Abstract
Five Arthrobacter isolates from clinical specimens were studied by phenotypic, chemotaxonomic, and genetic characterization. Two strains had characteristics consistent with those of Arthrobacter oxydans. One strain was related to A. citreus; however, DNA-DNA hybridization and phenotypic characteristics indicated that this strain belongs to a new species, for which the name Arthrobacter luteolus sp. nov. is proposed. Two strains were closely related to A. cumminsii by 16S rRNA gene sequencing, but DNA-DNA hybridization, peptidoglycan type, and some phenotypic features indicated that they should be assigned to a new species, for which the name Arthrobacter albus sp. nov. is proposed. The type strain of A. luteolus is CF25 (DSM 13067). The type strain of A. albus is CF43 (DSM 13068).
The genus Arthrobacter includes catalase-positive coryneform bacteria with an oxidative metabolism, the cell wall of which contains l-lysine as the diamino acid and cellular fatty acids of the branched type (6). Arthrobacter strains have only recently been recognized as opportunistic pathogens belonging to three species, A. cumminsii, A. woluwensis, and A. creatinolyticus, which hitherto were isolated only from humans (4, 7). Based on phenotypic, chemotaxonomic, and genetic studies, we describe the species identification of five Arthrobacter isolates. Two of them were assigned to the species A. oxydans, and the three others were assigned to two as-yet-undescribed species for which the names Arthrobacter luteolus sp. nov. and Arthrobacter albus sp. nov. are proposed.
Bacterial strains.
The five strains and the sites from which they were isolated are as follows: CF25, surgical wound; CF39, blood; CF43, blood; CF44, urine; and CF46, blood. Additional Arthrobacter strains were included for comparison in the study: seven A. cumminsii strains, including two laboratory isolates and the reference strains DSM 10493T, DSM 10494, CCUG 33745, CCUG 35863, and CCUG 35241, as well as the type strains A. woluwensis DSM 10495, A. oxydans DSM 20119, and A. citreus DSM 20133. They were inoculated on blood agar and incubated at 37°C.
Phenotypic characterization.
Most cultural, morphologic, and biochemical properties were investigated as described previously (6, 20). Flagellum staining was performed according to the method of Kodaka et al. (12). Gelatin hydrolysis was detected as outlined by Pitt and Dey (15). Oxidative acid production from carbohydrates was detected on phenol red agar slants with a low peptone content as described for acidification of ethylene glycol (20). Pyrrolidonyl peptidase, α-glucosidase, β-galactosidase, and N-acetylglucosaminidase were detected by diagnostic tablets (Rosco Diagnostica, Taastrup, Denmark). Susceptibility to desferrioxamine was tested by the method of Lindsay and Riley (13). API Coryne strips (bioMérieux, Marcy l'Etoile, France) were inoculated and interpreted according to the manufacturer's instructions. Carbohydrate assimilations were performed with the API 50 CH system using AUX medium (bioMérieux). Results were recorded after 5 days. API ZYM strips (bioMérieux) were read after 4 h of incubation at 37°C.
Antibiotic susceptibility.
To evaluate antibiotic susceptibility, MICs were determined using E-test strips (PDM, Solna, Sweden) on Mueller-Hinton blood agar incubated at 37°C for 24 h. Penicillin, ampicillin, cefotaxime, cephalothin, erythromycin, ciprofloxacin, gentamicin, and vancomycin were tested, and the results were interpreted according to the criteria established for staphylococci in 1997 by the National Committee for Clinical Laboratory Standards (14a).
Chemotaxonomic properties.
Cellular fatty acids (CFAs) were assayed by gas-liquid chromatography using a Delsi chromatograph as described previously (19). The amino acid composition of the peptidoglycan was studied by N. Weiss at the German Collection of Microorganisms and Cell Cultures (DSMZ) (Braunschweig, Germany) by a thin-layer chromatography method as outlined by Schleifer and Kandler (18).
