounts of ASO. Here, all three neuronal populations exhibited dose dependent suppression in target RNA uniformly across the population (57). This favors a model where low doses of ASOs target most cells modestly and evenly, rather than a subset of cells robustly. However, more work is needed to replicate this across cellular subtypes, and in human samples, as a limited data set in human patients suggested potentially more heterogeneity (58).
Given these principles and observations, it is not surprising that modulation of the ASO dosing paradigm can alter distribution. Enhancing convective forces either by increasing dose volume or applying percussive force improves distribution of ASOs up the neuraxis. Altering dose volume from 7% of total CSF to 17% of CSF changes the cord to cortex ratios, with higher cortical levels reached with higher dose volumes. Similarly, ASO bolus injection leads to wider and more efficient distribution than slow infusion (42). This is likely due to a higher maximal drug concentrations achieved in CSF (CMax) driving ASO into productive compartments, as has previously been demonstrated for liver (59). In the liver, ASO can be taken up into a productive compartment allowing ASO access to the target RNA, and into a non-productive compartment where ASO is unable to access the target RNA (59; 60). One can imagine a paradigm where simple changes in delivery technique can be used to achieve a desired distribution.
PRECLINICAL AND CLINICAL EXPERIENCE OF CENTRALLY ADMINISTERED ANTISENSE DRUGS
There are currently five approved antisense drugs for neurological diseases (Table 1). Four of the five are delivered systemically targeting either skeletal muscle or liver and have been previously reviewed (5; 14; 61; 62). The only approved antisense drug that is administered by intrathecal injection is nusinersen used for the treatment of spinal muscular atrophy. There are nine additional drugs in clinical trials for the treatment of amyotrophic lateral sclerosis, Huntington’s disease, Alzheimer’s disease, Parkinson’s disease, and Angelman syndrome, all delivered intrathecally. In addition, there are multiple drug discovery programs to identify intrathecally administered antisense drugs for the treatment of a both common, rare, and ultra-rare neurological diseases (56; 57; 63–70). Highlighting the efficiency and acceptance of intrathecally administered ASOs, a personalized antisense drug was recently identified and tested in a single patient with a unique form of Batten’s disease (71) and more recently a patient with ALS caused by mutation in fused in sarcoma (FUS) gene. A comprehensive review of antisense drugs used to treat neurological diseases is beyond the scope of this article and the reader is referred to several recent reviews on the topic (5; 14; 72). Here we will focus on intrathecal antisense drugs, examples that highlight lessons learned that translate across the platform.
Spinal Muscular Atrophy: Nusinersen
Spinal muscular atrophy is a severe pediatric neuromuscular disease characterized by muscle weakness and atrophy secondary to degeneration and death of neurons. Prior to therapeutic intervention, SMA was the most common genetic cause of infant mortality, with the life expectancy less than two years of age for the most severe form of the disease (73; 74). The disease is caused by mutations or deletions of the survival motor neuron 1 (SMN1) gene (75). Humans have a SMN1 paralog (SMN2) which was generated by an inverted duplication of the 5Q13 chromosomal region. SMN2 differs from SMN1 by approximately 5 nucleotides, one of which is a C to T transition in exon 7 that disrupts an exon splice enhancer and creates an exon splice suppressor resulting in skipping of exon 7 for the approximately 80% of SMN2 transcripts. The transcripts missing exon 7 produce a truncated protein that is rapidly degraded (76), whereas the 20% of the transcripts containing exon 7 make full length SMN protein identical to the protein derived from the SMN1 gene. Thus, SMA can be thought of as a SMN protein deficiency disease. This is further supported by the observation that patients with more copies of the SMN2 gene tend to have a milder form of the disease (77). SMA presents as a phenotypic spectrum that can roughly be classified as Type I SMA, Type II SMA and Type III SMA. Type I SMA being infantile onset, in which infants never gain the ability to sit and have short life-expectancy (< 2 years). Types II and III SMA have later ages of onset, usually in early childhood. Type II SMA children achieve the ability to sit but not walk independently and Type III SMA children gain the ability to walk, but often lose this ability as they develop (78).
Nusinersen is antisense drug that binds to a site in intron 7 of the SMN2 pre-mRNA, displacing hnRNPA1/A2 proteins which negatively regulate splicing of exon 7, resulting in full length SMN2 transcripts (23; 39). In multiple preclinical models, nusinersen enhances SMN2 exon 7 inclusion, improves muscle function and enhances survival (23; 79–83). These findings plus safety and tolerability studies in rodents and non-human primates supported advancing the drug into development. The drug was broadly studied in all SMA Types, demonstrating beneficial effects across the different patient populations (22; 84–89). Nusinersen has been approved for the treatment of SMA in over 40 different countries.
There are several important lessons from nusinersen that could have broader implications for antisense drugs being used to treat other neurological diseases. First, it was a collaborative project by the many stakeholders which effectively capitalized on shared interests in finding a therapy. Second, the studies demonstrated for the first time that SMA is a treatable disease. Third, studies in genetically diagnosed, pre-symptomatic patients demonstrated remarkable benefit, with many children achieving motor milestones that included sitting, standing, and walking within the normal age appropriate windows (89). These findings demonstrated that treating before symptoms develop may prevent or markedly minimize disease symptoms for a neurodegenerative disease, which was intuitive, but not previously proven. Finally, nusinersen was important for the ASO field as it demonstrated that the technology can create commercially successful drugs that have a major impact on patients’ lives.
Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS, also known as Lou Gehrig’s disease), is a severe neurodegenerative disease with progressive muscle weakness and paralysis leading to respiratory failure and death (90). The primary pathology observed in ALS is degeneration and death of upper and lower motor neurons and denervation-induced muscle atrophy. Although the cause of disease for the majority of cases is unknown, approximately 10 to 20% of cases are inherited. There are more than 50 genes that cause or contribute to ALS identified to date (91). Mutations in the superoxide dismutase 1 (SOD1) gene was the first example of a gene that causes familial forms of ALS and accounts for approximately 20% of familial ALS cases (92). More than 170 different ALS causing mutations have been identified. There are multiple molecular mechanism(s) proposed by which mutations in SOD1 protein cause ALS, but to date there is no agreement on a unifying mechanism (93). Most data implicate a toxic gain of function in the SOD1 protein rather than a loss of function (93; 94). Examining the function of more recently identified genes that contribute to the disease does not identify a common mechanism that contributes to ALS. Putative ALS-causing genes appear to impact several important areas of cell biology including RNA metabolism (e.g. TARDBP. FUS. hnRNPA1, MATR3, C9orf72, ANG), autophagy (OPTN, SQSTM1, TBK1), intracellular transport (DCTN1, TUB4A, PFN1) and proteostasis (SOD1, VCP), suggesting that perturbation of several biological pathways can lead to dysfunction and death of motor neurons.
