Abstract
Every type of nucleic acid in cells undergoes programmed chemical post-transcriptional modification. Generally, modification enzymes use substrates derived from intracellular metabolism, one exception is queuine (q)/queuosine (Q), which eukaryotes obtain from their environment; made by bacteria and ultimately taken into eukaryotic cells via currently unknown transport systems. Here, we use a combination of molecular, cell biology and biophysical approaches to show that in Trypanosoma brucei tRNA Q levels change dynamically in response to concentration variations of a sub-set of amino acids in the growth media. Most significant were variations in tyrosine, which at low levels lead to increased Q content for all the natural tRNAs substrates of tRNA-guanine transglycosylase (TGT). Such increase results from longer nuclear dwell time aided by retrograde transport following cytoplasmic splicing. In turn high tyrosine levels lead to rapid decrease in Q content. Importantly, the dynamic changes in Q content of tRNAs have negligible effects on global translation or growth rate but, at least, in the case of tRNATyr it affected codon choice. These observations have implications for the occurrence of other tunable modifications important for ‘normal’ growth, while connecting the intracellular localization of modification enzymes, metabolites and tRNAs to codon selection and implicitly translational output.
INTRODUCTION
Modifications are catalyzed by a remarkable array of different enzymes seemingly performing a myriad of chemical reactions, including deaminations, thiolations, methylations, isomerizations, amino acid additions, acetylations, etc. (1). In turn, modification enzymes utilize a variety of substrates, many of which are the products of central metabolism; yet the impact of metabolism on modifications remains largely unexplored.
Unique among modifications are transglycosylation reactions by the tRNA-guanine transglycosylase (TGT) family of enzymes, which catalyze incorporation of 7-deaza derivatives of guanosine in tRNAs (2). Formation of these modifications involves an isoenergetic reaction that breaks the glycosidic bond between the base and the sugar while replacing an encoded guanine for the nucleobase queuine (q) (and/or Q derivatives). In Archaea, archaeosine occurs at position 15 of tRNAs and is thought to play a structural role (3). In Bacteria and Eukarya (with the known exception of Saccharomyces cerevisiae, which lacks TGT), Q occurs at position 34 of the anticodon of a sub-set of tRNAs: tRNATyr, tRNAAsp, tRNAHis and tRNAAsn; all single-copy tRNAs reading two-codon boxes (4). At a molecular level, and because of its placement at the first anticodon position, the role of Q in decoding has been explored (5–8). The general principle that arises from such studies is that the role of Q in translation may well vary from organism to organism. In some, Q influences the accuracy of decoding the C-ending codons for histidine, aspartate, asparagine and tyrosine (6,7), while in other cases the presence of Q favors U-ending codons for the same amino acids (8,9). Further muddling matters is the fact that the Q base itself cannot be synthesized by eukaryotes and is salvaged from the external environment (10), suggesting that transport itself may limit intracellular Q levels.
At a more organismal level, in Drosophila, Q levels change during different developmental stages and presumably play an important function in codon selection during the developmental plan; an argument supported by strong evolutionary inferences (6); however, what triggers such changes in Q levels is not exactly clear. In Schizosaccharomyces pombe, Q34 is not only important for m5C formation at position 38 (11) but affects translational speed rather than codon choice (7). Beyond this, several reports invoke modification changes as a mechanism to deal with environmental stress via codon-biased translation (12–14). Cells may alter the modification levels of certain tRNAs to favor decoding of mRNAs for proteins involved in stress response. These mRNAs showed a marked bias for codons that can be accounted for by the increase in the particular modification (12,13,15). A corollary to codon-biased translation is the existence of mRNAs that are by themselves tunable; the so-called Modification Tunable Transcripts or MoTTs (16).
Trypanosomes undergo major metabolic reprogramming during their different developmental stages whereby in the insect vector they generate the bulk of their ATP via oxidative phosphorylation (17). In the mammalian stages, trypanosomes down-regulate mitochondrial function and rely on substrate-level phosphorylation (18). Changes in metabolism during development raise a possible connection between modifications and cellular metabolism, not solely as a response to stress but more importantly during ‘normal’ growth and maintenance of homeostasis. To explore these concepts, Trypanosoma brucei offers several advantages: (i) tRNA splicing is cytoplasmic (19) and tRNATyr is the only intron-containing tRNA in these organisms providing an important marker for where the tRNA has been, (ii) Q formation is a nuclear event, that only occurs after splicing, thus tRNATyr has to undergo retrograde transport to the nucleus to be modified (20) and (iii) the situation of Q as a micronutrient (9). Altogether the Q system in T. brucei may offer our best example of a dynamically changing modification that is tunable in response to nutrient changes.
