Abstract
A major Canada-wide outbreak of gastroenteritis due to Salmonella enterica serotype Enteritidis phage type (PT) 8 occurred in 1998, and this was traced to contaminated cheese in a commercial lunch pack product. Phage typing and pulsed-field gel electrophoresis linked the clinical and cheese isolates of serotype Enteritidis but failed to differentiate outbreak from nonoutbreak PT 8 strains. Further differentiation was made by biotyping based on melibiose fermentation.
Salmonella enterica serotype Enteritidis is one of the Salmonella serotypes most commonly associated with morbidity and mortality in humans (see the Salmonella dataset of the Communicable Disease Surveillance Centre of England and Wales. [http://www.phls.co.uk/facts/gastro/salm.htm]). In Canada the number of serotype Enteritidis clinical isolates received by the Laboratory Centre for Disease Control has risen steadily since the early 1980s, increasing from 307 in 1983 to 1,255 in 1994. Based on data submitted from provincial laboratories to the National Enteric Surveillance Program at the National Laboratory for Enteric Pathogens (NLEP) in Winnipeg, it is clear that serotype Enteritidis currently ranks as the second most common Salmonella serotype isolated from humans in Canada. In 1997 this serotype was involved in 4 community and 13 familial outbreaks (unpublished data). Serotype Enteritidis infections, particularly those due to phage type 8 (PT 8), are quite common in Canada and the United States and have been associated with the consumption of foods containing raw or lightly cooked eggs, which have caused outbreaks affecting large populations (6).
Epidemiological studies of serotype Enteritidis PT 8 infections are hampered by a lack of adequate subtyping procedures (17). Phage typing is often the initial method used for discrimination of isolates from serotype Enteritidis outbreaks. However, changes in phage type can occur as the result of plasmid or phage acquisition, changes in lipopolysaccharide expression, or spontaneous mutations affecting phage receptor sites (12). For this reason, a number of molecular typing and fingerprinting methods have been investigated for use with serotype Enteritidis, including pulsed-field gel electrophoresis (PFGE), random amplified polymorphic DNA analysis, plasmid profiling, and ribotyping (7, 9–12). Neither analysis of plasmid profiles nor selected ribotyping methods discriminate among serotype Enteritidis strains: PFGE gives a relatively higher level of discrimination, though it does not differentiate among PT 8 isolates (11).
On 23 March 1998, a provincewide outbreak of serotype Enteritidis was confirmed in Newfoundland based initially on the detection of 10 cases, most of which involved children (15). The NLEP was notified of the outbreak and subsequently subtyped the isolates as PT 8. At this time, a similar outbreak due to serotype Enteritidis PT 8 was recognized in Ontario. Eventually this outbreak proved to be Canada-wide in nature and affected over 800 persons, the majority of whom were children. Epidemiological and laboratory investigations were carried out in Newfoundland (15) and in the rest of Canada and included the testing of the food product and the processing environment from the manufacturer of the implicated food (A. Ellis, personal communication). Case control and trace-back studies implicated contaminated cheese in a commercial lunch pack product as the source of infection (15). Primary cases of infection and subsequent laboratory submissions decreased rapidly following the removal of the incriminated product from retail outlets on 31 March, although infections due to secondary cases continued for some time afterward (15).
Laboratory testing was performed to characterize serotype Enteritidis isolates obtained from clinical cases across the country and from incriminated food items. All isolates were maintained on nutrient agar and serologically identified as serotype Enteritidis according to the standard international scheme for serotyping Salmonella (14). Phage typing was performed by methods described previously (18) using 16 phages obtained from the Central Public Health Laboratory Service, London, England.
