Abstract
Micro RNAs (miRNAs) are processed from precursor RNA molecules with precisely defined secondary stem-loop structures. ARGONAUTE1 (AGO1) is the main executor component of miRNA pathway and its expression is controlled via the auto-regulatory feedback loop activity of miR168 in plants. Previously we have shown that AGO1 loading of miR168 is strongly restricted leading to abundant cytoplasmic accumulation of AGO-unbound miR168. Here, we report, that intrinsic RNA secondary structure of MIR168a precursor not only defines the processing of miR168, but also precisely adjusts AGO1 loading efficiency determining the biologically active subset of miR168 pool. Our results show, that modification of miRNA duplex structure of MIR168a precursor fragment or expression from artificial precursors can alter the finely adjusted loading efficiency of miR168. In dcl1-9 mutant where, except for miR168, production of most miRNAs is severely reduced this mechanism ensures the elimination of unloaded AGO1 proteins via enhanced AGO1 loading of miR168. Based on this data, we propose a new competitive loading mechanism model for miR168 action: the miR168 surplus functions as a molecular buffer for controlled AGO1 loading continuously adjusting the amount of AGO1 protein in accordance with the changing size of the cellular miRNA pool.
INTRODUCTION
RNA interference (RNAi) or RNA silencing is a widespread gene regulatory mechanism, playing roles in diverse biological processes in most eukaryotic organisms (1,2). The hallmark molecules of RNAi are the 21–24 nt long small RNAs (sRNA). A subclass of sRNAs are the micro RNAs (miRNAs) (3). They predominantly control endogenous gene expression (4,5) to coordinate developmental processes and stress responses (6,7).
During the biogenesis of miRNAs, a genome-encoded RNA, primary miRNA (pri-miRNA) with specific stem–loop secondary RNA structure is subjected to subsequent cleavages producing 21–24 nucleotide (nt) long miRNA duplexes (3,8). The stem–loops of plant pri-miRNAs exhibit extreme variability in length and structure compared to animal counterparts (9,10). In plants, miRNA precursors (pre-miRNAs) are typically excised initially from pri-miRNA transcripts by the action of DICER-LIKE1 (DCL1) with a cleavage event near the base of the stem. The next DCL1 cleavage occurs in 21–24 nucleotide distance from the end of the stem resulting in a miRNA duplex (3,11). In specific cases loop-to-base or bidirectional maturation can also take place by DCL1 (11–13).
The generated miRNA intermediate duplexes consist of a guide (miRNA) and a passenger (miRNA*) strand with 2-nt 3′ overhang and a 5′ phosphate at each strand. Plant pri-miRNAs produce miRNAs either from the 5′ or from 3′ arm of their precursors. Precise and efficient processing depends on pri-miRNA structural signals, like terminal loop structure, 14–15 bp paired stem proximal and base-pairing at defined region of stem distal to the miRNA duplex (3,14). The accurate and efficient processing of pri-miRNAs requires DCL1 co-factors, like the G-patch domain protein TOUGH (TGH), the zinc finger protein SERRATE (SE), and the dsRNA binding domain protein HYPONASTIC LEAVES1 (HYL1/DRB1)/ DOUBLE-STRANDED RNA-BINDING PROTEIN 1 (5,15). The miRNA/miRNA* duplexes are methylated by the sRNA methyltransferase HUA Enhancer 1 (HEN1) at the 3′ ends that aids their protection against exonucleases (16). The methylated miRNA/miRNA∗ duplexes, according to the previous model, were thought to be exported to the cytoplasm by the animal Exportin 5 (EXPO5) homologous protein HASTY (HST) (17).
Subsequently, miRNA/miRNA* duplexes are incorporated into the RNA-induced Silencing Complex (RISC), the effector of RNA silencing. During RISC assembly the guide miRNA strand is loaded onto ARGONAUTE protein, the central component of RISC, while the miRNA∗ is ejected and degraded (1). AGO1 is the major effector protein for miRNAs among the ten Arabidopsis encoded AGO proteins which are specialized for various RNAi pathways but often display functional redundancies (18). Recently, it has been shown that AGO1 containing RISC is mainly assembled in the nucleus and exported to the cytosol as a nucleoprotein complex by EXPO1 transporter (19). The miRNA-programmed AGO-RISC identifies its RNA targets via complementary base pairing and mediates their repression through cleavage (slicing) and/or translational inhibition (1).
Diverse intrinsic structural features of the miRNA/miRNA* duplexes determine their AGO loading specificities (20). Guide strand is selected according to the thermodynamic stability of the miRNA duplex ends (21). The 5′-nucleotide predominantly determines the sorting preference of miRNAs into specific AGO proteins (22,23). AGO1 exhibits strong preference for sRNAs having uridine at their 5′ end (5′U), consequently the majority of miRNAs fulfil this requirement ensuring their sorting into AGO1. Structural characteristics of the miR165/166 and miR390 miRNA duplexes target them specifically into AGO10 and AGO7, respectively (24,25). The AGO1 and AGO10 sorting properties of miR168 is also determined by metastable structural configurations of its precursor (26). Transient expression studies revealed that distinct secondary structural motifs of the miRNA/miRNA* duplexes control AGO1 and/or AGO2 sorting (27). Sorting of various miRNAs into different AGO-RISCs according to the structural and/or sequence characteristics therefore represents an important regulatory check-point of RNAi pathway.
However, recently another checkpoint of miRNA AGO loading was revealed by the identification of cytoplasmic protein unbound pool of miRNAs (28). This observation indicated that a highly controlled post-production regulatory mechanism is able to adjust the loading efficiency of particular miRNAs into the same AGO-RISC. This control mechanism can determine the biologically active portion of the produced miRNAs in the given cellular environment. Moreover, it was also suggested that RISC-loading efficiencies of distinct canonical 5′U miRNAs are predominantly controlled by their diverse precursor RNAs.
AGO1 protein is an absolutely essential trans factor of the miRNA pathway in Arabidopsis. Null ago1 mutations are lethal and hypomorphic ago1 mutants show severe developmental aberrancies (29). Moreover, over-accumulation of AGO1 also induces disturbances in miRNA levels and extremely drastic phenotypic/developmental alterations (30). Besides, AGO1 is also the central effector in siRNA-induced post-transcriptional gene silencing, antiviral immunity pathways and the target of virus RNAi suppressors (31,32).
The amount of functional AGO1 protein is precisely regulated through transcriptional and post-transcriptional feed-back loop mechanisms: (i) miR168-programmed AGO1-RISC controls the expression of AGO1 mRNA (33), (ii) AGO1 cleavage leads to production of secondary small interfering (si)RNAs that further repress AGO1 protein expression (34), (iii) AGO1 and MIR168 gene expressions are regulated co-transcriptionally (30), (iv) the existence of two MIR168 genes producing primary transcripts (pri-MIR168a and pri-MIR168b) with both distinct and overlapping functions gives an additional layer to the control of AGO1 (35), (v) miR168 is stabilized by AGO1 binding but insensitive to DCL1 levels (30). This complex multilayer regulation of AGO1 ensures a robust and precise functioning of the RNA silencing pathway itself.
Intriguingly, transgenic over-expression of miR168 induces limited phenotypic alterations such as delayed flowering and leaf serration (30). Recently, it has been shown that miR168 accumulates dominantly in a protein-unbound form in the cytoplasm suggesting a regulatory step at AGO1 loading (28). Why miR168 is expressed and maturated in such great quantities far exceeding its competence to be loaded into AGO1 and whether the unbound miR168 has any biological roles remains entirely elusive.
Given the importance of miRNA pathway self-regulation through AGO1 feed-back, here we investigated more in depth the effect of miR168/miR168* duplex structure on AGO1-loading efficiency. First, we have shown that intrinsic structural features precisely calibrate the loading efficiency of miR168/miR168* duplexes. Structural alterations of the miR168/miR168* duplex resulted in changes of loading efficiency, in both positive and negative manner that caused AGO1 protein amount changes and phenotypic alterations. Moreover, by altering the amount of competitor miRNA cellular pool we observed the establishment of a new AGO1 protein steady state level. In general, our data reveal a new RNAi regulatory action where not only the production rate but the finely tuned competitive loading rate into the RISC determines the biological activity of the miRNA in the given sRNA environment. Our findings suggests that the produced miR168/miR168* surplus coupled with structurally determined loading efficiency serves as a pivotal self-calibrator tool of the miRNA pathway.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana seeds were surface sterilized and after incubation for three days at 4°C, transgenic, mutant and wild type Columbia plants were germinated at 21°C on MS agar medium supplied with 1% sucrose with or without the presence of 50 μg/ml kanamycin, respectively. Seedlings were transferred into Jiffy peat blocks after seven days, and spent three weeks there before planting to pots filled with soil. Plants were grown under 8 h light 16 h darkness cycles at 21°C until planting, and then were moved to light room under 16 h light 8 h darkness at 21°C. Nicotiana benthamiana plants were grown under the light room conditions described above.
Plasmid constructs
All constructs were built using pGreen binary vector system (pGreen0029) and 35S cassette according to the instructions of the manufacturer (http://www.pgreen.ac.uk). cDNA was produced with RevertAid First strand cDNA synthesis Kit (Thermo Fisher Scientific) from A. thaliana RNA in all cases. Constructs of MIR168a, MIR168b, MIR156a and MIR171a contained the region 10–10 bp upstream and downstream from the position of miRNA stem-loop structure. For AMIRs, we used the modified hvu-MIR171 stem-loop described previously (36). PCR mutagenesis was applied to alter respective nucleotides in passenger strand of MIR168 stem–loop, and to change duplex part of hvu-MIR171 to miR168a and to its respective passenger strand. For the AGO1-sensor, a 558-bp-long cDNA fragment containing miR168 target site was amplified and fused to the 5′ part of GFP in-frame. Primers used to create constructs were presented in Additional file 2: Supplementary Table S1. All construct was introduced into Agrobacterium tumefaciens AGL1 strain with electroporation (360Ω, 25μF, 2,5kV; Biorad) in the presence of pSoup helper plasmid.
