Abstract
Histones, ubiquitous in eukaryotes as DNA-packing proteins, find their evolutionary origins in archaea. Unlike the characterized histone proteins of a number of methanogenic and themophilic archaea, previous research indicated that HpyA, the sole histone encoded in the model halophile Halobacterium salinarum, is not involved in DNA packaging. Instead, it was found to have widespread but subtle effects on gene expression and to maintain wild type cell morphology. However, the precise function of halophilic histone-like proteins remain unclear. Here we use quantitative phenotyping, genetics, and functional genomics to investigate HpyA function. These experiments revealed that HpyA is important for growth and rod-shaped morphology in reduced salinity. HpyA preferentially binds DNA at discrete genomic sites under low salt to regulate expression of ion uptake, particularly iron. HpyA also globally but indirectly activates other ion uptake and nucleotide biosynthesis pathways in a salt-dependent manner. Taken together, these results demonstrate an alternative function for an archaeal histone-like protein as a transcriptional regulator, with its function tuned to the physiological stressors of the hypersaline environment.
Graphical Abstract
Graphical Abstract.

The sole histone-like protein encoded by a hypersaline-adapted archaeon functions in transcriptional regulation.
INTRODUCTION
Phylogenetic analysis has shown that the histone fold domain originated in the Archaea (1–3). Histone proteins play a vital role in genome compaction and regulation of gene expression in eukaryotes (4). The four core eukaryotic histones (H3, H4, H2A, H2B) share a histone fold domain, which is involved in histone dimerization and DNA-binding (5–7). Proteins containing the histone fold are present in all known major archaeal lineages (7). Archaeal histone-like proteins have been most extensively characterized in species representing the euryarchaeal superphylum, with much work focusing on the thermophilic archaeal species Methanothermus fervidus (8) and Thermococcus kodakarensis (9–11). In vitro structural studies from these species demonstrate strong conservation between archaeal and eukaryotic histones in terms of histone fold, multimeric protein structure, and DNA wrapping (10,12–13). However, key differences from eukaryotes have also been noted (7,14): archaeal histones form extended polymeric structures called hypernucleosomes (10,13,15). These structural data help explain results from in vivo data. MNase digests yield fragments in multiples of 30–60 bp (11,15–16) and gene expression is significantly altered by histone binding (11,17). Like eukaryotic histones, archaeal histones can also hinder elongation (18) or inhibit the binding of site-specific transcription factors (TFs) through competition (19) to influence global transcription levels. These histones may act as the major chromatin protein. These studies led to the oft-noted hypothesis that many features of archaeal histone structure and function resemble those of eukaryotes in terms of genome compaction and gene expression, with some key differences (7,15).
Recent evidence in other model systems call for further testing of this hypothesis. For example, a deletion mutant of the sole histone of Methanosarcina mazei was viable, but exhibited reduced growth when exposed to radiation (20). Phylogenetics and molecular dynamics simulations in other model methanogens that encode multiple histone variants suggest various functions in the chromatin environment (21). In other model species, a dynamic variable set of bacterial-like chromatin and archaeal-specific proteins organize the genome rather than histones (22,23). Phylogenetic and proteomics evidence in hyperthermophiles suggests that chromatin compaction allows DNA stability to prevent unwanted transcription by promoter melting at high temperature (24). This hypothesis has been substantiated in vitro: plasmid DNA strand dissociation is prevented by histone binding at 90°C (25). Histone point mutants that cannot compact DNA exhibit differential expression of specific genomic regions (11). Previous work from our group demonstrated an alternative regulatory function for HpyA, the sole histone of the hypersaline-adapted species Halobacterium salinarum (26). HpyA is dispensable for cell viability but important for maintaining wild type gene expression and cell shape under optimum growth conditions. HpyA protein levels were too low to facilitate genome-wide DNA compaction (26). Together these findings reveal an expanding landscape of diverse histone functions across archaeal lineages. These functions are selected for by the diverse and sometimes extreme environments of archaea. However, the function of histone-like proteins in hypersaline-adapted archaea remains understudied relative to other archaeal lineages.
Halophilic archaea have adapted to survive extreme osmotic pressure (up to saturated NaCl) in their natural salt lake environments by counterbalancing with up to 4M potassium ions in the cytoplasm (27). Due to the resultant highly ionic cytoplasm, haloarchaeal proteins, including the histone protein, have evolved a negatively charged surface (28). This is in contrast to all other known species, where the positively charged surface of histones facilitates DNA-histone interactions (3,26). It has previously been observed in vitro that naked DNA under moderate salinity tends to spontaneously form structures similar to the beads-on-a-string observed with histone-bound DNA, with increasing compaction and even aggregation occurring at higher concentrations (29). These data call into question the need for protein-based genome compaction. In addition, proteomics data from our prior work demonstrated that the protein levels of HpyA are very low in Hbt. salinarum, and HpyA expression levels change little throughout growth (26). Based on these results and given the unusual chemistry of the haloarchaeal saturated salt cytoplasm, here we hypothesize that the non-canonical function of HpyA in gene regulation is linked to the unique hypersaline cytoplasmic environment of Hbt. salinarum.
We tested this hypothesis using a battery of in vivo quantitative phenotyping and functional genomics assays. Growth rate and cell morphology in low sodium was affected in the ΔhpyA deletion strain, confirming an association between the presence of the gene (and its product) and the effects of low sodium concentration. Protein-DNA binding assays (ChIP-seq) revealed reproducible, salt-dependent, genome-wide binding of HpyA at nearly 60 discrete sites—a binding pattern too sparse to coat or compact the genome. However, the high prevalence of binding within gene bodies suggests that the mechanism of regulation differs substantially from that of canonical TFs. Integration of DNA binding with transcriptomics data revealed direct regulation of iron uptake by HpyA. Global, indirect regulation of transport of other ions, biosynthesis of purines, and DNA replication and repair was also observed. Together, these results suggest that HpyA functions as a specific, direct transcriptional regulator of metal ion balance. HpyA thereby maintains growth rate and rod-shaped cell morphology during hypo-osmotic stress.
MATERIALS AND METHODS
Strains, media and general culturing
Strains used in this study have been described in Dulmage et al. (26), summarized in Supplementary Table S1. All strains were constructed from a Hbt. salinarum NRC-1 background with ura3 (encodes uracil biosynthesis functions) gene deleted to enable uracil counter-selection (30). Growth assays were carried out using strain MDK407 (Δura3) as the parent strain (control, referred to here as wild type, or WT) and KAD100 (Δura3ΔhpyA) as the ΔhpyA deletion strain.
For ChIP-seq, strains carrying the hpyA gene tagged at its C-terminus with the hemagglutinin (HA) epitope were used (26). The control strain was AKS134 (ΔhpyA deletion carrying the empty vector pMTF-cHA). The experimental strain was KAD128, which contained the pKAD17 plasmid expressing HpyA-HA driven by its native promoter (primers and plasmids given in Supplementary Table S1) (31). pKAD17 was generated by: (a) insertion of hpyA into the pMTF-cHA plasmid upstream of the HA tag sequence (between the NdeI and HindIII restriction sites); and (b) replacement by isothermal ligation of the Pfdx promoter of the plasmid with the Prpa200 native promoter sequence of HpyA at the KpnI site.
The media used for all experiments was Hbt. salinarum complete media (CM) containing 250 g/l NaCl, 20 g/l MgSO4•7H2O, 3 g/l trisodium citrate dihydrate, 2 g/l KCl, 10 g/l Bacteriological peptone (Oxoid). pH was adjusted to 6.8. Media were supplemented with 50 μg/ml uracil to compensate for the uracil auxotrophy of Δura3 parent and derivative strains. Reduced salt media was made identically except for NaCl, which was reduced to 199 g/l (3.4 M). For plasmid strains, 1 μg/ml mevinolin (AG Scientific) was added to liquid medium and 2.5 μg/ml to solid media to maintain selective pressure on the plasmid.