Targeting SOD1 as a potential treatment for ALS was the first use of antisense oligonucleotides for treating neurodegenerative diseases (44). A key factor in deciding to use SOD1 as a model system to learn about the behavior of ASOs delivered into the CNS was the overwhelming data supporting that the mutant SOD1 protein caused ALS through a toxic gain of function, providing a well validated target (93; 95–97). In addition, the initial study population was well defined and had a consistent natural history based on specific mutation, e.g., a good genotype to phenotype correlation (98; 99). Finally, several systemically dosed second-generation 2’-MOE modified ASOs had advanced in the clinic, providing comfort regarding safety and tolerability when applied systemically.
Preclinical studies documented dose-dependent reduction in SOD1 mRNA and protein in spinal cord and different brain regions following injection into the lateral ventricle of wild-type and transgenic rats (44). Based on the efficacy and safety of the ASO drug, ISIS 333611, it was advanced into a phase 1 clinical study in ALS patients with mutations in SOD1 (100). Because this was the first antisense drug to be delivered via the intrathecal route, the first study was primarily focused on safety. A 12- hour infusion of the drug into the intrathecal space was well tolerated. During the conduct of the study, significant advances in screening ASOs for CNS applications had been made resulting in more potent and well-tolerated drugs. In addition, preclinical data demonstrated that broader delivery within the CNS and better efficacy was achieved by bolus injection compared to constant infusion (42), therefore it was decided to terminate development of ISIS 333611 and advance a drug into development that took advantage of the improved screening.
Tofersen was identified following extensive screening in cell culture and transgenic rodents against the human SOD1 pre-mRNA. Tofersen was found to be more potent than ISIS 333611(46). It exhibited enhanced activity in mouse and rat SOD1 ALS models (46). Tofersen was advanced into a single and multiple ascending dose (MAD) Phase 1/2a study in ALS patients with pathogenic mutations in SOD1 gene. Forty-eight subjects participated in the study and received all planned doses, ranging from 20 to 100 mg in the MAD portion of the study (101). Patients in the MAD portion of the study received 5 doses on days 1, 15, 29, 57 and 85 by bolus intrathecal injection. Dose proportional plasma exposure was observed, but trough CSF concentrations (e.g. CSF concentration of the drug prior to administration of the next monthly dose) were less than dose proportional. The drug was generally well tolerated with most adverse events were ascribed to the underlying disease or the intrathecal procedure. A reduction from baseline in CSF SOD1 protein was observed in the 40, 60 and 100 mg dose groups, consistent with the drugs mechanism of action. In this short duration study, there was a lessening of decline in clinical measures of the disease in the 100 mg dose group which was significant in patients with fast progressing mutations. Consistent with decrease in rate of decline in clinical measures there was a decline in plasma and CSF phosphorylated neurofilament heavy chain and neurofilament light chain from baseline values. These encouraging data supported advancement of the drug into a pivotal Phase 3 study, which is currently ongoing (NCT02623699).
Hexanucleotide expansions in the first intron of the C9orf72 gene have recently been described as a pathogenic genetic change that leads to ALS and frontotemporal dementia (FTD) (3; 4). Mutations in C9orf72 account for approximately 8–10% of all ALS cases and 40% of familial cases. The size of the hexanucleotide repeat can range from fewer than 20 repeats in healthy controls to several thousand repeats in affected individuals (3; 4). The mechanism(s) by which the hexanucleotide expansion causes neurodegenerative diseases are not well understood. The protein product from the C9orf72 gene has a conserved DENN domain and can function as a guanine nucleotide exchange factor for Rab proteins (102). Most data support three potential non-exclusive mechanisms; RNA toxicity in which the expanded repeat sequesters RNA binding proteins, translation of the repeat RNA into peptides (RAN translation), and decrease in C9orf72 expression (3; 103–105). Toxicity from repeat containing RNAs or their RAN translation products appears to synergize with reduced C9orf72 protein produced from the affected C9orf72 allele (106).
ASOs designed to bind to the C9orf72 transcript containing the hexanucleotide expansion decreased the number of repeat-associated RNA foci, improved electrophysiological changes, decreased sensitivity to neurotoxins and repeat associated dipeptides in human fibroblasts and iPSCs (104; 107–109). In mice expressing human C9orf72 gene with a 500 nucleotide hexanucleotide expansion, one time treatment with an ASO targeting the C9orf72 transgene reduced dipeptides derived from the expansion and attenuated the behavioral effects (110).
The collective data support pathogenesis arising from an acquired toxicity combined with the reduction in C9orf72 expression observed in the clinical samples. Correspondingly, an “on disease mechanism” therapy would be to suppress the repeat containing RNAs without exacerbating reduction in protein coding C9orf72 mRNAs. Consistent with this, an antisense drug has been identified that selectively targets C9orf72 transcripts with repeat expansions, but which does not target the majority of C9orf72 protein-encoding RNAs, thereby preserving expression of C9orf72 protein from RNAs produced by the unaffected C9orf72 allele (107). This drug is currently in Phase 1/2 MAD clinical study (NCT03626012).
