Abstract
To facilitate the biochemical characterization of chromatin-associated proteins in the budding yeast Saccharomyces cerevisiae, we have developed a system to assemble nucleosomal arrays on immobilized templates using recombinant yeast core histones. This system enabled us to analyze the interaction of Isw2 ATP-dependent chromatin remodeling complex with nucleosomal arrays. We found that Isw2 complex interacts efficiently with both naked DNA and nucleosomal arrays in an ATP-independent manner, suggesting that ATP is required at steps subsequent to this physical interaction. We identified the second subunit of Isw2 complex, encoded by open reading frame YGL 133w (herein named ITC1), and found that both subunits of the complex, Isw2p and Itc1p, are essential for efficient interaction with DNA and nucleosomal arrays. Both subunits are also required for nucleosome-stimulated ATPase activity and chromatin remodeling activity of the complex. Finally, we found that ITC1 is essential for function of Isw2 complex in vivo, since isw2 and itc1 deletion mutants exhibit virtually identical phenotypes. These results demonstrate the utility of our in vitro system in studying interactions between chromatin-associated proteins and nucleosomal arrays.
The fundamental unit of chromatin is the nucleosome, composed of 147bp of DNA wrapped around an octamer of core histones H2A, H2B, H3, and H4 (29). Though required for genomic compaction, nucleosomes can inhibit processes dependent on protein-DNA interactions, including transcription. Therefore, chromatin remodeling is integral to the regulation of these processes. Two major classes of chromatin regulators have been identified in eukaryotic cells: histone-modifying enzymes and ATP-dependent chromatin remodeling factors (3, 22, 23, 30, 39, 40, 43, 46, 47, 50, 55). The significance of histone modifications in transcriptional regulation is highlighted by recent findings that a large number of previously identified transcriptional regulators possess acetylase and deacetylase activities (39, 41, 43). Acetylation of histone tails has been proposed to affect the higher-order folding of nucleosomal arrays (44) or the interaction of histone tails with DNA (54). Additionally, Strahl and Allis have proposed that histone modifications modulate the interactions of proteins with chromatin by serving as a code for recognition by specific proteins (39). However, the precise molecular mechanisms for the regulation of chromatin structure by covalent histone modifications remain to be determined.
The second type of chromatin regulators, ATP-dependent chromatin remodeling factors, use the energy of ATP hydrolysis to alter chromatin structure. They have been grouped into three classes, SWI/SNF, ISWI, and CHD1, according to their ATPase subunits (7, 20, 22, 32). Yeast SWI/SNF complex was originally identified as a positive regulator of a wide variety of genes (31, 53). Recent works suggest it may also have roles in the negative regulation of transcription (18, 42). The founding member of the ISWI class, Drosophila ISWI, is essential for cell viability (10). Deletion of the yeast ISW2 gene results in the mitotic transcriptional derepression of many genes normally induced during meiosis (13). Furthermore, the deletion of both yeast ISWI genes, ISW1 and ISW2, confers a synthetic stress-sensitive phenotype in combination with a chd1 mutation (45). In vitro, complexes in both the SWI/SNF (52) and ISWI (15, 25) classes induce sliding of nucleosomes along DNA templates. Therefore, nucleosome sliding may be one common mechanism underlying the functions of ATP-dependent chromatin remodeling factors. However, it is unknown exactly how these factors utilize the energy of ATP hydrolysis to alter chromatin structure.
In vitro chromatin assembly systems from Drosophila (2, 21) and Xenopus (1) extracts have been successfully applied to studying the effects of nucleosomes on various steps of transcription, including the interaction of transcription factors with chromatin templates. However, in vitro chromatin assembly systems using yeast core histones have not been widely available because of limited success in obtaining large quantities of purified core histones from Saccharomyces cerevisiae (28, 33, 38). In addition, native yeast histone preparations contain heterogeneous populations of modified core histones. To overcome these problems, we developed a nucleosome assembly system using recombinant yeast histones. Arrays of nucleosome core particles were assembled on DNA immobilized on magnetic beads to allow rapid detection of interacting proteins. In this text, for convenience we refer to arrays of nucleosome core particles composed of recombinant core histones assembled on DNA as nucleosomal arrays. We analyzed the interaction of the two-subunit Isw2 chromatin remodeling complex with chromatin and found that it interacts efficiently with both naked DNA and nucleosomal arrays in an ATP-independent manner. These results suggest that ATP is required subsequent to the physical interaction of the complex with nucleosomal arrays. Through mass spectrometry, we identified the second subunit of Isw2 complex as the product of open reading frame (ORF) YGL 133w and named it ITC1 (imitation switch 2 Complex subunit 1). We found that both subunits of the complex, Isw2p and Itc1p, are essential for efficient interaction with DNA and nucleosomal arrays, as well as for stimulation of the ATPase activity and chromatin remodeling activity of the complex. Finally, we demonstrate that isw2 and itc1 deletion mutants have virtually identical phenotypes, suggesting that both subunits are essential for Isw2 complex function in vivo, as predicted by in vitro experiments.
MATERIALS AND METHODS
Yeast strains.
All yeast strains used in this study are derived from W1588-4C. This strain is congenic to W303-1A, except that a weak rad5 mutation in the original W303 is repaired (56). To tag proteins with the FLAG epitope, we constructed a plasmid which serves as a template for PCR-based tagging by homologous recombination. Oligonucleotides encoding three copies of the FLAG epitope sequence followed by a termination codon were annealed, generating overhanging ends compatible with SacI at one end and Pstl at the other. The annealed fragment was then ligated into SacI/Pstl-digested pBlueScript SK(−) to create pBS-3FLAG. Subsequently, the NdeI-SpeI fragment of pUG6 (14), containing the KanMX marker flanked by loxP sites, was ligated into the EcoRI-XhoI sites of pBS-3FLAG (downstream of the FLAG sequence). The resulting plasmid, p3FLAG-KanMX, was then used as a template for generation of a PCR product that introduces three copies of the FLAG epitope just upstream of the termination codon of the ITC1 gene by homologous recombination, using 5′ AGTGGGCCAAACCTCAAGAACAGTAACACCTGCCCCAAATAGGGAACAAAAGCTGGAG 3′ as a 5′ primer and 5′ CAATTTACCAT CAGTTACAAAGGAAGTTTTTTATATATTACTATAGGGCGAATTG GGT 3′as a 3′ primer. The underlined bases indicate the sequence that anneals to p3FLAG-KanMX during PCR, while the remaining sequence corresponds to the site of insertion at the ITC1 locus. It should be noted that p3FLAG-KanMX was designed such that oligonucleotides used for FLAG tagging are also compatible with the pMPY vectors described previously (37) and thus can also be used for Myc- and hemagglutinin-epitope tagging, using these vectors as templates for PCR.