Cells were routinely streaked fresh from frozen stock onto solid medium. Individual colonies were picked from plates and inoculated into 5 ml CM (with additives when necessary) and allowed to grow for ∼4 days at 42°C at 225 rpm in a shaking incubator until stationary phase was reached. These starter cultures were diluted by sub-culturing to OD600 ∼0.02 into 50 ml of media indicated in the figures and grown until harvesting as described below.
Growth and microscopy
For growth curve phenotyping, 9 biological replicates of Δura3 (MDK407) and ΔhpyA (KAD100) strains were cultured in 125 ml flasks at 42°C in a shaking incubator. Optical density (OD) measurements were taken at time zero, then at 3–4 hour intervals following the initial lag phase of ∼12 h. Raw growth data are provided in Supplementary Table S2. Resultant growth curves were fit by logistic regression to calculate the maximum instantaneous growth rate (μmax) using the R package grofit (32). The code for analysis and visualization of these growth data are contained in https://github.com/amyschmid/HpyA_codes.
For microscopy, cultures of Δura3, ΔhpyA and ΔhpyA/pKAD17 (strain KAD128) were each grown to mid-exponential phase. 8 μl aliquots were placed on a thin, flat, agarose pad impregnated with 4.2 M NaCl as described (33). Cells were imaged at 100× using a Zeiss Axio Scope A1 microscope with a Pixelink PL-E421M camera. Images were analyzed for circularity using the MicrobeJ package within the ImageJ software (34). In this context, circularity is defined as the measure of deviation from a perfect circle, where 1 is a perfect circle and 0 is a polygon with 1 side infinitely longer than the other. Given that circularity distributions were skewed, adjusted bootstrap percentile corrected 95% confidence intervals were calculated by 1000-fold ordinary non-parametric bootstrap resampling of the median with replacement. The boot package in the R coding environment was used for these calculations.
ChIP-seq experiments
One biological replicate colony of AKS134 (Empty vector control) and four replicates of KAD128 (expressing HpyA-HA) were cultured as described above. The 50 mL cultures were grown in 125 ml flasks and their growth was monitored by OD600 until the time for harvesting (exponential phase: 36–50 h, OD ∼0.2–0.35, growth rate ∼0.059 h–1; stationary phase: ∼70–140 h, OD ∼1.4–1.7, growth rate ∼0.032 h–1). Strains were PCR-checked for the presence of the plasmid expressing hpyA-HA prior to each experiment (see Supplementary Table S1 for primers).
Harvested cells (45 ml) were immediately cross-linked using 1.4 ml 37% formaldehyde (final concentration = 1% v/v) and immuoprecipitated using Abcam HA-specific antibody (catalog #ab9110) as described in Wilbanks et al. (35), with certain modifications to the protocol: the cross-linking reaction was allowed to proceed for 20 min, and cell pellets were resuspended in 800 μl lysis buffer. Resulting DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and then ethanol precipitation. Library preparation and single-end sequencing was carried out by the Duke Center for Genomic and Computational Biology Sequencing and Genomic Technologies core facility using the Illumina HiSeq4000 instrument.
Analysis of ChIP-seq data
Gzipped FastQ files (accession: PRJNA703048, GEO: GSE182514) were analyzed using FastQC software. Outputs analyzed included read sequence quality, length distribution and presence of adapters. Adapters were trimmed from the reads using Trim Galore!, and these trimmed sequences were aligned to the Hbt. salinarum NRC-1 genome (RefSeq ID GCF_000006805.1, assembly ID ASM680v1) to generate a SAM file using Bowtie2 with default parameters. End-to-end alignment was suitable for trimmed reads (36). FastQC and Trim Galore! are available online at http://www.bioinformatics.babraham.ac.uk/projects/ (2015 version). The SAM files were converted to binary (BAM), sorted and indexed using SAMtools (37). Sorted BAM files were used for peak calling. WIG files for easy visualization were also generated using SAMtools, with coverage recorded every 10 bp. All code used to analyze ChIP-seq data are available in File S1 at https://github.com/amyschmid/HpyA_codes.
The sorted BAM files were used for peak-calling with MACS2 (38) version 2.1.1 callpeak function. Parameters were: nomodel, qval = 0.05 cutoff. Called peaks were combined across replicates using the multiBedIntersect function of the bedtools package (39). Only peaks detected in at least two biological replicate experiments were kept in downstream analyses. Genes within 500 bp of these reproducible peaks were annotated using the IRanges package in R (40). Resultant peaks were then manually curated to remove the following: (a) false positives caused by local variability in input control sequencing read depth; (b) local duplications and deletions associated with transposases and integrases; (c) one peak that was also detected in the HA tag-alone input control; (d) peaks located nearby redundant genes. Details of the code and dependencies for the entire workflow for peak calling and visualization are noted in the github repository https://github.com/amyschmid/HpyA_codes.
Resultant peaks regions (start to end of peak footprint) were then annotated based on their genomic context (details in Supplementary Table S3). Promoters were defined as the region from 500bp upstream of the translation start site [many halophile transcripts are leaderless (41)]. To classify binding peak center locations as ‘genic’ or ‘intergenic’ for the purpose of hypergeometric tests, the location of the center of the ChIP-seq peak (mid-point between start and stop chromosomal coordinates) relative to each type of genomic feature was tabulated. The code used to make this classification is in https://github.com/amyschmid/HpyA_codes. Operons were computationally predicted using the Operon-Mapper tool (42) and integrated with empirical predictions from Koide et al. (41). Classification of TrmB binding locations are given directly in reference (43) and significance of enrichment was computed using the hypergeometric test in R. Classification and computation of enrichment P-values for RosR binding locations [from reference (44)] were computed using BEDtools ‘fisher’ function (39).
RNA-seq experiments
Six biological replicate cultures of strains MDK407 (parent) and KAD100 (ΔhpyA) were cultivated as described above in either optimal salt (4.2 M NaCl) or low salt (3.4 M NaCl) media. Growth was monitored using OD600 until harvesting (exponential phase was defined as: ∼31–34 h of growth, OD ∼0.1–0.4 depending on the strain and medium).
A 4.2 ml aliquot of each culture was removed and centrifuged for 30 s at 21 000 × g in an Eppendorf tabletop centrifuge. The supernatant was discarded and the cell pellet was immediately plunged into liquid nitrogen and stored 1–7 days at −80°C. Extraction of RNA from these pellets was carried out using the Agilent Absolutely RNA Miniprep kit following the manufacturer's protocol, with an extended on-column DNase incubation of 45–60 min. Resultant RNA samples were checked for: (a) genomic DNA contamination using PCR with 200 ng input RNA and 35 amplification cycles using primers listed in Supplementary Table S1; (b) concentration using 260/280 nm ratio in a Nanodrop spectrophotometer; (c) quality using the Agilent Bioanalyzer RNA Nano 6000 chip (RNA Integrity Number (RIN) > 9.0). For each strain and condition, rRNA was removed from 3 replicates with NEBNext Bacteria rRNA Depletion Kit (New England Biolabs), while the other 3 were treated with NEBNext Depletion Core Reagent Set using custom probes targeted to Haloferax volcanii rRNA (Martinez-Pastor and Sakrikar, unpublished). These custom probes were designed using the NEBNext Custom RNA Depletion Design Tool (https://depletion-design.neb.com/). rRNA depletion was verified using the Bioanalyzer RNA chip. The NEBNext Ultra II Directional RNA Library Prep Kit for Illumina was used for preparing sequencing libraries, and cDNA libraries were quality-checked using the High-Sensitivity DNA Bioanalyzer chip. Paired-end sequencing was carried out at the Duke Center for Genomic and Computational Biology Sequencing core facility using the Novaseq6000 instrument (Illumina).