Given the early success targeting familial forms of ALS, there is increased interest in identifying targets for sporadic forms of the disease. A frequent histological observation in motor neurons in the cortex and spinal cord from autopsies of ALS patients is the presence of cytoplasmic inclusions which contain TAR DNA-binding protein 43 (TDP-43) (111). TDP-43 is an RNA binding protein involved in transcription, RNA processing and nuclear cytoplasmic transport. Another recently identified pathological feature identified in induced pluripotent stem cell (iPSC) derived motor neurons and autopsy samples from ALS patients is a defect in nuclear import, possibly through a nuclear pore defect (105; 112). These observations are being extensively studied and are not only providing insights into the pathogenesis of ALS but also identifying potential drug targets. As an example, the cytoplasmic retention of TDP-43 is associated with loss of nuclear TDP-43 resulting in changes in RNA metabolism (113; 114).
One transcript that is markedly decreased in ALS-derived iPSC and spinal cord tissues is the one encoding stathmin-2, a tubulin binding protein thought to play a role in neurite outgrowth by affecting microtubule dynamics. Investigation into the mechanism of stathmin-2 decrease in neurons revealed the presence of a cryptic splice and polyadenylation sites within intron 1 of the stathmin-2 pre-mRNA, adjacent to which are a trio of TDP-43 binding sites (115; 116). Loss of nuclear TDP-43 results in aberrant splicing and early polyadenylation of the stathmin-2 transcript, resulting in the decreased levels of the protein. Strategies to increase the expression of stathmin-2 could be of therapeutic utility for ALS patients.
A second target for ASO therapy in sporadic ALS is the RNA encoding the RNA binding protein ataxin-2, which was identified in a yeast screen as TDP-43 modifier (117). While a long trinucleotide expansion in the ataxin-2 gene is causative of ataxia (118), an intermediate CAG repeat length (27 to 33 repeats) in found at an enhanced rate in sporadic ALS patients (117). Genetic depletion or ASOs to decrease ataxin-2 expression increase survival in a mouse model of TDP-43 neurodegenerative disease, providing a therapeutic rationale for targeting ataxin-2 as a potential therapy for ALS (119).
Although still early, these results provide promise that disease modifying therapies for ALS patients will become available.
Alzheimer’s Disease
Given the high societal impact and unmet need, evaluating multiple therapeutic targets and strategies for the treatment of Alzheimer’s disease (AD) is warranted. Two therapeutic targets heavily researched as potential therapies for Alzheimer’s disease are amyloid precursor protein (APP) and tau (120). There are multiple approaches being pursued for modulating APP, including preventing processing of the protein and enhancing clearance of amyloid deposits, which to date have been disappointing in the clinic (120). Limited preclinical work has been conducted testing the efficacy of APP targeting ASOs in cell culture and mouse models of AD (121–124). Given the limited clinical success of agents that modulating the processing of APP or enhance clearance of amyloid plaques, preventing the synthesis of APP merits further investigation as a therapeutic strategy for Alzheimer’s disease.
Tau protein which is coded for by the microtubule-associated protein tau (MAPT) has been broadly implicated in contributing to pathology in Alzheimer’s, primary tauopathies and other neurodegenerative diseases (120; 125). Like most other proteinopathies associated with neurodegenerative diseases the precise mechanism by which the protein causes disease is not well understood but appears to be linked to intracellular misfolding and aggregation of the protein and cell to cell spread of pathogenic forms. It is worth noting that in contrast to amyloid deposits, the appearance of tau deposits is closely linked to onset of symptoms in AD patients (120). There are multiple drugs being tested that affect folding, prevent cell to cell spread, block synthesis, or enhance clearance of tau protein. ASO can be used to block the synthesis of tau protein or synthesize a potentially less pathogenic form of tau through alternative RNA splicing and are currently being explored as potential therapies (41; 126–129). An RNase H ASO targeting MAPT pre-mRNA decreased the amount of phosphorylated tau in brain tissue, preserving hippocampal neurons and enhancing survival in a mouse model expressing a pathogenic form of tau (41). In addition, lowering of tau protein using an ASO reduced sensitivity to seizures in mice (126), consistent with genetic manipulation of tau (130). IONIS-MAPTRx is an ASO that reduces the synthesis of tau protein through an RNase H mechanism of action is currently in a Phase1/2 clinical study in early Alzheimer’s disease patients (NCT03186989).
Parkinson’s Disease
Parkinson’s disease (PD) is a progressive movement disorder and is the second most common neurodegenerative disease affecting approximately 1% of the population over 60 (131). Parkinson’s disease can be idiopathic or familial, with both dominant and recessive inheritance patterns. Though palliative care exists, there is no disease-modifying therapy. The pathological hallmark of idiopathic PD and some familial PD is accumulation of alpha-synuclein into Lewy bodies and Lewy neurites, and subsequent loss of dopaminergic neurons (132). Alpha-synuclein is thought to be an underlying driver of PD, as duplication and triplication of SNCA, the gene that encodes alpha-synuclein, causes autosomal-dominant PD. Similarly, alpha-synuclein fibrils injected directly into the CNS can propagate and are directly toxic to dopaminergic neurons (133). Thus, it is not surprising that suppression of alpha-synuclein with ASOs has disease modifying benefits in multiple models (134; 135). Indeed, preventing production of alpha-synuclein can even reverse existing pathology and prevent dopaminergic cell death (135).
Dominantly inherited mutations in leucine-rich repeat kinase 2 (LRRK2) are the most common familial cause of PD (136). Patients with LRRK2 mutations are clinically and pathologically indistinguishable from idiopathic PD. ASO-mediated suppression of LRRK2 in an alpha-synuclein model of PD reduced the pathological propagation of alpha-synuclein, prevented motor deficits and dopaminergic cell loss (137). Given that ASOs to reduce CNS LRRK2 are delivered centrally with limited systemic exposure, ASOs do not have the same systemic on-target liabilities that small-molecule inhibitors of LRRK2 do. A LRRK2 targeting ASO is currently in an early stage clinical study (NCT03976349).
ASO drugs have the potential to be transformative disease-modifying therapies for PD. By targeting the primary underlying disease mechanism, either alpha-synuclein or LRRK2, dopaminergic cell loss can be prevented. Suppression of these targets is clearly beneficial after established pathology. PD is a particularly interesting case for disease modifying therapies, because good palliative therapies already exist. It is possible, that intervention in later, more established disease cases may have a greater opportunity for benefit than other neurodegenerative diseases, because combination therapy is possible. One can envision a scenario where disease pathology and progression are halted with an ASO, and established symptoms can be well managed with existing palliative care.