A null mutation of the ITC1 gene was created by replacing the coding region with the KanMX marker (14). Other mutants are described previously (45).
Expression, purification and reconstitution of recombinant yeast histone octamers.
Recombinant yeast histones were expressed and purified as described previously (29), with slight modifications. Each histone was purified individually as inclusion bodies and solubilized in unfolding buffer (29). Solubilized histones were then loaded onto tandemly connected Q-Sepharose and SP-Sepharose, each packed in an HR10/10 column (Amersham Pharmacia Biotech) and equilibrated in U buffer (29) containing 100 mM (for H2A and H2B) or 200 mM (for H3 and H4) NaCl. After washing, the Q-Sepharose column was detached, and core histones were eluted from the SP-Sepharose column by a linear salt gradient. The four core histones were denatured individually in unfolding buffer, mixed in equimolar ratios, and dialyzed against refolding buffer as described previously (29). Spectra/Por6 dialysis tubing (molecular size cutoff, 3.5 kDa; Spectrum Companies) was used in the reconstitution reaction. Reconstituted octamer was purified by gel filtration through a Superdex 200 column (Amersham Pharmacia Biotech) and stored at −20°C in 10 mM Tris-HCI (pH 7.6)–1 mM EDTA–2 M NaCl–0.05% NP-40–50% glycerol. A detailed protocol is available upon request.
Nucleosome spacing assays.
For nucleosome spacing assays (45), 0.3 μg of recombinant yeast histone octamer, approximately 1.8 μg of recombinant Nap 1p (nucleosome assembly protein 1) (12), 0.3 μg of lambda DNA, and approximately 45 fmol of Isw1 or Isw2 complex were used in each 30-μl reaction.
Assembly of nucleosomal arrays on an immobilized template.
pBlueScript SK(−) (Stratagene) was used as a template for the immobilized nucleosomal array. Approximately 6.6 × 108 (9.9 mg) Dynabeads M-280 (Dynal) were used for coupling with 20 μg of plasmid DNA linearized by EcoRI and ClaI as described previously (36). In a typical nucleosome assembly reaction, Dynabeads coupled with 1.5 μg of DNA were incubated with 1.5 μg of recombinant yeast histone octamer and approximately 9 μg of purified recombinant Nap1p in 10 mM HEPES-KOH (pH 7.6)–40 mM KCI–60 mM NaCl–5 mM MgCl2–0.5 mM EGTA–10% glycerol–0.1 μg of bovine serum albumin (BSA)/μl for 4 h at 30°C with constant mixing. Histone octamer was omitted from the reaction for naked DNA controls. The beads were washed six times with 1 ml of 25 mM HEPES-KOH (pH 7.6)–600 mM KCI–0.1 mM EDTA–0.5 mM EGTA–5 mM MgCl2–20% glycerol. To confirm the loading of histones on DNA, histones were eluted from the immobilized templates with 2 M NaCl and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining.
Purification of Isw2 complex and subunits.
Isw2 complex was purified as previously published by immunoaffinity purification with anti-FLAG M2 beads (Sigma) and a Source Q column (Amersham Pharmacia Biotech) (45). Monomer Isw2p and Itc1p were purified from itc1 and isw2 null mutants, respectively, using the same protocol.
Nucleosome-Isw2 interaction assay.
For a typical assay, immobilized nucleosomal arrays containing 25 ng of DNA were incubated with approximately 45 fmol of Isw2 complex, Isw2p, or Itc1p in 10 μl of 25 mM HEPES-KOH(pH 7.6)–1 mM EDTA–5 mM MgCl2–50 mM NaCl–45 mM KCl, 0.1 μg of BSA/μl–0.05% NP-40 at 30°C for 30 min at full speed on an Eppendorf 5436 shaker. The beads were concentrated on a magnetic particle concentrator (Dynal), and the supernatant was collected for analysis of unbound proteins. The beads were then washed in 100 μl of 25 mM HEPES-KOH (pH 7.6)–1 mM EDTA–5 mM MgCl2–50 mM NaCl–0.1 μg of BSA/μl–0.05% NP-40 for 1 min at 30°C at full speed on an Eppendorf 5436 shaker, and bound proteins were eluted by the addition of SDS-PAGE sample buffer to the beads. Bound and unbound proteins were detected by Western blotting using the anti-FLAG antibody M2 (Sigma).
ATPase assay.
ATPase assays were performed as described elsewhere (45). Standard reactions contained immobilized templates (25 ng of DNA or equivalent) and 45 fmol of Isw2 complex, Isw2p, or Itc1p in 5 μl. Reactions were performed at 30°C for 30 min on an Eppendorf 5436 shaker. Equivalent amounts of magnetic beads were used as a negative control.
RNA analysis.
Strains were grown in YEPD (2% Bacto Peptone, 1% yeast extract, 2% glucose) at 30°C and harvested during early log phase (optical density at 660 nm of 0.7). RNA was prepared using acid phenol extraction; 25 μg of total RNA was loaded per lane. The signals were quantified by a PhosphorImager (Molecular Dynamics).
RESULTS AND DISCUSSION
An in vitro nucleosome assembly system using recombinant yeast histones.