RNA-seq data analysis
For analysis of sequencing data, paired FastQ files were trimmed and checked for quality using Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/) and aligned to the genome using Bowtie2 (36). SAMtools was used to generate, sort, and index BAM files (37). The count function of HTSeq (45) was used to create a file assigning the number of reads to each gene (see File S1 within the Github repository for details). Outlier samples were removed from further analysis using Strong PCA (46) (https://github.com/amyschmid/HpyA_codes). The R package DESeq2 (47) was used to normalize counts and batch correct across replicates for each strain and genotype (using DEseq2 default parameters). Significant differential gene expression analysis using DESeq2 applied three pairwise contrasts: ΔhpyA versus WT in optimal salt, ΔhpyA versus WT in reduced salt, and reduced vs optimal salt in a WT background. For each contrast, reproducibility and quality was checked across replicates using dispersion, MA, and volcano plots. For each contrast, Benjamini-Hochberg (BH) adjusted (48) Wald test P < 0.05 (default within DESeq2) was used as the criterion for significant differential expression (results in Supplementary Table S4).
Averaged normalized counts across biological replicates for each strain and stress treatment were then mean and variance standardized and subjected to Kmeans clustering using the factoextra package in R, which also determines the best value for K (49). Resultant gene clusters were then subclustered using Kmeans and visualized using ggplot2 (50) and pheatmap (51) functions in R (https://github.com/amyschmid/HpyA_codes). This clustering procedure was carried out twice, once with genes differentially expressed in both reduced and optimal salt, and then excluding genes differentially expressed in optimal salt. Results of the clustering are given in Supplementary Table S5. For analysis of gene functional enrichments, the hypergeometric test P-value of enrichment for differentially expressed genes was calculated. Resultant P-values were BH-corrected for multiple hypothesis testing. The archaeal Clusters of Orthologous Genes (arCOG) functional ontology was used for functional assignments (52), results are listed in Supplementary Table S6.
RESULTS
HpyA is important for wild type growth and morphology in low salinity stress conditions
To test the hypothesis that HpyA plays a role in salt stress, we compared the growth rate of Hbt. salinarum Δura3 (parent strain, hereafter referred to as wild type or ‘WT’) to ΔhpyA cells in rich complete medium with salt concentrations supporting optimal growth (CM, 4.2 M NaCl) and CM with reduced salt (3.4 M NaCl). As expected from previous observations (26), instantaneous growth rate (μmax) under optimal salt of the WT strain was statistically indistinguishable from that of ΔhpyA (Figure 1A, Supplementary Table S2, Supplementary Figure S1). Reduced salt slows the instantaneous growth rate (μmax) of WT cultures to 89% of that in standard conditions. In contrast, ΔhpyA cultures show significant growth impairment in reduced salt relative to WT, growing at 67% of their standard rate. (Figure 1B; unpaired two-sample t-test P < 0.008).
Figure 1.
The ΔhpyA strain is impaired for growth under reduced salt conditions. (A) Spline-smoothed growth curves for the Δura3 parent (‘WT’, blue curves) compared to the ΔhpyA mutant (red curves) under optimal salt (dark colors) and reduced salt (light colors). For each strain under each condition, the mean of nine biological replicate growth curves is shown with surrounding shaded region representing the 99% confidence interval (CI, in some curves, shading is not visible because the mean line and CI overlap). (B) Dot plots of relative maximum instantaneous growth rate (μmax) for each of the WT and ΔhpyA strains. Each dot represents one of nine biological replicate trials measuring the μmax for each strain under reduced salt compared with its own growth in optimal conditions. Horizontal bars represent the median of each distribution.
Cell morphology of Hbt. salinarum changes from rod-shaped to circular in the presence of low salt due to disruption of charges in the glycoprotein surface layer (S-layer) (53–57). Our previous work demonstrated that the ΔhpyA strain exhibits similar circularity in standard conditions (26). To further test the hypothesis that HpyA plays a role in the salt stress response, we used phase contrast microscopy to visualize the combined effects of reduced salt and hpyA deletion on cell shape. From the images, we quantified circularity of individual cells (where 1 indicates a perfectly circular cell). In media containing optimal salt concentrations, WT cells are primarily rod-shaped, whereas the ΔhpyA cells are statistically significantly rounder (Figure 2, non-parameteric bootstrapped 95% confidence intervals of the medians of these distributions do not overlap, see Methods). In reduced salt, WT cell morphology was more circular: the median of the distribution was not significantly different from that of ΔhpyA in optimal salt. ΔhpyA morphology in reduced salt was the most circular of all strain-by-genotype combinations, indicating that this strain's morphology is strongly impacted by reduced salt.
Figure 2.

Circularity of Hbt. salinarum increases when hpyA is deleted under reduced salt. In dot plot, dots represent circularity measurements of individual cells. Horizontal bars are the median of the distribution in each strain under each condition. Shaded regions represent the 95% bias-corrected confidence interval from bootstrap resampling (see Materials and Methods). Below, representative micrograph images are shown for cells of WT, ΔhpyA, and complemented strain (ΔhpyA + hpyA-HA, i.e. pKAD17, Supplementary Table S1) cells in optimal and reduced salt media. Scale bar is 5 μm and consistent across images. Colors are as in Figure 1. Number of cells counted: WT in optimal salt, n = 363; WT in reduced salt, n = 383; ΔhpyA in optimal salt, n = 188; ΔhpyA in reduced salt, 187; complemented strain in optimal salt, n = 313; complemented strain in reduced salt, n = 360.
Growth and morphology defects are significantly complemented by expression of hpyA from its native promoter in trans on a plasmid (ΔhpyA + hpyA-HA, Figure 2). Whole genome resequencing of the ΔhpyA strain also demonstrated that: (a) second site suppressor mutations were absent; and (b) deletion of hpyA was complete through all chromosomal copies (Supplementary Table S1). Hbt. salinarum is highly polyploid (58), which necessitates validation that all gene copies have been deleted. These results indicate that ΔhpyA phenotypes are solely attributable to the deletion of hpyA.
Because cell shape differences can lead to alterations in light scattering in a spectrophotometer (59), as a control, we calculated CFU/mL by dilution plate counting. We found that OD600 measurements were well correlated with CFU counts for both strains and media preparations. In optimal salt, CFU to OD Spearman correlation of WT cultures was ρ = 0.81 (P = 0.00081), ΔhpyA ρ = 0.64 (P = 0.015). In reduced salt, WT correlation was ρ = 0.93 (P < 2.2 × 10–16), ΔhpyA ρ = 0.78 (P = 0.001; Supplementary Figure S2). This indicates that the ΔhpyA growth defect observed in reduced salt (as measured by optical density) is due to differences in growth and not an artefact of the shape change. Taken together, these batch culture (Figure 1) and single cell microscopy (Figure 2) quantitative phenotype data suggest that HpyA is important for maintaining wild type morphology and growth in response to hypo-osmotic salt stress.