16S rRNA gene sequence determination.
Determination of the 16S rRNA gene sequence was performed at the DSMZ by C. Sproër. Approximately 95% of the 16S rRNA gene sequence of the strains was determined by direct sequencing of PCR-amplified 16S ribosomal DNA. Genomic DNA extraction, PCR-mediated amplification of the 16S ribosomal DNA, and purification of the PCR products were carried out as previously described (16). Purified PCR products were sequenced using the ABI Prism dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Weiterstadt, Germany) according to the manufacturer's protocol. Sequencing products were electrophoresed using the Applied Biosystems 373A DNA sequencer. The resulting sequence data were put into the alignment editor ae2 (14), aligned manually, and compared with representative 16S rRNA gene sequences of organisms belonging to the Arthrobacter subgroup of the gram-positive bacteria (14). For comparison, 16S rRNA sequences were obtained from the EMBL database or the Ribosomal Database Project (14). The 16S rRNA gene similarity values were calculated by pairwise comparison of the sequences within the alignment. For construction of a phylogenetic dendrogram, operations of the PHYLIP package (version 3.5.1; J. Felsenstein, Seattle Department of Genetics, University of Washington) were used: pairwise evolutionary distances were computed from percent similarities by the correction method of Jukes and Cantor (10), and based on the evolutionary distance values, the phylogenetic tree was constructed by the neighbor-joining method (17). The root of the tree was determined by including the 16S rRNA gene sequence of Nocardioides simplex in the analysis.
DNA-DNA hybridization.
Hybridization was carried out at the DSMZ. DNA was isolated by chromatography on hydroxyapatite by the procedure of Cashion et al. (1). DNA-DNA hybridization was performed as described by De Ley et al. (2) with the modification described by Huss et al. (8) and Escara and Hutton (3), using a Gilford System model 2600 spectrometer equipped with a Gilford model 2527-R thermoprogrammer and plotter. Renaturation rates were computed with the TRANSFER.BAS program of Jahnke (9).
Results and discussion.
The five strains exhibited the general characteristics of the genus Arthrobacter. They were gram-positive coryneform bacteria with a nonfermentative metabolism. The CFAs were mainly of the branched type, with a predominant amount of anteiso 15:0 (Table 1). l-Lysine was the diamino acid of the peptidoglycan. Phenotypic, chemotaxonomic, and genetic studies based on 16S rRNA gene sequence determination and DNA-DNA hybridization allowed us to define three clusters among the five isolates.
TABLE 1.
Main CFA profile of human Arthrobacter isolates
| Strain | % of total CFAs
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| i 14:0 | 14:0 | i 15:0 | ai 15:0 | i 16:0 | 16:0 | i 17:0 | ai 17:0 | 18:0 | |
| CF25 | 0.7 | 0.6 | 15 | 72.0 | 2.5 | 1.6 | 0.4 | 4.6 | 0.3 |
| CF43 | 1.7 | tr | 8.9 | 61.8 | 10.1 | 1.9 | 0.8 | 11.8 | 0.7 |
| CF44 | 1.3 | tr | 4.5 | 61.3 | 11.3 | 2.3 | 0.5 | 16.5 | 0.7 |
| CF39 | 1.4 | 3 | 20.0 | 50.0 | 5.0 | 4.7 | 1.4 | 9.7 | tr |
| CF46 | 1.1 | 1.8 | 13.0 | 43.0 | 6.9 | 6.6 | 1.5 | 11.8 | 4.3 |
ai, anteiso; tr, trace.
Strains CF39 and CF46 had a peptidoglycan of the A3α type, l-Lys–l-Ser–l-Thr–l-Ala, found in A. oxydans (11). The 16S rRNA gene sequencing performed on CF46 showed a similarity of 99.5% to the type strain of A. oxydans, DSM 20119 (Table 2). DNA-DNA hybridization of both strains CF46 and CF39 with DSM 20119T resulted in homologies of 87.5 and 85%, respectively. CF39 was 92.5% related to CF46. Furthermore, the biochemical properties of both strains were consistent with those described for A. oxydans (4), and the API Coryne code was 3750004 (Arthrobacter spp.). These findings allow us to assign strains CF46 and CF39 to the species A. oxydans. The strains were susceptible to all antibiotics tested.
