Abstract
The telomere specific shelterin complex, which includes TRF1, TRF2, RAP1, TIN2, TPP1 and POT1, prevents spurious recognition of telomeres as double-strand DNA breaks and regulates telomerase and DNA repair activities at telomeres. TIN2 is a key component of the shelterin complex that directly interacts with TRF1, TRF2 and TPP1. In vivo, the large majority of TRF1 and TRF2 are in complex with TIN2 but without TPP1 and POT1. Since knockdown of TIN2 also removes TRF1 and TRF2 from telomeres, previous cell-based assays only provide information on downstream effects after the loss of TRF1/TRF2 and TIN2. Here, we investigated DNA structures promoted by TRF2–TIN2 using single-molecule imaging platforms, including tracking of compaction of long mouse telomeric DNA using fluorescence imaging, atomic force microscopy (AFM) imaging of protein–DNA structures, and monitoring of DNA–DNA and DNA–RNA bridging using the DNA tightrope assay. These techniques enabled us to uncover previously unknown unique activities of TIN2. TIN2S and TIN2L isoforms facilitate TRF2-mediated telomeric DNA compaction (cis-interactions), dsDNA–dsDNA, dsDNA–ssDNA and dsDNA–ssRNA bridging (trans-interactions). Furthermore, TIN2 facilitates TRF2-mediated T-loop formation. We propose a molecular model in which TIN2 functions as an architectural protein to promote TRF2-mediated trans and cis higher-order nucleic acid structures at telomeres.
INTRODUCTION
Telomeres are nucleoprotein structures that protect the ends of linear chromosomes by preventing them from activating DNA damage response and DNA double-strand break (DSB) repair pathways (1,2). Human telomeric DNA consists of ∼2–20 kb of TTAGGG repeats and a G-rich 3′ overhang (1,3). In humans, the shelterin complex, including TRF1, TRF2, RAP1, TIN2, TPP1 and POT1, forms complexes on telomeric DNA and shields telomeres from being spuriously recognized as double-strand DNA breaks (4–9). Among the shelterin proteins, TRF1 and TRF2 directly bind double-stranded telomeric DNA through the Myb/SANT domain, and POT1 directly binds to the single-stranded overhang through the OB domain (10–12). TRF1 and TRF2 differ at their N-termini (13), containing an acidic and a basic domain, respectively (13). TRF1 and TRF2 play distinct roles in promoting higher-order DNA structures at telomeres. TRF2 displays a unique activity in promoting T-loops, in which the 3′ single-strand overhang invades the upstream double-stranded telomeric region (14–17). The TRFH domain of TRF2 mediates the wrapping of dsDNA, which was proposed to be a critical step in T-loop formation (16). Furthermore, TRF2 recruits RAP1 to telomeres, and in turn, RAP1 enhances TRF2’s binding specificity for telomeric DNA (18–20). Consistent with its activity in promoting T-loop formation, TRF2 plays critical roles in regulating Mre11/Rad50/Nbs1-dependent ataxia-telangiectasia mutated (ATM) kinase signaling, classical nonhomologous end-joining (NHEJ) and alternative nonhomologous end-joining (alt-NHEJ) pathways at telomeres. In comparison, TRF1 promotes parallel pairing of telomeric DNA (21–23). TRF1 prevents DNA replication fork stalling at telomeres and contributes to the repression of telomere fragility (24).
TIN2 is the linchpin among the shelterin proteins and mediates the localization of three key DNA interacting proteins at telomeres (TRF1, TRF2 and POT1). TIN2 interacts with both TRF1 and TRF2 proteins and stabilizes them at telomeres (25,26). TRF2 and TIN2 form a complex with 2:1 stoichiometry (27). Deletion of TIN2 is embryonically lethal (28), and conditional knockout of TIN2 in mouse embryo fibroblasts (MEFs) leads to growth arrest. TIN2 depletion elicits a moderate level of chromosome-type fusions, a phenotype linked to TRF2 loss from telomeres. TIN2 loss also mimics the phenotypes of TPP1/POT1 deletion. TIN2 depletion leads to excess 3′ overhangs and fusion of duplicated chromatids that have been linked to POT1 or TPP1 deficiency (29). Consequently, TIN2 deletion elicits a DNA damage response involving both ataxia telangiectasia and Rad3-related (ATR) and ATM kinases (29).
Higher-order DNA structures play essential roles in telomere maintenance (30). T-loop formation plays a critical role in telomere protection as it enables cells to distinguish between native chromosome ends and aberrant double-stranded DNA breaks (15–17). STochastic Optical Reconstruction Microscopy (STORM) imaging of T-loops from MEFs showed that TRF2 is required for T-loop formation and maintenance (15). However, this study did not specifically test the impact of TIN2 toward T-loop formation as TIN2 loss leads to the removal of both TRF2 and POT1 from telomeres (29). A recent study evaluating the effect of TIN2 on T-loop formation was based on the comparison between the T-loop frequencies from cells overexpressing WT TRF2 and a TRF2 mutant missing its TIN2 and RAP1 binding sites (31). This study found no difference in T-loop frequencies in cells expressing wild-type TRF2 and TRF2 mutant containing deletions of its TIN2 and RAP1 binding sites. However, the authors acknowledged that the high expression levels of exogenous TRF2 and elongated 3′ overhangs after removing TIN2 might mask the defect in the T-loop formation due to the removal of TIN2 from telomeres.
Telomeric Repeat-containing RNA (TERRA) is expressed within a subset of subtelomeric regions in the proximity of the telomeric repeats (32–34). TERRA can be directly involved in telomere maintenance through association with telomeric chromatin forming telomeric R-loops. An R-loop is a three-stranded nucleic acid structure containing nascent RNA hybridized with its corresponding DNA template strand and the displaced ssDNA (35). It was recently shown that TERRA R-loops accumulate preferentially at short telomeres, contributing to the activation of DNA damage response (36,37). Telomeric R-loops support the telomerase-independent Alternative Lengthening of Telomeres (ALT) mechanism by promoting telomere recombination (38). TRF1 and TRF2 directly bind to TERRA. Furthermore, TRF2 promotes RNA invasion of double-stranded telomeric DNA to form telomeric R-loops (39,40). However, whether TIN2 directly enhances TRF2-mediated R-loop formation has not been investigated.
In human and mouse cells, TRF1, TRF2, TIN2 and RAP1 are approximately ten times more abundant than POT1 and TPP1 (41). The majority (∼90%) of the TRF1 and TRF2 proteins exist in complexes with TIN2 but lacking POT1 and TPP1. Consequently, investigating how TIN2 directly regulates TRF2–DNA binding is key to advancing our understanding of shelterin-mediated telomere maintenance. However, since the loss of TIN2 removes TRF1 and TRF2 from telomeres, it is technically challenging to identify all aspects of TIN2 functions from cell-based assays. Furthermore, despite the importance of higher-order DNA structures at telomeres, our understanding of how shelterin proteins other than TRF2 contribute to T-loop and telomeric R-loop formation is still limited. While ensemble-based bulk assays report the average properties of heterogeneous protein–DNA complexes, single-molecule imaging techniques can reveal protein-induced DNA conformations at specific DNA sequences, such as DNA compaction, DNA–DNA bridging and T-loop formation (16,42,43). Thus, there is a clear need to investigate the impact of TIN2 on TRF2–DNA binding at the single-molecule level using a defined in vitro system involving purified proteins. In this study, we focused on investigating whether or not TIN2 (TIN2S and TIN2L isoforms) regulates TRF2–DNA binding by applying single-molecule imaging platforms, including atomic force microscopy (AFM) (43–45) and fluorescence imaging of TRF2–TIN2 on telomeric DNA (42,46,47). The substrates include long native telomeric DNA purified from mouse liver tissues that contain same human telomeric DNA sequences and DNA tightropes containing regions with 270 TTAGGG repeats. We directly compared TRF2 and TRF2–TIN2 mediated telomeric DNA compaction, bridging and T-loop formation. These imaging platforms provide complementary results demonstrating that both TIN2S and TIN2L facilitate TRF2-mediated DNA compaction (cis-interactions), as well as dsDNA–ssDNA and dsDNA–ssRNA bridging (trans-interactions) in a telomeric sequence-dependent manner. Furthermore, TIN2 increases the TRF2-mediated T-loop formation frequency. This work sheds new light on the previously unknown biophysical functions of TIN2 and provides a new framework for future investigation of diverse functions of TIN2 in telomere maintenance.
MATERIALS AND METHODS
Protein purification
N-terminal His6-tagged TRF1 and TRF2 were purified using a baculovirus/insect cell expression system and an AKTA Explorer FPLC (GE Healthcare) as reported previously (48). Extensive use and reports of His-tagged TRF2 and HA-tagged TIN2 in the literature provide evidence that the tag does not interfere with protein activities or introduce DNA binding artifacts (8,16,27,49). TRF2 concentrations were determined using the Bradford assay. N-terminal HA-tagged TIN2L (HA-TIN2L, 1–451 aa) and TIN2S (HA-TIN2S, 1–354 aa) were expressed in the Sf9 insect cells using the pFastBac1 expression system (GenScript). The Sf9 cells infected with P2 virus were cultured in the Sf-900 I SFM medium (Gibco) for 3 days at 27°C and harvested at 72 h post-infection. Cell pellets were lysed in the cell lysis buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% glycerol, 1 mM PMSF, 12.5 μg/ml leupeptin, 6.25 μg/ml aprotinin and 2.5 μg/ml pepstain). HA-TIN2S and HA-TIN2L were purified using the HA-antibody agarose and stored in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 1% NP-40 and 1 mM EDTA. The concentrations of TIN2 proteins were determined by the BCA™ protein assay using BSA as the standard (ThermoFisher). The identities of purified HA-TIN2S and HA-TIN2L proteins were further established by the western Blot analysis using the HA antibody (GenScript A00168) and MALDI-TOF mass spectrometry analysis (UNC-Chapel Hill Proteomics Center). TIN2S and TIN2L were active in interacting with TRF1 based on electrophoresis mobility shift assays (EMSAs) using the telomeric DNA substrate containing three TTAGGG repeats (50). Mitochondrial single-stranded binding protein (mtSSB) was a gift from the Copeland laboratory (NIEHS) (51). T7 exonuclease was purchased from NEB.