Transient assay
Young leaves of six weeks old N. benthamiana were infiltrated with the respective mixture of Agrobacterium tumefaciens (AGL1) suspensions at 1.0 optical density of 600 nm [OD600] containing sensor, miRNA-producing and p14 constructs as described previously (37). P14, as a suppressor of siRNA pathway, was included in every experiment with uniform concentration (38), so it did not interfere with observation of relative signal reduction. Presence of p14 was checked with northern blot (Additional file 1: Supplementary Figure S1C) according to protocol described previously (38,39). To reduce the differences in miRNA production ability of the different constructs, normalized amounts were applied and mixtures were supplemented with empty pGreen0029 vector containing A. tumefaciens (AGL1). For detailed compilation of infiltration mixtures, see Additional file 2: Supplementary Table S2. Samples were taken at the third day post-infiltration; four discs of 1 cm diameter were pooled from patches of separate leaves for individual constructs. Samples were collected in parallel from both sides of same leaves. Every miRNA producing sensor construct combination was tested on four to five plants and each experiment was repeated at least three times.
Transgenic line production
Arabidopsis thaliana Columbia ecotype plants were transformed with the appropriate miR168 producing construct according to the standard floral dip protocol (40). Transformant T0 plants were selected on MS plates supplemented with 50 μg/ml kanamycin. Four weeks after planting their miR168 expression profile in young rosette leaves was analysed and plants exhibiting appropriate miR168 over-expression level were self-pollinated and used to generate transgenic lines. Homozygous lines were produced with self-pollination during two further generations. Selection was based on kanamycin resistance and consistent level of miR168 expression. To demonstrate the effect of over-expression, lines with comparable expression level were selected within one panel. To ensure the comparability of the used transgenic lines, different categories of over-expression levels were investigated, 10–35 times for MIR168a and MIR168-4bp, 2–6 times for MIR168a and MIR168-3mm and 8–19 times for MIR168a and AMIR1-2 lines.
Gel-filtration assay
Gel-filtration based size separation of crude extracts using Superdex-200 column was performed as described previously (41,42) with minor modifications. Optimized buffer of separation contained 50 mM Tris–HCl (pH 7.5), 10 mM NaCl, 5 mM MgCl2 and 4 mM DTT. 48 fractions of gel-filtration were divided into two and RNA was extracted with phenol-chloroform method from the odd samples, while even samples were used for protein purification using acetone precipitation. Crude extracts were prepared from 0.3 g plant material collected from leaves of N. benthamiana plants three days post-infiltration (Figure 5D, Additional file 1: Supplementary Figures S4D and S6B), from young leaves of Hordeum vulgare (Additional file 1: Supplementary Figure S6A), from 7-day-old Arabidopsis seedlings (Figures 2D, 3D, 4D and Additional file 1: Supplementary Figure S4A, B and C) and from young rosette leaves of 6 weeks old Arabidopsis thaliana plants (Figure 1D). In case of the given panel, all samples were collected in parallel and gel-filtration runs were carried out subsequently with same parameters.
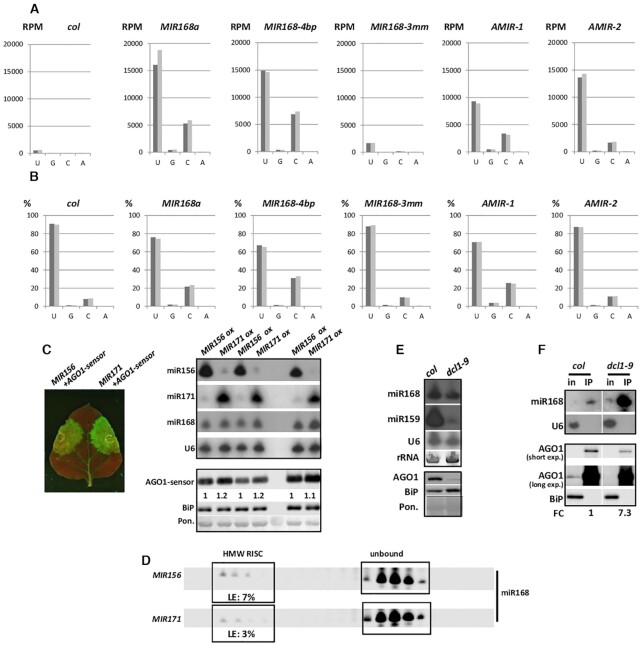
Figure 5.
HTS data analyses and miR168 loading rate under competitive circumstances. Distribution of 5′U/G/C/A miR168 species in HTS data of A. thaliana Col-0, MIR168a, MIR168-4bp, MIR168-3mm, AMIR-1 and AMIR-2 over-expressing plants displayed as (A) read per million (RPM) and (B) the percentage of whole miR168 homologous sequences. The two replicas were distinguished with different shades of grey. (C) MiR156, miR171, miR168 and AGO1-sensor content of ath-MIR156 and ath-MIR171 partial precursor infiltrated patches. U6, BiP and Ponceau staining were used as loading controls of RNA and protein blots, respectively. (D) MiR168 loading in case of transient ath-pri-MIR156 or ath-pri-MIR171 over-expression. Loading efficiency was calculated as the percentage of HMW RISC volume intensity referred to the sum of HMW RISC and unbound volume intensities. (E) MiR168, miR159 and AGO1 in wild type A. thaliana Columbia-0 (col) and dcl1-9 plants. U6, BiP and Ponceau staining were used as loading controls of RNA and protein blots, respectively. (F) MiR168 accumulation in AGO1 immuno-precipitated samples of col and dcl1-9 plants. Cytoplasmic contamination in IP samples at RNA and protein level was checked with U6 and BiP, respectively. Fold change (FC) was calculated as the volume intensity ratio of miR168 and AGO1 signal intensity of IP samples, and was reported on the basis of wild type Columbia (col).
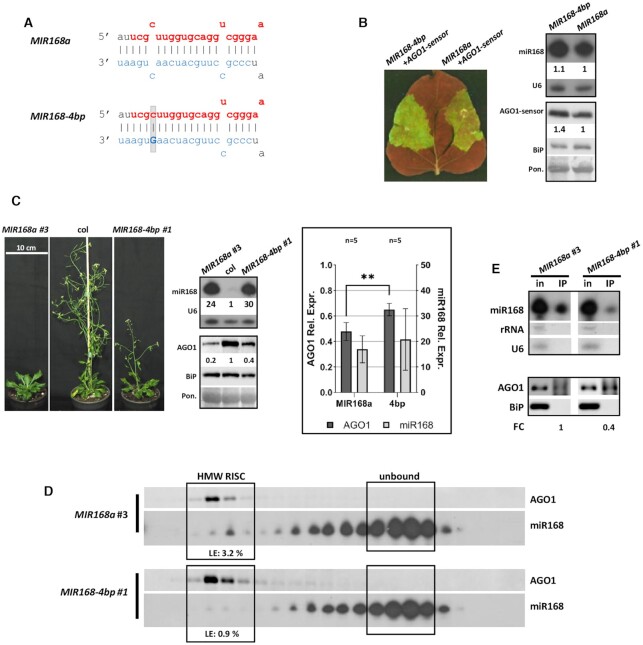
Figure 2.
Effect of MIR168a-4bp over-expression. (A) Structure of the modified duplex region in MIR168-4bp. Modified nucleotide is highlighted with a bold capital letter, grey background shows structural change. Mature miRNAs and star strands were marked with red and blue, respectively. (B) Transient over-expression of MIR168-4bp and wild type MIR168a. Blots demonstrate miR168 and AGO1-sensor content in infiltrated patches of N. benthamiana leaves. AGO1 part of the sensor fusion protein was detected with antibody raised against ath-AGO1. For the northern blot U6, while for the western blot, BiP and Ponceau staining were used as loading controls. (C) Phenotypes of nine weeks old MIR168-4bp and MIR168a lines, miR168 and AGO1 content of a transgenic line over-expressing MIR168-4bp and a wild type MIR168a line with comparably high over-expression rate of miR168. Statistical representation of five MIR168a (#: 3, 4, 2, 15, 16) and five MIR168-4bp (#: 1, 2, 6, 10, 14) lines. Asterisks represent significant difference (t-test, P = 0,05). (D) MiR168 distribution in gel-filtration fractions of the lines presented. High molecular weight RISC is identified by the presence AGO1 protein. Loading efficiency (LE) is calculated as described in Materials and Methods. (E) AGO1 IP of the investigated MIR168-4bp and the corresponding MIR168a over-expressing lines. Cytoplasmic contamination in IP samples at RNA and protein level was checked by detection of rRNA, U6 and BiP, respectively. Fold change (FC) of AGO1 associated miR168 was calculated as the volume intensity ratio of miR168 and AGO1 signals in IP samples and was presented on the basis of corresponding MIR168a line.
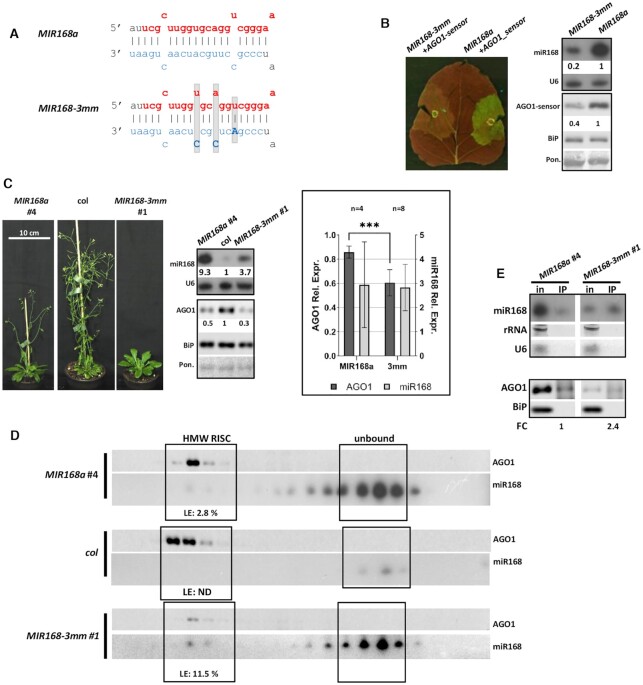
Figure 3.