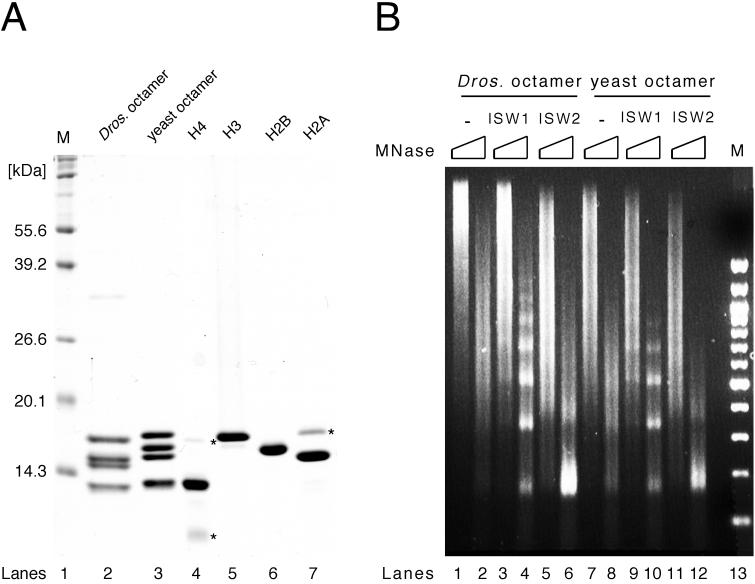
We sought to develop a system to assemble nucleosomal arrays using recombinant yeast histones in order to facilitate biochemical analyses of yeast chromatin remodeling factors. Using a published protocol (29) with minor modifications (see Materials and Methods for details), we expressed the four core histones separately in Escherichia coli and purified them individually (Fig. 1A). The four core histones were then mixed at equimolar ratios and renatured to form histone octamer. Reconstituted histone octamer was then separated from aggregate, H3/H4 tetramer, H2A/H2B dimer, and monomer histones by gel filtration through a Superdex 200 column. While minor contaminants were detectable by Coomassie blue staining in the purified monomer core histone fractions (Fig. 1 A, lanes 4 to 7), they were separated from reconstituted histone octamer in the Superdex column (lane 3). This procedure yields highly purified histone octamer containing stoichiometric amounts of all four core histones.
FIG. 1.
Reconstitution of recombinant yeast histone octamer. (A) Purified recombinant yeast core histones and histone octamer. Yeast core histones (H2A, H2B, H3, and H4) were expressed and purified individually from E. coli (lanes 4 to 7). Reconstituted recombinant yeast histone octamer (lane 3) is shown next to native Drosophila histone octamer (lane 2) for comparison. Proteins were separated by SDS-PAGE (15% gel) and stained with Coomassie brilliant blue R250. Lane M, size markers; ∗, minor contaminant present after purification of individual core histones. (B) Isw1 and Isw2 complexes facilitate the formation of regularly spaced nucleosomes assembled with recombinant yeast histone octamer. Nap1p-mediated nucleosome spacing assays using Drosophila (left) or yeast (right) histone octamer were performed on lambda DNA in the presence or absence of Isw1 and Isw2 complexes. All reactions contained ATP. Nucleosome spacing was analyzed by partial and extended MNase digestion. DNA was purified and separated by 1.3% agarose gel electrophoresis followed by ethidium bromide staining.
To assess the ability of yeast ISWI complexes to function with purified histone octamer, we compared the nucleosome spacing activities of Isw1 and Isw2 complexes using recombinant yeast histone octamer and native Drosophila octamer. With Drosophila octamer, we have previously shown that Isw1 and Isw2 complexes facilitate the regular spacing of nucleosomes deposited by recombinant histone chaperone Nap1p in vitro (45). This ATP-dependent spacing activity is revealed by the appearance of a discrete nucleosome ladder upon micrococcal nuclease (MNase) digestion of assembled nucleosomal arrays. Essentially identical results were obtained in nucleosome spacing assays using native Drosophila octamer and recombinant yeast octamer (Fig. 1B). As reported previously, digestion of nucleosomal arrays assembled in the absence of either ISWI complex resulted in a smear at both digestion time points, showing that nucleosomes are not regularly spaced (Fig. 1B, lanes 1, 2, 7, and 8). When nucleosomes are assembled in the presence of Isw1 complex, MNase digestion yields a discrete nucleosome ladder (lanes 3, 4, 9, and 10). On the other hand, MNase digestion of nucleosomal arrays assembled in the presence of Isw2 complex produces a strong mononucleosome signal with a less defined ladder as previously reported (lanes 5, 6, 11, and 12). These spacing activities are dependent on the presence of hydrolyzable ATP (data not shown). Therefore, we conclude that recombinant yeast histone octamer functions similarly to native Drosophila octamer in our biochemical assays.
The use of a recombinant system is particularly advantageous in yeast. Since S. cerevisiae has only two copies of the genes encoding each of the four core histones, genetic characterization of histones has been possible. These analyses have yielded a number of histone mutants that exhibit various defects in transcriptional regulation (16, 17, 24, 35, 51). Our system can be easily adapted to incorporate these mutants into histone octamers to test their effects on the stability of nucleosomal arrays and the interaction of these arrays with chromatin-associated proteins.
Development of a chromatin interaction assay.