Huntington’s Disease: Tominersen
Huntington’s disease (HD) is caused by expansion of a CAG repetitive sequence in the first intron of the huntingtin gene (HTT) (1). The CAG codon is translated into glutamine, resulting in an expanded polyglutamine track in the amino terminus of the HTT protein. Although the normal cellular function of HTT and the mechanism(s) by which the expanded CAG tract causes HD are not well understood, most data point to the mechanism being a dominant gain of function of the protein (138). At least three different antisense approaches have been investigated as potential treatment for HD; selective blocking translation of the RNA containing the CAG expansion, selective reduction of the mutant RNA and non-selective knockdown of wild-type and mutant HTT RNAs (43; 139–144). In general, blocking translation of the mutant HTT RNA (mHTT) has proven to be challenging due to the limited in vivo potency of the ASOs used. Allele-selective silencing through either RNase H or siRNA selective targeting of single nucleotide polymorphisms linked to the disease-causing allele would be a promising strategy, but these approaches are limited by 1) the limited sequence space available surrounding unique single nucleotide polymorphisms for designing ASOs which could impact potency and safety of the antisense drug and 2) because there is no polymorphism shared among all HD patients- a single drug would only be applicable to a sub-population of HD patients (145). The most advanced approach is non-allele selective reduction of huntingtin-encoding RNAs through an RNase H mechanism. Preclinical data in mouse models have demonstrated dose-dependent reduction of huntingtin expression yielding sustained, partial disease reversal including improvements in clinical phenotypes in several mouse model of HD and in an aggressive mouse model, stoppage of loss of brain mass (43; 45; 146).
A Phase 1 clinical study evaluating the safety and tolerability of tominersen (also known as IONIS-HTTRx or RG6042) has recently been completed (147). Tominersen is a chimeric 2’-MOE/DNA ASO designed to reduce both mutant and wild-type HTT expression through an RNase H mechanism of action. The first-in-human study was a randomized placebo-controlled dose escalation study in which subjects were administered four monthly doses of tominersen by bolus intrathecal injection. Five dose groups were analyzed, 10, 30, 60, 90 and 120 mg. The drug was well tolerated at all dose levels with adverse effects being mild or moderate in severity and none ascribed to the study drug. Dose-dependent trough CSF concentrations were observed which appeared to plateau at the highest dose groups. Importantly, a dose dependent reduction in CSF levels of mHTT protein were observed supporting the mechanism of action of the drug. No group-wise changes in clinical outcomes were observed, which was expected given the short treatment duration and small number of patients in each cohort (147). A global Phase 3 study of tominersen is currently enrolling, 801 subjects, who will be randomized to receive placebo, 120 mg tominersen every 2 months or 120 mg tominersen every 4 months (Generation HD1, NCT037662849).
Two allele selective antisense drugs have advanced into clinical studies, WVE-120101 and WVE-120102 (Table 1). However, there is limited publicly available information regarding the chemistry, design and preclinical data for these antisense drugs.
Spinocerebellar Ataxias
The spinocerebellar ataxias (SCAs) are a growing class of more than 30 diseases (148). They are progressive neurodegenerative diseases characterized predominantly by cerebellar dysfunction, which is often accompanied by broader CNS involvement. Patients typically suffer from incoordination, loss of balance, speech impairments and early mortality (149). There are no disease modifying therapies. The most prevalent diseases in the class are dominantly inherited SCAs caused by expansion mutations with a toxic gain of function. This group includes, but is not limited to, SCA1, SCA2, and SCA3 caused by CAG expansions in the ATXN1, ATXN2, and ATXN3 genes, respectively.
ASO-mediated suppression of the disease genes in rodent models robustly ameliorates symptoms. Transient suppression of Atxn1 in SCA1 knock-in mice results in sustained improvement of motor phenotype, improvement of MRS scores, normalization of SCA1 disease-associated genes and extension of survival (56). Suppression of ATXN2 in mouse models of SCA2 improves motor function, restores Purkinje cell neural networks and normalizes expression of SCA2-related proteins expressed in Purkinje cells (67). ASO-mediated lowering of ATXN3 results in reversal of nuclear ATXN3 accumulation, dose-dependent clearance of soluble and high molecular weight species, mitigation of motor deficits, repair of cerebellar network dysfunction, and gene expression changes (63; 150; 151).
In these disease models, partial and transient suppression of the disease gene lead to a sustained improvement in phenotype. Interestingly, this improvement of phenotype did not happen immediately, but once it did, it was sustained. This suggests a model where suppression of the disease-causing gene allows the system to recover, and once recovered, these slowly progressing diseases take time to become symptomatically detrimental again. This is similar to previous observations in models of HD, another CAG expansion disease (43; 152).
CONCLUSIONS
The approval of nusinersen as a treatment for SMA has helped validate ASOs as a viable therapeutic approach for the treatment of neurodegenerative diseases, neurodevelopmental disorders, and possibly other diseases of the CNS. Several additional clinical studies have provided proof of mechanism, e.g. modulation of the targeted protein and encouraging evidence of clinical benefit. The ongoing larger and longer-term studies are needed to provide robust evidence of clinical benefit, which will support the registration of additional antisense drugs. Like most technologies, ASO technology continues to evolve resulting in drugs with improved potency and safety profile. To help support use in large patient population more convenient methods of administration will be needed. In summary, ASOs may finally provide a therapeutic platform for developing drugs for the treatment of severe neurodegenerative and neurological diseases.
ACKNOWLEDGMENTS
We would like to thank Drs. Richard Smith (Center for Neurological Studies, La Jolla, CA), Dan Norris and Scott Henry (Ionis Pharmaceuticals) and our numerous colleagues who have contributed to the work cited in this review. We would like to thank the patients and their caregivers for participating in the clinical studies.
Footnotes
DISCLOSURE STATEMENT
C.F.B., and H.B.K., are employees of Ionis Pharmaceuticals and receive salary and stock options from the company. D.W.C. is a consultant for Ionis Pharmaceuticals.