DNA substrates
λ DNA and the pSXneo (T2AG3) plasmid DNA containing 1.6 kb telomeric TTAGGG repeats with a 23-bp nontelomeric sequence linking two (TTAGGG)135 regions (T270 DNA) were purchased from New England BioLabs (NEB) and Addgene, respectively (52). For AFM imaging, linear T270 DNA fragments were generated by digestion of T270 plasmid DNA at 37°C for 4 h using BglII in Buffer 4 (NEB) and further purification using the PCR DNA Purification Kit (Qiagen). The linearized T270 DNA for the DNA tightrope assay was ligated using the Quick Ligation™ Kit (NEB) at room temperature for 1 h followed by overnight incubation at 4°C. Ligated T270 DNA was purified using phenol-chloroform extraction. The biotinylated linear T270 DNA and control DNA substrates were generated through biotinylation of the gel-purified T270 fragment containing the (TTAGGG)270 region (pT270, 1.6 kb) or control nontelomeric DNA (noTel, 4.1 kb) using the 5′ EndTag Labeling DNA/RNA Kit (Vector Laboratories). Biotinylation efficiencies of the pT270 and noTel fragments were established using AFM imaging of DNA samples in the presence of streptavidin-coated quantum dots (strep-QDs, Invitrogen) (50). Single-stranded TTAGGG-12 DNA and UUAGGG-12 RNA containing 12 telomeric repeats and 5′ biotin labels were purchased from IDT.
To generate the model T-loop substrate, the linear T270 fragment (5.4 kb, 28 nM) was incubated with the T7 exonuclease (12.5 U) for 6 min on ice. The reaction was stopped by the addition of 11 mM EDTA. T7 exonuclease-treated linear T270 DNA was purified using the PCR DNA Purification Kit (Qiagen). Next, to mark the nontelomeric DNA end with QDs, the linear T270 DNA (2.8 nM) was incubated with a biotinylated primer (5′ biotin-TAT AGT GTC ACC TAA ATC GTA TGT GTA TGA TAC 3′, 11 nM) complementary to the nontelomeric 3′ overhang at 65°C for 5 min, followed by slow cooling to room temperature. Finally, strep-QDs (11 nM) were added to the sample to label the nontelomeric DNA end on the linear T270 DNA with a QD.
Purification of telomeric DNA from mouse liver tissues
Genomic DNA was isolated from C57B/6J mouse liver tissue using the Puregene DNA Isolation Kit (Qiagen) based on the standard protocol from the manufacturer. Approximately 0.5 g of mouse liver tissues yielded ∼2.5 mg of genomic DNA. Mouse genomic DNA (650 μg) was digested using a restriction enzyme cocktail containing HinfI, AluI, HphI and MnlI (NEB) overnight at 37°C (3,53,54). Region Specific Extraction (RSE) Purification Kit (Generation Biotech) was used to extract telomeric DNA from the digested genomic DNA (55). Digested genomic DNA was incubated with 2-, 5- or 10-fold molar excess of biotinylated 3′ telomeric overhang-complementary oligo (5′ biotin-ACT CCC CCT AAC CCT AAC CCT AAC CCT AAC CCT AAC CCT AAC CCT AAC CCT AA 3′) in the RSE H-buffer containing DNA polymerase and biotinylated dNTPs. The PCR reaction was carried out with the denaturation step at 92°C for 5 min, followed by the primer extension step at 64°C for 15 min. Streptavidin-coated magnetic beads were added to the PCR reaction at room temperature and incubated for another 20–30 min. Beads were washed twice with 200 μl of RSE Buffer before further incubation at 82°C for 15 min in a water bath, followed by slow cooling to room temperature. The concentrations of extracted telomeric DNA samples were measured using a NanoDrop Microvolume Spectrophotometer (Thermofisher).
Validation of the purity of telomeric DNA obtained using the RSE method
Detection of telomeric and genomic DNA containing the Alu repeat was performed using the GE Manifold spot blot apparatus based on a method described previously (54). For each experiment 7.5 ng of RSE-purified telomeric DNA was loaded with the corresponding genomic DNA (1, 5, 7.5, 10, 20 and 50 ng) in duplicates. The telomeric probes 5′-(TTAGGG)4–3′ and 5′-(CCCTAA)4–3′ and the Alu probe (5′-GGGAGGCCGAGGCGGGCGGA-3′) were radiolabeled as described previously (54). The membranes were incubated at 42°C for 30 min, in the hybridization buffer (Church Buffer: 0.25 M sodium phosphate buffer pH 7.2, 1 mM EDTA, 1% BSA, 7% SDS), and then overnight with the hybridization buffer containing the radiolabeled probe. The membranes were washed once in a buffer containing 2× SSC, once with 0.1% SDS and 0.1× SSC, and once more with 2× SSC before visualization on a Typhoon phosphorimager. Quantification of the signal intensities was done using ImageQuant software.
Direct capture of long telomeric DNA onto a surface for single-molecule imaging
For flow-stretching experiments, glass coverslips were first cleaned by sequential sonication in anhydrous ethanol, DI (deionized water) water, 2 M KOH, and DI water. The coverslips were then silanized by sonication in a mixture of APTES (1 ml) and acetone (50 ml) for 20 min. The silanization reaction was stopped by washing the coverslips with DI water. Coverslips were dried in a continuous stream of nitrogen gas. The first round of PEGylation was performed using mPEG-SVA (succinimidyl valeric acid PEG, MW 5000, 10 mg, Laysan Bio) and Biotin-PEG-SVA (MW 5000, Laysan Bio) mixed in a ratio of 80:1 in 0.1 M NaHCO3 buffer (70 μl per coverslip sandwich) and incubation of 3–5 h. The second round of overnight PEGylation was done using 7 mg of Methyl-PEG4-NHS (Thermoscientific, Catalogue number: 22341) dissolved in 0.1 M NaHCO3 buffer (pH 8.5, 70 μl per coverslip sandwich). After PEGylation, the streptavidin solution (0.1 mg/ml, 30 μl) was introduced into the flow cell (30 μl volume), and the reaction was incubated for 10 min. To directly capture the long telomeric DNA, the enzyme cocktail (HinfI, AluI, HphI and Mnll) digested genomic DNA was first purified using phenol-chloroform extraction (54). Next, purified genomic DNA was incubated with biotinylated oligo complementary to the 3′ overhang (oligo to overhang ratio at 10:1) at 68°C for 5 min, followed by slow cooling to room temperature. The resulting biotinylated telomeric DNA was stained with YOYO1 (dye-to-DNA base pair ratio at 1:10) overnight at 4°C. YOYO1-stained DNA substrates were introduced into the flow cell containing a streptavidin-coated glass coverslip for flow-stretching experiments.
AFM imaging and analysis
TRF2 (80 nM dimer) without or with TIN2 (TIN2S or TIN2L, 80, 120 and 200 nM) was incubated with the linear pT270 DNA (10.5 nM) in TRF2 Imaging Buffer (20 mM HEPES [pH 7.5], 100 mM NaCl) at room temperature for 10 min. All DNA–protein samples were diluted 10-fold in TRF2 Imaging Buffer before being deposited on to a freshly prepared 1-(3-aminopropyl)silatrane (APS)-treated mica (SPI Supply) surface for 30 s (56). The APS-treated mica surface was washed with DI water and dried under a stream of nitrogen gas. All images were collected in the tapping mode using Pointprobe® PPP-FMR probes (Nanosensors, spring constants at ∼2.8 N/m) on an MFP-3D-Bio AFM (Asylum Research). All images were captured at a scan rate of 1–2 Hz, a scan size of 1–3 μm × 1–3 μm and a resolution of 512 × 512 pixels.
The DNA tightrope assay
The oblique angle Total Internal Reflection Microscopy (TIRFM) based DNA tightrope assay was carried out according to protocols described previously (42,46,57–60). Briefly, ligated DNA substrates were introduced into the flow cell using a syringe pump at a flow rate of 300 μl/min to stretch the DNA between poly-L-lysine (2.5 mg/ml, MW > 30 000 kDa, Wako Chemicals) treated silica beads on a PEGylated coverslip surface. The primary HA antibody (Abcam) was conjugated to red QDs (655 nm) to form HA-Ab-QDs using the SiteClick™ QD antibody labeling kit (Life Technologies) following the manufacturer’s protocols. HA-tagged TIN2S or TIN2L was conjugated to QDs by incubating proteins with HA-Ab-QDs at a 1:1 molar ratio for 20 min at room temperature. All protein–QD samples were diluted 100-fold using the imaging buffer (20 mM HEPES [pH 7.9], 100 mM NaCl, 0.3 mM MgCl2, 1 mM EDTA, 0.5 mM DTT and 1 mg/ml BSA) to a final concentration of 10 nM proteins in the flow cell. 1× Blocking Reagent (Sigma, catalog number 11096176001) was added to the imaging buffer to reduce nonspecific binding.
Statistical analysis
Data sets from AFM, TIRFM imaging and the DNA tightrope assay were from at least two to three independent experiments. The numbers reported are mean ± SEM, unless stated otherwise. The statistical significance level was set at P < 0.05 based on the Student’s t-test.
RESULTS
Flow-stretching long telomeric DNA for single-molecule fluorescence imaging
One technical barrier in studying DNA binding by the shelterin system using single-molecule fluorescence imaging techniques is the availability of long native telomeric DNA (>2 kb) with high purity. In general, purification of telomeres from cultured cells or tissues was achieved through restriction digestion of genomic DNA using a cocktail of high-frequency cutters and subsequent separation of telomeric DNA from the digested genomic DNA fragments (54,61). The separation step is typically performed using gel filtration, gel electrophoresis or streptavidin-coated magnetic bead pulldown of biotinylated DNA (14,54). The major drawback of using gel filtration and electrophoresis methods for the separation step is that the final products include other long DNA fragments with repetitive sequences. While the magnetic bead pulldown method was successful in purifying telomeric DNA from HeLa cells (∼2–20 kb) (54), the yield of telomeric DNA from cultured cells or tissues using this method is relatively low (Kaur, unpublished data). This low pulldown efficiency is likely due to reduced accessibility of the biotin on the complementary strand and a large shearing force on the longer telomeric DNA during the incubation of DNA with beads. To overcome this technical barrier, we optimized a new DNA target capture technology, the Region-Specific Extraction (RSE, Generation Biotech) system, for purifying longer telomeric DNA (Figure 1A). The RSE method has been shown to enrich DNA fragments with lengths >20 kb achieving an accuracy of over 99% (55). We hybridized a complementary capture primer to the 3′ overhang of the DNA, followed by extension of the primer using PCR reactions containing biotinylated-dNTP and purification using streptavidin-coated magnetic beads. PCR extended primers containing multiple biotinylated nucleotides provide extra stability and accessibility to streptavidin-coated magnetic beads. First, to validate the specificity and efficiency of the RSE system, PCR extension and capture steps of RSE were performed on λ DNA in the absence (mock treatment) or presence of a primer complementary to the overhang. From 8 μg of λ DNA, we recovered ∼20 ng DNA for the mock treatment and ∼650 ng for the λ DNA annealed to the complementary primer. These experiments established that the RSE system could capture long DNA with lengths comparable to telomeric DNA from mouse liver tissues.