Effect of MIR168-3mm over-expression. (A) Structure of the modified duplex region in MIR168-3mm in comparison with wild type MIR168a duplex. Modified nucleotides of star strand are highlighted with bold capital letters, grey background indicates structural changes. Mature miRNAs and star strands were marked with red and blue, respectively. (B) Transient over-expression of MIR168-3mm and wild type MIR168a precursor fragment constructs. Blots demonstrate miR168 and AGO1-sensor content of infiltrated patches of N. benthamiana leaves. AGO1 part of the sensor fusion protein was detected with antibody raised against ath-AGO1. For the northern blot U6, while for the western blots, BiP and Ponceau staining were used as loading controls. (C) Phenotypes of nine weeks old MIR168-3mm and a MIR168a line. MiR168 and AGO1 content of a transgenic line over-expressing MIR168-3mm and a wild type ath-pri-MIR168a line with a comparable over-expression rate of miR168. Statistical presentation of four MIR168a (#: 11–14) and eight MIR168-3mm (#: 1–8) lines. Asterisks label significant difference (t-test, P = 0,01). (D) MiR168 distribution in gel-filtration fractions of the investigated lines and wild type Columbia. High molecular weight (HMW) RISC is presented with AGO1. Loading efficiency (LE) is calculated as described in Methods. (E) AGO1 IP of the investigated MIR168-3mm and the corresponding MIR168a over-expressing line. Cytoplasmic contamination in IP samples at RNA and protein level was checked by detection of rRNA, U6 and BiP, respectively. Fold change (FC) of AGO1 associated miR168 was calculated as the volume intensity ratio of miR168 and AGO1 signals in IP samples and was reported on the basis of corresponding MIR168a line.
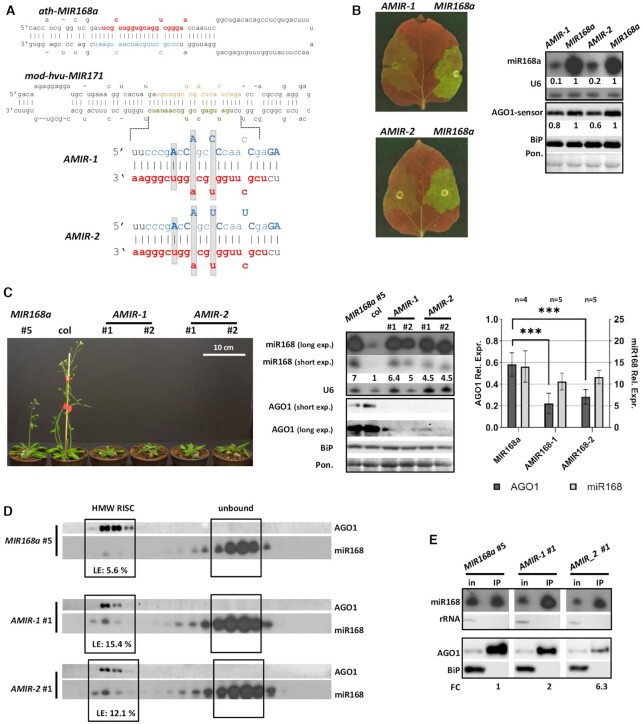
Figure 4.
Over-expression of miR168 producing AMIR constructs. (A) Structure of the modified duplex region implemented into hvu-MIR171 backbone in case of AMIR constructs. Modified nucleotides of star strands compared to wild type MIR168a duplex are highlighted with bold capital letters, grey background shows structural changes. Mature miR168, miR168*, miR171 and miR171* strands were marked with red, blue, green and brown, respectively. (B) Transient over-expression of AMIR constructs and wild type MIR168a precursor fragment. Blots demonstrate miR168 and AGO1-sensor content of infiltrated patches of N. benthamiana leaves. For the northern blot U6, while for the western blot, BiP and Ponceau staining were used as loading controls. (C) Phenotype of eight weeks old MIR168a, AMIR-1 and AMIR-2 lines. MiR168 and AGO1 content of transgenic lines over-expressing AMIR precursors and a MIR168a line with a comparable over-expression rate of miR168. Statistical analysis of four MIR168a (#: 2, 6, 8, 9), five AMIR-1 (#: 3, 4, 5, 6, 7) and five AMIR-2 (#: 5, 6, 7, 8, 9) lines. Asterisks display significant difference (t-test, P = 0,01). (D) MiR168 distribution in gel-filtration fractions of MIR168a #5, AMIR-1 #1 and AMIR-2 #1 lines. High molecular weight RISC is presented with AGO1. Loading efficiency (LE) is calculated as described in Materials and Methods. (E) AGO1 IP of the investigated AMIR-1, AMIR-2 and the corresponding MIR168a over-expressing line. Cytoplasmic contamination in IP samples at RNA and protein level was checked by detection of rRNA and BiP, respectively. Fold change (FC) of AGO1 associated miR168 was calculated as the volume intensity ratio of miR168 and AGO1 signals in IP samples and was reported on the basis of corresponding MIR168a line.
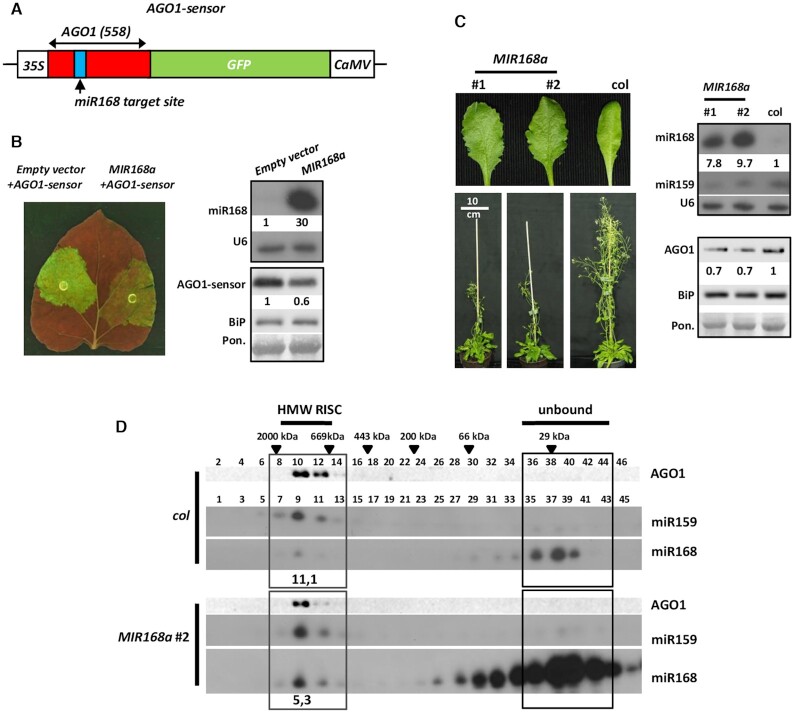
Figure 1.
Over-expression of wild type ath-pri-MIR168a in transient and transgenic systems. (A) Schematic representation of the AGO1-sensor construct. AGO1-derived part of the sensor is indicated with red, miR168 target site is marked with blue rectangle. (B) Transient over-expression of ath-pri-MIR168a (MIR168a on figures) precursor fragment in the presence of AGO1-sensor. Left panel shows the GFP fluorescence in leaves after co-infiltration of AGO1-sensor construct containing Agrobacteria and either empty vector (pGreen0029) or MIR168a. Right panels indicate the RNA levels of miR168 and the protein levels of the AGO1-sensor in the infiltrated patches. AGO1 part of the sensor fusion protein was detected with antibody raised against ath-AGO1. For the northern blot U6, while for the western blot BiP (Lumenal binding proteins) and Ponceau staining were used as loading controls. (C) Phenotypes of nine weeks old MIR168a precursor fragment over-expressing transgenic lines. Right panels show miR168, miR159 and AGO1 levels in over-expressing and control plants. U6, BiP and Ponceau staining were used as loading controls, respectively. (D) MiR168 distribution among gel-filtration fractions of wild type A. thaliana Columbia (col) and MIR168a over-expressing plants. High molecular weight (HMW) RISCs were detected with AGO1 western and miR159 northern blots. Even fractions were used for protein extraction, odd fractions for RNA purification. Black triangles show positions of known molecular weight markers amongst gel-filtration fractions.
RNA extraction and protein sample preparation
For miRNA and protein analyses indicated amount of agro-infiltrated leaves, 0.05–0.2 g of A. thaliana seedlings (Figures 1C, 2C, 3C and 4C and Additional file 1: Supplementary Figure S3B) or young rosetten leaves (Figure 5E and Additional file 1: Supplementary Figure S2B) were collected, homogenized in an ice-cold mortar in 355 μl of extraction buffer (0.1 M glycine–NaOH, pH 9.0, 100 mM NaCl, 10 mM EDTA, 2% sodium dodecyl sulfate and 1% sodium lauroylsarcosine) and divided into two aliquots. To one part (60 μl) an equal amount of 2× Laemmli buffer was added and was centrifuged 5 min after boiling for 5 min. The remaining part was supplemented with 355 μl extraction buffer and was used for RNA extraction with the standard phenol–chloroform method. This method ensures the comparability of protein and RNA samples within one panel.
miRNA detection and quantitation
For small RNA northern blot analyses 4 μg of total RNA or samples of gel-filtration were fractionated on denaturing 12% polyacrylamide gels containing 8 M urea and transferred to Hybond NX membrane (GE Healthcare) with semi-dry blotting (Biorad). Membranes were chemically cross-linked (43) and probed with radiolabelled locked nucleic acid (LNA) oligonucleotide probes (Exiqon, Vedbaek, Denmark) or DNA probes as described (44). To detect AGO1-derived siRNAs probes described previously were used simultaneously; AGGAGCTCCCAGTCAGGCAAT, AGCCTACACTGTCTGAGGTGA, AGGCTTTCAAGTTTCCAATGA, GTCCTACCCAGGTACCAGAAC (45). Signal was detected using X-ray film or phosphorimager screen (Amersham). In gel-filtration replicates (Additional file 1: Supplementary Figure S4A, B and C) miR168 was hybridized with biotinylated LNA oligonucleotide and detected using Chemiluminescent Nucleic Acid Detection Module Kit (ThermoFischer, Cat.: 89880).