In the present work, we explore whether or not Q formation in tRNA is rapidly tuned in response to transient nutrient changes, even under a situation of ‘normal’ growth, defined here as environmental changes that do not affect growth rate. We show that the levels of Q in tRNA can change in response to changes in certain amino acids, importantly, these Q changes have negligible effect on growth rate or global translation. We also show that part of the tuning mechanism rests on tRNA intracellular transport dynamics. Lastly, the outcome of such responses, to variations in amino acid levels in the media, is reflected in the importance of Q34 to efficiently decode the U-ending codons for Tyr.
MATERIALS AND METHODS
Culture conditions and generation of cell lines
Procyclic form (29-13 cells) (21) of T. brucei were grown at 27°C in SDM-79 media. Bloodstream form of T. brucei (Lister 427 strain) was grown at 37°C with 5% CO2 in HMI-9 media that contained 10% fetal bovine serum as previously described (22). All the dual-luciferase constructs were cloned separately in a pABPURO vector using the HindIII site that resulted in nine constructs (DL1–DL9). All the nine constructs were linearized using NotI restriction enzyme and separately electroporated into PF (29-13) cells as described (21). All the dual-luciferase cell-lines were selected using puromycin. The growth of cells was monitored using a hemocytometer.
APB gel electrophoresis and northern hybridization
APB gels required 50 milligrams of 3-aminophenylboronic acid (Sigma) per 10 ml of the polyacrylamide mix (8% polyacrylamide and 8 M urea). The total RNA was isolated using the guanidine isothiocyanate protocol (23). Deacylated tRNAs were prepared by incubation of total RNA in 100 mM Tris (pH 9.0) for 30 min at 37°C. Oxidation controls were prepared by RNA incubation in 50 mM NaOAc (pH 4.5) and 2.5 mM NaIO4 for 2 h at 37°C in the dark and quenched with 2 mM glucose for 30 min at 37°C in the dark. All RNA samples were ethanol precipitated and 10 μg of RNA was resuspended in RNA urea loading dye to load onto the APB polyacrylamide gel. The electrophoresis was carried out for 4 hours at 80 Volts. The northern hybridization was performed on zeta-probe membrane according to manufacturer's instructions (Bio-Rad). The blots were scanned using a Typhoon FLA 9000 scanner and the Q levels were quantified using an ImageQuant TL software (GE healthcare). The membranes were hybridized with the following probes:
tRNATyrGTGGTCCTTCCGGCCGGAATCGAA, tRNAHisGGGAAGACCGGGAATCGAAC, tRNAAspCGGGTCACCCGCGTGACAGG,
tRNAAsnAACCAACGACCTGTAGGTTAACAGC
Sucrose density gradient sedimentation
The PF cells were grown to mid-log phase (1 × 107 cells/ml) in SDM-79 media and washed twice in 1× PBS. The cells were transferred to the new flasks containing 50 ml of normal, low, and high tyrosine media (1 × 106 cells/ml as starting concentration). After 48 hours cycloheximide (100 μg/ml) was added to the flasks and centrifuged. The cell pellets were washed twice in ice-cold 1× PBS supplemented with cycloheximide. The cell pellets were lysed in 790 μl of 1× polysome buffer (10 mM Tris–HCl (pH7.5), 10 mM MgCl2, 300 mM KCl, 0.2% NP-40, 1× protease inhibitor, cycloheximide (100 μg/ml) and 2 mM DTT) for 10 min on ice. The cells were centrifuged at 15 000 g for 10 min and the supernatant was transferred to a freshly prepared 10–50% sucrose gradient prepared in polysome buffer. The gradients were centrifuged using the SW41Ti rotor (Beckman) at 36 600 RPM for 2 h and passed through a gradient fractionator (ISCO UA-6 UV with type 11 optical unit). The peak was visualized using the peak chart software (Brandel) and collected manually in a 1.5 ml Eppendorf tubes. The data was analyzed using peak chart software (Brandel) and plotted using GraphPad Prism8.
Extraction of queuine and queuosine from cells and media
An extraction method for Q/q was optimized through Q/q spiking studies involving the use of cell lysates produced from cells cultured in Q/q free media. A methanol extraction methodology was selected because it had minimal matrix effects and had highest overall Q/q recovery. Cells were washed twice with 1 mL of PBS. 500 μl of ice-cold MeOH–H2O (80:20) was added directly onto cells, scraped, transferred to an Eppendorf tube, vortexed and the homogenate was flash frozen in liquid nitrogen for 2 min followed by thawing in an ice bath for 10 min. The sample was centrifuged at 5800 g at 4°C for 5 min and the supernatant was transferred into a new tube. The pellet was re-extracted twice more with 250 μl of ice-cold MeOH–H2O (80:20) and the combined extracts were dried using SpeedVac at 0.1 Vac, 40°C for 2 h.