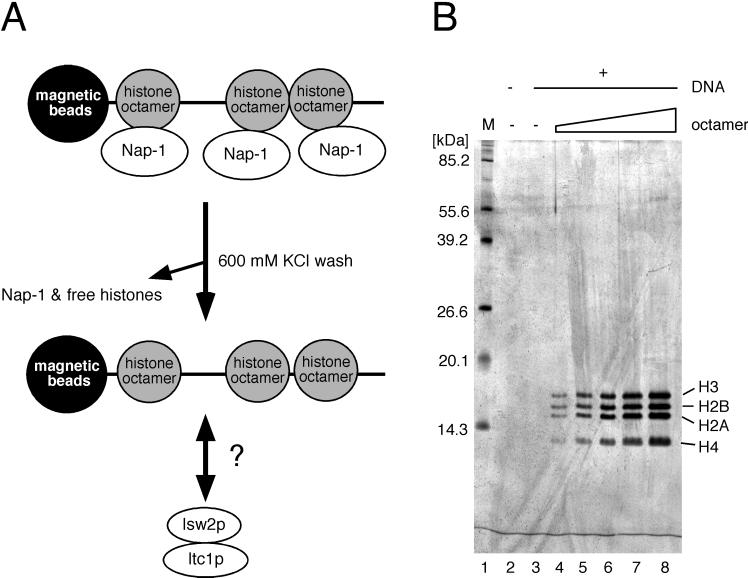
Previously, glycerol gradient fractionation has been widely used to assess the association of proteins with nucleosomes (6). This method, however, is time-consuming and requires large quantities of starting materials. To overcome these problems, we developed a system to study interactions between nucleosomes and proteins. We use recombinant Nap1p histone chaperone to deposit yeast histone octamer onto DNA immobilized on magnetic beads, followed by removal of free histones and Nap1p by a high (600 mM) salt wash (Fig. 2A). This protocol yields nucleosomal arrays composed of highly purified core histones and permits quantitative loading of nucleosomes as assessed by silver staining (Fig. 2B). This system can be used to study the interactions of chromatin-associated proteins with nucleosomal arrays.
FIG. 2.
Assay for protein-nucleosomal array interactions. (A) Schematic representation of the assay. Nucleosomes assembled on immobilized templates with Nap1p are washed with high salt to remove free histones and Nap1p. Nucleosomal arrays are then incubated with Isw2 complex to analyze their interactions. (B) Quantitative loading of recombinant yeast histone octamer onto immobilized templates. Increasing amounts of recombinant yeast histone octamer (0, 0.5, 1.0, 1.5, 2.0, and 3.0 μg) and Nap1p (9 μg) were mixed with a fixed amount of immobilized template (1.5 μg of DNA). Proteins were eluted from immobilized templates, assembled with no (lane 3) or increasing amounts of (lanes 4 to 7) histone octamer. Lane 2 shows elution from the beads alone. Proteins were separated by SDS-PAGE (15% gel) and silver stained. Histone H4 commonly does not stain as strongly as H2A, H2B, and H3. Lane M, size markers.
Isw2 complex interacts with nucleosomal arrays in an ATP-independent manner.
We applied our in vitro system to analyze the interaction of Isw2 complex with nucleosomal arrays in order to begin dissecting its mechanism of chromatin remodeling. One of the unique biochemical properties of the ISWI class of ATP-dependent chromatin remodeling complexes is that their ATPase activities are strongly stimulated by nucleosomes but not as efficiently by naked DNA or free histones (nucleosome-stimulated ATPase activity) (4, 27, 45, 46, 49). This property is distinct from the ATPase activity of the SWI/SNF class of chromatin remodeling factors, which is equally well stimulated by naked DNA and nucleosomes (9, 11, 26). The specificity of their ATPase activity for nucleosomes implies that ISWI complexes recognize structural features of nucleosomes that are absent from naked DNA and free histones. However, this recognition process is not understood at the molecular level. It is also unknown at which step(s) during chromatin remodeling ISWI complexes utilize the energy of ATP hydrolysis. One possibility is that ISWI complexes interact with nucleosomal arrays in an ATP-dependent manner. Alternatively, ISWI complexes may interact preferentially with nucleosomal arrays over naked DNA in an ATP-independent manner. The ATPase activity of the complex is then stimulated subsequent to this interaction.
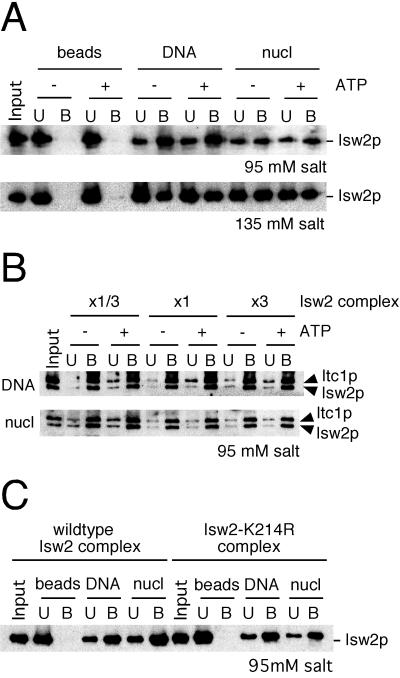
We first tested whether purified Isw2 complex requires ATP to stably interact with nucleosomal arrays. Native Isw2 complex was incubated with beads alone, immobilized DNA, or immobilized nucleosomal arrays in the presence or absence of ATP. Each reaction contained approximately 3.5 molecules of Isw2 complex per immobilized template (3 kb), or an estimated 4.5 nucleosomes for each molecule of the complex, assuming an average of 185 bp DNA per nucleosome. As shown in Fig. 3A, Isw2 complex does not bind detectably to the beads alone, whereas it binds with substantial affinity to both naked DNA and nucleosomal arrays at 95 mM salt. Unexpectedly, this interaction is not significantly affected by the presence of ATP. The addition of ATPγS, a nonhydrolyzable ATP analog, also does not affect Isw2-nucleosome interactions (data not shown). Similar results were found at 135 mM salt, a condition under which the interaction of Isw2 complex with the nucleosomal array was slightly impaired. In order for this assay to be meaningful, the concentration of Isw2 complex used in the interaction assay must be within a linear range. Otherwise, it is possible that Isw2 complex has saturated a limited number of interaction sites on the immobilized templates. To address this point, we performed the interaction assay with three different concentrations of Isw2 complex (15, 45, and 135 fmol in 10-μl reactions) in the presence and absence of ATP. To load equivalent amounts of protein in Western blotting, the samples from the 45- and 135-fmol reactions were diluted three- and nine fold, respectively. As shown in Fig. 3B, Isw2 complex efficiently interacted with both naked DNA and nucleosomal arrays under all conditions, and the signals from all three concentrations of Isw2 complex were similar. This result indicates that the concentration of Isw2 complex in our standard condition (45 fmol in 10 μl) is within a linear range of Isw2-template interactions. These data further suggest that the interactions between Isw2 complex and templates are ATP independent over a range of Isw2 concentrations.