Figure 1.
Purification of long telomeric DNA from the mouse liver tissue using the region-specific extraction (RSE) method. (A) Schematics of the RSE method for purifying mouse telomeric DNA. (B) An AFM image of genomic DNA purified from the mouse liver tissue. XY scale bar = 1 μm. (C) Agarose gel electrophoresis showing purified mouse genomic DNA and genomic DNA digested with a cocktail of four restriction enzymes. (D) Dot-Blot assay validating the purity of mouse telomeric DNA obtained from the RSE method. Various amounts of undigested genomic DNA and the telomeric DNA purified using the RSE method were loaded on to a membrane that was hybridized with either a radiolabeled telomere (left panel) or Alu repeat DNA probe (right panel). Telomeric DNA preparations using 2×, 5× and 10 × molar excess of the complementary capture primers relative to the estimated telomeric fragments were tested.
Next, we applied the RSE method to purify telomeric DNA from mouse liver tissues. We first used AFM imaging to validate long DNA fragments in the genomic DNA preparation (Figure 1B). We then digested genomic DNA purified from mouse liver tissues using a restriction enzyme cocktail containing HinfI, AluI, HphI and Mnll, which do not cut telomeric DNA (Figure 1C) (54). AFM imaging and agarose gel electrophoresis validated that this restriction enzyme cocktail digested most of the genomic DNA into smaller DNA fragments (<0.5 kb, Figure 1C). To use the RSE method, we hybridized a capture primer to the 3′ overhang of the telomeric DNA in the digested genomic DNA pool, followed by primer extension using PCR reactions containing biotinylated-dNTPs (Materials and Methods). For the primer annealing step, we used a primer/telomeric DNA fragment ratio of ∼2:1 (2×), 5:1 (5×) or 10:1 (10×). Mouse telomeric DNA fragments annealed to these biotinylated primers at the 3′ overhang were later captured by the streptavidin-coated magnetic beads (Figure 1A). For mice with a total nuclear genome content of 2.8 × 109 bp per cell, telomeric DNA (∼60 kb) constitutes ∼0.17% of the total genome. From 125 μg of mouse genomic DNA, under all three purification conditions, the RSE system generated ∼0.18 μg of purified telomeric DNA, achieving a yield of ∼85%. The higher yield for telomeric DNA is likely due to longer overhangs at telomeres (hundreds of nt) compared to λ DNA (12 nt).
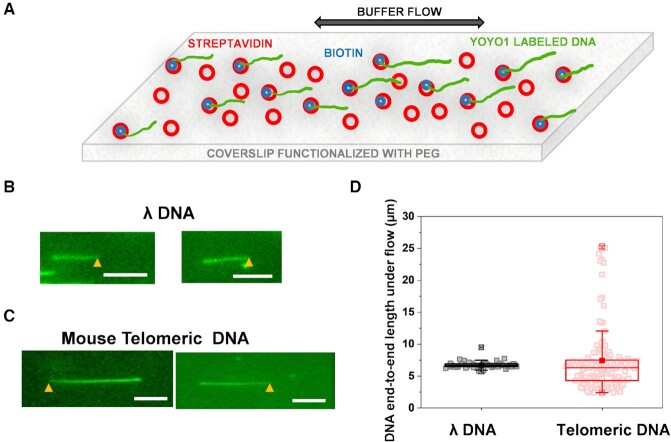
We performed the Dot-Blot assay to estimate the purity of telomere DNA obtained using the RSE system (54). We loaded various amounts of genomic DNA, and 7.5 ng of purified telomere DNA, followed by hybridization with a radiolabeled telomeric or Alu repeat DNA probe as the negative control (Figure 1D). Alu repeats are short interspersed nuclear elements (SINEs) that comprise ∼13% and 18% of the primate and rodent genomes, respectively (62,63). Based on signal intensities quantified for the telomeric DNA probe, using 5 × complementary primers for the RSE protocol led to the highest yield of the telomeric DNA. Notably, there were no significant signals detected using the Alu probe from the telomeric DNA purified using the RSE method. These results established that the RSE method enabled us to purify telomeric DNA with high purity. To ensure that the telomeric DNA was intact through the RSE purification process, the end-to-end length of the flow-stretched control λ DNA and purified telomeric DNA was measured using objective-based TIRFM (Figure 2) (42,46,47). YOYO1-stained DNAs were anchored onto streptavidin-coated PEGylated glass slides and stretched under a flow rate of 250 μl/min (Figure 2A–C) (64). The DNA molecules stretched under the buffer flow recoiled back as the flow stopped. These results established that there were no significant nonspecific interactions between DNA molecules and the flow chamber surface. Furthermore, the large majority of flow-stretched telomeric DNA molecules were anchored at one end only. This result indicated that the capture primer predominantly targeted the 3′ overhang of the telomeric DNA. Annealing of the capture primer at the double-stranded telomeric DNA region was not significant. The end-to-end length of λ DNA at its maximum extension under this flow rate was measured at 6.7 μm (± 0.6 μm, N = 52, Figure 2D). Meanwhile, the purified telomeric DNA displayed a wider length distribution at 7.4 μm (± 5.2 μm, N = 133, Figure 2D). Based on the end-to-end lengths of flow-stretched λ DNA (48.5 kb) under the same experimental conditions, the end-to-end lengths of telomeric DNA were consistent with results from previous terminal restriction fragment (TRF) analysis of samples from the same mouse strain (C57B/6J) (65).
Figure 2.
Flow-stretching of λ DNA and mouse telomeric DNA purified using the RSE method. (A) Schematics of flow stretching of YOYO1-stained DNA that was anchored onto the surface at one end through biotin-streptavidin interactions. (B and C) Fluorescence images of flow stretched λ DNA anchored through the biotinylated primer annealed to the overhang (B), and RSE-purified mouse telomeric DNA anchored onto the surface through biotins on the primer annealed to the 3′ overhang (C); scale bar: 5 μm. The yellow arrows represent the DNA anchoring points. (D) Box plots (range 25–75 percentile) of the length of the flow-stretched λ and telomeric DNA purified using the RSE method. λ DNA: N = 52, 6.7 μm ± 0.6 μm (mean ± STD); telomeric DNA: N = 133, 7.4 μm ± 5.2 μm (mean ± STD). Mean  , max and min
, max and min  , 99%, and 1%
, 99%, and 1%  are represented by these symbols in the box plot.
are represented by these symbols in the box plot.
Next, to streamline the single-molecule imaging platform, we directly anchored the telomeric DNA from the digested mouse genomic DNA pool onto the flow chamber surface. Specifically, after digestion of mouse genomic DNA using the restriction enzyme cocktail and removing enzymes using phenol extraction, a biotinylated oligo was annealed to the 3′ overhang of telomeric DNA. The digested genomic DNA sample containing telomeric DNA fragments with a biotin-labeled primer annealed to the 3′ overhang was then introduced into the flow chamber containing a streptavidin-coated coverslip surface (Materials and Methods). The lengths of flow-stretched DNA captured on the surface (8.0 μm ± 3.7 μm) were consistent with what was measured for the telomeric DNA purified using the RSE method (7.4 μm ± 5.2 μm, Figure 2D). In stark contrast, there was no long DNA attached to the streptavidin-coated coverslip following the same experimental procedure but with biotinylated Alu probe (Supplementary Figure S1A). Thus, this direct surface capture platform enables the anchoring of long telomeric DNA onto a surface from the digested mouse genomic DNA pool. The ability to specifically anchor individual long telomeric DNA molecules on to a surface at DNA ends opens new possibilities for investigating shelterin protein assembly on physiologically relevant long telomeric DNA substrates.
TRF2 recruits TIN2 to telomeric DNA and forms stable protein–DNA complexes on DNA
Next, to study the interaction between TRF2 and TIN2, we utilized the oblique angle TIRFM imaging to monitor QD-labeled proteins on DNA tightropes anchored between micron-sized silica beads (23,58–60,66). In this imaging platform, DNA molecules are stretched using hydrodynamic flow inside a flow cell. Anchoring of stretched DNA between two poly-L-lysine coated microspheres forms DNA tightropes. Uniquely, DNA tightropes created using tandemly ligated DNA allow us to directly correlate DNA binding events with the underlying specific DNA sequences or structures along DNA tightropes (23,42,46). Specifically, to study telomere binding proteins, we ligated linear T270 DNA fragments containing 270 TTAGGG repeats (LT270 DNA) to form DNA tightropes with telomeric regions at defined spacing (Figure 3A). The lengths of the LT270 DNA tightropes are typically in the range of ∼2.1–22 μm, corresponding to ligation of ∼2–12 of T270 fragments (5.4 kb) (23). Three TIN2 isoforms have been identified in human cell lines (67–69), including TIN2S (354 AAs), TIN2L (451 AAs) and TIN2M (420 AAs). TIN2S, TIN2L and TIN2M share the same TRF1, TRF2 and TPP1 binding domains and localize to telomeres (67,69,70). We purified the full-length His-tagged TRF2 using the insect cell/baculovirus protein expression system and obtained HA-tagged TIN2 (TIN2S and TIN2L, Supplementary Figure S2A) (23,50,71). The DNA binding activity of purified TRF2 and formation of TRF2–TIN2S and TRF2–TIN2L complexes on DNA were validated by electrophoresis mobility shift assays (EMSAs) (Supplementary Figure S2B) (72). To monitor the recruitment of TRF2 at the telomeric sequences, we previously conjugated His-tagged TRF2 to the strep-QDs through the biotinylated multivalent chelator tris-nitrilotriacetic acid (BTTris-NTA) linker (TRF2–QDs) (23). QD-labeling of TRF2 does not significantly affect its telomeric DNA binding specificity (23). In addition, no strep-QDs were observed on DNA tightropes in the presence of BSA (1 mg/ml) and strep-QDs (10 nM), indicating that QD labeling is specific to His-tagged proteins (Supplementary Figure S2C). We observed that TRF2 bound specifically to the telomeric DNA on LT270 DNA tightropes and could transit between slow diffusion over telomeric and fast diffusion over nontelomeric regions (23). To monitor the recruitment of TIN2 onto DNA by TRF2 in this study, we conjugated HA-tagged TIN2 to primary HA antibody-coated QDs (HA–Ab–QDs) using the antibody sandwich method (TIN2S–QDs and TIN2L–QDs, Figure 3B). Without TRF2, there was no significant binding of TIN2S–QDs or TIN2L–QDs on LT270 DNA tightropes (Figure 3C). To monitor the recruitment of TIN2 on to DNA tightropes by TRF2, after establishing LT270 DNA tightropes in the flow cell, we introduced unlabeled TRF2 (25 nM dimer) together with TIN2–QDs (TIN2S or TIN2L, 10 nM) using a syringe pump. After the buffer flow was stopped, videos of TIN2–QDs on unstained LT270 DNA tightropes were recorded at 20 frames/s (Figure 3D). For both TRF2–TIN2S–QDs and TRF2–TIN2L–QDs, approximately 86% of TIN2–QD complexes remained on DNA tightropes after 10 or 30 s (N = 567). Furthermore, ∼86% of TIN2–QDs were static, indicating stable TRF2–TIN2 binding to DNA. The histograms of the pairwise distance between adjacent TIN2S–QDs and TIN2L–QDs displayed distinct peaks centered at 1.5 (± 0.6) μm and 1.4 (± 0.5) μm (Figure 3E), respectively. The average center-to-center spacing between (TTAGGG)270 regions on LT270 DNA tightropes is ∼1.5 μm, assuming DNA is stretched to ∼90% of its contour length under hydrodynamic flow during attachment to microspheres (58). Thus, the distance between adjacent TIN2–QDs (nearest neighbors) was consistent with the expected spacing between telomeric regions on the LT270 tightropes (Figure 3A). Furthermore, we expect approximately 6 (TTAGGG)270 regions per 10 μm of LT270 DNA tightropes. We measured the densities of QDs on LT270 DNA tightropes with single or multiple complexes (static or mobile). The QD densities were 3.8 (N = 23 tightropes) and 3.4 (N = 74 tightropes) complexes per 10 μm of DNA, respectively, for TIN2S– and TIN2L–QDs. These results indicated that close to half of the individual (TTAGGG)270 regions contained TRF2–TIN2. Furthermore, TRF2–QD and TIN2–QD co-localized on LT270 DNA tightropes (Figure 3F). In summary, these results demonstrate that TRF2 loads TIN2 (TIN2S and TIN2L) specifically at telomeric regions and forms stable complexes on telomeric DNA.