HpyA binds genome-wide in a salt-specific manner
To determine which genes are potential targets of transcriptional regulation by HpyA, we performed genome-wide DNA binding location analysis using chromatin immunoprecipitation coupled to sequencing (ChIP-seq). For this purpose, we generated an ΔhpyA strain expressing in trans HpyA translationally fused at its C-terminus to the hemagglutinin (HA) epitope tag. This fusion construct was driven by its native promoter (see Methods and Supplementary Table S1 for strain details). As described above, expression of hpyA-HA in trans complemented the circularity defect of ΔhpyA, demonstrating that HA tag and plasmid-based expression does not interfere with wild type function of HpyA (Figure 2). Based on the ΔhpyA phenotypes observed (Figures 1 and 2), the ChIP-seq experiments were performed at both physiological and reduced salt concentrations in both mid-exponential and stationary phase. HpyA binding was enriched relative to the background input control at a total of 59 discrete genomic locations (ChIP-seq peaks) across all conditions tested (Supplementary Table S3). These 59 peaks were consistently detected in reduced salt across growth phases and biological replicate experiments (Figure 3A and B), but only five of these peaks remained bound in optimal salt conditions. Of the low salt peaks, 35 were detected exclusively during exponential growth phase, 14 exclusively during stationary phase, and nine across both growth phases (Figure 3C). HpyA protein levels do not change significantly across the growth curve (26,60), suggesting that such differential binding occurs at the level of HpyA activity. Because HpyA binds DNA primarily under low salt conditions, these results corroborate the growth and morphological impairments of ΔhpyA cells observed in early log phase under reduced salt conditions (Figure 2).
Figure 3.

ChIP-seq of HpyA shows salt and growth phase dependent binding patterns. (A) Chromosome-wide binding pattern (measured as read-depth of IP/Input) of HpyA-HA in optimal salt and exponential growth phase (red), optimal salt and stationary phase (pink), reduced salt and exponential phase (dark blue), reduced salt and stationary phase (light blue). (B) Reproducible peaks detected across at least two of four biological replicates for each condition – shorter peaks represent those found in two replicates only, while peaks at full height were detected in at least three replicates for that particular condition. Note that peaks shown in tag-alone control have been removed from the other conditions and from further analysis. (C) Venn diagram indicating the number of peaks detected in the different conditions. Circles are not scaled by number of peaks.
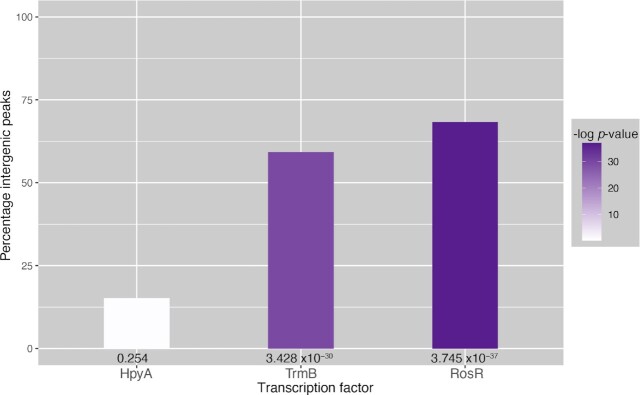
HpyA binding peaks were located nearby 86 genes (within the gene coding region or 500 bp upstream of the gene start in the promoter region). Few of the HpyA binding sites are located within non-coding regions of the genome (15.2%, P = 0.253). In contrast, other previously characterized Hbt. salinarum TFs bind in a sequence-specific manner with significant preference non-coding regions [Figure 4, (43,44)]. Binding of HpyA is also not statistically enriched for binding withing gene coding regions (P = 0.615). This high number was as expected because, like many archaeal genomes, the Hbt. salinarum genome is dense with coding sequences (86%). Taken together these DNA binding results suggest that, unlike canonical histone proteins of eukaryotes and other archaeal species, HpyA binds in a salt-specific manner to a restricted set of sites genome-wide. However, unlike canonical TFs, HpyA binds apparently without preference for coding vs non-coding regions.
Figure 4.
HpyA binds without preference for coding vs non-coding regions. Height of the bar graph corresponds to percentage of ChIP-seq peaks in non-coding (intergenic) regions of the genome. Colors of the bars are shaded by negative log10P-value of enrichment of peak locations in non-coding regions (see scale at right). Actual P-values of enrichment calculated by hypergeometric test for each TF are also written below each bar. HpyA binding locations (left) are compared with those for characterized TFs TrmB and RosR in Hbt. salinarum (43,44).
HpyA functions primarily as an activator of genes encoding ion transport and metabolic proteins
Based on the quantitative phenotyping and ChIP-seq data, we reasoned that HpyA may regulate gene expression in response to salt stress. To test this hypothesis, we performed transcriptome profiling experiments in WT versus ΔhpyA strains in both optimal and reduced salt using RNA-seq (see Methods). In the WT strain, over one-third of the genes in the transcriptome were significantly differentially expressed during exponential growth phase in reduced salt compared to optimal salt conditions (P < 0.05; 882 genes; 37% of genome; Supplementary Table S4). Of the 37 genes previously identified by microarray analysis (61), 22 genes were also identified as significantly differentially expressed in the current dataset. For these 22 genes, the fold-change in expression was strongly and significantly correlated across the two datasets (ρ = 0.86, P < 2.2 × 10–16). Our results therefore recapitulate but also extend previous observations that Hbt. salinarum mounts a strong, reproducible, and global regulatory response to hypo-osmotic stress.
To determine the extent of HpyA’s regulatory reach, gene expression ratios (ΔhpyA:WT) were calculated during mid-exponential growth in optimal salt and reduced salt conditions (in two separate DEseq2 analyses, see Methods). A total of 168 differentially expressed genes (DEG) were detected, 143 of which were significantly altered in reduced salt and 46 in optimal salt in ΔhpyA vs WT (Figure 5A, Supplementary Table S4). Of these, 121 genes were uniquely differentially expressed in response to low salt. These genes are significantly enriched for a wide variety of functions critical to maintaining cell growth and physiology in adverse conditions, especially ion transport and nucleotide metabolism (hypergeometric test P < 0.05 enrichment in arCOG categories (52), Figure 5B, Supplementary Table S6). Across both optimum and reduced salt conditions, the expression of 21 genes was significantly affected by hpyA deletion. These genes encode predicted functions in DNA recombination, replication, and repair pathways including RadA, DNA topoisomerase VI, and RPA family proteins (Supplementary Table S4, Supplementary Table S6).
Figure 5.
HpyA regulates gene expression in a salt-dependent manner. (A) Venn diagram illustrates the number of genes differentially expressed due to knockout of hpyA in different conditions. Genes with significant ΔhpyA: WT ratios in optimal salt are shown in red, genes with significant ΔhpyA : WT ratios low salt in pink, genes with significant low : optimal salt ratios in WT in blue. (B) arCOG enrichment of differentially expressed genes. X-axis shows the number of differentially expressed genes in each category that are annotated in the arCOG ontology, y-axis lists the arCOG category functions and short-hand single letter designations. Categories enriched in low salt are listed in pink text, categories enriched across conditions in black. Bars are shaded by Benjamini-Hochberg corrected (48) P-values of significance of enrichment according to the scale shown in the legend.
To determine the role of HpyA in the activation or repression of these genes, we performed K-means clustering analysis of normalized read count data for gene expression across the four conditions tested (ΔhpyA in low salt, ΔhpyA in optimal salt, WT in low salt, WT in optimal salt, details in Materials and Methods). We first analyzed the expression of the 21 genes that are differentially regulated the ΔhpyA strain in both optimal and reduced salt conditions. These 21 genes fall into two clear categories—10 genes downregulated in the ΔhpyA strain and 11 genes upregulated (Figure 6A, Supplementary Table S5). As noted above, genes across these two clusters are significantly enriched for DNA recombination, replication, and repair functions (eight genes). HpyA binding was detected in ChIP-seq near only one of these genes (ssb, encoding single-stranded DNA binding protein, Supplementary Table S7). HpyA binding was not detected for the 20 other genes in this cluster, indicating indirect regulation by HpyA.