TABLE 2.
Levels of 16S rRNA sequence similarity between A. oxydans (CF46), A. luteolus sp. nov. (CV25), A. albus sp. nov. (CF43), and other Arthrobacter species and related taxa
| Species | % 16S rRNA similarity with:
|
||
|---|---|---|---|
| CF46 | CF25 | CF43 | |
| Arthrobacter agilis DSM 20550T | 95.5 | 96.4 | 95.0 |
| Arthrobacter atrocyaneus DSM 20127T | 93.9 | 94.6 | 94.5 |
| Arthrobacter aurescens DSM 20116T | 97.4 | 97.7 | 94.7 |
| Arthrobacter citreus DSM 20133T | 95.7 | 98.7 | 94.6 |
| Arthrobacter creatinolyticus JCM 10102T | 94.7 | 94.9 | 95.9 |
| Arthrobacter crystallopoietes DSM 20117T | 96.0 | 97.0 | 96.0 |
| Arthrobacter cumminsii DSM 10493T | 94.4 | 95.5 | 99.5 |
| Arthrobacter globiformis DSM 20124T | 97.4 | 96.0 | 96.0 |
| Arthrobacter histidinolovorans DSM 20115T | 97.7 | 96.6 | 94.9 |
| Arthrobacter ilicis DSM 20138T | 96.8 | 97.0 | 94.0 |
| Arthrobacter nicotianae DSM 20123T | 96.4 | 94.4 | 95.4 |
| Arthrobacter nicotinovorans DSM 420T | 98.0 | 96.9 | 95.2 |
| Arthrobacter oxydans DSM 20119T | 99.5 | 96.4 | 94.9 |
| Arthrobacter pascens DSM 20545T | 98.0 | 95.7 | 95.6 |
| Arthrobacter polychromogens DSM 20136T | 99.1 | 96.4 | 94.8 |
| Arthrobacter protophormiae DSM 20168T | 95.7 | 94.7 | 95.0 |
| Arthrobacter ramosus DSM 20546T | 98.0 | 95.7 | 95.6 |
| Arthrobacter rhombi CCUG 38813T | 95.3 | 95.0 | 95.6 |
| Arthrobacter sulfureus DSM 20167T | 95.7 | 95.0 | 94.4 |
| Arthrobacter ureafaciens DSM 20126T | 96.9 | 96.8 | 95.6 |
| Arthrobacter uratoxydans DSM 20647T | 95.0 | 94.3 | 94.6 |
| Arthrobacter woluwensis DSM 10495T | 96.2 | 95.7 | 94.9 |
| Renibacterium salmoninarum ATCC 33209T | 94.8 | 94.2 | 94.3 |
| Micrococcus luteus DSM 20030T | 95.9 | 94.9 | 94.7 |
| Micrococcus lylae DSM 20315T | 95.8 | 94.5 | 94.6 |
| Rothia dentocariosa ATCC 17931 | 94.1 | 93.6 | 93.5 |
| Dermatophilus congolensis ATCC 14637T | 92.9 | 92.2 | 92.1 |
| Stomatococcus mucilaginosus DSM 20746T | 94.4 | 94.3 | 93.9 |
| Kocuria kristinae DSM 20032T | 94.0 | 94.7 | 95.7 |
| Kytococcus sedentarius DSM 2074T | 92.0 | 92.4 | 93.6 |
| Jonesia denitrificans DSM 20603T | 91.5 | 91.0 | 92.5 |
| Nocardioides simplex DSM 20130T | 89.3 | 88.6 | 87.8 |
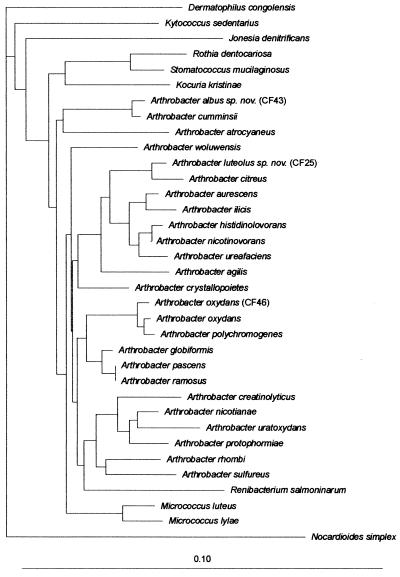
Strain CF25 represented the second cluster. It had a cell wall of the A3α type, l-Lys–l-Thr–l-Ala2, which is present in A. citreus (11). The 16S rRNA gene sequencing revealed the highest similarity to A. citreus DSM 20133T, at a level of 98.7% (Table 2). However, when DNA-DNA hybridization was performed, it appeared that CF25 had a homology of only 44.0% with the type strain of A. citreus. Colonies grown on solid media at 37°C were slightly yellow. Urease, Simmons citrate, and pyrrolidonyl peptidase were negative but gelatin, DNase, and tyrosine