Figure 3.
TRF2 loads TIN2 specifically at telomeric regions on LT270 DNA tightropes. (A) Schematics of the ligated T270 DNA (LT270) substrate for generating DNA tightropes. (B) QD-labeling scheme: HA-tagged TIN2 conjugated to HA antibody-coated QDs (HA-Ab-QDs). (C) TIN2-QDs do not bind to LT270 DNA tightropes in the absence of TRF2. DNA was stained with YOYO1 (left panel) after the introduction of TIN2L-QDs (right panel) into the flow cell. (D) QD-labeled TIN2 on LT270 DNA tightropes in the presence of unlabeled TRF2. Representative fluorescence images (top panels) and kymographs (bottom panels) of red (655 nm) HA-Ab-QD-labeled TIN2S (TIN2S-QD, left panel) and TIN2L (TIN2L-QD, right panel). (E) Histograms representing the distance between two adjacent TIN2S-QD or TIN2L-QD complexes on LT270 DNA tightropes. Fitting the data with Gaussian functions shows peaks centered at 1.5 μm ± 0.6 μm (mean ± STD) for TRF2-TIN2S-QD (N = 538, R2> 0.98), and 1.4 μm ± 0.5 μm for TRF2-TIN2L-QD (N = 288, R2> 0.98). (F) Co-localization of TRF2-QD and TIN2-QD on L270 DNA tightropes; N = 42 observations.
TIN2 facilitates DNA compaction and DNA–DNA bridging by TRF2
Previous single-molecule studies of TRF2 and shelterin proteins using DNA substrates ranging from 32 to 270 TTAGGG repeats demonstrated that TRF2 alone can compact telomeric DNA (8,13,23,43). After confirming that TRF2 loads TIN2 specifically at telomere regions, we went further to investigate how TIN2 affects DNA binding by TRF2 at the single-molecule level. We anchored individual YOYO1-stained and biotinylated control λ or telomeric DNA molecules on to the flow chamber surface using the biotin-streptavidin linkage (Figure 4A). The telomeric DNA was purified using the RSE method or from the digested mouse genomic DNA pool. Before and after introducing TRF2 without or with TIN2, we recorded videos and positions of stretched DNA under back-and-forth buffer flow (Figure 4). Videos of DNA before adding proteins were used to establish their original extended DNA end-to-end lengths. After introducing TRF2 alone (200 nM, unlabeled) into the flow chamber, we recorded videos of the same sets of DNA molecules under the same buffer flow conditions (Figure 4B). The extended end-to-end lengths of λ DNA under buffer flow after incubation with TRF2 were only slightly reduced by 2.3% (± 3.8%) from their original lengths (Figure 4E). The change was not statistically significant. In stark contrast, a direct comparison of extended telomeric DNA end-to-end lengths before and after introducing TRF2 (200 nM) into the flow chamber showed that TRF2 significantly reduced the telomere DNA lengths by 22.1% (±25.3%, Figure 4B and E). Notably, upon incubation with TRF2, QD-labeled TRF2 co-localized with beadlike structures showing higher YOYO1 intensities on DNA tightropes that represented folded or compacted DNA (N = 35 events, Figure 4B). These structures are consistent with previous reports of multiprotein TRF2–DNA complexes (43,73). The beadlike DNA structure upon TRF2 binding demonstrated that the long telomeric DNA compaction was heterogeneous and possibly was mediated through the cooperative binding of multiple TRF2 molecules. In addition, comparison experiments using telomeric DNA purified through the RSE method and from direct anchoring of the telomeric DNA from the digested genomic DNA pool showed the same trend of DNA end-to-end length reduction upon the addition of TRF2 (Supplementary Figure S1B).
Figure 4.
TIN2 facilitates TRF2-mediated compaction of long mouse telomeric DNA. (A) Model depicting the formation of compacted DNA structures on incubating end-anchored telomeric DNA with TRF2 and TIN2. YOYO1-stained mouse telomeric DNA was anchored at one end through the linkage between the biotin on the primer annealed to the 3′ overhang and streptavidin-coated glass coverslip surface. (B–D) Examples of telomeric DNA compaction mediated by QD-labeled TRF2 (B), unlabeled TRF2–TIN2S and TRF2–TIN2L (C), and unlabeled TRF2 and QD-labeled TIN2S (D). (B) His-tagged TRF2 was labeled with the strep-QD through the BTTris-NTA linker. (D) HA–TIN2S was labeled with the HA–Ab–QD using the antibody sandwich method. White arrows: buffer flow directions. Black arrows: DNA anchoring points. Green-blue arrows: compacted beadlike structures. The numbers are total lengths of the video in seconds; scale bar: 5 μm. (E) Box plots (range 25–75 percentile) comparing compaction of λ DNA by TRF2 (N = 43, 2.3% ± 3.8%) and telomeric DNA by TRF2 (N = 91, 22.1% ± 25.3%), TRF2–TIN2S (N = 55, 56.3% ± 27.8%) and TRF2–TIN2L (N = 46, 65.0% ± 21.8%). % compaction is reported as mean ± STD. TRF2: 100 nM dimer; TIN2: 100 nM. Mean  , max and min
, max and min  , 99% and 1%
, 99% and 1%  are represented by these symbols in the box plot; P < 10–5 *, P < 10–11***.
are represented by these symbols in the box plot; P < 10–5 *, P < 10–11***.
Since anchoring of telomeric DNA after the RSE purification or from the digested genomic DNA pool provided similar results, we carried out additional experiments with TRF2–TIN2 using the latter method. Next, to investigate whether TIN2 facilitates TRF2-mediated telomeric DNA compaction (cis-interactions), after anchoring telomeric DNA from the digested genomic DNA pool, we introduced both TRF2 (100 nM dimer) and TIN2 (100 nM, TIN2S or TIN2L) into the flow cell (Figure 4C and D). Strikingly, from their original lengths without proteins, the telomeric DNA end-to-end lengths were reduced by 56.3% (± 27.8%) and 65.0% (± 21.8%), respectively, for TRF2–TIN2S and TRF2–TIN2L. These levels of DNA compaction by TRF2-TIN2 on telomeric DNA were 2 to 3-fold higher than what was observed for TRF2 alone (22.1% ± 25.3%, Figure 4E). Clearly, the addition of TIN2 shifted the protein–DNA complexes to the higher DNA compaction mode (Supplementary Figure S3A). In comparison, the extended DNA end-to-end lengths of λ DNA under buffer flow after incubation with both TRF2 (200 nM) and TIN2 (100 nM, TIN2S or TIN2L) were only slightly reduced by 1.2% (± 1.6%) and 7.8% (± 8.6%), respectively, from their original lengths (Supplementary Figure S3B). Furthermore, the extent of DNA compaction for individual DNA molecules was constant during the duration of the back-and-forth flow-stretching (for 30 s to 1 min, Figure 4B and C), indicating that TRF2–TIN2 mediated DNA compaction was stable. It is worth noting that after adding either TRF2 alone or TRF2–TIN2 in the flow chamber, the directions of the flow-stretched λ-DNA and telomeric DNA molecules changed with buffer flow direction switching. This observation indicated that DNA molecules did not nonspecifically interact with the flow chamber surface upon the addition of proteins (Figure 4B and C). In addition, QD-labeled TIN2S localized to compacted DNA regions with beadlike structures (N = 50 events, Figure 4D). Since TIN2 does not directly bind to DNA, these results suggested that TRF2–TIN2 together compact telomeric DNA.
To further establish the structure of TRF2-TIN2 mediated DNA–DNA compaction, we applied AFM imaging of telomeric DNA in the presence of TRF2–TIN2. We used the linear DNA substrate containing 270 TTAGGG repeats (pT270 DNA) that was gel-purified to remove the nontelomeric regions after double-digestion of the T270 plasmid. To investigate the function of TIN2, we carried out AFM imaging of samples after incubation of the linear pT270 DNA (10.5 nM) with TRF2 (80 nM dimer) and increasing concentrations of TIN2 (either TIN2S or TIN2L, 0, 80, 120 and 200 nM, Figure 5). We identified protein complexes using AFM heights of molecules on DNA. Since dsDNA alone displayed an AFM height <0.5 nm (47), structures with AFM heights >0.8 nm were categorized as protein complexes on DNA (Figure 5A and B). Consistent with previous observations (13,43), TRF2 alone significantly (P < 0.05) compacted the pT270 DNA from its original length of 605 nm (± 2 nm) to 550 nm (± 10 nm, Figure 5C). With a fixed TRF2 concentration (80 nM dimer) and increasing TIN2 concentrations (80, 120 and 200 nM), the pT270 DNA lengths were significantly (P < 0.05) shortened compared to TRF2 alone (Figure 5B and C). In addition, TIN2 alone (200 nM) did not significantly bind to pT270 DNA and induce telomeric DNA compaction. Collectively, consistent with previous studies using shorter telomeric DNA (13,43), imaging of flow-stretched DNA demonstrated that TRF2 compacts physiologically relevant long telomeric DNA (cis-interactions). Significantly, both TIN2S and TIN2L facilitate TRF2-mediated telomeric DNA compaction.