Figure 6.
HpyA-dependent regulon shows diverse expression patterns in the conditions tested. (A) Clustering heatmap of genes differentially expressed in response to hpyA deletion across both optimal and low salt conditions. Each column corresponds to the genotype in each condition and rows represent the averaged normalized counts for each gene. Each row is self-standardized for normalization. Genes labeled with certain colors to the right of the heat map represent gene functional categories (see legend for colors). Dots next to genes represent monocistronic genes, vertical bars indicate differentially expressed operons. (B) Clustering heatmap of genes differentially expressed in response to hpyA deletion in low salt conditions alone. (C) Normalized reads for 3 selected direct targets of HpyA. Each box corresponds to a particular gene target indicated in the heatmap. In each panel, ChIP-seq data are shown in the top box, RNA-seq data in the middle, genomic context at bottom. ChIP-seq y-axes represent the ratio of IP to input control (whole cell extract, or WCE). RNA-seq y-axes represent read depth for WT in reduced salt (grey traces) and KO in reduced salt (black). Genonmic context images include the differentially expressed gene(s) (black arrows) and neighboring genes (grey arrows).
A separate clustering analysis of the 122 genes differentially expressed only in reduced salt in ΔhpyA yielded two main patterns (Figure 6B, Supplementary Table S5). In cluster 1, genes are elevated in expression in the ΔhpyA background under reduced salt relative to WT, whereas cluster 2 genes are downregulated. Cluster 2 includes 64% of genes differentially expressed in low salt, suggesting HpyA functions as an activator in the majority of cases in reduced-salt conditions.
To more clearly observe the gene expression patterns and the function of differentially expressed genes, we further divided these two main clusters, resulting in a total of 4 sub-clusters (Figure 6B, Supplementary Table S5). Subcluster 1.1 contains 11 genes whose expression pattern is downregulated in WT in reduced salt but upregulated in ΔhpyA. This cluster includes 3 genes predicted to encode ion transport proteins (chloride, iron, and other metals). Subcluster 1.2 contains 28 genes that are upregulated in reduced salt in WT but more heavily upregulated in reduced salt in the knockout strain. The function of genes in subcluster 1.2 are varied and not statistically enriched for a particular function. However, notable among genes in cluster 1.2 include transcription factor B (TFB), four amino acid biosynthesis genes, and HelA ATP-dependent DNA helicase (Figure 6A, Supplementary Table S5). ChIP-seq enrichment for HpyA binding was not detected nearby any of the genes in subclusters 1.1 and 1.2, suggesting indirect regulation (Figure 6B, Supplementary Table S7). HpyA is therefore necessary but not sufficient for repression of cluster 1 in low salt conditions.
Cluster 2 contains many genes encoding transporters (21 genes across both subclusters 2.1 and 2.2). Notably, genes encoding known metal cation transporters exhibit tight clustering with their cognate transcriptional regulators SirR and VNG0147C (62,63) (Supplementary Table S5). Subcluster 2.1 consists of 37 genes modestly upregulated in reduced salt in the WT but strongly downregulated in reduced salt in the ΔhpyA mutant. Interestingly, ChIP-seq enrichment for HpyA binding was detected at four sites nearby genes in this subcluster (Supplementary Table S7). Three of these four sites are nearby genes involved in maintenance of iron levels (Figure 6C). These encode the siderophore (iron chelator) biosynthesis and transport operon, the Suf iron-sulfur cluster biosynthesis and transport system, and a putative oxidoreductase (VNG0405C). Surprisingly, the siderophore biosynthesis operon is bound at both the 5′ and 3′ ends by HpyA, which is associated with significant activation of this operon in low salt conditions (Figure 6C, top panel). These results indicate that HpyA is required for direct activation of iron uptake under low sodium.
In subcluster 2.2, 46 genes are downregulated or constitutive across optimal and reduced salt in the WT, but more heavily downregulated in ΔhpyA in reduced salt. All 8 differentially expressed nucleotide metabolism genes are found within this subcluster, and all encode de novo purine biosynthesis enzymes. However, only one of the 46 genes of subcluster 2.2 is a direct target of HpyA (VNG0161G, encoding glutamate dehydrogenase). This suggests that HpyA regulates purine biosynthesis and other functions in this subcluster in an indirect manner.
Together, these transcriptome profiling data integrated with ChIP-seq binding locations suggest an important role for HpyA as specific, direct activator of iron uptake, and an indirect global regulator of ion transport and nucleotide biosynthesis during hypo-osmotic stress.
DISCUSSION
Here, we integrate quantitative phenotyping and functional genomics data to demonstrate that the sole histone-like protein encoded in the hypersaline adapted archaeal species Hbt. salinarum directly activates iron uptake transporters under hypo-osmotic stress. At other sites in the genome, HpyA also functions as an indirect, global activator of genes encoding functions central to cellular physiology in low ionic strength medium. These transcriptional effects enable cells to maintain rod-shaped cellular morphology and growth in hypo-osmotic conditions.
Two of the five operons under the direct transcriptional control of HpyA encode transmembrane ABC transporters that are predicted to import iron. One operon (VNG0524G-VNG0527C) encodes a putative iron-sulfur (Fe-S) cluster assembly system of the Suf family. The predicted encoded proteins exhibit moderate identity to the well-characterized E. coli Fe-S assembly proteins SufC, SufB, and SufD (45%, 56% and 28%, respectively (64)). The other operon (gabT/bdb/iucABC, VNG6210-VNG6216) encodes siderophore biosynthesis and uptake. Siderophores are high-affinity iron binding chelators that are secreted from the cell and then imported via a dedicated ABC transporter (65). In Hbt. salinarum and many bacteria, in addition to the ABC transporter, this operon includes a novel L-2,4-diaminobutyrate decarboxylase (DABA DC; encoded by gabT) and a DABA aminotransferase (encoded by bdb) for siderophore biosynthesis (65). Because amino acids are precursors for DABA biosynthesis, down-regulation of iucABC in the ΔhpyA mutant strain may also explain the indirect differential expression of amino acid biosynthesis genes during hypo-osmotic stress.
Hbt. salinarum is a facultative anaerobe capable of aerobic and anaerobic respiratory metabolism (66). Across the tree of life, including Hbt. salinarum, iron is an essential cofactor for the function of respiratory complexes in the oxygen-accepting electron transport chain (67). Because reduced salinity increases oxygen saturation in the medium, these conditions would favor aerobic respiratory metabolism over anaerobic metabolism, increasing the cellular demand for iron (60). Indeed, we observe that these iron transport systems are induced in an HpyA-dependent manner under low salt conditions (Figure 6B). Low levels of iron transport expression in the ΔhpyA strain would therefore be expected to lead to low intracellular iron levels. Low intracellular iron has also been observed previously for strains deleted for idr2, which encodes a DtxR family iron-dependent TF in Hbt. salinarum. This TF also functions as a direct activator of the iucABC siderophore biosynthesis and transport operon, and intracellular iron levels are low in the Δidr2 strain due to dysregulation of iucABC (62,68). Idr2 is a member of a complex network of TFs that regulate the response to iron imbalance (62,68) and the current study suggests that HpyA is also involved in regulation of iron uptake. This mode of transcriptional regulation by HpyA explains the ΔhpyA growth impairment observed in low sodium conditions tested here (Figure 1).