For the analysis of gel-filtration blots, images were acquired with ChemiDoc equipment in Colorimetric mode. Volume intensity of the four most prominent RISC loaded and unbound fractions were measured and summarized. Loading efficiency (LE) was calculated as RISC loaded volume intensity divided with the total sum of RISC loaded plus unbound volume intensities and was represented as a percentage. Within one panel miR168 expression of all different constructs was detected with same exposure time and images of different miRNAs were produced with subsequent probing of the same membrane after washing. Between the two hybridizations, membranes were checked for activity with phosphorimager screens (Amersham).
Northern blotting
To detect p14 6 μg of same RNA as was used to detect miR168 was separated on formaldehyde agarose gels, blotted onto Hybond NX membrane (GE Healthcare) with capillary transfer, UV crosslinked and subjected to hybridization with radiolabeled PCR product of p14 at 65ºC as described previously (39). Probe was labelled using DecaLabel DNA labelling Kit of Thermo Fisher Scientific according to the manufacturer's instructions. Image was created with phosphorimager screen (Amersham).
Western blotting
5 μl extract of infiltrated N. benthamiana leaves, or 20 μl of Arabidopsis samples were separated on 10 or 8% sodium dodecylsulphate–polyacrylamide gel, blotted overnight to PVDF Transfer Membrane (Hybond-P; GE Healthcare, Freiburg, Germany) using wet tank transfer and subjected to western blot analysis. Membranes were blocked using 5% non-fat dry milk in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST) for 60 min. Blots were cut into two, and respective parts were incubated with anti-BiP (Agrisera, AS09 481) for 1 h or with anti-AGO1 (Agrisera, AS09 527) for 2.5 h at a dilution of 1:7500 in 1% non-fat dried milk in 1× PBST. AGO1-sensor was also detected on Supplementary Figure S1A with anti-EGFP (Agrisera, AS132700) at a dilution of 1:7500. After washing in PBST, blot was incubated with secondary goat anti-rabbit IgG HRP conjugated antibody (Agrisera, AS09 602) for 1 h at a dilution of 1:10 000 in 1× PBST with agitation. Blots were developed with High Clarity Western ECL (Biorad), exposure was made using ChemiDoc (Biorad) equipment in signal accumulation mode.
Immuno-precipitation
For crude extracts, 0.4 g of seedlings (Figures 2E, 3E, 4E and Additional file 1: Supplementary Figure S5A, B and C) or same amount of small rosette leaves (Figure 5F and Additional file 1: Supplementary Figure S5D) were homogenized in four volume of lysis buffer (10 mM Tris–HCl pH 7.6; 1 mM EDTA; 150 mM NaCl; 10% glycerol; 0.5% Nonidet P-40; 5 mM NaF; 1 mM dithiothreitol; 0.5 mM Na3VO4; 1 mM phenylmethylsulfonyl fluoride), and centrifuged three times at 4°C in fresh tubes to get rid of cellular debris. 100–100 μl of extracts were used to purify RNA with the phenol-chloroform method and protein by adding an equal volume of Laemmli buffer (2×). RNA was dissolved in 20 μl nuclease-free water. 1 ml extract was pre-incubated with 3 μl anti-AGO1 HRP conjugated antibody (Agrisera) for 2 h at 4°C with agitation, then applied to Dynabeads Kit according to the manufacturer. Immuno-precipitated fraction was eluted in 20 μl, from which 10 μl was used to purify RNA. Immuno-precipitated RNA samples were dissolved in 20 μl nuclease-free water. To the remaining 10 μl equal volume of Laemmli buffer was added, and used as a protein sample. Input RNA samples for northern blot were diluted 40 times before loading, and the same volume of input and immune-precipitated samples were applied on the gel. Northern and western blots were carried out as described above. Volume intensities of miR168 blots were measured with ImageLab 5.2.1. software of ChemiDoc equipment (Biorad). MiR168 signal volume intensities of immuno-precipitated samples were referred to AGO1 level in corresponding IP samples. Fold change values (FC on figures) were presented relative to the point of reference (35S::MIR168a plants on Figures 2E, 3E, 4E and Additional file 1: Supplementary Figure S5A, B and C; col on Figure 5F and Additional file 1: Supplementary Figure S5D).
High-throughput sequencing (HTS)
To create cDNA libraries for sequencing, high quality RNA samples were purified with the phenol–chloroform method from 0.3 g of bulked seedlings of respective homozygote lines. 20 μg of the samples were loaded onto separate poly-acrylamide gels, the 21–22 nt enriched small RNA fraction was isolated and libraries were prepared only from this fraction using the Truseq Small RNA Library Preparation Kit (Illumina, San Diego, CA, USA) and the modified protocol described earlier (46). Sequencing was carried out on HiScanSQ by UD-Genomed (Debrecen, Hungary) with a 50 bp, single-end chemistry (8 samples/sequencing lane). QIAGEN CLC Genomics Workbench 20 was used for sequence analysis. First, raw sequences were subjected to quality control, adapters and stop sequence were trimmed and reads within the 15–30 nucleotide size range were used for further analysis. Trimmed read number of different libraries varied between 1.5 and 7.7 million, and were used as a base of RPM calculations. The size-selected reads were mapped to the wild type ath-pri-MIR168a, ath-pri-MIR168b and to the respective modified precursor sequence using the Map Reads to Reference tool. The created alignments were extracted to new sequence lists, and with the help of Microarray and Small RNA Analysis tool of CLC Genomics Workbench, reads of individual sequences were counted and exported to a single Excel file. The selected complementary reads were used for further analysis. For analysis of AGO1 derived siRNAs the whole cDNA sequence of ath-AGO1 mRNA was used as reference.
Semi-quantitative RT-PCR
2 μg of DNase I (NEB) treated RNA samples were used to cDNA synthesis with RevertAid First Strand cDNA Synthesis Kit (ThermoFischer). 100 ng of the samples was used to amplify ath-AGO5 (CATTCCGTGCCCGCTACTACAT; ATTAGCAATAAAACATAACCTC) and ath-CPH (GGCGAGAAAGGAATGGGAAA; GTTCTTGGCGGTGAAATCA).
RESULTS
Over-expression of miR168 from ath-MIR168 precursor fragments induces limited AGO1 down-regulation
First, we investigated the AGO1 controlling efficiency of miR168, over-produced from the wild type A. thaliana MIR168a precursor fragment containing the hairpin structure and 10–10 base pairs up- and downstream. Due to the limited sensitivity of AGO1 antibody to detect endogenous AGO1 in Agrobacterium-infiltrated N. benthamiana leaves we built a sensor construct expressing an AGO1-GFP fusion protein (AGO1-sensor; Figure 1A). AGO1-sensor was expressed transiently in Nicotiana benthamiana leaves in the presence or absence of 35S::pri-MIR168a (MIR168a) precursor binary construct (Figure 1A). To eliminate siRNA mediated transgene induced RNAi, viral p14 silencing suppressor construct was added to Agrobacterium infiltration mix. The expression of p14 mRNA was checked by northern blot analyses in the experiments (Additional file 2: Supplementary Table S2, Additional file 1: Supplementary Figure S1C) (38,47).
The robust miR168 over-expression, however, resulted in only a moderate decrease in the GFP signal of AGO1-sensor compared to control infiltration with empty vector (Figure 1B, Additional file 1: Supplementary Figure S1A). The AGO1-sensor showed also moderate down-regulation. To further investigate the regulatory efficiency of miR168 on AGO1 accumulation we produced transgenic A. thaliana (Columbia) plants over-expressing both wild type ath-MIR168a and ath-MIR168b precursor fragments (MIR168a and MIR168b). In accordance with a previous work (30), transgenic over-expression of MIR168a caused only minor changes in the phenotype. The overall look and fertility of the transgenic plants resembled to the wild-type ones. We observed only minor developmental alterations, like serrated rosette leaves and delayed flowering in transgenic lines (Figure 1C, Additional file 1: Supplementary Figure S2A). The severity of the phenotypes correlated with miR168 accumulation levels.
In MIR168a over-expressing transgenic lines we observed only a moderate down-regulation of AGO1 protein level compared to wild type plant (Figure 1C, Additional file 1: Supplementary Figure S2B). Similarly to previous results (30), we detected slightly decreased accumulation of miR159 in parallel with the reduced AGO1 level (Figure 1C). Over-expression of MIR168b induced similar phenotypic alterations and also associated with moderate AGO1 protein down-regulation in young leaves (Additional file 1: Supplementary Figure S2A and B). Since MIR168a and MIR168b transgenic plants exhibited similar phenotypes and AGO1 down-regulation properties, only MIR168a lines were used in following experiments.
The limited impact of miR168 over-expression on AGO1 protein levels, detected in transient and stable transformant systems, correlated very well and suggested that miR168 inefficiently programs AGO1-RISC complexes. To investigate the loading efficiency of miR168 into AGO1-RISC we employed size separation gel-filtration assay (42). Crude extracts of young leaves were loaded onto size-separating column and the collected fractions were analysed for their miRNA and AGO1 protein content. In line with our previous data, miR159 was present predominantly in high molecular weight (HMW) AGO1-RISC containing complexes. In contrast, miR168 accumulated mainly in protein-unbound form in the same sample and only the minority of miR168 was loaded into HMW AGO1-RISC (28,42) (Figure 1D).
Similarly, as was described previously the elevated miR168 level resulted only in a moderate increase in HMW AGO1-RISC loading of miR168 (Figure 1D). This observation indicates that the massive excess of miR168 matured from over-produced wild type MIR168a precursor fragment is not able to incorporate into AGO1-RISC efficiently. This restricted loading efficiency of miR168 resulted only in a moderately reduced AGO1 protein level. The majority of the produced miR168 accumulated in fractions representing protein-unbound miRNAs. This phenomenon is characteristic of miR168 since transient or transgenic over-expression of miR159 and miR171 results in almost total or very efficient loading of HMW AGO1-RISC, respectively (28). In MIR168a over-expressing plants, miR159 preserved its well-loading feature (Figure 1D). This observation confirms that not the unavailability of free, unloaded AGO1 proteins limits the incorporation of miR168 into AGO1-RISC.