Figure 5.
Facilitation of TRF2-mediated telomeric DNA compaction by TIN2 revealed by AFM imaging. (A and B) Representative AFM images of pT270 fragment (1.6 kb) with (A) TRF2 alone (80 nM dimer), and (B) TRF2 and increasing concentrations of TIN2S and TIN2L. XY scale bar = 100 nm. (C) DNA contour lengths of the linear pT270 fragments alone, with TRF2 only, or with TRF2 and TIN2. DNA only: N = 75, 605 ± 2 nm; +TRF2: N = 119, 550 ± 10 nm; +TRF2 + TIN2S (80 nM): N = 138, 520 ± 12 nm; +TRF2 + TIN2S (120 nM): N = 261, 514 ± 10 nm; +TRF2 + TIN2S (200 nM): N = 168, 386 ± 18 nm; +TRF2 + TIN2L (80 nM): N = 118, 537 ± 12 nm; +TRF2 + TIN2L (120 nM): N = 141, 532 ± 11 nm; +TRF2 + TIN2L (200 nM): N = 140, 479 ± 16 nm. The error bars represent SEM from two independent data sets.
TIN2 facilitates TRF2-mediated double-stranded telomeric DNA–DNA bridging
In addition to DNA compaction (cis-interactions) (Figure 5), AFM imaging also revealed that compared to TRF2 alone, TRF2 and TIN2 together significantly increased the percentage of molecules that contained multiple strands of pT270 DNA bridged by protein complexes (Supplementary Figure S4). To further establish whether or not TIN2 facilitates TRF2-mediated trans-interactions (DNA–DNA bridging) in solution, we anchored LT270 DNA tightropes between micron-sized silica beads and introduced free biotinylated pT270 (270 TTAGG repeats, pT270) or the control nontelomeric dsDNA fragments (noTel, Materials and Methods, Figure 6A) into the flow cell. To visualize the pT270 DNA and noTel bridged on to DNA tightropes by proteins, we labeled ends of these two DNA substrates with red (655 nm) strep-QDs (QDpT270 and QDnoTel, Materials and Methods). AFM imaging revealed that 60.7% (± 3.1%) of the pT270 and 67.7% (± 6.7%) of the noTel DNA fragments were labeled with strep-QDs (50). Previously, we also established that the QDpT270 DNA fragments do not nonspecifically pair with LT270 DNA tightropes without proteins (50). Thus, QD signals on DNA tightropes enable us to directly observe protein-mediated bridging of the linear QDpT270 dsDNA fragments from solution to DNA tightropes.
Figure 6.
TIN2 enhances TRF2-mediated double-stranded telomeric DNA–DNA bridging. (A) Schematics depicting the strategy to monitor protein meditated double-stranded DNA–DNA bridging using the DNA tightrope assay. DNA tightropes were incubated with TRF2 (25 nM dimer) without or with TIN2 (10 nM) before introducing QD-labeled linear double-stranded pT270 fragment (QDpT270, 1.6 kb, 50 nM) and TRF2 (25 nM dimer) either without or with TIN2 (10 nM). (B) Representative fluorescence images (top panels) and kymographs (bottom panels) of the QDpT270 DNA bridged onto LT270 DNA tightropes by TRF2 alone, TRF2-TIN2S and TRF2-TIN2L. (C) Histograms and fitting using Gaussian function of the spacing between QDpT270 DNA fragments bridged on to the LT270 tightropes by TRF2 alone (N = 215, 1.43 μm ± 0.02 μm, R2= 0.93), TRF2-TIN2S (N = 229, 1.45 μm ± 0.04 μm, R2= 0.96) and TRF2-TIN2L (N = 212, 1.64 μm ± 0.02 μm, R2= 0.98) (Gaussian peak ± STD). (D) Intensities (arbitrary units: a.u.) of QDpT270 on LT270 DNA tightropes mediated by TRF2 (N = 183), TRF2-TIN2S (N = 148) and TRF2-TIN2L (N = 180) with their standard deviations from two independent data sets. P < 0.05 *, P < 10–36 ***.
To evaluate whether or not TIN2 facilitates DNA–DNA bridging by TRF2, we first introduced unlabeled TRF2 (25 nM dimer) without or with TIN2 (TIN2S or TIN2L, 10 nM) into the flow cell and incubated for 5 min to allow proteins binding to LT270 DNA tightropes. Then, we introduced QDpT270 DNA fragments (50 nM) along with TRF2 (25 nM dimer), either without or with TIN2 (TIN2S or TIN2L, 10 nM). Under all experimental conditions, we observed QDpT270 bound to LT270 DNA tightropes that were long-lasting. All QDpT270 DNA molecules stayed on DNA tightropes until the end of the 10–120 s observational windows (Figure 6B). The average spacing between QDpT270 signals mediated by TRF2 alone (1.43 ± 0.02 μm), TRF2-TIN2S (1.45 ± 0.04 μm) and TRF2-TIN2L (1.64 ± 0.02 μm) on the DNA tightropes was consistent with the distance between adjacent telomeric regions (Figure 6C). Under the same experimental conditions, when both TRF2 and TIN2 (TIN2S or TIN2L) were present, the QD intensities from the QDpT270 fragments bridged to individual telomeric regions on LT270 tightropes were significantly stronger than when TRF2 alone was present (Figure 6D). Even though only one QD was conjugated to each pT270 DNA (50), the heterogeneity in the intensity from individual QDs prevented us from precisely determining the number of QDs in each cluster. However, these results strongly suggested that compared to TRF2 alone, when both TRF2 and TIN2 were present, higher numbers of linear QDpT270 DNA fragments were bridged to individual telomere regions on LT270 tightropes. In comparison, there were no significant QDpT270 DNA or QDnoTel DNA signals on nontelomeric DNA tightropes either in the presence of TRF2 alone, TRF2–TIN2S or TRF2–TIN2L. In summary, results from AFM imaging and the DNA tightrope assay show that both TIN2S and TIN2L facilitate TRF2-mediated telomeric dsDNA-dsDNA bridging (trans-interactions).
TRF2 facilitates the bridging of telomeric single-stranded DNA and RNA to double-stranded DNA
Results from both in vitro and cell-based experiments support the notion that TRF2 is the crucial player in T-loop formation, in which the 3′ overhang invades the double-stranded telomeric DNA region (13,15,16). Furthermore, TRF2 directly binds to TERRA (39). To investigate whether or not TIN2 contributes to the recruitment of single-stranded telomeric DNA or RNA to the double-stranded telomeric DNA regions, we applied the DNA tightrope assay with QD-labeled single-stranded DNA (TTAGGG-12) or RNA (UUAGGG-12) containing 12 telomeric repeats in the flow chamber (Figure 7A and B). To visualize the ssDNA and ssRNA recruited to the DNA tightrope, biotinylated TTAGGG-12 and UUAGGG-12 were conjugated to strep-QDs. Without proteins, there was no significant QDTTAGGG-12 or QDUUAGGG-12 signals observed on LT270 DNA tightropes. In stark contrast, with the introduction of TRF2 (25 nM dimer) and single-stranded DNA or RNA (QDTTAGGG-12 or QDUUAGGG-12, 50 nM) in the flow cells, QD signals decorated LT270 tightropes (Figure 7A and B). These results indicated that TRF2 recruited single-stranded TTAGGG-12 and UUAGGG-12 to LT270 DNA tightropes. The histograms of the pairwise distance between adjacent QD-labeled single-stranded telomeric DNA or RNA were consistent with the spacing between adjacent T270 regions (Figure 7C). Thus, consistent with previous observations (13,15,16,39), these results demonstrated that TRF2 recruits telomeric single-stranded DNA and RNA to the double-stranded telomeric regions. Next, we introduced TRF2 (25 nM dimer), TIN2 (10 nM, TIN2S or TIN2L) and ssDNA or ssRNA (QDTTAGGG-12 and QDUUAGGG-12, 50 nM) into the flow chamber with LT270 DNA tightropes. The spacing between adjacent QD-labeled TTAGGG-12 and UUAGGG-12 observed on LT270 tightropes was again consistent with the distance between telomeric regions on LT270 DNA (Figure 7C). In the presence of TRF2–TIN2 (TIN2S or TIN2L), the intensities of the QDTTAGGG-12 and QDUUAGGG-12 signals at individual telomeric regions on the LT270 DNA tightropes were significantly greater than what was observed for TRF2 alone (Figure 7D). These results indicate that compared to TRF2 alone, TRF2 -TIN2 together bridged higher numbers of TTAGGG-12 ssDNA or UUAGGG-12 ssRNA molecules to individual telomeric regions on LT270 tightropes. In contrast, there were no significant QDTTAGGG or QDUUAGGG signals on nontelomeric DNA tightropes either in the presence of TRF2 alone, TRF2–TIN2S or TRF2–TIN2L. Thus, results from the DNA tightrope assay establish that both TIN2S and TIN2L facilitate TRF2-mediated bridging of telomeric single-stranded DNA and RNA to double-stranded DNA.
Figure 7.
TIN2 enhances TRF2-mediated bridging of telomeric ssDNA and ssRNA to LT270 dsDNA tightropes. LT270 DNA tightropes were incubated with TRF2 (25 nM dimer) without or with TIN2 (10 nM) before introducing QD-labeled single-stranded TTAGGG-12 (QDTTAGGG-12), or UUAGGG-12 (QDUUAGGG-12, 50 nM) along with TRF2 (25 nM dimer) either without or with TIN2 (10 nM). (A and B) Representative fluorescence images (top panels) and kymographs (bottom panels) of the QDTTAGGG-12 DNA (A) and QDUUAGGGG-12 RNA (B) bridged onto LT270 DNA tightropes by TRF2 alone, TRF2-TIN2S and TRF2-TIN2L. (C) Histograms and fitting using Gaussian function (R2> 0.94) of the spacing between QDTTAGGG-12 and QDUUAGGG-12 on LT270 tightropes, mediated by TRF2, TRF2-TIN2S and TRF2-TIN2L proteins. QDTTAGGG-12: TRF2 alone (N = 172, 1.72 ± 0.01 μm), TRF2-TIN2S (N = 202, 1.54 ± 0.01 μm), TRF2-TIN2L (N = 175, 1.38 ± 0.01 μm). QDUUAGGG-12: TRF2 alone (N = 223, 1.55 ± 0.01 μm), TRF2-TIN2S (N = 322, 1.35 ± 0.01 μm), TRF2-TIN2L (N = 227, 1.58 ± 0.03 μm). (D) Comparing intensities (arbitrary unit: a.u.) of QDTTAGGG-12 and QDUUAGGG-12 bridged onto LT270 DNA tightropes mediated by TRF2 (NTTAGGG= 153, NUUAGGG= 162), TRF2-TIN2S (NTTAGGG= 108, NUUAGGG= 137) and TRF2-TIN2L (NTTAGGG= 225, NUUAGGG= 76) with their standard deviations from two independent data sets; P < 10–12–10–24 ***.