The remaining three operons under direct HpyA regulation encode central metabolic functions (Supplementary Table S7). HpyA activates glutamate dehydrogenase and acyl-coA ligase enzymes, encoded by the gdhB/alkK operon. These enzymes control the entry of glutamate into the TCA cycle via the conversion of glutamate to 2-oxoglutarate. Glutamate is also a key precursor for biosynthesis of many metabolites, including purines and other amino acids (69,70). Direct control of this operon may explain the indirect transcriptional dysregulation of these pathways in the ΔhpyA mutant strain. HpyA activates an oxidoreductase gene (NAD-dependent epimerase predicted to act on nucleotide-sugar substrates; VNG0405C) and glycerol dehydrogenase gene and its associated operon (VNG0161G / VNG0162G), also encoding key components of core metabolism. The gene encoding a single-stranded DNA binding protein (ssb) is the only direct target predicted to be regulated by HpyA under both optimal and low salt conditions, and repressed rather than activated. Although the precise relationship between these HpyA regulatory targets and the ΔhpyA growth defect remains unclear, current knowledge of metabolism in Hbt. salinarum suggests that, in the ΔhpyA mutant, disruption in the levels of key metabolic intermediates (glycerol, glutamate) may contribute to the growth impairment of this strain under low salt conditions.
Dysregulation of import and/or efflux of other ions (divalent metal cations, chloride, and other transporters) in the ΔhpyA mutant may also explain the cell shape change in this strain (Figure 2). The proteinaceous surface layer (S-layer) is a key cell shape determinant of Hbt. salinarum (57,71). The S-layer is pliable and allows for changes in cell shape under physical pressure and low salinity (26,53). This shape change is exacerbated in ΔhpyA (Figure 2), which we hypothesize is due to dysregulation of ion transport expression. Iron has also been shown to impact cell morphology in the related haloarchaeal species Haloferax volcanii, although the underlying mechanism remains unknown (72). Expression of other pathways, for example, the S-layer (encoded by csg) and glycosylation enzymes (VNG0140G; Figure 6 and Supplementary Table S4) is reduced in the ΔhpyA mutant under low salt conditions. However, these appear to play a more minor role in the ΔhpyA morphology defect given that: (a) these genes are indirect targets of HpyA regulation; and (b) overall S-layer glycosylation is unaffected in strains deleted of hpyA (26). Taken together, these data suggest that HpyA salt-dependent regulation of ionic balance is a major contributor to maintenance of wild type cell morphology and growth in reduced sodium environments.
Apart from these cases of direct regulation by HpyA, the majority of differentially expressed genes are located >500 bp away from HpyA binding sites. This can be explained in a number of ways. Several TFs are differentially expressed in the ΔhpyA strain relative to WT in low salt (Supplementary Table S4, Figure 6). Therefore, the proximate cause of indirect differential gene expression can be inferred based on prior knowledge of the global gene regulatory network (GRN) in this organism (44,73–74). For example, the general TF, TfbB, is differentially expressed in ΔhpyA in low salt (cluster 1.1, Figure 6), and most of the genes in this cluster are indirectly regulated. TfbB is a direct regulator of several of the genes in this cluster, including cysK (74). Hbt. salinarum encodes 7 paralogs of transcription factor B (TFB) (75). Together, TFB and TATA binding protein recruit RNA polymerase to core promoters to initiate transcription (76). The TFB network in Hbt. salinarum is highly interconnected: for example, TfbB directly activates TfbG, which in turn regulates other genes indirectly regulated by HpyA (e.g. metal transporter VNG1744H, Supplementary Table S4). Other indirect regulation by HpyA can be attributed to metal-responsive TFs. For example, SirR and VNG0147C, members of cluster 2.1, have previously been experimentally characterized as regulators of operons encoding metal transporters, specifically manganese uptake (ZurA) and the heavy metal efflux (ZntA), respectively (62,63). Aside from indirect regulation as part of a transcriptional network, we note that our data do not exclude the possibility that HpyA may function as a co-regulator, perhaps by binding DNA through interaction with another TF. Hence, we propose that HpyA may, in part, achieve its global, indirect regulatory effect via its regulation of genes encoding other TFs and/or through protein-protein interaction with other sequence-specific TFs.
In addition to transcriptional regulation of ion balance, HpyA may play other functional roles during hypo-osmotic stress. More than 40 HpyA binding sites were detected with no corresponding significant change in gene expression in the ΔhpyA knockout (Supplementary Tables S3, S7). HpyA prefers to bind neither coding nor non-coding genomic regions, setting it apart from characterized haloarchaeal TFs that function by canonical, sequence-specific DNA binding to promoter regions [TrmB (43) or RosR (44), Figure 4]. We provide evidence of direct regulation both among targets bound in promoter and genic regions (Supplementary Table S7). Direct regulation of expression via binding in gene bodies has also been reported for E. coli regulator RutR (77). The mechanism of RutR binding is complex and includes DNA bending, specific cis-regulatory sequence binding, and interaction with DNA-wrapping proteins (78). Future biochemical studies on HpyA are therefore needed to elucidate its specific DNA binding mechanism. Our data do not rule out that non-canonical binding modes of HpyA could also influence other aspects of the transcription cycle, including elongation or termination. Bacterial nucleoid associated proteins (NAPs) bind DNA to regulate gene expression, remodel chromatin by bending or wrapping, and/or protect the nucleoid during stress (79,80). For example, the E. coli transcription regulator CRP can function both as a canonical TF (site-specific gene regulation) for some genes, and as a DNA-bending chromatin remodeler at other genomic sites (79,80). These newly-discovered and expanding roles for DNA binding proteins calls for a broader perspective on the function of transcriptional regulators (79,80). Likewise, further research is needed to explore such functional roles for HpyA.
Taken together, the results presented here strongly suggest that HpyA functions as a direct activator of iron regulatory genes and a global indirect regulator of diverse pathways. This function is markedly different than other characterized H3/H4-like histones in archaea and eukaryotes.
DATA AVAILABILITY
Raw sequencing data (ChIP-seq, RNA-seq, and ΔhpyA whole genome resequencing) are freely available through the National Center for Biotechnology Information (NCBI) accession PRJNA 703048. ChIP-seq and RNA-seq metadata and additional information can be found on the Gene Expression Omnibus (GEO) through accession GSE182514. All code and input data are available on GitHub at https://github.com/amyschmid/HpyA_codes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank current and former Schmid lab members: Cynthia Darnell, Rylee Hackley, Mar Martinez-Pastor, Angie Vreugdenhill, Peter Tonner, Sungmin Hwang and Preeti Bhanap for technical assistance with experimental methods and analysis, and for comments on the manuscript. We thank former Schmid lab member Keely Dulmage for generation of the pKAD plasmids. Saaz Sakrikar is grateful to his graduate thesis committee members (David Macalpine, Amy Grunden, Richard Brennan and Raluca Gordan) for mentorship and comments on the manuscript. We thank Deyra Rodriguez and New England Biolabs for providing reagents and advice for rRNA depletion and RNA library preparation. We thank Antoine Hocher and Tobias Warnecke for scientific discussions during the preparation of the manuscript. We thank the Duke Sequencing and Genomic Technologies Core Facility for their technical expertise in generating the sequencing data reported here.
Contributor Information
Saaz Sakrikar, Biology Department, Duke University, Durham, NC, 27708, USA; University Program in Genetics and Genomics, Duke University, Durham, NC, 27708, USA.