PFGE was performed using the method of Böhm and Karch (2) at the time of the outbreak and confirmed later using the Centers for Disease Control and Prevention (CDC) standard method for Salmonella spp. (4). Strains were included or excluded from the outbreak on the basis of laboratory and epidemiological information acquired during the outbreak, and these strains were chosen for PFGE on that basis. Electrophoresis was carried out in 1% agarose gels made by using pulsed-field certified agarose (Bio-Rad Laboratories Canada, Inc., Mississauga, Ontario, Canada) with the method of Böhm and Karch (2) and in SeaKem Gold agarose (Mandel Scientific Co., Ltd., Guelph, Ontario, Canada) with the CDC standard method (4). The PFGE buffer was 0.5× Tris-borate-EDTA made from 5× Tris-borate-EDTA buffer concentrate (Sigma-Aldrich Canada, Ltd., Oakville, Ontario, Canada). Switch times for the Böhm and Karch method were 2 to 40 s throughout a 27-h run, and those for the CDC method were 2.2 to 54.2 s for a 21-h run. Gels were run at a temperature of 14°C and a voltage of 6 V/cm in a CHEF-DR III PFGE unit (Bio-Rad). Gels were stained in 0.5 μg of ethidium bromide per ml, and the DNA was visualized with an Alpha Imager 2000 (Alpha Innotech Corp., San Leandro, Calif.). All isolates tested were analyzed using XbaI, and selected isolates were analyzed using SpeI as the second enzyme; however, the second enzyme did not differentiate isolates that were identical when XbaI was used. PFGE patterns differing by more than three bands were given a different letter designation (A to E), while those differing by three bands or less were given numerical designations within each letter designation (A1, A2, and A5, etc.). PFGE results were interpreted in accordance with the standards described previously (4, 16).
Antimicrobial susceptibility assays were performed using the agar dilution method based on breakpoints as described in the NCCLS guidelines (13). Those antibiotics tested included kanamycin, ampicillin, tetracycline, chloramphenicol, ciprofloxacin, sulfadiazine, streptomycin, nitrofurantoin, and trimethoprim-sulfamethoxazole. Plasmid profiles were determined using a modification of the plasmid DNA extraction method of Birnboim and Doly (1). The DNA was electrophoresed for 2.5 h through 1% agarose at 100 V in 1× Tris-agarose-EDTA buffer. The gels were stained with ethidium bromide and the molecular weight of the plasmids was determined by comparison with plasmids of known molecular weight. Biotyping was done based on the ability of serotype Enteritidis to ferment melibiose at 37°C in 18 to 24 h (5, 8).
Serotype Enteritidis isolates from across Canada were submitted to the NLEP at the time of the outbreak for characterization. These included 373 clinical isolates, 16 isolates from incriminated cheese portions of the lunch pack, 15 from chicken portions of the lunch pack which were suspected to have been involved, and 53 clinical and animal isolates that were not associated with the outbreak but were included to serve as controls. The ability to ferment melibiose was determined for all suspected outbreak strains and a subset of control strains. Of the 373 clinical isolates tested, 343 (92%) did not ferment melibiose in 24 h (Table 1). Similarly, 14 of 16 (87.5%) of the cheese isolates did not ferment melibiose. In contrast, all of the chicken isolates and most control strains, including all 29 PT 8, 12 PT 4, and 8 of 11 PT 13a strains tested, were melibiose positive. Epidemiological information coupled with the results of melibiose fermentation suggested that some of the 373 clinical isolates were unrelated to the outbreak and were derived from sporadic cases of gastroenteritis. This strongly supported the correlation of the melibiose-negative phenotype with exposure to the lunch packs (A. Ellis, personal communication). It is also possible, however, that some infections due to serotype Enteritidis melibiose-fermenting strains could have arisen from contaminated lunch packs.
TABLE 1.