FIG. 3.
Isw2 complex interacts with DNA and nucleosomal arrays in an ATP-independent manner. (A) Interaction of Isw2 complex with immobilized templates. The interaction assay was performed by incubating Isw2 complex with beads alone, immobilized DNA, or immobilized nucleosomal arrays (nucl) in the presence or absence of ATP at 95 or 135 mM salt. The supernatant was collected, beads were washed, and bound proteins were eluted from the beads in SDS-PAGE sample buffer. Equivalent amounts of unbound (U) and bound (B) fractions were separated by SDS-PAGE, and Isw2p was detected by Western blotting. “Input” indicates the amount of the fractions used in each reaction. (B) Quantitative interaction of Isw2 complex with templates. Threefold serial dilutions of Isw2 complex were used in the interaction assay in the presence and absence of ATP. x1/3, x1, and x3 correspond to 15, 45, and 135 fmol of Isw2 complex. For Western blotting, the samples from x1 and x3 reactions were diluted three- and ninefold, respectively. The standard reaction uses 45 fmol of Isw2 complex. In this experiment, Isw2 complex with FLAG-tagged Isw2p and Itc1p was used. (C) Interactions of wild-type and catalytically inactive (Isw2-K214R) Isw2 complexes with immobilized templates. The experiment was performed under standard conditions.
We sought to confirm the ATP independence of the interactions between Isw2 complex and nucleosomal arrays through an alternate approach. To this end, Isw2 complex containing a catalytically inactive form of Isw2p (Isw2-K214R) was purified and tested in the interaction assay. The K214R mutation lies in the putative ATP-binding pocket of Isw2p and reduces the ATPase activity of the complex to background levels, without affecting its subunit composition (data not shown). This mutation completely inactivates the ability of Isw2 complex to repress early meiotic genes during mitotic growth in vivo (13). As shown in Fig. 3C, the mutant Isw2 complex interacts with naked DNA and nucleosomal arrays as efficiently as the wild-type complex. From these data, we conclude that the initial interaction of Isw2 complex with nucleosomal arrays and naked DNA does not require ATP.
Interaction of Isw2 complex with both DNA and nucleosomal arrays was somewhat unexpected since naked DNA does not effectively stimulate the ATPase activity of the complex. Our preliminary results show that Isw2 complex does not efficiently interact with high-density nucleosomal arrays or with nucleosome core particles (M. E. Gelbart and T. Tsukiyama, unpublished data). It is therefore possible that Isw2 complex interacts with linker regions first, and its ATPase activity is stimulated at subsequent steps involving recognition of nucleosomal structures. This model is consistent with the observed ATP-independent interaction of Isw2 complex with the immobilized templates and the requirement of the energy of ATP hydrolysis for chromatin remodeling activities of the complex (45). Furthermore, an earlier report shows that Drosophila ISWI protein does not interact with nucleosome core particles whereas it efficiently interacts with mononucleosomes containing linker DNA (5).
Both Isw2p and Itc1p subunits are essential for efficient interaction of Isw2 complex with nucleosomal arrays.
Next, we tested whether the second subunit of Isw2 complex, Itc1p (previously referred to as p140 [45]), is required for the interaction of the complex with nucleosomal arrays. This question is particularly interesting since monomeric Drosophila ISWI protein is partially functional in nucleosome remodeling at high concentrations in vitro (8, 15, 19). ITC1 is encoded by ORF YGL133W, as determined by mass spectrometry (data not shown). This ORF is predicted to encode a 1,264-amino-acid protein with an estimated molecular mass of 145 kDa. Though this ORF has no known function, it is homologous to another uncharacterized yeast ORF, YPL216w (4, 19). Both of these ORFs share an N-terminal WAC (WSTF/Acf1/cbp146) motif also present in Drosophila Acf1 (ATP-utilizing chromatin assembly and remodeling factor 1), human WSTF (Williams Syndrome transcription factor), human WCRF180 (Williams syndrome transcription factor-related chromatin remodeling factor 180)/BAZ1A (bromodomain adjacent to zinc finger domain 1A)/human ACF1, and mouse cbp146 (4, 19, 27, 34). Acf1 is the second subunit of Drosophila ACF, an ISWI-containing chromatin remodeling complex (19). WCRF180/BAZ1A/hACF1 is the second subunit of the human WCRF/hACF complex as well as the highly related HuCHRAC complex, both containing the ISWI homolog, hSNF2h, as the catalytic subunit (4, 27, 34). These data indicate that the WAC domain may be responsible for functions of ISWI complexes that have been evolutionarily conserved.