Amy K Schmid, Biology Department, Duke University, Durham, NC, 27708, USA; University Program in Genetics and Genomics, Duke University, Durham, NC, 27708, USA; Center for Genomics and Computational Biology, Duke University, Durham, NC, 27708, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSF MCB [1651117, 1936024, 1615685 to A.K.S.]; NIH T32 training grant [5T32GM007754 to the Duke University Program in Genetics and Genomics]. Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Malik H.S., Henikoff S.. Phylogenomics of the nucleosome. Nat. Struct. Mol. Biol. 2003; 10:882–891. [DOI] [PubMed] [Google Scholar]
- 2. Brunk C.F., Martin W.F.. Archaeal histone contributions to the origin of eukaryotes. Trends Microbiol. 2019; 27:703–714. [DOI] [PubMed] [Google Scholar]
- 3. Sandman K., Reeve J.N.. Archaeal histones and the origin of the histone fold. Curr. Opin. Microbiol. 2006; 9:520–525. [DOI] [PubMed] [Google Scholar]
- 4. Talbert P.B., Henikoff S.. Histone variants -- ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010; 11:264–275. [DOI] [PubMed] [Google Scholar]
- 5. Mariño-Ramírez L., Kann M.G., Shoemaker B.A., Landsman D. Histone structure and nucleosome stability. Expert Rev. Proteomics. 2005; 2:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arents G., Moudrianakis E.N.. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:10489–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laursen S.P., Bowerman S., Luger K.. Archaea: the final frontier of chromatin. J. Mol. Biol. 2021; 433:166791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sandman K., Krzycki J.A., Dobrinski B., Lurz R., Reeve J.N.. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:5788–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Čuboňová L., Katano M., Kanai T., Atomi H., Reeve J.N., Santangelo T.J.. An archaeal histone is required for transformation of Thermococcus kodakarensis. J. Bacteriol. 2012; 194:6864–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mattiroli F., Bhattacharyya S., Dyer P.N., White A.E., Sandman K., Burkhart B.W., Byrne K.R., Lee T., Ahn N.G., Santangelo T.J.et al.. Structure of histone-based chromatin in Archaea. Science. 2017; 357:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanders T.J., Ullah F., Gehring A.M., Burkhart B.W., Vickerman R.L., Fernando S., Gardner A.F., Ben-Hur A., Santangelo T.J.. Extended archaeal histone-based chromatin structure regulates global gene expression in Thermococcus kodakarensis. Front Microbiol. 2021; 12:681150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Decanniere K., Babu A.M., Sandman K., Reeve J.N., Heinemann U.. Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus. J. Mol. Biol. 2000; 303:35–47. [DOI] [PubMed] [Google Scholar]
- 13. Henneman B., Brouwer T.B., Erkelens A.M., Kuijntjes G.J., van Emmerik C., van der Valk R.A., Timmer M., Kirolos N.C.S., van Ingen H., van Noort J.et al.. Mechanical and structural properties of archaeal hypernucleosomes. Nucleic. Acids. Res. 2021; 49:4338–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henneman B., van Emmerik C., van Ingen H., Dame R.T.. Structure and function of archaeal histones. PLoS Genet. 2018; 14:e1007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maruyama H., Harwood J.C., Moore K.M., Paszkiewicz K., Durley S.C., Fukushima H., Atomi H., Takeyasu K., Kent N.A.. An alternative beads-on-a-string chromatin architecture in Thermococcus kodakarensis. EMBO Rep. 2013; 14:711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tomschik M., Karymov M.A., Zlatanova J., Leuba S.H.. The archaeal histone-fold protein HMf organizes DNA into bona fide chromatin fibers. Structure. 2001; 9:1201–1211. [DOI] [PubMed] [Google Scholar]
- 17. Sanders T.J., Marshall C.J., Santangelo T.J.. The role of archaeal chromatin in transcription. J. Mol. Biol. 2019; 431:4103–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie Y., Reeve J.N.. Transcription by an archaeal RNA polymerase is slowed but not blocked by an archaeal nucleosome. J. Bacteriol. 2004; 186:3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilkinson S.P., Ouhammouch M., Geiduschek E.P.. Transcriptional activation in the context of repression mediated by archaeal histones. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weidenbach K., Glöer J., Ehlers C., Sandman K., Reeve J.N., Schmitz R.A.. Deletion of the archaeal histone in Methanosarcina mazei Gö1 results in reduced growth and genomic transcription. Mol. Microbiol. 2008; 67:662–671. [DOI] [PubMed] [Google Scholar]
- 21. Stevens K.M., Swadling J.B., Hocher A., Bang C., Gribaldo S., Schmitz R.A., Warnecke T.. Histone variants in archaea and the evolution of combinatorial chromatin complexity. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:33384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hocher A., Rojec M., Swadling J.B., Esin A., Warnecke T.. The DNA-binding protein HTa from Thermoplasma acidophilum is an archaeal histone analog. eLife. 2019; 8:e52542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maruyama H., Prieto E.I., Nambu T., Mashimo C., Kashiwagi K., Okinaga T., Atomi H., Takeyasu K.. Different Proteins Mediate Step-Wise Chromosome Architectures in Thermoplasma acidophilum and Pyrobaculum calidifontis. Front Microbiol. 2020; 11:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hocher A., Borrel G., Fadhlaoui K., Brugère J.-F., Gribaldo S., Warnecke T.. Growth temperature is the principal driver of chromatinization in archaea. 2021; bioRxiv doi:08 July 2021, preprint: not peer reviewed 10.1101/2021.07.08.451601. [DOI] [PMC free article] [PubMed]
- 25. Soares D., Dahlke I., Li W.-T., Sandman K., Hethke C., Thomm M., Reeve J.N.. Archaeal histone stability, DNA binding, and transcription inhibition above 90°C. Extremophiles. 1998; 2:75–81. [DOI] [PubMed] [Google Scholar]
- 26. Dulmage K.A., Todor H., Schmid A.K.. Growth-phase-specific modulation of cell morphology and gene expression by an archaeal histone protein. mBio. 2015; 6:e00649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner G., Hartmann R., Oesterhelt D. Potassium uniport and ATP synthesis in Halobacterium halobium. Eur. J. Biochem. 1978; 89:169–179. [DOI] [PubMed] [Google Scholar]
- 28. Ng W.V., Kennedy S.P., Mahairas G.G., Berquist B., Pan M., Shukla H.D., Lasky S.R., Baliga N.S., Thorsson V., Sbrogna J.et al.. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eickbush T.H., Moudrianakis E.N.. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978; 13:295–306. [DOI] [PubMed] [Google Scholar]
- 30. Peck R.F., DasSarma S., Krebs M.P.. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 2000; 35:667–676. [DOI] [PubMed] [Google Scholar]
- 31. Dulmage K.A. Large-scale Effectors of Gene Expression and New Models of Cell Division in the Haloarchaea. 2015; Duke University; PhD Thesis. [Google Scholar]
- 32. Kahm M., Hasenbrink G., Lichtenberg-Fraté H., Ludwig J., Kschischo M.. grofit: fitting biological growth curves with R. 2010. 2010; 33:21. [Google Scholar]
- 33. Darnell C.L., Zheng J., Wilson S., Bertoli R.M., Bisson-Filho A.W., Garner E.C., Schmid A.K.. The ribbon-helix-helix domain protein CdrS regulates the tubulin homolog ftsZ2 to control cell division in archaea. mBio. 2020; 11:e01007-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ducret A., Quardokus E.M., Brun Y.V.. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016; 1:16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilbanks E.G., Larsen D.J., Neches R.Y., Yao A.I., Wu C.-Y., Kjolby R.A.S., Facciotti M.T.. A workflow for genome-wide mapping of archaeal transcription factors with ChIP-seq. Nucleic Acids Res. 2012; 40:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koide T., Reiss D.J., Bare J.C., Pang W.L., Facciotti M.T., Schmid A.K., Pan M., Marzolf B., Van P.T., Lo F.Y.et al.. Prevalence of transcription promoters within archaeal operons and coding sequences. Mol. Syst. Biol. 2009; 5:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taboada B., Estrada K., Ciria R., Merino E.. Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics. 2018; 34:4118–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmid A.K., Reiss D.J., Pan M., Koide T., Baliga N.S.. A single transcription factor regulates evolutionarily diverse but functionally linked metabolic pathways in response to nutrient availability. Mol. Syst. Biol. 2009; 5:282–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tonner P.D., Pittman A.M.C., Gulli J.G., Sharma K., Schmid A.K.. A regulatory hierarchy controls the dynamic transcriptional response to extreme oxidative stress in archaea. PLoS Genet. 2015; 11:e1004912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen X., Zhang B., Wang T., Bonni A., Zhao G.. Robust principal component analysis for accurate outlier sample detection in RNA-Seq data. BMC Bioinformatics. 2020; 21:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B (Methodological). 1995; 57:289–300. [Google Scholar]