Phage typing, PFGE, and biotype profiles of serotype Enteritidis isolates
| Source | No. of isolates tested | Phage type | PFGE pattern | Melibiose fermentation |
|---|---|---|---|---|
| Human | 106 | 8 | A1 | − |
| Human | 237 | 8 | NTa | − |
| Human | 20 | 8 | A1 | + |
| Human | 8 | 8 | NT | + |
| Human | 1 | 8 | D1 | + |
| Human | 1 | 8 | E1 | + |
| Cheese | 14 | 8 | A1 | − |
| Cheese | 2 | 8 | A1 | + |
| Chicken | 15 | 13a | NT | + |
| Human, control | 20 | 8 | A1 | + |
| Human, control | 1 | 8 | A5 | + |
| Human, control | 1 | 8 | A6 | + |
| Animal, control | 7 | 8 | A1 | + |
| Human, control | 12 | 4 | A2 | + |
| Human, control | 7 | 13a | A1 | + |
| Human, control | 3 | 13a | A1 | − |
| Human, control | 1 | 13a | B1 | + |
| Human, control | 1 | UTb | C1 | + |
NT, not tested.
UT, untypeable.
Phage typing demonstrated that 373 clinical and all 16 cheese isolates were PT 8 (Table 1). In contrast, the 15 chicken isolates were found to be PT 13a. Based on the phage pattern, the chicken isolates were considered unrelated to the outbreak. Though PT 8 strains can be converted to PT 13a by acquisition of a plasmid (3), plasmid analysis did not suggest any consistent differences between the PT 8 and PT 13a strains that would indicate that this had occurred. The sporadic control isolates included PT 8, PT 13a, and PT 4 isolates and isolates with untypeable patterns (Table 1).
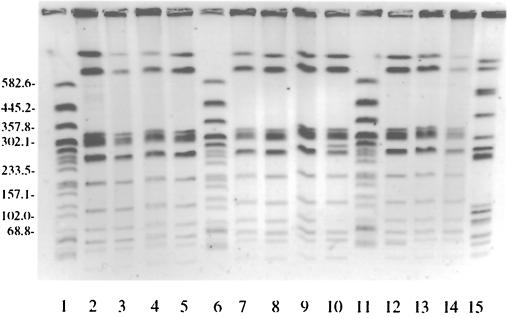
PFGE analysis did not discriminate well among serotype Enteritidis isolates (Fig. 1). The A1 pattern was found in both melibiose-fermenting and non-melibiose-fermenting clinical isolates, as well as in many of the clinical PT 8 and PT 13a human sporadic isolates collected in 1999. The A1 pattern was also present in isolates from cheese regardless of their ability to ferment melibiose. Two PT 8 clinical isolates had very different PFGE patterns (D1 and E1 [Table 1]), indicating that they were not part of the outbreak. A single PT 13a control strain and an untypeable control strain each had unique PFGE patterns (Table 1; Fig. 1, lanes 12 and 15, respectively). None of the chicken isolates were tested using PFGE because phage typing had already shown that they were not responsible for the outbreak. PFGE was not a reliable means of differentiating among serotype Enteritidis strains, although it was used to exclude a small number of isolates from the outbreak. Interestingly, the PFGE profiles (A2 pattern) of 12 PT 4 strains differed by only two bands from the most common PFGE profiles (A1 pattern) of the PT 8 strains examined (Table 1), suggesting that there is little diversity among Canadian serotype Enteritidis strains.
FIG. 1.
Characterization of selected S. enterica serotype Enteritidis strains by PFGE following digestion with the restriction endonuclease XbaI. Lanes 2 to 4, 8, and 9, outbreak isolates (all PT 8, melibiose negative); lanes 5 and 7, cheese isolates (PT 8, melibiose negative); lanes 10, 12, 13, 14, and 15, nonoutbreak control isolates (PT4, PT13a, PT8, PT8, and PT untypeable, respectively; all melibiose positive). Escherichia coli CDC strain G5244 was used as a molecular size marker (lanes 1, 6, and 11). Molecular sizes are indicated in kilobases.