hydrolysis were positive. Nitrate reduction was positive. There was no fermentation of sugars, but glucose was oxidatively acidified, as were sucrose, maltose, and xylose. The following carbohydrates were used with the API 50 CH strips: glucose, ribose, d-xylose, d-fructose, d-mannose, rhamnose, cellobiose, maltose, sucrose, trehalose, xylitol, l-fucose, gluconate, and 5-keto-gluconate. The other substrates of the API 50 CH strips were not utilized. By the API ZYM system, only alkaline phosphatase, esterase, esterase-lipase, leucine arylamidase, acid phosphatase, phosphoamidase, and α-glucosidase were detected. The strain was resistant to desferrioxamine. The API Coryne code was 3110004, corresponding to Rhodococcus spp. with a probability of 94.1%. The strain was susceptible to all antibiotics tested except cephalosporins. Although CF25 is yellow pigmented and has the same peptidoglycan type as A. citreus, it is clearly distinct from this species on the basis of DNA-DNA hybridization, notwithstanding a close relationship of the 16S rRNA gene sequences (Table 2). Phenotypic characteristics also distinguish CF25 from A. citreus. Strain CF25 grows well at 37°C, while A. citreus does not. A. citreus does not assimilate any carbohydrate in the API 50 CH system, while CF25 utilizes several of them. Other distinctive biochemical reactions are listed in Table 3. These findings suggest that CF25 belongs to a new species within the genus Arthrobacter, for which the name A. luteolus is proposed. It is closely related to A. citreus (Fig. 1).
TABLE 3.
Distinctive characteristics of Arthrobacter species of human origina
| Speciesb | nc | Mot | 20° | 42°C | Des | Gel | DNa | Twe | Tyr | Citr | NO3 | Glu | Mnt | Pyr | β-Gal | α-Glu | Aga |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. cumminsii | 7 | − | v | + | S | + | +w | − | − | − | − | − | − | + | − | − | − |
| A. albus (CF43, CF44) | 2 | − | − | + | R | +w | − | − | − | − | − | − | − | + | − | − | − |
| A. woluwensis | 1 | − | + | + | R | + | + | + | + | + | − | + | + | + | + | + | + |
| A. oxydans (CF39, CF46) | 3 | − | + | v | R | + | + | v | + | + | + | + | + | − | + | + | − |
| A. luteolus (CF25) | 1 | + | + | + | R | + | + | − | + | − | + | + | − | − | − | + | − |
| A. citreus | 1 | +w | + | − | R | + | + | + | + | − | + | + | + | − | + | + | − |
Mot, motility; 20°C and 42°C, growth at 20°C and 42°C; Des, desferrioxamine; Gel, gelatin, DNa, DNase; Twe, Tween 80; Tyr, decomposition of tyrosine; Citr, citrate; NO3, nitrate, Glu and Mnt, acid from glucose and mannitol; Pyr, pyrrolidonyl peptidase; β-Gal, β-galactosidase; α-Glu, α-glucosidase; Aga, N-acetylglucosaminidase; +, positive; +w, weak reaction; −, negative; v, variable, S, susceptible; R, resistant.