Altogether, the observation that MIR168a precursor fragment mediated miR168 over-accumulation is not associated with drastically enhanced AGO1-RISC loading implies that AGO1 loading efficiency of miR168 is strictly regulated.
Modification of miR168/miR168* duplex structure of MIR168a precursor fragment can further reduce AGO1 loading efficiency
According to gel-filtration experiments, miRNAs processed from structurally different precursors, can be incorporated into AGO1-RISC to variant extents (28) (Figure 1D). In silico analyses of miR168/miR168* duplex structure encoded by genomes of various plant species revealed a dominantly conserved nucleotide mismatch at the fourth nucleotide of the duplex. Structural features may affect AGO1-loading efficiency. To test whether this mismatch has any impact on AGO-loading we created a construct in which base pairing at position fourth was introduced by the modification of the miR168* strand only (MIR168-4bp precursor fragment construct, Figure 2A, Additional file 1: Supplementary Figure S2C).
Transient over-expression of MIR168-4bp by agro-infiltration resulted in higher GFP signal coupled with elevated AGO1-sensor protein level compared to wild type MIR168a precursor fragment over-expression (Figure 2B, Additional file 1: Supplementary Figure S1A and B). This observation indicates that despite the high production rate the biological activity of MIR168-4bp derived miR168 species was reduced.
To corroborate these observations, we generated multiple independent MIR168-4bp stable transformants. MIR168-4bp and wild type MIR168a over-expressing lines having comparable miR168 level were selected and their phenotype and AGO1 content were analysed. The selected MIR168-4bp lines showed less delay in flowering and higher AGO1 protein level than the corresponding MIR168a lines suggesting reduced control of AGO1 level (Figure 2C, Additional file 1: Supplementary Figure S3B–E). Although the severity of delayed flowering phenotype and AGO1 down-regulation showed correlation with miR168 level, even extreme over-expression of MIR168-4bp derived miR168 could not provoke greater effect than observed by the MIR168a control line (Additional file 1: Supplementary Figure S3B–E). In gel-filtration experiments MIR168-4bp over-expressing plants showed reduced HMW-RISC loading efficiency of miR168 relative to MIR168a over-expressing plants (Figure 2D, Additional file 1: Supplementary Figure S4A). This observation was further confirmed by AGO1 immuno-precipitation experiments detecting relatively decreased accumulation of miR168 in MIR168-4bp versus MIR168a transgenic plants (Figure 2E, Additional file 1: Supplementary Figure S5A).
All together, these data suggest that establishment of the miR168/miR168* duplex base-pairing at the fourth nucleotide does not inhibit the production of miR168 but decreases its AGO1-RISC loading capacity leading to a new AGO1 protein steady state level compared to wild type precursor fragment (MIR168a) over-expression.
AGO1 loading efficiency can be enhanced by modifying the miR168/miR168* duplex structure of MIR168a precursor fragment
In contrast to miR168, Arabidopsis miR171 exhibits a higher AGO1-RISC loading efficiency (28). Remarkably, in barley hvu-miR168 is also inefficiently loaded, while hvu-miR171 efficiently loaded into HMW AGO1-RISC (Additional file 1: Supplementary Figure S6A). These observations suggest that the loading properties of miRNAs are precisely set and conserved through evolution. In our previous work we have successfully used partial hvu-MIR171 precursor to produce efficient artificial miRNAs (amiRs) (36).
Next, we attempted to increase the loading efficiency of miR168 into AGO1-RISC by remodelling the secondary structure of miR168/miR168* duplex to mimic that of hvu-miR171/miR171* duplex. In hvu-MIR171 the miRNA duplex contains three mismatches at the 4th, 9th and 12th positions and the mature miRNA originates from the 3′ arm of the stem-loop structure. In contrast, ath-MIR168a miRNA duplex has two mismatches at the 4th and 15th positions and the mature miR168 originates from 5′ arm of the precursor (Additional file 1: Supplementary Figure S2C). Mutations were introduced into the passenger strand of ath-MIR168a precursor fragment to create the three mismatches in the appropriate positions, producing MIR168-3mm precursor fragment construct (Figure 3A, Additional file 1: Supplementary Figure S2C).
The transient test of MIR168-3mm revealed that despite the profoundly reduced production rate of miR168, it exhibits increased capacity in down-regulating AGO1-sensor compared to MIR168a construct (Figure 3B; Additional file 1: Supplementary Figure S1). In accordance with this, leaf patches agro-infiltrated with MIR168-3mm showed remarkable GFP signal reduction and low level of AGO1-sensor accumulation compared to MIR168a infiltrated leaves (Figure 3B, Additional file 1: Supplementary Figure S1A and B). Transgenic plants containing MIR168-3mm construct showed only moderate miR168 over-expression. However, this moderate over-expression of miR168 resulted in more pronounced delay in flowering and greater AGO1 down-regulation compared to transgenic lines expressing miR168 more abundantly from the wild type MIR168a construct (Figure 3C, Additional file 1: Supplementary Figure S3B and C).
To test whether the enhanced activity of MIR168-3mm originated miR168 species is caused by their enhanced RISC-loading efficiency, gel-filtration experiments were performed. We found indeed that, MIR168-3mm derived miR168 incorporates more efficiently into HMW-RISC bringing about the enhanced down-regulation of AGO1 protein (Figure 3D, Additional file 1: Supplementary Figure S4B). Immuno-precipitation of AGO1 from MIR168-3mm transgenic line also confirmed the tendency of enhanced loading efficiency of miR168 compared to the MIR168a line (Figure 3E, Additional file 1: Supplementary Figure S5B).
These data imply that intrinsic structural features of wild type miR168/miR168* duplex restrictively regulate AGO1-loading of miR168.
MiR168 produced from hvu-MIR171 stem-loop based artificial constructs exhibits increased AGO1 down-regulation capacity
MIR168-3mm construct exhibits hvu-MIR171 specific features in miR168/miR168* duplex region but the backbone is derived from MIR168a precursor fragment. We wanted to further investigate the role of structural features defining miR168 AGO1-loading capacity in other experimental system. For this we built different artificial miR168 precursor (AMIR) constructs based on the modified version of barley hvu-MIR171 precursor fragment (36). Two variants of artificial AMIR constructs were created to express miR168 from hvu-MIR171 backbone. To retain the hvu-MIR171 stem-loop structure we changed the orientation of miR168 guide strand and modified the star strand in order to keep the distribution of the three mismatches within the duplex in the same positions as in hvu-miR171 duplex (Figure 4A and Additional file 1: Supplementary Figure S2C). AMIR-1 and AMIR-2 differ only in the identity of mismatched nucleotides in the duplex at 4th and 9th positions.
These miR168 producing AMIR168 constructs (AMIR-1, -2) were expressed transiently in N. benthamiana leaves by agro-infiltration in the presence of AGO1-sensor. Both constructs induced a higher reduction of the GFP signal and the AGO1-sensor level compared to MIR168a (Figure 4B and Additional file 1: Supplementary Figure S1A and B). Moreover, as small RNA northern blot results indicated, the increased AGO1 down-regulation in these cases were associated with less amount of miR168 over-expression compared to MIR168a (Figure 4B and Additional file 1: Supplementary Figure S1A and B).
Gel-filtration experiments of crude extracts originated from AMIR-1 and AMIR-2 agro-infiltrated leaf patches confirmed elevated HMW-RISC loading ability of miR168 compared to transient over-expression of MIR168a (Additional file 1: Supplementary Figure S6B). Complete loading of miR159 showed the existence of functional AGO1-RISCs in HMW fractions of leaves expressing AMIR constructs. These membranes were also used to detect miR168* strands using probes specific for every AMIR constructs. Signals detected in the low molecular weight, protein-unbound fractions suggest that AMIR originated miR168 species exist at least partly in duplex form (Additional file 1: Supplementary Figure S6B) confirming previous results (28).
To further investigate how the altered stem-loop structures affect miR168 HMW-RISC loading efficiency, AMIR-1 and -2 stable transgenic lines were generated. Over-expressing lines producing similar amount of miR168 were selected for further studies. AMIR-1 and AMIR-2 displayed more pronounced phenotypic alterations (including delayed flowering time, reduced rosette diameter) compared to MIR168a line. (Figure 4C, Additional file 1: Supplementary Figure S2A and B). The severity of phenotypes of the analysed AMIR-1 and AMIR-2 lines correlated with the level of over-produced miR168 and in the cases of plants expressing extremely high level of miR168 resembled to that of hypomorph ago1–25 and ago1–27 mutants (Additional file 1: Supplementary Figure S2A). Although the amount of miR168 over-expression was moderately lower, the AGO1 protein content of AMIR lines was severely reduced compared to MIR168a lines (Figure 4C).
Next, size separation of protein complexes with gel-filtration experiments were performed from seedlings of selected AMIR-1 and AMIR-2 transgenic lines and a control MIR168a line. AMIR-1 and AMIR-2 lines displayed enhanced miR168 accumulation in AGO-RISC containing fractions in spite of the reduced accumulation of AGO1 protein (Figure 4D, Additional file 1: Supplementary Figure S4C). Immuno-precipitation experiments from seedlings of AMIR-1 and AMIR-2 over-expressing lines confirmed that miR168 incorporation into AGO1-RISC is increased in both cases (Figure 4E, Additional file 1: Supplementary Figure S5C).
These data indicate that production of miR168 from alternative stem-loop structures could increase the AGO1-RISC loading efficiency leading to its enhanced biological activity.
High-throughput sequencing analysis of transgenic plants identifies canonical miR168 species
Over-production of miR168 from modified precursor fragments can be associated with the misprocessing of the miR168/miR168* duplex structure resulting in the differential accumulation of canonical and potentially non-canonical mi168 species. The non-canonical miR168 species may be loaded into different AGOs, at different rates to alternatively program the RISC effectors leading to the distorted auto-regulation of RNAi. To get a comprehensive understanding about how the miR168 species are maturated in our stable transgenic lines, we performed a high-throughput sequencing (HTS) analyses of sRNA pools of the selected transgenic lines.