LITERATURE CITED
- 1.Huntington’sDiseaseCollaborativeResearchGroup. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes Cell 72:971–83 [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Zoghbi HY. 2007. Trinucleotide repeat disorders. Annu Rev Neurosci 30:575–621 [DOI] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72:245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72:257–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooke ST, Witztum JL, Bennett CF, Baker BF. 2018. RNA-Targeted Therapeutics. Cell Metab 27:714–39 [DOI] [PubMed] [Google Scholar]
- 6.Osen-Sand A, Catsicast M, Staple JK, Jones KA, Ayala G, et al. 1993. Inhibition of axonla growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature 364:445–8 [DOI] [PubMed] [Google Scholar]
- 7.Hooper ML, Chiasson BJ, Robertson HA. 1994. Infusion into the brain of an antisense oligonucleotide to the immediate-early gene c-fos suppresses production of Fos and produces a behavioral effect. Neuroscience 63:917–24 [DOI] [PubMed] [Google Scholar]
- 8.Dragunow M, Lawlor P, Chiasson B, Robertson H. 1993. c-fos antisense generates apomorphine and amphetamine- induced rotation. Neuroreport 5:305–6 [DOI] [PubMed] [Google Scholar]
- 9.Weiss B, Zhou L-W, Zhang S-P, Qin Z-H. 1993. Antisense oligodeoxynucleotide inhibits D2 dopamine receptor- mediated behavior and D2 messenger RNA. Neuroscience 55:607–12 [DOI] [PubMed] [Google Scholar]
- 10.Wahlestedt C, Golanov E, Yamamoto S, Yee F, Ericson H, et al. 1993. Antisense oligodeoxynucleotides to NMDA-R1 receptor channel protect cortical neurons from excitotoxicity and reduce focal ischaemic infarctions. Nature 363:260–3 [DOI] [PubMed] [Google Scholar]
- 11.Bennett CF, Swayze EE. 2010. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Ann. Rev. Pharmacol. Toxicol 50:259–93 [DOI] [PubMed] [Google Scholar]
- 12.Cerritelli SM, Crouch RJ. 2009. Ribonucelase H: the enzymes in eukaryotes. FEBS J. 276:1494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Lima WF, Crooke ST. 1999. Properties of cloned and expressed human RNase H1. J Biol Chem 274:28270–8 [DOI] [PubMed] [Google Scholar]
- 14.Bennett CF. 2019. Therapeutic Antisense Oligonucleotides Are Coming of Age. Annu Rev Med 70:307–21 [DOI] [PubMed] [Google Scholar]
- 15.Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, et al. 2018. Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N Engl J Med 379:22–31 [DOI] [PubMed] [Google Scholar]
- 16.Scott LJ. 2020. Givosiran: First Approval. Drugs 80:335–9 [DOI] [PubMed] [Google Scholar]
- 17.Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, et al. 2018. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med 379:11–21 [DOI] [PubMed] [Google Scholar]
- 18.Alterman JF, Godinho B, Hassler MR, Ferguson CM, Echeverria D, et al. 2019. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat Biotechnol 37:884–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazenave C, Stein CA, Loreau N, Thuong NT, Neckers LM, et al. 1989. Comparative inhibition of rabbit globin mRNA translation by modified antisense oligodeoxynucleotides. Nucl.Acids Res 17:4255–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominski Z, Kole R. 1993. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA 90:8673–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, et al. 2001. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet 10:1547–54 [DOI] [PubMed] [Google Scholar]
- 22.Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, et al. 2017. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med 377:1723–32 [DOI] [PubMed] [Google Scholar]
- 23.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82:834–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward AJ, Norrbom M, Chun S, Bennett CF, Rigo F. 2014. Nonsense-mediated decay as a terminating mechanism for antisense oligonucleotides. Nucleic Acids Res 42:5871–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nomakuchi TT, Rigo F, Aznarez I, Krainer AR. 2016. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat Biotechnol 34:164–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers TA JRW, Burckin T, Bennett CF, Freier SM. 2001. Fully modified 2’-MOE oligonucleotides redirect polyadenylation. Nucl.Acids Res 29:1293–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, et al. 2004. Micro-RNA-143 regulates adipocyte differentiation. J. Biol. Chem 279:52361–5 [DOI] [PubMed] [Google Scholar]
- 28.Liang XH, Sun H, Shen W, Wang S, Yao J, et al. 2017. Antisense oligonucleotides targeting translation inhibitory elements in 5’ UTRs can selectively increase protein levels. Nucleic Acids Res 45:9528–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang X- H, Shen W, Sun H, Migawa MT, Vickers TA, Crooker ST. 2016. Specific increase in translation of proteins by antisense oligonucleotides targeting uORFs. Nat Biotechnol. In Press [DOI] [PubMed] [Google Scholar]
- 30.Choi WY, Giraldez AJ, Schier AF. 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318:271–4 [DOI] [PubMed] [Google Scholar]
- 31.Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. 2017. Pharmacology of Antisense Drugs. Annu Rev Pharmacol Toxicol 57:81–105 [DOI] [PubMed] [Google Scholar]
- 32.Eckstein F. 2000. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 10:117–21 [DOI] [PubMed] [Google Scholar]
- 33.Iversen PL. 2008. Morpholinos. In Antisense Drug Technology: Principles, Strategies, and Applications, ed. Crooke ST:565–82. Boca Raton: Taylor and Francis. Number of 565–82 pp. [Google Scholar]
- 34.Koshkin AA, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. 1998. LNA (locked nucleic acid): an RNA mimic forming excedingly stable LNA:LNA duplexes. J. Am. Chem. Soc 120:13252–3 [Google Scholar]
- 35.Obika S, Nanbu D, Hari Y, Andoh J, Morio K, et al. 1998. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2’-O, 4’-C-methyleneribonucleosides. Tetrahedron Lett. 39:5401–4 [Google Scholar]
- 36.Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, et al. 2010. Synthesis and biophysical evaluation of 2’,4’-constrained 2’O-methoxyethyl and 2’,4’-constrained 2’O-ethyl nucleic acid analogues. J Org Chem 75:1569–81 [DOI] [PubMed] [Google Scholar]
- 37.Freier SM, Altmann K-H. 1997. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucl.Acids Res 25:4429–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen W, Liang XH, Sun H, Crooke ST. 2015. 2’-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res 43:4569–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigo F, Hua Y, Chun SJ, Prakash TP, Krainer AR, Bennett CF. 2012. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat Chem Biol 8:555–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang XH, Sun H, Shen W, Crooke ST. 2015. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res 43:2927–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, et al. 2017. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigo F, Chun SJ, Norris DA, Hung G, Lee S, et al. 2014. Pharmacology of a central nervous system delivered 2’-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther 350:46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, et al. 2012. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74:1031–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, et al. 2006. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest 116:2290–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southwell AL, Kordasiewicz HB, Langbehn D, Skotte NH, Parsons MP, et al. 2018. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci Transl Med 10 [DOI] [PubMed] [Google Scholar]