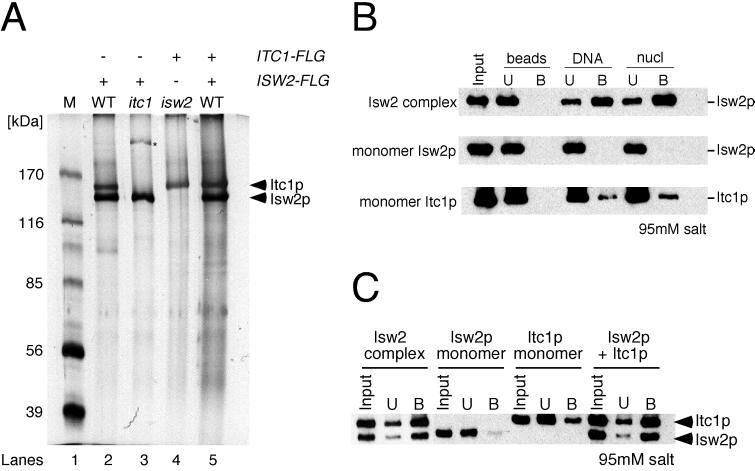
FLAG purification of Isw2p and Itc1p from itc1 (ygl133w) and isw2 deletion mutants yielded monomeric Isw2p and Itc1p, respectively (Fig. 4A, lanes 3 and 4), confirming the identity of the ITC1 gene. These data also suggest that neither subunit is present in other major complexes. As shown in Fig. 4B, monomeric Isw2p showed little to no detectable binding to naked DNA or nucleosomal arrays. Monomeric Itc1p exhibited minor binding to both naked DNA and nucleosomes, but the efficiency was much lower than that of the native Isw2 complex. These results demonstrate that both subunits are necessary for efficient interaction of the complex with DNA and nucleosomal arrays. For additional confirmation, we mixed purified monomeric Isw2p and Itc1p to determine whether the reconstituted complex functioned like the native complex in the interaction assay. As shown in Fig. 4C, the reconstituted complex interacted with nucleosomal arrays as efficiently as the native Isw2 complex. In this experiment, native Isw2 complex was purified from a strain in which both Isw2p and Itc1p were FLAG tagged to simultaneously detect both subunits (Fig. 4A, lane 5). ATP also did not affect interaction of monomer Isw2p with templates (data not shown).
FIG. 4.
Both Isw2p and Itc1p of Isw2 complex are required for efficient interactions. (A) Purified Isw2 subunits used in analyses. Lanes 2 and 5 show a silver stain of wild-type (WT) Isw2 complex, purified from strains in which only Isw2p and both subunits, respectively, were FLAG tagged. Lane 3 shows a silver stain of the complex from an itc1 deletion background in which Isw2p was FLAG tagged. Lane 4 shows protein purified from an isw2 deletion background in which Itc1p was FLAG tagged. Lane M, size markers; ∗, minor contaminant present in some but not all preparations. (B) Interaction assay using native Isw2 complex, monomeric Isw2p, and monomeric Itc1p. Monomeric Isw2p and Itc1p were purified from itc1 and isw2 deletion mutants, respectively. Unbound (U) and bound (B) proteins were separated by SDS-PAGE and detected by Western blotting. (C) Interaction of reconstituted Isw2 complex with immobilized nucleosomal arrays. Unbound (U) and bound (B) proteins were separated by SDS-PAGE and detected by Western blotting. “Isw2p + Itc1p” denotes Isw2 complex reconstituted in vitro by mixing monomeric Isw2p and Itc1p at equimolar ratios and incubating the mixture for 30 min on ice. In this experiment, Isw2 complex with FLAG-tagged Isw2p and Itc1p was used.
Both Isw2p and Itc1p subunits are essential for biochemical activities of Isw2 complex.
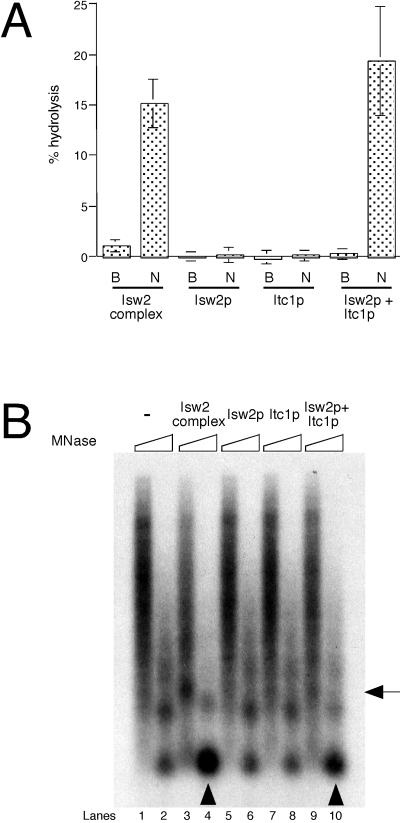
The requirement of Itc1p for the biochemical activities of Isw2 complex was further supported by ATPase assays (Fig. 5A). Monomeric Isw2p and Itc1p showed no detectable ATPase activity in response to stimulation with immobilized nucleosomal arrays, whereas the reconstituted complex was as active as the native Isw2 complex. This evidence suggests that efficient physical interaction of Isw2 complex with nucleosomal arrays may be needed to stimulate Isw2 ATPase activity. In addition, we examined the chromatin remodeling activity of native and reconstituted Isw2 complex, as well as monomeric Isw2p and Itc1p, by incubating them with preassembled nucleosomal arrays on immobilized templates (Fig. 5B). Extensive MNase digestion of the template in the absence of additional factors yielded mono- and dinucleosome signals (lane 2), showing that canonical nucleosomes were formed in this system. In contrast, limited digestion yielded a smear (lane 1), revealing that nucleosomes are not regularly spaced. Nucleosomal arrays incubated with ATP and native Isw2 complex yielded a much stronger mononucleosome signal upon extensive MNase digestion (lane 4), whereas limited digestion revealed a shift in the position of the dinucleosome signal (lane 3). These changes observed on immobilized templates are similar to those observed on free naked DNA (Fig. 1B) (45). Previously, we proposed that this increase in mononucleosome signal may be due to facilitation of nucleosome assembly by Isw2 complex (45). However, our data showed that the mononucleosome signal still increases upon action of Isw2 complex on preassembled nucleosomal arrays in the absence of free histones. This result suggests that Isw2 complex may function by altering histone-DNA interactions within the nucleosome as proposed previously (48). In contrast to Isw2 complex, monomeric Isw2p and Itc1p did not induce detectable structural changes in nucleosomal arrays (lanes 5 to 8). MNase digestion of nucleosomal arrays incubated with reconstituted Isw2 complex revealed changes in chromatin structure similar to but less prominent than those observed with native Isw2 complex (lanes 9 and 10). This result suggests that simple mixing of monomeric Isw2p and Itc1p is not sufficient to reconstitute full biochemical activity of the native complex.
FIG. 5.