Samples originated from bulked seedlings of the representative lines were used in two replica experiments. Following the quality check and adapter and stop oligo sequence trimming (Additional file 1: Supplementary Figure S7A) reads between 15–26 nt were initially analysed for their size distribution (Additional file 1: Supplementary Figure S8A). In the investigated samples the 21 nt reads exhibited the highest abundance, but reads between 15 and 24 nt long were also represented in high proportion. In contrary to total read size distribution, reads mapped to miR168 producing precursors revealed the high dominance of 21 nt long small RNAs. 81–98% of the sequences were find to be between 20 and 22 nt, with the lowest proportion observed in case of MIR168a over-expressing plants (Additional file 1: Supplementary Figure S8B).
Investigation of the reads per million (RPM) data revealed, in accordance with the small RNA northern blot data, that all the investigated precursor fragments over-produced miR168 compared to the wild type plant (Figure 5A; Additional file 1: Supplementary Figure S5E). In addition to the dominant over-production of 5′U miR168 we also experienced the over production of 5′C and the presence of minor quantity 5′G and 5′A miR168 species in the transgenic plants (Figure 5A). The differential accumulation of various miR168 species in transgenic lines can raise the possibility that misprocessing of miR168 can be an important factor influencing AGO1 loading efficiency.
To investigate this possibility, we calculated the relative accumulation of different 5′ end miR168 species (Figure 5B). Detailed investigation of iso-miRNA168 species revealed, that the majority of 5′U reads represent the canonical miR168 species except AMIR-2 where one nt truncated version of miR168 accumulated to the highest level (Additional file 1: Supplementary Figure S7B). The 5′C iso-miRNA content of MIR168-4bp and MIR168-3mm over-expressors was similar to that of wild type and MIR168a precursor fragment over-expressing transgenic plants exhibiting one nt truncation at the 5′ end. In contrary, in AMIR-2 and especially AMIR-1 over-expressors the dominant version of 5′C iso-miRNAs were one nt longer at the 5′ end. (Additional file 1: Supplementary Figure S7B). Since, AMIR-1 and AMIR-2 exhibit very similar features it is unlikely that the observed differences in the accumulation of iso-miR168 species play important role in enhanced AGO loading. 5′G and A miR168 iso-miRNAs were strongly under-represented in the samples, less than 4 and 0.6% of total miR168 reads, respectively. Due to the small quantity of these sRNAs it is unlikely that they contribute considerably to the investigated phenomenon.
MIR168a and AMIR-1 lines revealed a very similar distribution of 5′U and 5′C miR168 species, even detecting slightly less 5′U miR168 in AMIR-1 line compared to MIR168a (Figure 5B). This observation suggests that the generated 5′C miR168 species do not interfere with or take part in differential AGO1 loading at a high extent since at the level of comparable expression AMIR-1 derived miR168 is more efficient in AGO1 loading than that of MIR168a (Figure 4E, Additional file 1: Supplementary Figure S5C). AMIR-2 line, which has slightly more 5′U but less 5′C miR168 relative to MIR168a line, behaved very similarly to AMIR-1 (Figure 4E, Additional file 1: Supplementary Figure S5C). This finding also indicates that the ratio of 5′U and 5′C iso-miRNAs may not have profound effect on the AGO1 loading efficiency.
In the case of MIR168-3mm we detected elevated 5′U miR168 ratio relative to 5′C miR168, very similar to that of was detected in AMIR-2 line (Figure 5B). However, MIR168-3mm was associated with more efficient AGO1 loading at less than 20% miR168 over-expression level compared to AMIR-1–2 and MIR168a lines (Figure 5A). MIR168-4bp mediated over-production of miR168, exhibiting very inefficient AGO loading, was associated with higher 5′C miR168 ratio (Figure 5B). However, due to the high over-expression level in MIR168-4bp the absolute content of 5′U miR168 was comparable to MIR168a and AMIR-2, and higher than MIR168-3mm and AMIR-1 (Figure 5A.). This finding further supports the assumption that the altered 5′C miR168 content is not the major component that determines the loading efficiency changes in the investigated transgenic lines.
Furthermore, by comparing analysis of miR168 species in previously published AGO1 immuno-precipitation data (22) (Additional file 1: Supplementary Figure S8C) with 5′C miR168 ratio in input total RNA of our Columbia sample, we found, that in wild type Arabidopsis the 5′C miR168 ratio in input total RNA and AGO1 precipitated samples is very similar (7–8%). This observation suggests that most of the produced 5′C miR168 species are biologically active and follow the same AGO loading rules as 5′U miR168. To further investigate the potential alternative function of 5′C miR168 we tested the accumulation of AGO5, an AGO protein preferentially associated with 5′C miRNAs. We found, in accordance with published data (48), that AGO5 does not express at detectable level in the investigated tissue type under our conditions (seedlings) (Additional file 1: Supplementary Figure S8D). These data indicate that the activity of this AGO protein does not interfere with our results. Moreover, we found that AGO1 mRNA associated siRNAs do not exhibit higher level accumulation in our HTS data and northern blot analysis in plants expressing the manipulated precursor constructs indicating that the activity of this pathway has not been altered in the used transgenic lines (Additional file 1: Supplementary Figure S5E and F).
Altogether these data suggest that mainly the structural features of miRNA duplex govern the loading efficiency changes of miR168 in our study. However, we cannot exclude the possibility that, to a lesser extent, the altered production of miR168 species can also contribute to the phenomenon in some cases.
Competing miRNAs can affect the loading of miR168 into AGO1-RISC
Our findings suggest that loading efficiency of miR168 into AGO1-RISC is tuned-down by structural elements located within miRNA duplex. Importantly, low loading properties of miR168 seem to be conserved (Figure S6A and B; (42)), suggesting its biological relevance. We hypothesized that the high excess of unbound miR168 may act as a balancer continuously adjusting the required physiological level of AGO1 in response to external or endogenous stimuli. Modification in the secondary structure of the duplex can shift this adjusted loading balance leading to miR168 over/under loading into AGO1-RISC. The general existence of high excess of miR168 pool unbound to AGO1 suggests that the calibrated loading efficiency of miR168 into AGO1-RISC represents a flexible adaptive regulatory system.
To test this hypothesis, first we investigated whether modulating the small RNA content of the cellular environment can competitively alter the loading of miR168 into AGO1-RISC. Previously we demonstrated that miR171 is well-loaded, while miR156 exhibits very inefficient AGO1 loading ability since it accumulates predominantly in protein-unbound form in A. thaliana (28).
In transient assays, MIR156 and MIR171 precursor fragments were massively over-expressed in the presence of Ago1-sensor to find out whether the endogenous miR168 can be competitively sequestered from AGO1 loading. Robust over-expression of miR171 resulted in a higher GFP signal and slightly but consistently increased AGO1-sensor level compared to miR156 co-infiltrated control (Figure 5C, Additional file 1: Supplementary Figure S4D). These findings suggest that the miR171 efficiently outcompetes miR168 from AGO1 loading that subsequently results in de-repression of AGO1-sensor. Size separation gel-filtration assays confirmed that the over-expression of well-loaded miR171 reduced the loading efficiency of endogenous miR168 into HMW AGO1-RISC compared to low-loaded miR156 infiltration control (Figure 5D, Additional file 1: Supplementary Figure S4D). These observations indicate that AGO1 loading of miR168 can respond to alterations in the small RNA pool of the given cellular environment in a competitive manner.
The competitive loading model of miR168 predicts that the decrease of AGO1 protein amount should be achieved by increasing the competitiveness of miR168 inversely to the cellular miRNA pool. For this we took advantage of dcl1-9 mutant, in which the production of most miRNAs is severely impaired (49). Intriguingly, miR168 level was reported to be relatively unaffected in this mutant plant (30). To avoid potential deteriorating effects of the presence of unloaded AGO1-RISCs in these mutants, it is expected that AGO1 protein level is adjusted to the available miRNA pool by the action of miR168/AGO1 regulatory loop. We speculated that in dcl1-9 background, the competitiveness of miR168 would be elevated due to the stoichiometric imbalance, the relatively high level of miR168 in comparison to the competitor miRNA pool. We confirmed that in contrast to miR159, miR168 content was only slightly affected in dcl1-9 mutant, while AGO1 protein level was remarkably down-regulated (Figure 5E). Since the low level of AGO1 made it technically difficult to use gel-filtration assays, immuno-precipitation experiments were carried out. Supporting our hypothesis, the relative miR168 content in AGO1 immuno-precipitates of dcl1-9 mutants increased (Figure 5F, Additional file 1: Supplementary Figure S5D).
These data indicate that in dcl1-9 mutant miR168 loading into AGO1 is enhanced. This new miR168 loading kinetics in turn generates a lower AGO1 protein level adjusted to the suppressed miRNA content of the cells.
DISCUSSION
Plant miRNAs play fundamental roles in plant growth and development, as well as in adaptation to biotic, abiotic stresses and other physiological processes via controlling the expression of target transcription factors and stress-response linked proteins (6). The multi-layered regulatory roles render the miRNA pathway to be one of the most pliable and versatile controlling mechanisms. According to these attributes, the miRNA pathway is fine tuned in an environment responsive manner involving transcriptional control of the miRNA encoding genes, tissue‐specific expression of biogenesis co-factors, post‐translational modifications, control of miRNA stability and processing (50).
The secondary structure of the pre-miRNA, encompassing the miRNA/miRNA* duplex region, plays a pivotal role in determining the efficiency of miRNA biogenesis (14,15,51,52). Moreover, structural motifs of the precursors can also contribute to specification of AGO proteins determining the sorting of miRNAs in proper executor complexes (20,26). The central executor component of miRNA pathway, AGO1, is feedback regulated by the conservative AGO1-competent miR168 family (48). The importance of miR168 driven auto-regulatory loop was demonstrated by the over-expression of miR168-resistant version of AGO1 mRNA inducing various developmental defects and eventually the death of the plants (33). These data revealed that unbalanced over-accumulation of AGO1 protein imposes severe danger to the proper functioning of the plant.
Previously, the utilization of size-separation gel-filtration method of plant crude extracts revealed an extraordinary property of miR168. It was shown that only a small subset of mature miR168 was present in HMW-RISC while the majority of mature miR168 accumulated in the low molecular weight fractions in protein-unbound form (42). Notably, the accumulation of unbound miR168 seems to be conserved, however the significance of the free miR168 pool remains elusive.