Antibiotic susceptibility testing was performed on 20 clinical isolates chosen randomly from the 106 PT 8, PFGE A1 isolates and on 5 randomly chosen cheese isolates. It was determined that 15 of the 20 clinical isolates and 5 of 5 cheese isolates were resistant to nitrofurantoin: these isolates were susceptible to all other antibiotics tested. These 15 clinical and 5 cheese isolates carried a 38-MDa serovar-specific plasmid. One of the clinical PT 8 non-melibiose-fermenting strains also possessed an additional 60-MDa plasmid. Epidemiological investigations indicated that the patient matched the epidemiological profile of the outbreak. The relevance of this is unknown, since the plasmid could have been acquired during the outbreak. The three control strains tested possessed 2.1- and 2.8-MDa plasmids in addition to the 38-MDa plasmid.
Because serotype Enteritidis strains belonging to PT 8 are common in North America, multiple epidemiologic, phenotyping, and genotyping methods are required to distinguish outbreak from nonoutbreak strains. In this outbreak, phage typing was helpful for the initial characterization of isolates but could not be used to definitively identify these isolates as part of the outbreak. The use of PFGE did not greatly increase discrimination among strains. Although 95% of all Salmonella species ferment melibiose, all outbreak strains were found to be non-melibiose fermenters. This finding is striking when compared with the 5% non-melibiose fermenters that would normally be expected among PT 8 strains in general. It is also interesting that both melibiose fermenters and non-melibiose fermenters were found in the cheese, which suggests that the melibiose-fermenting PT 8, PFGE A1 cheese isolates may not have been prevalent or widely distributed in the snack foods made with the cheese. The fortuitous finding that the outbreak strain was deficient in its ability to ferment melibiose allowed for rapid and inexpensive screening of isolates. Together with phage typing and PFGE data, results from the melibiose assay supported the epidemiological findings and established a link between the clinical and cheese isolates. Although the potential for cross contamination existed between various ingredients used in the incriminated lunch product which contained both cheese and chicken, all serotype Enteritidis isolates from chicken had a phage type and biotype profile different from those of the outbreak cases and the incriminated cheese samples; thus, the chicken was excluded as the source of the outbreak. A lack of human infection due to PT 13a may have been due to a reduction of viable organism numbers in the chicken portions to levels below the infectious dose during processing. However, quantitative assessments of the relative levels of contamination of cheese and chicken portions of the lunch pack were not done. Differences in virulence between these two phage types seem unlikely given the fact that both could cause sporadic cases in humans (Table 1). However, it remains possible that the outbreak PT 8 strain had virulence or growth capabilities that made it more likely to cause an outbreak.
While PFGE was able to discriminate between different phage types, it failed to distinguish the outbreak PT 8 strains from the nonoutbreak PT 8 strains. Antibiogram and plasmid profiling were unable to discriminate beyond the serotype level.
Both epidemiological and bacterial typing methods were required for characterizing serotype Enteritidis PT 8 as strains involved in this major Canada-wide outbreak of salmonellosis due to contaminated cheese. Isolates could be characterized as outbreak strains on the basis of melibiose fermentation and phage typing, while PFGE excluded a minority of isolates. Epidemiological investigations were required to validate the laboratory findings and will be reported more fully elsewhere. These investigations led to the identification and removal of the vehicle of infection, and this was confirmed both by the identification of the outbreak strain in the product and by the rapid decline in the epidemic curve upon removal of the food from the market. Cooperation between reference laboratories and epidemiologists involved in the Canadian enteric surveillance system was pivotal in the rapid recognition and containment of this outbreak. It is crucial that reference laboratory centers maintain the capacity to perform a wide range of both phenotypic and genotypic methods to support outbreak investigations of salmonellosis.
Acknowledgments
We thank provincial public health laboratories, federal laboratories, veterinary laboratories, and the Canadian Food Inspection Agency for providing serotype Enteritidis isolates. We also thank Abdool Ali, Irene Martin, Annie Savoie, Maritia Gully, Russell Easy, Shelley Johnson, and Craig Murphy for technical assistance. We acknowledge the cooperation of Andrea Ellis and the staff epidemiologists of the Bureau of Infectious Diseases during and after the outbreak.