- 46.McCampbell A, Cole T, Wegener AJ, Tomassy GS, Setnicka A, et al. 2018. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J Clin Invest 128:3558–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thrane AS, Rangroo Thrane V, Nedergaard M. 2014. Drowning stars: reassessing the role of astrocytes in brain edema. Trends in neurosciences 37:620–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinker T, Stopa E, Morrison J, Klinge P. 2014. A new look at cerebrospinal fluid circulation. Fluids and barriers of the CNS 11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oreskovic D, Klarica M. 2014. A new look at cerebrospinal fluid movement. Fluids and barriers of the CNS 11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casaca-Carreira J, Temel Y, Hescham SA, Jahanshahi A. 2017. Transependymal Cerebrospinal Fluid Flow: Opportunity for Drug Delivery? Molecular neurobiology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jessen NA, Munk AS, Lundgaard I, Nedergaard M. 2015. The Glymphatic System: A Beginner’s Guide. Neurochem Res 40:2583–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iliff JJ, Wang M, Liao Y, Plogg BA, Peng W, et al. 2012. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med 4:147ra11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazur C, Powers B, Zasadny K, Sullivan JM, Dimant H, et al. 2019. Brain pharmacology of intrathecal antisense oligonucleotides revealed through multimodal imaging. JCI insight 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagemann TL, Powers B, Mazur C, Kim A, Wheeler S, et al. 2017. Antisense suppression of glial fibrillary acidic protein as a treatment for Alexander disease. Annals of neurology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fitzsimmons BL, Zattoni M, Svensson CI, Steinauer J, Hua XY, Yaksh TL. 2010. Role of spinal p38alpha and beta MAPK in inflammatory hyperalgesia and spinal COX-2 expression. Neuroreport 21:313–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedrich J, Kordasiewicz HB, O’Callaghan B, Handler HP, Wagener C, et al. 2018. Antisense oligonucleotide-mediated ataxin-1 reduction prolongs survival in SCA1 mice and reveals disease-associated transcriptome profiles. JCI insight 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohan A, Fitzsimmons B, Zhao HT, Jiang Y, Mazur C, et al. 2018. Antisense oligonucleotides selectively suppress target RNA in nociceptive neurons of the pain system and can ameliorate mechanical pain. Pain 159:139–49 [DOI] [PubMed] [Google Scholar]
- 58.Ramos DM, d’Ydewalle C, Gabbeta V, Dakka A, Klein SK, et al. 2019. Age-dependent SMN expression in disease-relevant tissue and implications for SMA treatment. J Clin Invest 129:4817–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geary RS, Wancewicz EV, Matson JE, Pearce M, Siwkowski A, et al. 2009. Effect of dose and plasma concentration on liver uptake and pharmacological activity of a 2’-methoxethyl modified chimeric antisense oligonucleotide targeting PTEN. Biochem. Pharmacol 78:284–91 [DOI] [PubMed] [Google Scholar]
- 60.Koller E, Vincent TM, Chappell A, De S, Manoharan M, Bennett CF. 2011. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39:4795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gertz MA, Mauermann ML, Grogan M, Coelho T. 2019. Advances in the treatment of hereditary transthyretin amyloidosis: A review. Brain Behav 9:e01371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korinthenberg R. 2019. A new era in the management of Duchenne muscular dystrophy. Developmental medicine and child neurology 61:292–7 [DOI] [PubMed] [Google Scholar]
- 63.McLoughlin HS, Moore LR, Chopra R, Komlo R, McKenzie M, et al. 2018. Oligonucleotide therapy mitigates disease in spinocerebellar ataxia type 3 mice. Ann Neurol 84:64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raymond GJ, Zhao HT, Race B, Raymond LD, Williams K, et al. 2019. Antisense oligonucleotides extend survival of prion-infected mice. JCI insight 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen X, Kilikevicius A, O’Reilly D, Prakash TP, Damha MJ, et al. 2018. Activating frataxin expression by single-stranded siRNAs targeting the GAA repeat expansion. Bioorg Med Chem Lett 28:2850–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. 2015. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518:409–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, et al. 2017. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544:362–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lieberman AP, Yu Z, Murray S, Peralta R, Low A, et al. 2014. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell reports 7:774–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niu C, Prakash TP, Kim A, Quach JL, Huryn LA, et al. 2018. Antisense oligonucleotides targeting mutant Ataxin-7 restore visual function in a mouse model of spinocerebellar ataxia type 7. Sci Transl Med 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahashi K, Katsuno M, Hung G, Adachi H, Kondo N, et al. 2015. Silencing neuronal mutant androgen receptor in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet [DOI] [PubMed] [Google Scholar]
- 71.Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, et al. 2019. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med 381:1644–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verhaart IEC, Aartsma-Rus A. 2019. Therapeutic developments for Duchenne muscular dystrophy. Nature reviews. Neurology 15:373–86 [DOI] [PubMed] [Google Scholar]
- 73.Finkel RS, McDermott MP, Kaufmann P, Darras BT, Chung WK, et al. 2014. Observational study of spinal muscular atrophy type 1 and implications for clinical trials. Neurology 83:974–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rudnik-Schoneborn S, Berg C, Zerres K, Betzler C, Grimm T, et al. 2009. Genotype-phenotype studies in infantile spinal muscular atrophy (SMA) type I in Germany: implications for clinical trials and genetic counselling. Clin Genet 76:168–78 [DOI] [PubMed] [Google Scholar]
- 75.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–65 [DOI] [PubMed] [Google Scholar]
- 76.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, et al. 2007. Refined characterization of the expression and stability of the SMN gene products. Am J Pathol 171:1269–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, et al. 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 16:265–9 [DOI] [PubMed] [Google Scholar]
- 78.Kostova FV, Williams VC, Heemskerk J, Iannaccone S, Didonato C, et al. 2007. Spinal muscular atrophy: classification, diagnosis, management, pathogenesis, and future research directions. J Child Neurol 22:926–45 [DOI] [PubMed] [Google Scholar]