Previously, we demonstrated that the transient or transgenic over-expression of various miRNA precursors can lead to complete (miR159), efficient (miR171) or limited (miR168) HMW-RISC loading in the same cellular environment (28). These miRNAs are matured from precursors having different secondary structures raising the possibility that structural motifs of miRNA precursors can affect the AGO1 loading rate. To study the biological function of this low-calibrated loading efficiency of miR168 into AGO1-RISC, the wild type ath-MIR168a and ath-MIR168b precursor fragments were over-expressed in both transient and stable transgenic systems. Drastic enhancement of miR168/AGO1-RISC loading and AGO1 down-regulation could not be achieved in transgenic lines. Majority of the over-expressed miR168 were sorted into the free pool and only a smaller subset was loaded into the RISC. This finding suggested that restricted miR168 loading equilibrium into AGO1 has an important biological role. We hypothesized that structural motifs of MIR168a precursor can be responsible for the restricted loading of miR168 into AGO1-RISC.
To test this hypothesis, a series of alternative, but miR168 producing precursor fragments were created by modifying the wild type MIR168a precursor miRNA duplex region or by expressing miR168 from heterologous constructs containing hvu-MIR171 precursor backbone. The secondary structure of miRNA/miRNA* duplex regions of these constructs were manipulated by introducing modifications into the star strand only, leaving the guide strand intact. In the transformation experiments several transgenic lines have been generated with various expression levels of miR168 (Additional file1: Supplementary Figure S2A, B and S3). The distribution of various construct lines amongst the over-expression rate categories was different raising the possibility that structural modifications in duplex may also affect the efficiency of biogenesis (Additional file1: Supplementary Figure S3A). Since our work focuses on the AGO loading efficiency of miR168, over-produced from modified precursor fragments, transgenic lines displaying similar expression properties have been selected to allow the precise comparison in the experiments. According to this, high, intermediate and low MIR168a precursor fragment over-expressing lines were used for the analyses of MIR168-4bp, AMIR168-1-2 and MIR168-3mm, respectively.
The majority of these modifications resulted in enhanced down-regulation of AGO1-sensor in transient assays or AGO1 level in transgenic plants. Gel-filtration and immuno-precipitation experiments indicated that increased AGO1 down-regulation is a consequence of enhanced AGO1-RISC loading of miR168. Both AMIR-1 and AMIR-2 have similar effect on AGO1 down-regulation. This observation suggests that the identity of mismatched nucleotides within a duplex do not have strong influence on AGO1 loading rather the structure of the duplex is important. Intriguingly, in the case of MIR168-4bp the introduced modification resulted in even less efficient miR168/AGO1 loading showing that the loading balance can be altered in both directions.
The over-expressed miR168 species were produced mainly in correct size and no elevated secondary siRNA production was detected from AGO1 mRNA in the experiments. However, we experienced altered 5′U/C ended miR168 ratio during the over-expression of various modified miR168 precursor fragments. Alternative maturation of miR168 may affect loading and downstream effects on AGO1 feed-back loop. The similar AGO loading properties of 5′C or 5′U miR168 species in the wild type plant (col on Figure 5B; Additional file 1: Supplementary Figure S8C) and association of various AGO1 loading efficiencies with similar 5′U/C miR168 ratios renders this possibility less likely. Still, we cannot absolutely exclude the role of altered production of miR168 isoforms in differential AGO sorting or loading as for example in the floral tissue where AGO5 is dominantly expressed (33).
Moreover, we also demonstrated that loading of miR168 into AGO1-RISC is an environmentally responsive regulatory process. The drastic suppression of the overall miRNA level in dcl1-9 biogenesis mutant triggered adjustment of AGO1 content to a lower level through the enhanced loading of miR168 into AGO1. Furthermore, miR168 can be outcompeted from RISC by massive over-expression of an efficiently AGO1-competent miRNA suggesting that miRNAs compete for RISC loading. Combining previous findings with our new results we propose a refined model for miR168 mediated regulation of AGO1 homeostasis (Figure 6).
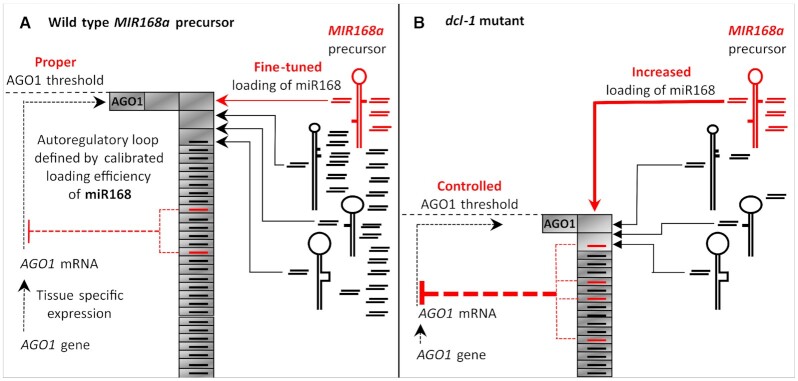
Figure 6.
Proposed model for calibrated loading efficiency based miR168-AGO1 autoregulatory loop. (A) Action of MIR168a precursor in wild type plant. AGO1 mRNA, possessing miR168 recognition site, is expressed by tissue specific manner under the control of AGO1 gene promoter (dotted black line with arrowheads). Unloaded AGO1 proteins (gray rectangles) are continuously translated from the mRNA. MiRNA precursors are also expressed in a tissue specific manner (schematically represented by black hairpin structures), including MIR168a precursor (red), which are subjected to subsequent cleavages to produce miRNA duplexes (short paired lines) determining the composition of the miRNA pool. The mature miRNA strand of the miRNA duplexes can associate with AGO1 proteins with different efficiencies (lines with arrowheads) generating miRNA loaded AGO1 pool (grey rectangles (AGO1) with lines (miRNAs)). MiRNA duplexes not able to load into AGO1 can accumulate in protein-unbound forms (paired lines on the right side of the precursor structures). The AGO1 loading of miR168 is finely calibrated by structural features of the precursor RNA allowing only a subset of the miR168 pool to be loaded into AGO1 (grey rectangles with red line) and majority of miR168 accumulates in duplex form, unbound to protein (red paired lines on the right side of the precursor structures). Due to this highly sensitive autoregulatory loop, the defined miR168/AGO1 complexes negatively regulate AGO1 mRNA (red dashed lines) determining the proper physiological AGO1 threshold (schematically represented by the column of miRNA loaded AGO1 proteins (gray rectangles with lines). (B) Regulatory action of miR168/AGO1 autoregulatory loop in RNAi defective mutants. In dcl1-9 mutant the production of endogenous miRNAs is strongly inhibited (represented by the absence of black paired lines) except the biogenesis of miR168. Because of the lack of AGO1-competent miRNAs there is a danger of over-accumulation of unloaded AGO1 proteins leading to interference with proper cell functions. However, in the absence of efficiently AGO1-competent miRNA species the extensively produced miR168 is able to load into AGO1 proteins with higher efficiency imposing strong control on AGO1 mRNA. Due to this regulatory mechanism, a new, controlled, AGO1 equilibrium is formed in balance with reduced miRNA content of the cell.
According to this ‘competitive’ model, the efficiently processed MIR168a precursor produces a high excess of miR168 but only a small subset of this is loaded into AGO1-RISC. The unincorporated miR168 species accumulates as miR168/miR168* duplexes in the cytoplasm. Balance of AGO1-RISC loaded and unbound miR168 is determined by structural features of the precursor encompassing the miR168/miR168* duplex region. Other miRNA precursors such as MIR159a or MIR171a, for example, possess structural features enabling their more efficient AGO1 loading (28). The amount of AGO1-RISC associated miR168 is determined by competition of miR168 with the AGO1-competent small RNA pool of the cell for the limiting free AGO1 proteins. The low-calibrated AGO1-RISC loading of miR168 could fine tune the proper physiological level of AGO1 relative to given small RNA population of the cell (Figure 6A).
The fundamental requirement of this regulatory mechanism is the continuous presence of biologically active miR168 excess competent to be incorporated into AGO1-RISC. This requirement seems to be guaranteed by uncoupling the biogenesis of miR168 from the canonical miRNAs since miR168 production is insensitive to many mutations affecting miRNA pathway, such as dcl1-9 (30) (Figure 5E). The insensitivity of miR168 production to disorders in miRNA pathway can ensure the continuous miR168 driven control of AGO1 level eliminating the risk of over-loading AGO1 with unwanted sRNA species. Indeed, we found that in dcl1-9 mutant where the level of endogenous miRNA pool is severely lowered, less AGO1 protein is present due to enhanced incorporation of miR168 into AGO1 (Figure 5F). This finding suggests that the suppressed miRNA pool results in less loaded AGO1. The availability of free AGO1 proteins enables the more efficient loading of the miR168 excess. The enhanced loading of miR168 in turn reinforces the feedback regulatory loop bringing down the AGO1 protein level to a new equilibrium (Figure 6B). The observation that MIR168 and AGO1 expression is co-regulated (30) is in line with this model since increase in AGO1 protein level requires the increase in miR168 level to maintain the proper balance between AGO1 loaded and unbound miR168 species.
Previously, we showed that various virus infections on different host plants induce drastic miR168 induction in the infected leaves which is usually associated with strong down-regulation of AGO1 level (42,53). The efficiency of the virus mediated AGO1 control can be explained by the observation that virus infection also resulted in drastic AGO1 mRNA induction. The AGO1 mRNA excess can continuously produce AGO1 proteins enabling the loading of miR168 in higher extent and the establishment of a strong feedback loop.
The striking variability of RNA stem-loop shape and size of plant miRNA precursors (13,54) compared to stereotypic animal counterparts (55) suggests that structural features can play a dominant role in the biogenesis and action of plant miRNAs. The calibrated loading action of miR168 governed by RNA structural motifs is potentially applicable to other miRNAs. This hypothesis is supported by the observation that many miRNAs exhibit the presence of AGO-loaded as well AGO-unbound forms (28). It is not clear, whether miRNA duplexes accumulated in the cytoplasm in protein-unbound form represent a biologically active pool of AGO1 loading competent molecules or they are superfluous by-products of the miRNA biogenesis. Recently it has been shown that miRNA loading of AGO1 predominantly takes place in the nucleus (19) suggesting that the loading rate of miR168 is calibrated in the nucleus. However, it cannot be excluded that the cytoplasmic miRNA duplexes represent a biologically active reservoir.