TIN2 facilitates TRF2-mediated T-loop formation in vitro
While the DNA tightrope assay clearly showed that TIN2 enhances TRF2-mediated bridging of double-stranded telomeric DNA and single-stranded DNA to LT270 tightropes, it did not address whether or not TIN2 facilitates the T-loop formation. To directly evaluate the impact of TIN2 on T-loop formation in vitro, we generated a model T-loop substrate based on the established protocols and validated the presence of the 3′ overhang using AFM imaging (14). First, the ends of the linear T270 DNA (5.4 kb) containing 270 TTAGGG (1.6 kb) repeats at one end were resected using T7 exonuclease. AFM imaging of the T7 exonuclease treated T270 substrate after incubation with mitochondrial SSB (mtSSB) revealed that approximately 88% (N = 100) DNA molecules were bound by mtSSB, and among which 86% DNA molecules with mtSSB bound only at one or both ends without internal DNA binding (Supplementary Figure S5A). This establishes that the majority of T7 exonuclease treated linear T270 DNA contained ssDNA overhangs. Furthermore, after T7 exonuclease treatment, the double-stranded DNA contour length decreased from 1744 (± 80) nm to 1652 (± 110) nm (Supplementary Figure S5B), indicating that the average overhang was ∼150 nt at each DNA end. Next, to differentiate the telomeric and nontelomeric DNA ends, we annealed a biotinylated primer to the 3′ overhang of the nontelomeric end and labeled it with strep-QDs (Figure 8A). The majority (72%) of the QDs observed on the T7 exonuclease treated T270 DNA fragment were located at DNA ends, demonstrating the specificity of this DNA end labeling strategy. To benchmark the T-loop formation efficiency by TRF2, we incubated increasing concentrations of TRF2 with the model T-loop substrate. T-loops, positioned at the opposite side of the nontelomeric end marked by the QD, were observed as lasso-like structures with TRF2 bound at the base of the loop (Figure 8B). The T-loop formation efficiency is TRF2 concentration-dependent, with the highest efficiency of T-loop formation observed at 25 nM TRF2 dimer concentration (Figure 8C).
Figure 8.
TIN2 facilitates TRF2-mediated T-loop formation. (A) Blocking and marking of the 3′ overhang at the genomic DNA end on the linear T270 DNA substrate (5.4 kb). Position distribution of strep-QDs on the linear T270 DNA after treatment with T7 exonuclease and annealing of a biotinylated primer to the 3′ overhang at the genomic DNA end. Insert: an AFM image of T7 exonuclease-treated linear T270 DNA with a strep-QD at one DNA end; XY scale bar = 100 nm. (B) AFM images of T-loops formed on the linear T270 DNA with the nontelomeric end labeled with QDs in the presence of TRF2 (25 nM dimer); XY scale bar = 100 nm. (C) Percentages of T-loop formation on the linear T270 DNA with a 3′ overhang at different TRF2 concentrations. TRF2 dimer concentrations tested: 12.5 nM (N = 832, 3.1%), 18 nM (N = 1114, 3.5%), 20 nM (N = 445, 7.0%), 25 nM (N = 332, 10.2%), 30 nM (N = 276, 5.4%), 35.5 nM (N = 269, 6.7%), 40 nM (N = 88, 4.5%). (D) AFM images of T-loop formed in the presence of TRF2-TIN2L on the linear T270 DNA with a 3′ overhang; XY scale bar = 100 nm. (E) Percentages of T270 DNA molecules with loop formation at a fixed TRF2 concentration (25 nM dimer) and increasing concentrations of TIN2L. No overhang control DNA: T270 DNA without T7 exonuclease treatment. All other data sets: T270 DNA with T7 exonuclease treatment. The data were pooled from at least three independent experiments. N = 912–2463 DNA molecules for each condition.
To evaluate the effect of TRF1 on TRF2 mediated T-loop formation, we incubated the linear T270 DNA containing a 3′ overhang with TRF2 (25 nM dimer) and TRF1 (25 nM dimer) in a time-dependent sequential manner. On addition of TRF1 with a time delay of 30 s to the reaction mixture of TRF2 and DNA with 3′ overhangs, the percentage of T-loop formation remained almost the same as when TRF2 alone was present (Supplementary Figure S6). In contrast, when adding TRF1 30 s before or together with TRF2 to the DNA reaction mixture, the percentage of T-loop formation was significantly (P < 0.05) reduced. This result suggests that TRF1 and TRF2 compete during binding to telomeric DNA and TRF1 impairs the ability of TRF2 toward T-loop formation.
To evaluate if TIN2 facilitates TRF2 T-loop formation, we incubated the linear T270 DNA with a 3′ overhang generated through T7 exonuclease treatment with TRF2 (25 nM dimer) and increasing concentrations of TIN2L (3, 5 and 7.5 nM). Concentrations of TIN2L >7.5 nM led to large protein–DNA complexes covering the whole telomeric regions on the T270 DNA and prohibited the analysis of T-loop structures. With both TRF2 and TIN2L present, the percentages of the linear T270 DNA with T-loops significantly increased from 8.3% (± 0.3%) for TRF2 alone to 8.7% (± 0.8%) at 3 nM TIN2L, 15.0% (± 1.1%) at 5 nM TIN2L, to 22.5% (± 0.3%) at 7.5 nM TIN2L (Figure 8E). Furthermore, the T-loops formed in the presence of TRF2 or TRF2–TIN2L at all three TIN2 concentrations displayed contour lengths consistent with the length of the telomeric region (1.6 kb) on the T270 DNA (Supplementary Figure S7). In stark contrast, on the control linear T270 DNA without a 3′ overhang, TRF2 (25 nM dimer) as well as TRF2 (25 nM dimer) combined with TIN2L (7.5 nM) did not induce significant populations of molecules with loops (1.3% ± 0.5% and 2.9% ± 0.8%, respectively, Figure 8E). These results demonstrated that TRF2-TIN2L mediated telomeric DNA loop structure was 3′ overhang specific. The AFM volumes of protein complexes at the base of the loop (>10 000 nm3, Supplementary Figure S7) when both TRF2 and TIN2 were present were significantly larger compared to the volumes of TRF2 dimer (∼99.6 nm3) and TIN2 alone (TIN2S: ∼41.3 nm3; TIN2L: ∼41.9 nm3) in solution (50). While the exact numbers of TRF2 and TIN2 molecules in the complexes were technically challenging to determine based on AFM imaging, these results support a model in which TIN2 forms multiprotein complexes with TRF2 and facilitates TRF2-mediated T-loop formation in a 3′ overhang dependent manner.
DISCUSSION
It has been established that at telomeres, the large majority (∼90%) of the TRF1 and TRF2 proteins exist in complexes with TIN2 but lacking POT1 and TPP1 (41). Consequently, understanding how TIN2 regulates TRF2–DNA binding is a critical step in advancing our understanding of TIN2 in telomere maintenance. In this study, complementary results from AFM imaging and the DNA tightrope assay shed new light on TIN2’s role in regulating TRF2 function to promote higher-order nucleic acid structures at telomeres (Figure 9).
Figure 9.
TIN2 facilitates TRF2-mediated trans- and cis-interactions on telomeric DNA. Results from AFM imaging and the DNA tightrope assay demonstrate that TRF2 facilitates TRF2-mediated telomeric DNA compaction, T-loop formation, and bridging of dsDNA, ssDNA and ssRNA (TERRA) to duplex DNA. ssDNA is relevant in the context of the 3′ overhang.
Mechanism of stabilization of TRF2 at telomeres by TIN2
TIN2 binds TRF1 and TRF2 through different domains (25) and stabilizes both TRF1 and TRF2 at telomeres (25,29). The depletion of TIN2 using siRNA or conditional knockout in MEFs leads to decreased localization of TRF1 and TRF2 at telomeres. Single-molecule imaging from this study sheds new light on the molecular mechanism underlying stabilization of TRF2 by TIN2 at telomeres. Consistent with results from cell-based assays, we observed that TRF2 loads both TIN2S and TIN2L specifically at telomeric regions and forms long-lasting protein complexes on LT270 DNA tightropes. AFM imaging revealed that in the presence of TIN2, large protein complexes consisting of multiple copies of TRF2 and TIN2 form on telomeric DNA and induce DNA compaction (top left panel of Figure 9). These results suggest that protein–protein and protein–DNA interaction networks mediated by TIN2 stabilize TRF2 at telomeres.
TIN2 facilitates TRF2-mediated dsDNA-dsDNA bridging and T-loop formation
Applying super-resolution fluorescence imaging of the native chromatin in MEFs nuclei, Dokasani et al. established that TRF2 is required for the formation and/or maintenance of the T-loop structure, while the deletion of TPP1, POT1 and RAP1 does not affect the frequency of T-loop formation (15). Furthermore, super-resolution fluorescence imaging of telomere structures with an active and ongoing ATM-dependent DDR further revealed that ATM-dependent DDR at telomeres correlates with the telomere structural change from the looped to the linear configurations (17). A ‘three-state’ model that includes the closed-state, intermediate-state and uncapped-state has been proposed to explain telomere-mediated chromosome end protection (74). TRF2 overexpression in TIN2 knock out cells leads to a significant decrease in the chromosome-type telomere fusion but only a slight reduction in the phosphorylation of Chk2 (29). These results suggest that TRF2 alone without TIN2 elicits a cell phenotype that is consistent with intermediate-state telomeres that are linearized DDR + chromosome ends (29). It was proposed that TIN2 might contribute to the formation of the closed-state telomeres (74). However, how TIN2 functions together with TRF2 to promote the closed-state telomeres is largely unknown. Single-molecule imaging in this study revealed previously unknown activities of TIN2 in facilitating TRF2-mediated bridging of telomeric ssDNA (a surrogate for 3′ overhangs) and dsDNA to telomeric dsDNA (bottom panel of Figure 9). Consistent with these activities, AFM imaging revealed that, in a concentration-dependent manner, TIN2 increases the frequency of TRF2-mediated T-loop formation (top right panel of Figure 9). In the presence of TRF2 and TIN2, large multiprotein complexes with AFM volumes >15 000 nm3 formed at the base of T-loop structures. Collectively, results from our single-molecule imaging strongly suggest that multiprotein TRF2–TIN2 complexes promote T-loop formation with higher efficiency than TRF2 alone.