Quantification of free queuine and queuosine by LC-MS/MS
Queuine standard was provided by Professor Vincent Kelly (Trinity College Dublin, Ireland) and dissolved in ultrapure water to prepare 1 mM stock solution. A queuosine stock solution (1 mM in water) was provided by Professor Valerie de Crecy-Lagard (University of Florida, USA). Both stock solutions were aliquoted and stored at –20°C. Calibration standards (1, 0.3, 0.1, 0.03, 0.01, 0.003, 0.001 and 0.0003 μM), and three quality controls (QCs; 0.8, 0.02 and 0.0008 μM) were prepared freshly in ultrapure water on the day of analysis. A typical 7-point calibration curve is shown in the Supplementary Figure 2C and D. The cell and media extracts were reconstituted in 100 μl of water and analyzed using a LC/MS/MS system consisting of an AB SCIEX ExionLC system (Foster, CA, USA) coupled to an AB SCIEX Triple Quad 5500 + mass spectrometer (Foster, CA, USA). The system operated in ESI positive mode using Multiple Reaction Monitoring (MRM). The analyte separation was achieved on a Waters XSelect HSS T3 column (100 × 4.6 mm, 3.5 μm, Milford, MA, USA) at 45°C with the flow rate of 1.0mL/min. The injection volume was 5 μl. Mobile phase A was water with 0.1% formic acid; mobile phase B was acetonitrile with 0.1% formic acid. The mobile gradient was set as follows: 2% B to 3% B from 0 to 3 min, ramped to 98% B at 4 min and hold for 2 min, decrease from 98% to 2% B in 0.5 min and then equilibrated for 3.5 min at 2% B. Mass spectrometric parameters were set as follows: curtain gas 30; collision gas 9; ion spray voltage 5500; temperature 550; ion source gas 1, 50; ion source gas 2, 50 and entrance potential 10. The MRM transition settings are shown in Supplementary Table S1. Supplementary Figure S2 shows chromatograms from q and Q standards (A), and cell extract (B).
Dual luciferase reporter assay
The PF cells harboring DL1-DL9 constructs were grown to mid-log phase (1 × 107 cells/ml) in SDM-79 media and washed twice in 1× PBS. The cells were transferred to the new flasks containing 50 ml of normal, low, and high tyrosine media (1 × 106 cells/ml as starting concentration). The cells were harvested after 48 h and washed twice in ice-cold 1× PBS. The cell pellets were transferred to a new 1.5 ml Eppendorf and stored at –20°C until used for assays. The assays were carried out as per the manufacturer's recommended protocol using the Firefly/Renilla dual-luciferase reporter assay system (Promega E1960) (24). The cells were resuspended in 70 μl 1× Passive lysis buffer (Promega) and allowed to lyse on ice for 15 min. Cells were centrifuged and 20 μl of supernatant was added to the individual wells of a 96-well plate (white wall clear bottom, Corning 3610). 100 μl of LAR II was added to each sample and incubated for 5 min. It was followed up by luminometer detection using Spectramax i3X multimode microplate reader (10 seconds integration time and height 1 mm). After firefly luciferase detection, the Renilla luciferase activity was measured using Stop and Glo substrate. 100 μl of Stop and Glo substrate was added to each sample and incubated for 5 min that was followed up the luminometer detection (10 s integration time and height 1 mm). Each data point represents the technical duplicates of biological duplicates. The data was normalized for +1 frameshifting % by calculating the ratio of Firefly luciferase activity to that of Renilla luciferase activity in a test codon relative to its in-frame control as previously described (25). Statistical significance was determined by a two-tailed pairwise student t-test.
Fluorescence in situ hybridization (FISH)
1 × 107 cells were harvested and washed with PBS. Cells were resuspended in 4% paraformaldehyde/PBS solution and fixed to poly-l-lysine coated microscope slides for 30 min. Non-adhered cells were removed by washing with PBS and remaining cells were dehydrated by a series of increasing ethanol concentrations (50, 80 and 100%, for 3 min each). Subsequently, the permeabilized cells were pre-hybridized with hybridization solution (2% BSA, 5× Denhardt′s solution, 4× SSC, 5% dextran sulphate, 35% deionized formamide, 10 U/ml RNase inhibitor), for 2 h. The slides were then incubated overnight at room temperature in a humid chamber in the presence of 10 ng/μl Cy3-labeled oligonucleotide probe, in the afore-mentioned hybridization solution. Afterward, slides were washed for 10 min, once with 4× SSC with 35% deionized formamide, followed by one wash each with 2× SSC and 1× SSC. Finally, the slides were mounted with mounting medium supplemented with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI). Images were taken with confocal microscope Olympus FluoView™ FV1000 and analyzed using Fluoview and ImageJ (NIH) software. FISH data were quantified as described elsewhere (26). In brief, six cells per micrograph were selected randomly. Fluorescence intensities were measured using ImageJ with a plot profile analysis along a 3.5 μm line drawn across the nucleus with overhangs covering the cytoplasm. The results are expressed as the average value of relative fluorescence intensity ± SD.