In summary, we used an artificial system built for comparative analyses of AGO1 loading efficiency of miR168 derived from differently altered precursor fragments. This comparative analysis, carried out in transient and stable transgenic lines, helped us to provide a biologically relevant explanation for the existence of the recently discovered AGO unbound miR168 species, and refine the model of RNA silencing autoregulation. According to this, (i) the excessive processing of ready-to-load miR168 species, (ii) the structural properties embedded in miR168/miR168* duplex structure, and (iii) the amount of competing cellular miRNAs are defining the AGO1-RISC loading efficiency of miR168. This competition-based mechanism precisely and dynamically correlates the amount of AGO1 protein according to cellular needs. The presence of unbound cytoplasmic pool was demonstrated for many miRNAs (28) indicating that regulatory action of competition based loading efficiency can be valid for other miRNAs as well.
Since we investigated solely the role of precursor fragment stem-loop structures further experiments will be necessary to investigate this phenomenon in the complex context of miRNA biogenesis. Moreover, it will also be important to fine map the requirements of precursor stem–loop structure for governing AGO1 loading by extensive comprehensive structural–function analyses. In the future it will be also important to identify and characterize protein cofactors playing a role in RNA structure-based communication between miRNA biogenesis and action.
DATA AVAILABILITY
HTS data were deposited at SRA database under the BioProject accession number PRJNA640279.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tibor Csorba and Péter Gyula for the critical review of the manuscript and Erzsébet Poldán for her valuable assistance in the laboratory.
Contributor Information
Ágnes Dalmadi, Hungarian University of Agriculture and Life Sciences, Institute of Genetics and Biotechnology, Páter Károly Street 1, Gödöllő 2100, Hungary.
Fabio Miloro, Hungarian University of Agriculture and Life Sciences, Institute of Genetics and Biotechnology, Páter Károly Street 1, Gödöllő 2100, Hungary.
Jeannette Bálint, National Agricultural Research and Innovation Centre, Agricultural Biotechnology Institute, Szent-Györgyi Albert Street 4, Gödöllő 2100, Hungary.
Éva Várallyay, Hungarian University of Agriculture and Life Sciences, Institute of Plant Protection, Ménesi Street 44, Budapest 1118, Hungary.
Zoltán Havelda, Hungarian University of Agriculture and Life Sciences, Institute of Genetics and Biotechnology, Páter Károly Street 1, Gödöllő 2100, Hungary.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research, Development and Innovation Office [NKFI K116602, K134914]. Funding for open access charge: National Research, Development and Innovation Office [NKFI-K134914].
Conflict of interest statement. None declared.
REFERENCES
- 1. Borges F., Martienssen R.A.. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015; 16:727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Treiber T., Treiber N., Meister G.. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019; 20:5–20. [DOI] [PubMed] [Google Scholar]
- 3. Rogers K., Chen X.M.. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013; 25:2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel D.P. Metazoan microRNAs. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J.L., Mei J., Ren G.D.. Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci. 2019; 10:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li S., Castillo-Gonzalez C., Yu B., Zhang X.. The functions of plant small RNAs in development and in stress responses. Plant J. 2017; 90:654–670. [DOI] [PubMed] [Google Scholar]
- 7. Brant E.J., Budak H.. Plant small non-coding RNAs and their roles in biotic stresses. Front. Plant Sci. 2018; 9:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Achkar N.P., Cambiagno D.A., Manavella P.A.. miRNA biogenesis: a dynamic pathway. Trends Plant Sci. 2016; 21:1034–1044. [DOI] [PubMed] [Google Scholar]
- 9. Bologna N.G., Voinnet O.. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014; 65:473–503. [DOI] [PubMed] [Google Scholar]
- 10. Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C.. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005; 138:2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bologna N.G., Schapire A.L., Zhai J., Chorostecki U., Boisbouvier J., Meyers B.C., Palatnik J.F.. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013; 23:1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu H., Zhou Y., Castillo-Gonzalez C., Lu A., Ge C., Zhao Y.T., Duan L., Li Z., Axtell M.J., Wang X.J.et al.. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 2013; 20:1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bologna N.G., Mateos J.L., Bresso E.G., Palatnik J.F.. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009; 28:3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu J., Li C., Peng X., Zhang X.. RNA architecture influences plant biology. J. Exp. Bot. 2021; 72:4144–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Z., Ma Z., Castillo-Gonzalez C., Sun D., Li Y., Yu B., Zhao B., Li P., Zhang X.. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature. 2018; 557:516–521. [DOI] [PubMed] [Google Scholar]
- 16. Ji L., Chen X.. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012; 22:624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S.. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:3691–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma Z., Zhang X.. Actions of plant Argonautes: predictable or unpredictable. Curr. Opin. Plant Biol. 2018; 45:59–67. [DOI] [PubMed] [Google Scholar]
- 19. Bologna N.G., Iselin R., Abriata L.A., Sarazin A., Pumplin N., Jay F., Grentzinger T., Dal Peraro M., Voinnet O.. Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the pMicroRNA pathway. Mol. Cell. 2018; 69:709–719. [DOI] [PubMed] [Google Scholar]
- 20. Iki T. Messages on small RNA duplexes in plants. J. Plant Res. 2017; 130:7–16. [DOI] [PubMed] [Google Scholar]
- 21. Eamens A.L., Smith N.A., Curtin S.J., Wang M.B., Waterhouse P.M.. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. 2009; 15:2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C.et al.. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008; 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y.. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008; 49:493–500. [DOI] [PubMed] [Google Scholar]
- 24. Zhu H.L., Hu F.Q., Wang R.H., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X.R.. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011; 145:242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Endo Y., Iwakawa H.O., Tomari Y.. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 2013; 14:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iki T., Clery A., Bologna N.G., Sarazin A., Brosnan C.A., Pumplin N., Allain F.H.T., Voinnet O.. Structural flexibility enables alternative maturation, ARGONAUTE sorting and activities of miR168, a global gene silencing regulator in plants. Mol Plant. 2018; 11:1008–1023. [DOI] [PubMed] [Google Scholar]
- 27. Zhang X.M., Niu D.D., Carbonell A., Wang A.R., Lee A., Tun V., Wang Z.H., Carrington J.C., Chang C.E.A., Jin H.L.. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 2014; 5:5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dalmadi A., Gyula P., Balint J., Szittya G., Havelda Z.. AGO-unbound cytosolic pool of mature miRNAs in plant cells reveals a novel regulatory step at AGO1 loading. Nucleic. Acids. Res. 2019; 47:9803–9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morel J.B., Godon C., Mourrain P., Beclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H.. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002; 14:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaucheret H., Mallory A.C., Bartel D.P.. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006; 22:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Csorba T., Kontra L., Burgyan J.. viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology. 2015; 479-480:85–103. [DOI] [PubMed] [Google Scholar]
- 32. Carbonell A., Carrington J.C.. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015; 27:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaucheret H., Vazquez F., Crete P., Bartel D.P.. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev. 2004; 18:1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mallory A.C., Vaucheret H.. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009; 10:521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One. 2009; 4:e6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kis A., Tholt G., Ivanics M., Varallyay E., Jenes B., Havelda Z.. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016; 17:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vargason J.M., Szittya G., Burgyan J., Hall T.M.. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003; 115:799–811. [DOI] [PubMed] [Google Scholar]
- 38. Szadeczky-Kardoss I., Csorba T., Auber A., Schamberger A., Nyiko T., Taller J., Orban T.I., Burgyan J., Silhavy D.. The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res. 2018; 46:4632–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merai Z., Kerenyi Z., Molnar A., Barta E., Valoczi A., Bisztray G., Havelda Z., Burgyan J., Silhavy D. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005; 79:7217–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J.. Protocol: streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods. 2009; 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lakatos L., Szittya G., Silhavy D., Burgyan J.. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004; 23:876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Varallyay E., Valoczi A., Agyi A., Burgyan J., Havelda Z.. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010; 29:3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pall G.S., Hamilton A.J.. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008; 3:1077–1084. [DOI] [PubMed] [Google Scholar]
- 44. Varallyay E., Burgyan J., Havelda Z.. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008; 3:190–196. [DOI] [PubMed] [Google Scholar]
- 45. Iki T., Clery A., Bologna N.G., Sarazin A., Brosnan C.A., Pumplin N., Allain F.H.T., Voinnet O.. Structural flexibility enables alternative maturation, ARGONAUTE sorting and activities of miR168, a global gene silencing regulator in plants. Molecular Plant. 2018; 11:1008–1023. [DOI] [PubMed] [Google Scholar]
- 46. Czotter N., Molnar J., Pesti R., Demian E., Barath D., Varga T., Varallyay E.. Use of siRNAs for diagnosis of viruses associated to woody plants in nurseries and stock collections. Methods Mol. Biol. 2018; 1746:115–130. [DOI] [PubMed] [Google Scholar]
- 47. Merai Z., Kerenyi Z., Molnar A., Barta E., Valoczi A., Bisztray G., Havelda Z., Burgyan J., Silhavy D.. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005; 79:7217–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mallory A., Vaucheret H.. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010; 22:3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Park W., Li J., Song R., Messing J., Chen X.. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002; 12:1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Manavella P.A., Yang S.W., Palatnik J.. Keep calm and carry on: miRNA biogenesis under stress. Plant J. 2019; 99:832–843. [DOI] [PubMed] [Google Scholar]
- 51. Meyers B.C., Simon S.A., Zhai J.. MicroRNA processing: battle of the bulge. Curr. Biol. 2010; 20:R68–R70. [DOI] [PubMed] [Google Scholar]
- 52. Moro B., Chorostecki U., Arikit S., Suarez I.P., Hobartner C., Rasia R.M., Meyers B.C., Palatnik J.F.. Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 2018; 46:10709–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Varallyay E., Havelda Z.. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 2013; 14:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cuperus J.T., Fahlgren N., Carrington J.C.. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011; 23:431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HTS data were deposited at SRA database under the BioProject accession number PRJNA640279.