The nonhuman type strains A. oxydans DSM 20119T and A. citreus DSM 20133T were included in the strains tested. Tests for A. citreus were performed at 30°C.
Number of strains analyzed.
FIG. 1.
Unrooted tree showing the phylogenetic positions of A. oxydans (CF46), A. luteolus sp. nov. (CF25), and A. albus sp. nov. (CF43) within the genus Arthrobacter and related taxa. The scale bar indicates 10 nucleotide substitutions per 100 nucleotides.
The third cluster consisted of two strains: CF43 and CF44. Their cell wall was of the A4α type, l-Lys–l-Ala–l-Glu, similar to that of the A. nicotianae group (11). 16S rRNA gene sequencing of CF43 showed the highest similarity, 99.5%, to A. cumminsii (Table 2). However, the cell wall of A. cumminsii is markedly different (5). DNA-DNA hybridization was carried out on strains CF43 and CF44 and the type strain of A. cumminsii, DSM 10493. CF43 and CF44 displayed a similarity of 88.1% to each other, suggesting that the two strains are members of the same species. The two strains showed only 54.5 and 58.6% homology, respectively, to the type strain of A. cumminsii. The two strains shared many phenotypic characteristics with A. cumminsii. Gram staining showed small coryneform bacteria. The colonies were white and opaque and reached 1 mm after 48 h of incubation at 37°C, while the colonies of A. cumminsii are grayish and less opaque. Urease, nitrate reduction, esculin hydrolysis, tyrosine degradation, and DNase were negative. Gelatin was delayed and weakly positive by the gelatin agar method, but with the API Coryne system, CF43 liquefied gelatin after 1 day, whereas CF44 remained negative even after 5 days. There was no susceptibility to desferrioxamine, unlike with A. cumminsii. Pyrrolidonyl peptidase was positive. No sugar acidification occurred within 5 days, and none of the carbohydrates included in the API 50 CH panel were utilized. The enzymatic profile obtained by API ZYM strips was similar to that of A. cumminsii (4). The API Coryne codes were 6102004 (Brevibacterium spp.) for strain CF43 and 6100004 (Corynebacterium afermentans or Corynebacterium coyleae) for CF44. The strains were susceptible to the antibiotics tested. These findings justify assigning strains CF43 and CF44 to a new species within the genus Arthrobacter, for which the name A. albus is proposed. It is phylogenetically related to A. cumminsii (Fig. 1).
The small number of strains available for most named species of Arthrobacter of human origin does not allow establishment of a definitive profile of their phenotypic properties, especially for those represented by only one strain. Nevertheless, a provisional scheme of distinctive characteristics has been developed based on our clinical isolates and reference strains (Table 3). Although these results have to be confirmed in the future, some of the proposed tests seem to have a discriminant value. Susceptibility to desferrioxamine is specific to A. cumminsii. A strong N-acetylglucosaminidase activity is found only in A. woluwensis. Tyrosine clearing and α-glucosidase are regularly observed in the glucolytic species, and within this group, only A. luteolus does not acidify mannitol and is β-galactosidase negative.