- 49. Kassambara M., Mundt F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyse. R package version 1.0.7 ed. 2020; https://CRAN.R-project.org/package=factoextra. [Google Scholar]
- 50. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016; NY: Springer Verlag. [Google Scholar]
- 51. Kolde R. pheatmap: Pretty Heatmap. R package version 1.0.12 ed. 2019; https://CRAN.R-project.org/package=pheatmap. [Google Scholar]
- 52. Makarova K.S., Wolf Y.I., Koonin E.V.. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life. 2015; 5:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eun Y.J., Ho P.Y., Kim M., LaRussa S., Robert L., Renner L.D., Schmid A., Garner E., Amir A.. Archaeal cells share common size control with bacteria despite noisier growth and division. Nat Microbiol. 2018; 3:148–154. [DOI] [PubMed] [Google Scholar]
- 54. Vauclare P., Natali F., Kleman J.P., Zaccai G., Franzetti B.. Surviving salt fluctuations: stress and recovery in Halobacterium salinarum, an extreme halophilic Archaeon. Sci. Rep. 2020; 10:3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dyall-Smith M. Dyall-Smith M. The Halohandbook: Protocols for Haloarchaeal Genetics. 2009; https://haloarchaea.com/wp-content/uploads/2018/10/Halohandbook_2009_v7.3mds.pdf. [Google Scholar]
- 56. Guan Z., Naparstek S., Calo D., Eichler J.. Protein glycosylation as an adaptive response in Archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ. Microbiol. 2012; 14:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Todor H., Dulmage K., Gillum N., Bain J.R., Muehlbauer M.J., Schmid A.K.. A transcription factor links growth rate and metabolism in the hypersaline adapted archaeon Halobacterium salinarum. Mol. Microbiol. 2014; 93:1172–1182. [DOI] [PubMed] [Google Scholar]
- 58. Zerulla K., Soppa J.. Polyploidy in haloarchaea: advantages for growth and survival. Front Microbiol. 2014; 5:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stevenson K., McVey A.F., Clark I.B.N., Swain P.S., Pilizota T.. General calibration of microbial growth in microplate readers. Sci. Rep. 2016; 6:38828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmid A.K., Reiss D.J., Kaur A., Pan M., King N., Van P.T., Hohmann L., Martin D.B., Baliga N.S.. The anatomy of microbial cell state transitions in response to oxygen. Genome Res. 2007; 17:1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coker J.A., DasSarma P., Kumar J., Muller J.A., DasSarma S.. Transcriptional profiling of the model Archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature. Saline Syst. 2007; 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martinez-Pastor M., Lancaster W.A., Tonner P.D., Adams M.W.W., Schmid A.K.. A transcription network of interlocking positive feedback loops maintains intracellular iron balance in archaea. Nucleic Acids Res. 2017; 45:9990–10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaur A., Pan M., Meislin M., Facciotti M.T., El-Gewely R., Baliga N.S.. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 2006; 16:841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Outten F.W. Recent advances in the Suf Fe-S cluster biogenesis pathway: beyond the Proteobacteria. Biochim. Biophys. Acta. 2015; 1853:1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Burrell M., Hanfrey C.C., Kinch L.N., Elliott K.A., Michael A.J.. Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol. Microbiol. 2012; 86:485–499. [DOI] [PubMed] [Google Scholar]
- 66. Andrei A.S., Banciu H.L., Oren A.. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol. Lett. 2012; 330:1–9. [DOI] [PubMed] [Google Scholar]
- 67. Gonzalez O., Gronau S., Pfeiffer F., Mendoza E., Zimmer R., Oesterhelt D. Systems analysis of bioenergetics and growth of the extreme halophile Halobacterium salinarum. PLoS Comput. Biol. 2009; 5:e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schmid A.K., Pan M., Sharma K., Baliga N.S.. Two transcription factors are necessary for iron homeostasis in a salt-dwelling archaeon. Nucleic Acids Res. 2011; 39:2519–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Falb M., Muller K., Konigsmaier L., Oberwinkler T., Horn P., von Gronau S., Gonzalez O., Pfeiffer F., Bornberg-Bauer E., Oesterhelt D. Metabolism of halophilic archaea. Extremophiles. 2008; 12:177–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Todor H., Gooding J., Ilkayeva O.R., Schmid A.K.. Dynamic metabolite profiling in an archaeon connects transcriptional regulation to metabolic consequences. PLoS One. 2015; 10:e0135693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mescher M.F., Strominger J.L.. Structural (shape-maintaining) role of the cell surface glycoprotein of Halobacterium salinarium. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:2687–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. de Silva R.T., Abdul-Halim M.F., Pittrich D.A., Brown H.J., Pohlschroder M., Duggin I.G.. Improved growth and morphological plasticity of Haloferax volcanii. Microbiology (Reading). 2021; 167: 10.1099/mic.0.001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bonneau R., Facciotti M.T., Reiss D.J., Schmid A.K., Pan M., Kaur A., Thorsson V., Shannon P., Johnson M.H., Bare J.C.et al.. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007; 131:1354–1365. [DOI] [PubMed] [Google Scholar]
- 74. Facciotti M.T., Reiss D.J., Pan M., Kaur A., Vuthoori M., Bonneau R., Shannon P., Srivastava A., Donohoe S.M., Hood L.E.et al.. General transcription factor specified global gene regulation in archaea. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:4630–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Baliga N.S., Goo Y.A., Ng W.V., Hood L., Daniels C.J., DasSarma S.. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors?. Mol. Microbiol. 2000; 36:1184–1185. [DOI] [PubMed] [Google Scholar]
- 76. Martinez-Pastor M., Tonner P.D., Darnell C.L., Schmid A.K.. Transcriptional regulation in archaea: from individual genes to global regulatory networks. Annu. Rev. Genet. 2017; 51:143–170. [DOI] [PubMed] [Google Scholar]
- 77. Shimada T., Ishihama A., Busby S.J.W., Grainger D.C.. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008; 36:3950–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nguyen Le Minh P., Bervoets I., Maes D., Charlier D. The protein-DNA contacts in RutR*carAB operator complexes. Nucleic Acids Res. 2010; 38:6286–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Holowka J., Zakrzewska-Czerwinska J.. Nucleoid associated proteins: the small organizers that help to cope with stress. Front Microbiol. 2020; 11:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dorman C.J., Schumacher M.A., Bush M.J., Brennan R.G., Buttner M.J.. When is a transcription factor a NAP?. Curr. Opin. Microbiol. 2020; 55:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data (ChIP-seq, RNA-seq, and ΔhpyA whole genome resequencing) are freely available through the National Center for Biotechnology Information (NCBI) accession PRJNA 703048. ChIP-seq and RNA-seq metadata and additional information can be found on the Gene Expression Omnibus (GEO) through accession GSE182514. All code and input data are available on GitHub at https://github.com/amyschmid/HpyA_codes.