TIN2 facilitates TRF2-mediated association of TERRA to telomeric dsDNA
TERRA plays key roles in telomere maintenance by regulating telomerase activity, homologous recombination (HR) and heterochromatin structure (75). Tightly regulated telomeric R-loop formation promotes HR without severely impacting DNA replication at telomeres. Despite the importance of TERRA in regulating telomere functions, how TERRA specifically associates with chromosome ends is not well understood. TRF1 and TRF2 bind to TERRA in vitro (39). Results from enzyme-linked immunosorbent assay (ELISA) showed that TERRA binding affinity by TRF2 is similar to its affinity for single-stranded telomeric G-rich DNA. Human TERRA forms G-quadruplex structures both in vivo and in vitro (76–78), and TRF2 binds to TERRA through G-quadruplex dependent interactions (79). TRF2 mutant lacking the B domain (TRF2ΔB) decreases the TERRA localization at telomeres. In our experimental setup, TERRA (UUAGGG-12) in solution was bridged onto the telomeric regions by TRF2. TIN2 further increases the efficiency of ssRNA–dsDNA bridging, manifested by higher intensities from more UUAGGG-12 molecules bridged to each telomeric region on LT270 DNA tightropes. Results from our single-molecule studies provide direct experimental evidence for TERRA acting in trans (Figure 9) and supporting the notion that TERRA expressed from one chromosome participates in telomere function at multiple chromosome ends (80). TERRA has been proposed to serve as a scaffold for telomere-binding proteins to regulate telomere histone modifications and the telomere length (39). TERRA acts as a scaffold to recruit other RNA binding proteins, such as translocated in liposarcoma (TLS), also known as fused in sarcoma (FUS) protein to telomeres (81). Furthermore, TERRA expression and recruitment to telomeres are regulated by multiple factors that include DNA methylation at CpG island promoters, cell cycle, DDR at telomeres, telomere lengths and DNA repair factors such as RAD51 (75,82,83). Based on these previous observations, our results are consistent with a model in which TERRA recruited by TRF2–TIN2 links other biological signals from cells to telomeres by serving as a scaffold to introduce additional proteins and functions at telomeres (84).
The N-terminal TRFH domain of TIN2 (residues 2–20) binds to a short motif (residues 350–366) on the intrinsically disordered regions (IDRs) between the TRFH and Myb domains on TRF2 (85). We speculate that binding of TIN2 to TRF2 could regulate the dynamics TRF2 dimers and stability of TRF2-mediated DNA–DNA and DNA–RNA bridging. Consistent with this model, our recent single-molecule study showed that TIN2 increases the lifetimes of TRF1-mediated DNA–DNA bridging (50). Furthermore, TIN2L, but not TIN2S, is phosphorylated at its C-terminal domain by the casein kinase 2 (CK2) and the C-terminal domain of TIN2L stabilizes TRF2-TIN2 in vivo (86). Consistent with these results, single-molecule experiments in this study show that TIN2L is more efficient than TIN2S in promoting TRF2-mediated DNA–DNA and DNA–RNA bridging.
Importantly, the novel RSE-based telomeric DNA purification method enables us to investigate the structure and dynamics of shelterin proteins on native telomeric DNA purified from mouse tissues. Recently, long-read genome sequencing technologies generated for the first-time telomere-to-telomere assembly and uncovered the sequence heterogeneity in the human telomeric regions (87). Only a small number of degenerative telomeric (variant) sequences were found in between TTAGGG repeats. Even though the full sequences of telomeres from the mouse genome are not available for comparison, we do not expect the possible difference between small numbers of mouse and human telomeric variant sequences to affect the interpretation of results from our single-molecule studies.
In summary, this study based on single-molecule imaging using purified proteins and defined DNA substrates revealed a unique role of TIN2 in promoting TRF2-mediated higher-order DNA structures, including DNA compaction, T-loop formation and bridging of TERRA to telomeric dsDNA. Previous cell-based assays clearly demonstrated the existence of shelterin subcomplexes at telomeres, such as TRF1 or TRF2 complexes with TIN2 but without TPP1 and POT1 (41). This study on TRF2–TIN2 and the previous one on the full shelterin complex support a model in which shelterin assembly are heterogeneous, with shelterin subcomplexes and full complexes displaying distinct biophysical properties and contributing toward different functions at telomeres (8).
DATA AVAILABILITY
All data are available in the main text or the supplementary materials.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Opresko group at the University of Pittsburgh, the Smith group at New York University, and Johannes Dapprich at Generation Biotech for technical support.
Author Contributions: P.K. and R.B. designed and carried out the experiments, analyzed the data, and wrote the paper. H.P., A.C.D., C.M., R.B., J.H., Z.M., M. L. and C.Y. carried out experiments and/or analyzed the data. J.P., R.C.S., R.R., P. L.O. and H.W. designed the experiments and/or wrote the paper.
Contributor Information
Parminder Kaur, Physics Department, North Carolina State University, Raleigh, NC, 27695, USA; Center for Human Health and the Environment, North Carolina State University, Raleigh, NC 27695, USA.
Ryan Barnes, Department of Environmental and Occupational Health, University of Pittsburgh, UPMC Hillman Cancer Center, PA 15213, USA.
Hai Pan, Physics Department, North Carolina State University, Raleigh, NC, 27695, USA.
Ariana C Detwiler, Department of Environmental and Occupational Health, University of Pittsburgh, UPMC Hillman Cancer Center, PA 15213, USA.
Ming Liu, Physics Department, North Carolina State University, Raleigh, NC, 27695, USA.
Chelsea Mahn, Physics Department, North Carolina State University, Raleigh, NC, 27695, USA.
Jonathan Hall, Center for Human Health and the Environment, North Carolina State University, Raleigh, NC 27695, USA; Toxicology Program, North Carolina State University, Raleigh, NC, 27695, USA.
Zach Messenger, Toxicology Program, North Carolina State University, Raleigh, NC, 27695, USA.
Changjiang You, Department of Biology/Chemistry, Universität Osnabrück, Osnabrück 49076, Germany.
Jacob Piehler, Department of Biology/Chemistry, Universität Osnabrück, Osnabrück 49076, Germany.
Robert C Smart, Center for Human Health and the Environment, North Carolina State University, Raleigh, NC 27695, USA; Toxicology Program, North Carolina State University, Raleigh, NC, 27695, USA.
Robert Riehn, Physics Department, North Carolina State University, Raleigh, NC, 27695, USA.
Patricia L Opresko, Department of Environmental and Occupational Health, University of Pittsburgh, UPMC Hillman Cancer Center, PA 15213, USA.
Hong Wang, Physics Department, North Carolina State University, Raleigh, NC, 27695, USA; Center for Human Health and the Environment, North Carolina State University, Raleigh, NC 27695, USA; Toxicology Program, North Carolina State University, Raleigh, NC, 27695, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM123246 to H.W., R.R., and P.L.O.]; P30 ES025128 Pilot Project Grants to H.W. and P.K. through the Center for Human Health and the Environment at NCSU. Funding for open access charge: National Institutes of Health [R01GM123246].
Conflict of interest statement. None declared.
REFERENCES
- 1. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 2. Muraki K., Nyhan K., Han L., Murnane J.P.. Mechanisms of telomere loss and their consequences for chromosome instability. Front. Oncol. 2012; 2:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wright W.E., Tesmer V.M., Huffman K.E., Levene S.D., Shay J.W.. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997; 11:2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004; 116:273–279. [DOI] [PubMed] [Google Scholar]
- 5. Songyang Z., Liu D. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit. Rev. Eukaryot. Gene Expr. 2006; 16:103–118. [DOI] [PubMed] [Google Scholar]
- 6. Verdun R.E., Karlseder J.. Replication and protection of telomeres. Nature. 2007; 447:924–931. [DOI] [PubMed] [Google Scholar]
- 7. Giraud-Panis M.J., Pisano S., Poulet A., Le Du M.H., Gilson E.. Structural identity of telomeric complexes. FEBS Lett. 2010; 584:3785–3799. [DOI] [PubMed] [Google Scholar]
- 8. Erdel F., Kratz K., Willcox S., Griffith J.D., Greene E.C., de Lange T. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep. 2017; 18:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Palm W., Hockemeyer D., Kibe T., de Lange T.. Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 2009; 29:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Broccoli D., Chong L., Oelmann S., Fernald A.A., Marziliano N., van Steensel B., Kipling D., Le Beau M.M., de Lange T.. Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression and conserved protein domains. Hum. Mol. Genet. 1997; 6:69–76. [DOI] [PubMed] [Google Scholar]
- 11. Broccoli D., Smogorzewska A., Chong L., de Lange T.. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997; 17:231–235. [DOI] [PubMed] [Google Scholar]
- 12. Lei M., Podell E.R., Cech T.R.. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004; 11:1223–1229. [DOI] [PubMed] [Google Scholar]
- 13. Poulet A., Pisano S., Faivre-Moskalenko C., Pei B., Tauran Y., Haftek-Terreau Z., Brunet F., Le Bihan Y.V., Ledu M.H., Montel F.et al.. The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA. Nucleic Acids Res. 2012; 40:2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 15. Doksani Y., Wu J.Y., de Lange T., Zhuang X.. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013; 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benarroch-Popivker D., Pisano S., Mendez-Bermudez A., Lototska L., Kaur P., Bauwens S., Djerbi N., Latrick C.M., Fraisier V., Pei B.et al.. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol. Cell. 2016; 61:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Ly D., Low R.R.J., Frolich S., Bartolec T.K., Kafer G.R., Pickett H.A., Gaus K., Cesare A.J.. Telomere loop dynamics in chromosome end protection. Mol. Cell. 2018; 71:510–525. [DOI] [PubMed] [Google Scholar]
- 18. Janouskova E., Necasova I., Pavlouskova J., Zimmermann M., Hluchy M., Marini V., Novakova M., Hofr C.. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015; 43:2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y., Rai R., Zhou Z.R., Kanoh J., Ribeyre C., Yang Y., Zheng H., Damay P., Wang F., Tsujii H.et al.. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 2011; 18:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arat N.O., Griffith J.D.. Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere. J. Biol. Chem. 2012; 287:41583–41594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Griffith J., Bianchi A., de Lange T.. TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol. 1998; 278:79–88. [DOI] [PubMed] [Google Scholar]
- 22. Bianchi A., Stansel R.M., Fairall L., Griffith J.D., Rhodes D., de Lange T.. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999; 18:5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin J., Countryman P., Buncher N., Kaur P., Longjiang E., Zhang Y., Gibson G., You C., Watkins S.C., Piehler J.et al.. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014; 42:2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye J.Z., Donigian J.R., van Overbeek M., Loayza D., Luo Y., Krutchinsky A.N., Chait B.T., de Lange T.. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 2004; 279:47264–47271. [DOI] [PubMed] [Google Scholar]
- 26. Chen Y., Yang Y., van Overbeek M., Donigian J.R., Baciu P., de Lange T., Lei M.. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008; 319:1092–1096. [DOI] [PubMed] [Google Scholar]