- 79.Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, et al. 2010. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 24:1634–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, et al. 2011. Peripheral SMN restoration is essential for long-term rescue of a severe SMA mouse model. Nature 478:123–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, et al. 2011. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 3:72ra18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sahashi K, Hua Y, Ling KK, Hung G, Rigo F, et al. 2012. TSUNAMI: an antisense method to phenocopy splicing-associated diseases in animals. Genes Dev 26:1874–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bogdanik LP, Osborne MA, Davis C, Martin WP, Austin A, et al. 2015. Systemic, postsymptomatic antisense oligonucleotide rescues motor unit maturation delay in a new mouse model for type II/III spinal muscular atrophy. Proc Natl Acad Sci U S A 112:E5863–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, et al. 2016. Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology 86:890–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, et al. 2016. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388:3017–26 [DOI] [PubMed] [Google Scholar]
- 86.Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, et al. 2018. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med 378:625–35 [DOI] [PubMed] [Google Scholar]
- 87.Montes J, Dunaway Young S, Mazzone ES, Pasternak A, Glanzman AM, et al. 2019. Nusinersen improves walking distance and reduces fatigue in later-onset spinal muscular atrophy. Muscle Nerve 60:409–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Darras BT, Chiriboga CA, Iannaccone ST, Swoboda KJ, Montes J, et al. 2019. Nusinersen in later-onset spinal muscular atrophy: Long-term results from the phase 1/2 studies. Neurology 92:e2492–e506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Vivo DC, Bertini E, Swoboda KJ, Hwu W-L, Crawford TO, et al. 2019. Nusinersen intitated in infants during the presymptomatic stage of spinal muscular atrophy: interim efficacy and safety results from the Phase 2 NURTURE study. . Neuromuscular Disorders 29:842–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown RH, Al-Chalabi A. 2017. Amyotrophic Lateral Sclerosis. N Engl J Med 377:162–72 [DOI] [PubMed] [Google Scholar]
- 91.Taylor JP, Brown RH Jr., Cleveland DW. 2016. Decoding ALS: from genes to mechanism. Nature 539:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, et al. 1993. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- 93.Bruijn LI, Miller TM, Cleveland DW. 2004. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci 27:723–49 [DOI] [PubMed] [Google Scholar]
- 94.Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, et al. 1998. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281:1851–4 [DOI] [PubMed] [Google Scholar]
- 95.Bruijn LI, Cleveland DW. 1996. Mechanisms of selective motor neuron death in ALS: insights from transgenic mouse models of motor neuron disease. Neuropathol. Appl. Neurobiol 22:373–87 [DOI] [PubMed] [Google Scholar]
- 96.Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, et al. 2006. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312:1389–92 [DOI] [PubMed] [Google Scholar]
- 97.Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, et al. 1997. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18:327–38 [DOI] [PubMed] [Google Scholar]
- 98.Cudkowicz ME, McKenna-Yasek D, Sapp PE, Chin W, Geller B, et al. 1997. Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol 41:210–21 [DOI] [PubMed] [Google Scholar]
- 99.Bali T, Self W, Liu J, Siddique T, Wang LH, et al. 2017. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry 88:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miller TM, Pestronk A, David W, Rothstein J, Simpson E, et al. 2013. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol 12:435–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miller TM, Cudkowicz ME, Shaw PJ, Graham D, Fradette S, et al. 2019. Safety PK, PD, and exploratory efficacy in single and multiple dose study of a SOD1 antisense oligonucleotide (BIIB067) administered to participants with ALS. In American Academy of Neurology. Philadelphia, PA [Google Scholar]
- 102.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, et al. 2016. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 35:1656–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, et al. 2013. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339:1335–8 [DOI] [PubMed] [Google Scholar]
- 104.Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, et al. 2013. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80:415–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, et al. 2016. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu Q, Jiang J, Gendron TF, McAlonis-Downes M, Jiang L, et al. 2020. Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat Neurosci 23:615–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, et al. 2013. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 110:E4530–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, et al. 2013. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5:208ra149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, et al. 2017. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci Transl Med 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, et al. 2016. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron 90:535–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neumann M, Sampathu DM, Kwong LK, Truax AC, Miscenyi MC, et al. 2006. Ubiquinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–3 [DOI] [PubMed] [Google Scholar]
- 112.Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, et al. 2018. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci 21:228–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ling JP, Pletnikova O, Troncoso JC, Wong PC. 2015. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349:650–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14:459–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Melamed Z, Lopez-Erauskin J, Baughn MW, Zhang O, Drenner K, et al. 2019. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci 22:180–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, et al. 2019. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci 22:167–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, et al. 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scoles DR, Pulst SM. 2018. Spinocerebellar Ataxia Type 2. Adv Exp Med Biol 1049:175–95 [DOI] [PubMed] [Google Scholar]
- 119.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, et al. 2017. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544:367–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Long JM, Holtzman DM. 2019. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 179:312–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Erickson MA, Niehoff ML, Farr SA, Morley JE, Dillman LA, et al. 2012. Peripheral administration of antisense oligonucleotides targeting the amyloid-beta protein precursor reverses AbetaPP and LRP-1 overexpression in the aged SAMP8 mouse brain. J Alzheimers Dis 28:951–60 [DOI] [PubMed] [Google Scholar]