Both Isw2p and Itc1p are required for biochemical activities of Isw2 complex. (A) ATPase assays using native Isw2 complex, monomeric Isw2p, monomeric Itc1p, and reconstituted Isw2 complex. Assays were done in the presence of buffer (B) or immobilized nucleosomal arrays (N). (B) Chromatin remodeling assay performed on preassembled nucleosomal arrays on immobilized templates. After incubation of the templates with ATP and the fractions indicated above, limited (lanes 1, 3, 5, 7, and 9) and extensive (lanes 2, 4, 6, 8, and 10) MNase digestion was performed. Nucleosomal arrays containing 25 ng of DNA and 45 fmol of Isw2 complex, Isw2p, or Itc1p were used. Arrowheads denote mononucleosome signals increased by native and reconstituted Isw2 complex. The arrow on the right indicates a dinucleosome signal that is shifted upward by the native Isw2 complex (lane 3). DNA was visualized by Southern blotting.
Biochemical characterization of monomeric Drosophila ISWI protein has been reported previously. Nucleosome-stimulated ATPase activity was detected when 1.3 pmol of E. coli-expressed ISWI and nucleosomes containing 360 ng of DNA were used in a 10-μl reaction (8) and when 0.14 to 0.28 pmol of baculovirus-expressed ISWI and nucleosomes containing 25 ng of DNA were used in a 5-μl reaction (15). Therefore, requirement of both Isw2p and Itc1p subunits in all of our assays (DNA and nucleosome binding, ATPase, and chromatin remodeling) was somewhat unexpected. To compare our results with previous reports, we performed the ATPase assay using 0.2 pmol of Isw2p and nucleosomes containing 25 ng of DNA in a 5-μl reaction, a condition identical to that in the study by Hamiche et al. (15). Even under this condition, we did not detect any nucleosome-stimulated ATPase activity of monomeric Isw2p (data not shown). This result implies that Isw2p may be different from Drosophila ISWI protein and lacks biochemical activities as a monomer. However, it should be noted that monomeric Drosophila ISWI protein is extremely labile, and its biochemical activities can easily be lost upon freeze-thaw cycles or prolonged storage (C. Wu and H. Xiao, personal communications). This implies that minor differences in the folding properties of monomeric Drosophila ISWI protein and Isw2p may account for the observed differences in their biochemical activities. It is also possible that biochemically active monomer Isw2p needs to be purified under special conditions yet to be determined. While active in some biochemical assays, Drosophila ISWI exhibits significantly higher specific activities when incorporated into complexes according to two reports. Hamiche et al. reported that 3 to 5 fmol of NURF and 0.14 to 0.28 pmol of monomeric ISWI exhibited comparable ATPase activities (15). Additionally, Ito et al. showed that 2.2 but not 0.22 pmol of monomeric Drosophila ISWI was active in the nucleosome spacing assay, and 22 fmol of recombinant ACF exhibited a comparable activity (19). In contrast, Drosophila CHRAC and E. coli-expressed monomeric ISWI exhibited comparable specific activities in ATPase, nucleosome spacing, and nucleosome disruption assays (8, 25). The basis for the differences among these reports remains unknown.
Itc1p is essential for functions of Isw2 complex in vivo.
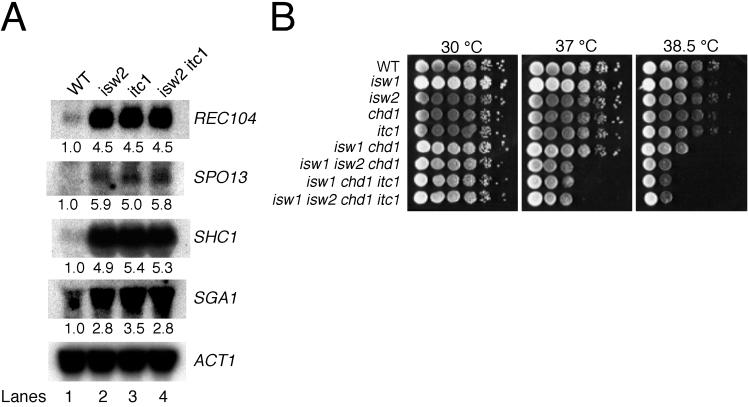
We next tested whether the results of our in vitro experiments are relevant to the physiological functions of Isw2 complex. One of the consequences of an isw2 null mutation is that many genes induced during meiosis are derepressed relative to wild type under mitotic conditions in haploid (13). To test the requirement of ITC1 for Isw2 complex function in vivo, we prepared RNA from vegetatively growing wild-type, isw2, itc1, and isw2 itc1 strains and assayed the expression of several meiotic genes by Northern blotting. As demonstrated previously, expression of REC104, SPO13, SHC1, and SGA1 was increased in isw2 mutants relative to wild-type strains (Fig. 6A, lanes 1 and 2). Each of these genes was derepressed in both itc1 and isw2 itc1 mutants to levels indistinguishable from those of the isw2 single mutant (Fig. 6A, lanes 2 to 4). The observation that isw2 and itc1 null mutants exhibit similar levels of transcriptional derepression indicates that Itc1p is essential for the in vivo function of Isw2 complex, as predicted by in vitro assays. Furthermore, the isw2 itc1 mutant phenotype is no more severe than that of either single mutant, suggesting that both Isw2p and Itc1p function primarily as part of Isw2 complex.
FIG. 6.
Isw2 and Itc1 function in the same pathways. (A) Deletion of ISW2 or ITC1 results in derepression of meiotic genes. Northern blot analysis was performed to compare expression levels of meiotic genes in vegetatively growing cells. The genotype of cells used is listed above each lane; the gene probed is indicated at the right. Numbers below the lanes represent levels of transcription in mutants relative to wild-type cells. ACT1 was used as a loading control. WT, wild type. (B) isw2 and itc1 mutations confer temperature-sensitive growth defects in combination with isw1 and chd1 mutations. Tenfold serial dilutions of saturated wild-type and mutant liquid cultures were spotted onto rich medium (YEPD) and incubated at 30, 37, or 38.5°C.