The pathologic significance of Arthrobacter isolates in humans has yet to be assessed. In our series, one strain, A. albus CF43, was clinically relevant, as it was isolated from several cultures of blood from a surgery patient with severe phlebitis. The other strains were isolated from either a superficial body site, urine, or one single blood culture bottle, and their role in the disease was not proved. Nevertheless, they were not considered to be environmental contaminants since, except in blood cultures, numerous colonies were present at primary isolation. There is a need to increase the number of human isolates belonging to new Arthrobacter species, to define more accurately their phenotypic characteristics. Greater recognition of coryneform bacteria in clinical samples may contribute to better understanding of their role in human disease.
Description of A. luteolus sp. nov.
Cells of A. luteolus (lu-te'-o-lus. N.L. adj. meaning yellowish, because of the yellow-pigmented colonies) are gram-positive coryneform bacteria. No spores are formed. They are motile by peritrichous flagella. Growth is obligately aerobic. Colonies are slightly yellow, smooth, and ∼1.5 mm in diameter after 24 h of incubation at 37°C on blood agar. Catalase is positive. Nitrate is reduced. No urease is detected. Gelatin and tyrosine are hydrolyzed, but not esculin. Simmons citrate agar is alkalinized. There is no susceptibility to desferrioxamine. Tween esterase is negative, but DNase is positive. Pyrrolidonyl peptidase is not detected. Acid is produced oxidatively from glucose, maltose, sucrose, and xylose, but not from mannitol and lactose. Glycerol, ribose, d-xylose, d-glucose, d-fructose, d-mannitol, rhamnose, cellobiose, maltose, saccharose, trehalose, xylitol, l-fucose, and 5-keto-gluconate are utilized. Erythritol, d-arabinose, l-arabinose, l-xylose, adonitol, β-methylxyloside, galactose, l-sorbose, dulcitol, inositol, mannitol, sorbitol, α-methyl-d-mannoside, α-methyl-d-glucoside, N-acetylglucosamine, amygdalin, arbutin, esculin, salicin, lactose, melibiose, inulin, melezitose, d-raffinose, starch, glycogen, geniobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, d-arabitol, l-arabitol, and 2-keto-gluconate are not utilized. The following enzymatic activities are detected on API ZYM strips: alkaline and acid phosphatase, esterase, esterase-lipase, leucine arylamidase, trypsin, phosphoamidase, and α-glucosidase. Not present are lipase, valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetylglucosaminidase, α-mannosidase, and α-fucosidase. The major CFA is anteiso C15:0, and the peptidoglycan type is l-Lys–l-Thr–l-Ala2. The type strain, CF25, has been deposited in the DSMZ as strain DSM 13067. It was isolated from an infected surgical wound.
Description of A. albus sp. nov.
Cells of A. albus sp. nov. (al'-bus. L.N. adj. meaning white, because of the white colonies of the organism) are small coryneform bacteria, nonmotile and without spore formation. Colonies are white and are 1 mm in diameter after 48 h of incubation at 37°C on blood agar. Metabolism is obligately aerobic. Catalase is positive. Urease and esculin are not hydrolyzed, and nitrates are not reduced. Gelatin is slowly and weakly liquefied, and there is no DNase activity. Tyrosine is not hydrolyzed. Simmons citrate is negative. Pyrrolidonyl peptidase is positive. The organism is resistant to desferrioxamine. No acid is produced from carbohydrates, and there is no utilization of these substrates in the API 50 CH system. The following enzymatic activities are detected: alkaline and acid phosphatase, phosphoamidase, esterase, esterase-lipase, leucine arylamidase, and trypsin. The following activities are not present: lipase, valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetylglucosaminidase, α-mannosidase, and α-fucosidase. The main CFA is anteiso C15:0, and the peptidoglycan type is l-Lys–l-Ala–l-Glu. The strains were isolated from human clinical specimens. The type strain, CF43, has been deposited in the DSMZ as strain DSM 13068. It was isolated from human blood.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the following strains are deposited in the EMBL Data Library under the indicated accession numbers: CF25 (DSM 13067T), no. AJ243422; CF43 (DSM 13068T), no. AJ243421; and CF46 (DSM 13066), no. AJ243423.