- 27. Lim C.J., Zaug A.J., Kim H.J., Cech T.R.. Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat. Commun. 2017; 8:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiang Y.J., Kim S.H., Tessarollo L., Campisi J., Hodes R.J.. Telomere-associated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway. Mol. Cell. Biol. 2004; 24:6631–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takai K.K., Kibe T., Donigian J.R., Frescas D., de Lange T.. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell. 2011; 44:647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan J., Lan L.. The DNA secondary structures at telomeres and genome instability. Cell Biosci. 2020; 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Timashev L.A., De Lange T.. Characterization of t-loop formation by TRF2. Nucleus. 2020; 11:164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 33. Schoeftner S., Blasco M.A.. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008; 10:228–236. [DOI] [PubMed] [Google Scholar]
- 34. Azzalin C.M., Lingner J.. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015; 25:29–36. [DOI] [PubMed] [Google Scholar]
- 35. Groh M., Gromak N.. Out of balance: R-loops in human disease. PLoS Genet. 2014; 10:e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Porro A., Feuerhahn S., Delafontaine J., Riethman H., Rougemont J., Lingner J.. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014; 5:5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Graf M., Bonetti D., Lockhart A., Serhal K., Kellner V., Maicher A., Jolivet P., Teixeira M.T., Luke B.. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell. 2017; 170:72–85. [DOI] [PubMed] [Google Scholar]
- 38. Rippe K., Luke B.. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015; 22:853–858. [DOI] [PubMed] [Google Scholar]
- 39. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M.. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee Y.W., Arora R., Wischnewski H., Azzalin C.M.. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 2018; 25:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takai K.K., Hooper S., Blackwood S., Gandhi R., de Lange T.. In vivo stoichiometry of shelterin components. J. Biol. Chem. 2010; 285:1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin J., Countryman P., Chen H., Pan H., Fan Y., Jiang Y., Kaur P., Miao W., Gurgel G., You C.et al.. Functional interplay between SA1 and TRF1 in telomeric DNA binding and DNA-DNA pairing. Nucleic Acids Res. 2016; 44:6363–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaur P., Wu D., Lin J., Countryman P., Bradford K.C., Erie D.A., Riehn R., Opresko P.L., Wang H.. Enhanced electrostatic force microscopy reveals higher-order DNA looping mediated by the telomeric protein TRF2. Sci. Rep. 2016; 6:20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang Y., Wang H., Erie D.A.. Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods. 2003; 29:175–187. [DOI] [PubMed] [Google Scholar]
- 45. Wang H., Nora G.J., Ghodke H., Opresko P.L.. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem. 2011; 286:7479–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Countryman P., Fan Y., Gorthi A., Pan H., Strickland J., Kaur P., Wang X., Lin J., Lei X., White C.et al.. Cohesin SA2 is a sequence-independent DNA-binding protein that recognizes DNA replication and repair intermediates. J. Biol. Chem. 2018; 293:1054–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pan H., Jin M., Ghadiyaram A., Kaur P., Miller H.E., Ta H.M., Liu M., Fan Y., Mahn C., Gorthi A.et al.. Cohesin SA1 and SA2 are RNA binding proteins that localize to RNA containing regions on DNA. Nucleic Acids Res. 2020; 48:5639–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A.. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002; 277:41110–41119. [DOI] [PubMed] [Google Scholar]
- 49. Schmutz I., Mensenkamp A.R., Takai K.K., Haadsma M., Spruijt L., de Voer R.M., Choo S.S., Lorbeer F.K., van Grinsven E.J., Hockemeyer D.et al.. TINF2 is a haploinsufficient tumor suppressor that limits telomere length. Elife. 2020; 9:e61235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pan H., Kaur P., Barnes R., Detwiler A.C., Sanford S.L., Liu M., Xu P., Mahn C., Tang Q., Hao P.et al.. Structure, dynamics, and regulation of TRF1-TIN2-mediated trans- and cis-interactions on telomeric DNA. J. Biol. Chem. 2021; 297:101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaur P., Longley M.J., Pan H., Wang H., Copeland W.C.. Single-molecule DREEM imaging reveals DNA wrapping around human mitochondrial single-stranded DNA binding protein. Nucleic Acids Res. 2018; 46:11287–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hanish J.P., Yanowitz J.L., de Lange T.. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA. 1994; 91:8861–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rogalla P., Kazmierczak B., Rohen C., Trams G., Bartnitzke S., Bullerdiek J.. Two human breast cancer cell lines showing decreasing telomeric repeat length during early in vitro passaging. Cancer Genet. Cytogenet. 1994; 77:19–25. [DOI] [PubMed] [Google Scholar]
- 54. Parikh D., Fouquerel E., Murphy C.T., Wang H., Opresko P.L.. Telomeres are partly shielded from ultraviolet-induced damage and proficient for nucleotide excision repair of photoproducts. Nat. Commun. 2015; 6:8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dapprich J., Ferriola D., Mackiewicz K., Clark P.M., Rappaport E., D’Arcy M., Sasson A., Gai X., Schug J., Kaestner K.H.et al.. The next generation of target capture technologies - large DNA fragment enrichment and sequencing determines regional genomic variation of high complexity. BMC Genomics. 2016; 17:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shlyakhtenko L.S., Gall A.A., Filonov A., Cerovac Z., Lushnikov A., Lyubchenko Y.L.. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003; 97:279–287. [DOI] [PubMed] [Google Scholar]
- 57. Liu L., Kong M., Gassman N.R., Freudenthal B.D., Prasad R., Zhen S., Watkins S.C., Wilson S.H., Van Houten B.. PARP1 changes fromthree-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic acids Res. 2017; 45:12834–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kad N.M., Wang H., Kennedy G.G., Warshaw D.M., Van Houten B.. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol. Cell. 2010; 37:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dunn A.R., Kad N.M., Nelson S.R., Warshaw D.M., Wallace S.S.. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011; 39:7487–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hughes C.D., Wang H., Ghodke H., Simons M., Towheed A., Peng Y., Van Houten B., Kad N.M.. Real-time single-molecule imaging reveals a direct interaction between UvrC and UvrB on DNA tightropes. Nucleic Acids Res. 2013; 41:4901–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mazzucco G., Huda A., Galli M., Piccini D., Giannattasio M., Pessina F., Doksani Y.. Telomere damage induces internal loops that generate telomeric circles. Nat. Commun. 2020; 11:5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deininger P. Alu elements: know the SINEs. Genome Biol. 2011; 12:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maquat L.E. Short interspersed nuclear element (SINE)-mediated post-transcriptional effects on human and mouse gene expression: SINE-UP for active duty. Philos. Trans. R. Soc. Lon. Ser. B Biol. Sci. 2020; 375:20190344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Komazin-Meredith G., Mirchev R., Golan D.E., van Oijen A.M., Coen D.M.. Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proc. Natl. Acad. Sci. USA. 2008; 105:10721–10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hemann M.T., Greider C.W.. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000; 28:4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pan H., Bilinovich S.M., Kaur P., Riehn R., Wang H., Williams D.C. Jr. CpG and methylation-dependent DNA binding and dynamics of the methylcytosine binding domain 2 protein at the single-molecule level. Nucleic Acids Res. 2017; 45:9164–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaminker P.G., Kim S.H., Desprez P.Y., Campisi J.. A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell Cycle. 2009; 8:931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith S. The long and short of it: a new isoform of TIN2 in the nuclear matrix. Cell Cycle. 2009; 8:797–798. [DOI] [PubMed] [Google Scholar]
- 69. Pike A.M., Strong M.A., Ouyang J.P.T., Greider C.W.. TIN2 functions with TPP1/POT1 to stimulate telomerase processivity. Mol. Cell. Biol. 2019; 39:e00593-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim S.H., Kaminker P., Campisi J.. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999; 23:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nora G.J., Buncher N.A., Opresko P.L.. Telomeric protein TRF2 protects Holliday junctions with telomeric arms from displacement by the Werner syndrome helicase. Nucleic Acids Res. 2010; 38:3984–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Opresko P.L., Fan J., Danzy S., Wilson D.M., Bohr V.A. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005; 33:1230–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stansel R.M., de Lange T., Griffith J.D.. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001; 20:5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cesare A.J., Karlseder J.. A three-state model of telomere control over human proliferative boundaries. Curr. Opin. Cell Biol. 2012; 24:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bettin N., Oss Pegorar C., Cusanelli E.. The emerging roles of TERRA in telomere maintenance and genome stability. Cells. 2019; 8:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Collie G.W., Haider S.M., Neidle S., Parkinson G.N.. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010; 38:5569–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Randall A., Griffith J.D.. Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J. Biol. Chem. 2009; 284:13980–13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu Y., Suzuki Y., Ito K., Komiyama M.. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. USA. 2010; 107:14579–14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Biffi G., Tannahill D., Balasubramanian S.. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012; 134:11974–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Montero J.J., Lopez de Silanes I., Grana O., Blasco M.A.. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016; 7:12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kondo K., Mashima T., Oyoshi T., Yagi R., Kurokawa R., Kobayashi N., Nagata T., Katahira M.. Plastic roles of phenylalanine and tyrosine residues of TLS/FUS in complex formation with the G-quadruplexes of telomeric DNA and TERRA. Sci. Rep. 2018; 8:2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Feretzaki M., Renck Nunes P., Lingner J.. Expression and differential regulation of human TERRA at several chromosome ends. RNA. 2019; 25:1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Feretzaki M., Pospisilova M., Valador Fernandes R., Lunardi T., Krejci L., Lingner J.. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. 2020; 587:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lalonde M., Chartrand P.. TERRA, a multifaceted regulator of telomerase activity at telomeres. J. Mol. Biol. 2020; 432:4232–4243. [DOI] [PubMed] [Google Scholar]
- 85. Hu C., Rai R., Huang C., Broton C., Long J., Xu Y., Xue J., Lei M., Chang S., Chen Y.. Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex. Cell Res. 2017; 27:1485–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nelson N.D., Dodson L.M., Escudero L., Sukumar A.T., Williams C.L., Mihalek I., Baldan A., Baird D.M., Bertuch A.A.. The C-terminal extension unique to the long isoform of the shelterin component TIN2 enhances its interaction with TRF2 in a phosphorylation- and dyskeratosis congenita cluster-dependent fashion. Mol. Cell. Biol. 2018; 38:e00025-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A.et al.. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020; 585:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.