A second phenotype of isw2 null mutants is synthetic temperature sensitivity in combination with isw1 chd1 null mutations (45). As with isw2 mutants, deletion of the ITC1 gene alone had no obvious effect on growth or viability under the conditions we tested. We then deleted the ITC1 gene in an isw1 chd1 null background and tested the temperature sensitivity of the resulting triple mutant. As shown in Fig. 6B, serial dilutions of isw1 isw2 chd1, isw1 chd1 itc1, and isw1 isw2 chd1 itc1 mutant cultures plated on rich media do not show significant growth defects at 30°C. However, all three mutants show comparable growth defects at 37 and 38.5°C. These data further support our conclusion that ISW2 and ITC1 act in the same pathways. The epistatic relationship of ISW2 and ITC1 observed here is also consistent with our observation that Itc1p and Isw2p do not form any other major complexes in vivo.
This report describes the development of a novel biochemical system to analyze interactions between chromatin proteins and nucleosomal arrays, which was then used to begin dissecting the steps of ATP-dependent chromatin remodeling by Isw2 complex. We found that the interaction of Isw2 complex with naked DNA and nucleosomal arrays is ATP independent and that Itc1p is essential for Isw2 complex function in vitro and in vivo. Furthermore, we have demonstrated the utility of the newly developed chromatin interaction assay in analyzing the interactions of proteins with nucleosomal arrays.
ACKNOWLEDGMENTS
We are grateful to Tom Fazzio, Jesse Goldmark, Cedar McKay, and Jay Vary for helpful discussions, encouragement, and critical reading of the manuscript. We also thank C. Wu and H. Xiao for information regarding biochemical activity of monomeric Drosophila ISWI protein.
This work was supported in part by a Pew Charitable Trust Biomedical Scholars Fellowship and National Institutes of Health grant GM58465 to T.T. and by a grant from the Swiss National Fund for Scientific Research to T.J.R.
REFERENCES
- 1.Almouzni G, Mechali M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988;7:665–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker P B, Tsukiyama T, Wu C. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 3.Belotserkovskaya R, Berger S L. Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit Rev Eukaryot Gene Expr. 1999;9:221–230. doi: 10.1615/critreveukargeneexpr.v9.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 4.Bochar D A, Savard J, Wang W, Lafleur D W, Moore P, Cote J, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci USA. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm A, Langst G, Kehle J, Clapier C R, Imhof A, Eberharter A, Muller J, Becker P B. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 2000;19:4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulger M, Ito T, Kamakaka R T, Kadonaga J T. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc Natl Acad Sci USA. 1995;92:11726–11730. doi: 10.1073/pnas.92.25.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 8.Corona D F, Langst G, Clapier C R, Bonte E J, Ferrari S, Tamkun J W, Becker P B. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 9.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 10.Deuring R, Fanti L, Armstrong J A, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley S L, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun J W. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 11.Du J, Nasir I, Benton B K, Kladde M P, Laurent B C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J Biol Chem. 1992;267:20980–20986. [PubMed] [Google Scholar]
- 13.Goldmark J P, Fazzio T G, Estep P W, Church G M, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 14.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 16.Han M, Kim U J, Kayne P, Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschhorn J N, Bortvin A L, Ricupero-Hovasse S L, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 21.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 22.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 24.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 25.Langst G, Bonte E J, Corona D F, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 26.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 27.LeRoy G, Loyola A, Lane W S, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 28.Lorch Y, Kornberg R D. Isolation of the yeast histone octamer. Proc Natl Acad Sci USA. 1994;91:11032–11034. doi: 10.1073/pnas.91.23.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luger K, Rechsteiner T, Richmond T J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 30.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 31.Peterson C L. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 32.Peterson C L. SWI/SNF complex: dissection of a chromatin remodeling cycle. Cold Spring Harbor Symp Quant Biol. 1998;63:545–552. doi: 10.1101/sqb.1998.63.545. [DOI] [PubMed] [Google Scholar]
- 33.Pilon J, Terrell A, Laybourn P J. Yeast chromatin reconstitution system using purified yeast core histones and yeast nucleosome assembly protein-1. Protein Expr Purif. 1997;10:132–140. doi: 10.1006/prep.1996.0716. [DOI] [PubMed] [Google Scholar]
- 34.Poot R A, Dellaire G, Hulsmann B B, Grimaldi M A, Corona D F, Becker P B, Bickmore W A, Varga-Weisz P D. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Recht J, Osley M A. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandaltzopoulos R, Becker P B. Solid phase DNase I footprinting: quick and versatile. Nucleic Acids Res. 1994;22:1511–1512. doi: 10.1093/nar/22.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider B L, Steiner B, Seufert W, Futcher A B. pMPY-ZAP: a reusable polymerase chain reaction-directed gene disruption cassette for Saccharomyces cerevisiae. Yeast. 1996;12:129–134. doi: 10.1002/(sici)1097-0061(199602)12:2<129::aid-yea891>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 38.Schultz M C, Hockman D J, Harkness T A, Garinther W I, Altheim B A. Chromatin assembly in a yeast whole-cell extract. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 40.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 41.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 42.Sudarsanam P, Iyer V R, Brown P O, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suka N, Carmen A A, Rundlett S E, Grunstein M. The regulation of gene activity by histones and the histone deacetylase RPD3. Cold Spring Harbor Symp Quant Biol. 1998;63:391–399. doi: 10.1101/sqb.1998.63.391. [DOI] [PubMed] [Google Scholar]
- 44.Tse C, Sera T, Wolffe A P, Hansen J C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 47.Tyler J K, Kadonaga J T. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- 48.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 49.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 50.Wade P A, Wolffe A P. Transcriptional regulation: SWItching circuitry. Curr Biol. 1999;9:R221–R224. doi: 10.1016/s0960-9822(99)80134-1. [DOI] [PubMed] [Google Scholar]
- 51.Wechser M A, Kladde M P, Alfieri J A, Peterson C L. Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 53.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 55.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 56.Zhao X, Muller E G, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]