REFERENCES
- 1.Cashion P, Holder-Franklin M A, McCully J, Franklin M. A rapid method for base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- 2.De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 3.Escara J F, Hutton J R. Thermal stability and renaturation of DNA in dimethylsulphoxide solutions: acceleration of renaturation rate. Biopolymers. 1980;19:1315–1327. doi: 10.1002/bip.1980.360190708. [DOI] [PubMed] [Google Scholar]
- 4.Funke G, Hutson R A, Bernard K A, Pfyffer G E, Wauters G, Collins M D. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J Clin Microbiol. 1996;34:2356–2363. doi: 10.1128/jcm.34.10.2356-2363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funke G, Pagano-Niederer M, Sjödén B, Falsen E. Characteristics of Arthrobacter cumminsii, the most frequently encountered Arthrobacter species in human clinical specimens. J Clin Microbiol. 1998;36:1539–1543. doi: 10.1128/jcm.36.6.1539-1543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funke G, von Graevenitz A, Clarridge III J E, Bernard K A. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou X-G, Kawamura Y, Sultana F, Shu S, Hirose K, Goto K, Ezaki T. Description of Arthrobacter creatinolyticus sp. nov., isolated from human urine. Int J Syst Bacteriol. 1998;48:423–429. doi: 10.1099/00207713-48-2-423. [DOI] [PubMed] [Google Scholar]
- 8.Huss V A R, Festl H, Schleifer K H. Studies on the spectrometric determination of DNA hybridization from renaturation rates. J Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- 9.Jahnke K-D. Basic computer program for evaluation of spectroscopic DNA renaturation data from GILFORD System 2800 spectrometer on a PC/XT/AT type personal computer. J Microbiol Methods. 1992;15:61–73. [Google Scholar]
- 10.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 11.Koch C, Schumann P, Stackebrandt E. Reclassification of Micrococcus agilis (Ali-Cohen 1889) to the genus Arthrobacter as Arthrobacter agilis comb. nov. and emendation of the genus Arthrobacter. Int J Syst Bacteriol. 1995;45:837–839. doi: 10.1099/00207713-45-4-837. [DOI] [PubMed] [Google Scholar]
- 12.Kodaka H, Armfield A Y, Lombard G L, Dowell V R., Jr Practical procedure for demonstrating bacterial flagella. J Clin Microbiol. 1982;16:948–952. doi: 10.1128/jcm.16.5.948-952.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsay J A, Riley T V. Susceptibility to desferrioxamine: a new test for the identification of Staphylococcus epidermidis. J Med Microbiol. 1991;35:45–48. doi: 10.1099/00222615-35-1-45. [DOI] [PubMed] [Google Scholar]
- 14.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.National Committee for Clinical Laboratory Standards. Minimum inhibitory concentration (MIC) interpretive standards (μg/ml) for organisms other than Haemophilus spp., Neisseria gonorrhoeae, and Streptococcus spp. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 15.Pitt T L, Dey D. A method for the detection of gelatinase production by bacteria. J Appl Bacteriol. 1970;33:687–691. doi: 10.1111/j.1365-2672.1970.tb02251.x. [DOI] [PubMed] [Google Scholar]
- 16.Rainey F A, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol. 1996;46:1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wauters G, Driessen A, Ageron E, Janssens M, Grimont P A D. Propionic acid-producing strains previously designated as Corynebacterium xerosis, C. minutissimum, C. striatum, and CDC group I2 and group F2 coryneforms belong to the species Corynebacterium amycolatum. Int J Syst Bacteriol. 1996;46:653–657. [Google Scholar]
- 20.Wauters G, Van Bosterhaut B, Janssens M, Verhaegen J. Identification of Corynebacterium amycolatum and other nonlipophilic fermentative corynebacteria of human origin. J Clin Microbiol. 1998;36:1430–1432. doi: 10.1128/jcm.36.5.1430-1432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]