- 122.Kumar VB, Farr SA, Flood JF, Kamlesh V, Franko M, et al. 2000. Site-directed antisense oligonucleotide decreases the expression of amyloid precursor protein and reverses deficits in learning and memory in aged SAMP8 mice. Peptides 21:1769–75 [DOI] [PubMed] [Google Scholar]
- 123.Coulson EJ, Barrett GL, Storey E, Bartlett PF, Beyreuther K, Masters CL. 1997. Down-regulation of the amyloid protein precursor of Alzheimer’s disease by antisense oligonucleotides reduces neuronal adhesion to specific substrata. Brain Research 770:72–80 [DOI] [PubMed] [Google Scholar]
- 124.Denman RB. 1997. Ribozyme and antisense RNAs inhibit coupled transcription translation by binding to rabbit polyribosomes. Biochem Biophys Res Commun 230:226–31 [DOI] [PubMed] [Google Scholar]
- 125.Holmes BB, Diamond MI. 2014. Prion-like properties of Tau protein: the importance of extracellular Tau as a therapeutic target. J Biol Chem 289:19855–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.DeVos SL, Goncharoff DK, Chen G, Kebodeaux CS, Yamada K, et al. 2013. Antisense reduction of tau in adult mice protects against seizures. J Neurosci 33:12887–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schoch KM, DeVos SL, Miller RL, Chun SJ, Norrbom M, et al. 2016. Increased 4R-Tau Induces Pathological Changes in a Human-Tau Mouse Model. Neuron 90:941–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sud R, Geller ET, Schellenberg GD. 2014. Antisense-mediated Exon Skipping Decreases Tau Protein Expression: A Potential Therapy For Tauopathies. Molecular therapy. Nucleic acids 3:e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ong AAL, Tan J, Bhadra M, Dezanet C, Patil KM, et al. 2019. RNA Secondary Structure-Based Design of Antisense Peptide Nucleic Acids for Modulating Disease-Associated Aberrant Tau Pre-mRNA Alternative Splicing. Molecules 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, et al. 2007. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science 316:750–4 [DOI] [PubMed] [Google Scholar]
- 131.Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, et al. 2017. Parkinson disease. Nat Rev Dis Primers 3:17013- [DOI] [PubMed] [Google Scholar]
- 132.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. 2003. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiology of aging 24:197–211 [DOI] [PubMed] [Google Scholar]
- 133.Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, et al. 2012. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338:949–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Uehara T, Choong C- J, Nakamori M, Hayakawa H, Nishiyama K, et al. 2019. Amido-bridged nucleic acid (AmNA)-modified antisense oligonucleotides targeting α-synuclein as a novel therapy for Parkinson’s disease. Scientific reports 9:7567- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cole TA, Zhao H, Collier TJ, Sandoval I, Sortwell CE, et al. 2019. Alpha-synuclein antisense oligonucleotides as a disease-modifying therapy for Parkinson’s disease. bioRxiv:830554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martin I, Kim JW, Dawson VL, Dawson TM. 2014. LRRK2 pathobiology in Parkinson’s disease. Journal of neurochemistry 131:554–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao HT, John N, Delic V, Ikeda-Lee K, Kim A, et al. 2017. LRRK2 Antisense Oligonucleotides Ameliorate α-Synuclein Inclusion Formation in a Parkinson’s Disease Mouse Model. Molecular Therapy - Nucleic Acids 8:508–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saudou F, Humbert S. 2016. The Biology of Huntingtin. Neuron 89:910–26 [DOI] [PubMed] [Google Scholar]
- 139.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, et al. 2009. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nature Biotech. 27:478–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yu D, Pendergraff H, Liu J, Kordasiewicz H, Cleveland DW, et al. 2012. Single-Stranded RNAs Act Through RNAi, Allele-Selectively Target Expanded CAG Repeats, and Potently Inhibit Huntingtin Expression. Cell In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Southwell AL, Skotte NH, Kordasiewicz HB, Ostergaard ME, Watt AT, et al. 2014. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther 22:2093–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, et al. 2011. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther 19:2178–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, et al. 2009. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr Biol 19:774–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Evers MM, Pepers BA, van Deutekom JC, Mulders SA, den Dunnen JT, et al. 2011. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS One 6:e24308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kay C, Collins JA, Skotte NH, Southwell AL, Warby SC, et al. 2015. Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol Ther 23:1759–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stanek LM, Yang W, Angus S, Sardi PS, Hayden MR, et al. 2013. Antisense oligonucleotide-mediated correction of transcriptional dysregulation is correlated with behavioral benefits in the YAC128 mouse model of Huntington’s disease. J Huntingtons Dis 2:217–28 [DOI] [PubMed] [Google Scholar]
- 147.Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, et al. 2019. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N Engl J Med 380:2307–16 [DOI] [PubMed] [Google Scholar]
- 148.Sullivan R, Yau WY, O’Connor E, Houlden H. 2019. Spinocerebellar ataxia: an update. Journal of neurology 266:533–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun YM, Lu C, Wu ZY. 2016. Spinocerebellar ataxia: relationship between phenotype and genotype - a review. Clinical genetics 90:305–14 [DOI] [PubMed] [Google Scholar]
- 150.Moore LR, Rajpal G, Dillingham IT, Qutob M, Blumenstein KG, et al. 2017. Evaluation of Antisense Oligonucleotides Targeting ATXN3 in SCA3 Mouse Models. Molecular therapy. Nucleic acids 7:200–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Moore LR, Keller L, Bushart DD, Delatorre RG, Li D, et al. 2019. Antisense oligonucleotide therapy rescues aggresome formation in a novel spinocerebellar ataxia type 3 human embryonic stem cell line. Stem Cell Res 39:101504- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lu XH, Yang XW. 2012. “Huntingtin holiday”: progress toward an antisense therapy for Huntington’s disease. Neuron 74:964–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, et al. 2013. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol 74:637–47 [DOI] [PubMed] [Google Scholar]
- 154.Charleston JS, Schnell FJ, Dworzak J, Donoghue C, Lewis S, et al. 2018. Eteplirsen treatment for Duchenne muscular dystrophy: Exon skipping and dystrophin production. Neurology [DOI] [PubMed] [Google Scholar]
- 155.Frank DE, Schnell FJ, Akana C, El-Husayni SH, Desjardins CA, et al. 2020. Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology [DOI] [PMC free article] [PubMed] [Google Scholar]