REFERENCES
- 1.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böhm H, Karch H. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30:2169–2172. doi: 10.1128/jcm.30.8.2169-2172.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D J, Baggesen D L, Platt D J, Olsen J E. Phage type conversion in Salmonella enterica serotype Enteritidis caused by the introduction of a resistance plasmid of incompatibility group X (IncX) Epidemiol Infect. 1999;122:19–22. doi: 10.1017/s0950268898001794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Atlanta, Ga: The National Molecular Subtyping Network for Foodborne Disease Surveillance, Centers for Disease Control and Prevention; 1998. [Google Scholar]
- 5.Ewing W H. Edwards and Ewings's identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1986. pp. 181–340. [Google Scholar]
- 6.Hennessey T W, Hedberg C W, Slutsker L, White K, Besser-Wiek J, Moen M, Feldman J, Coleman W, Edmonson L, MacDonald K, Osterholm M the Investigation Team. A national outbreak of Salmonella enteritidis infections from ice cream. N Engl J Med. 1996;334:1281–1286. doi: 10.1056/NEJM199605163342001. [DOI] [PubMed] [Google Scholar]
- 7.Hilton A C, Penn C W. Comparison of ribotyping and arbitrarily primed PCR for molecular typing of Salmonella enterica and relationships between strains on the basis of three molecular markers. J Appl Microbiol. 1998;85:933–940. doi: 10.1111/j.1365-2672.1998.tb05256.x. [DOI] [PubMed] [Google Scholar]
- 8.Kauffman F. The bacteriology of Enterobacteriaceae. Baltimore, Md: The Williams & Wilkins Co.; 1966. [Google Scholar]
- 9.Laconcha I, López-Molina N, Rementeria A, Audicana A, Perales I, Garaizar J. Phage typing combined with pulsed-field gel electrophoresis and random amplified polymorphic DNA increases discrimination in the epidemiological analysis of Salmonella enteritidis strains. Int J Food Microbiol. 1998;40:27–34. doi: 10.1016/s0168-1605(98)00007-5. [DOI] [PubMed] [Google Scholar]
- 10.Landeras E, Mendoza M C. Evaluation of PCR-based methods and ribotyping performed with a mixture of PstI and SphI to differentiate strains of Salmonella serotype Enteritidis. J Med Microbiol. 1998;47:427–434. doi: 10.1099/00222615-47-5-427. [DOI] [PubMed] [Google Scholar]
- 11.Liebisch B, Schwarz S. Molecular typing of Salmonella enterica subsp. enterica serovar Enteritidis isolates. J Med Microbiol. 1996;44:52–59. doi: 10.1099/00222615-44-1-52. [DOI] [PubMed] [Google Scholar]
- 12.Mendoza M C, Landeras E. Molecular epidemiological methods for differentiation of Salmonella enterica serovar Enteritidis strains. In: Saeed A M, Gast R E, Potter M E, Wall P G, editors. Salmonella enterica serovar Enteritidis in humans and animals: epidemiology, pathogenesis, and control. Ames: Iowa University Press; 1999. pp. 125–140. [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4:17, (2) M100-S7. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 14.Popoff M Y, LeMinor L. Antigenic formulas of the Salmonella serovars. 6th ed. Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella, Institute Pasteur; 1992. [Google Scholar]
- 15.Ratnam S, Stratton F, O'Keefe C, Roberts A, Coats R, Yetman M, Khakhria R, Hockin J. Salmonella Enteritidis outbreak due to contaminated cheese—Newfoundland. Can Communicable Dis Rep. 1999;25:1–4. [PubMed] [Google Scholar]
- 16.Tenover F C, Arbeit R, Goering R. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect Control Hosp Epidemiol. 1997;18:426–439. doi: 10.1086/647644. [DOI] [PubMed] [Google Scholar]
- 17.Usera M A, Popovic T, Bopp C A, Strockbine N A. Molecular subtyping of Salmonella enteritidis phage type 8 strains from the United States. J Clin Microbiol. 1994;32:194–198. doi: 10.1128/jcm.32.1.194-198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward L R, de Sa J D H, Rowe B. A phage typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–304. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]