RESULTS
Queuosine levels differ between developmental stages of T. brucei
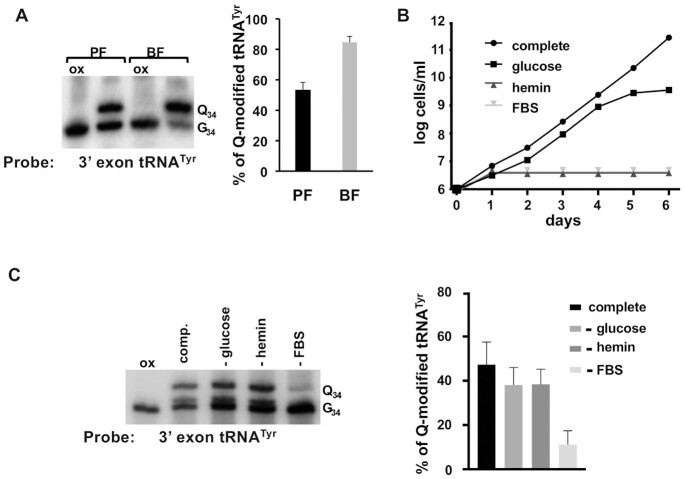
In Drosophila melanogaster changes in the Q content of tRNAs have been associated with developmental progression via effects on accuracy driven codon usage (6). Since trypanosomes undergo distinct life cycle stages that include major metabolic reprogramming, we explored the possibility of similar changes in the Q content of tRNAs. To test this, we used N-acryloyl-3-aminophenylboronic acid (APB) polyacrylamide gel electrophoresis. cis-Diol moieties, such as those present in Q as well as the 3’ end of RNA, have affinity for boronic acid, in the case of tRNA the presence of Q causes a pronounced band shift in APB gels allowing easy detection of Q-containing tRNAs by northern blots (27). To establish the steady-state levels of Q in the different developmental stages of T. brucei, we isolated total RNA from procyclics (PF, insect forms) and bloodstream (BF, mammal form) stages of trypanosomes grown in culture. The RNA from PF and BF were separated by APB gel electrophoresis, followed by northern blots using a tRNATyr-specific radioactive oligonucleotide probe (Figure 1). A fraction of the same total RNA sample was treated with sodium m-periodate (ox), which oxidizes the vicinal diols found in Q leading to the formation of dialdehydes; this prevents a band shift during APB gel electrophoresis, thus serving as a negative control. We found that in PF trypanosomes ∼50% of tRNATyr has Q, similar to previous results (20), whereas in BF ∼86% of tRNATyr contains Q (Figure 1A), supporting differential Q modification levels in PF and BF stage trypanosomes.
Figure 1.
Differences in the steady-state levels of Queuosine in tRNA between developmental stages of T. brucei cannot be explained by changes in general nutrients. (A) total RNA from procyclic form (PF, insect stage) and bloodstream form (BF, mammal stage) T. brucei was analyzed by APB-gel northern blot, using a radioactive oligonucleotide probe specific for tRNATyr, to determine the levels of Q-containing (Q34) vs. unmodified tRNA (G34) (left panel) and quantification of the bands in (right panel), where percent Q-tRNA was calculated by dividing the intensity of the Q34 band by the sum of Q34 and G34 and multiplied by 100. (B) shows a growth curve of PF T. brucei comparing complete media to similar media lacking glucose, hemin or fetal bovine serum (FBS). The graph shows the log of cells per ml of media from cumulative cell counts over 6 days as indicated. (C) APB-gel northern blot as in (A) to assess Q levels in tRNATyr, where ‘comp.’ denotes complete media as compared to media lacking glucose (-glucose), hemin (-hemin) or fetal bovine serum (–FBS) (left panel) and quantification of the band intensities from the left panel to determine Q levels as in. In all blots, ‘ox’ refers to an oxidized control (negative control); oxidation of the cis-diols from Q prevents the observed band shift and indicates the presence of Q on the shifted untreated samples. All graphs are representative of at least 3 different independent experiments.
One of the salient features for the growth of T. brucei in the insect vector vs. the mammalian host is the availability of different nutrients. In the insect, parasites (PF) encounter low levels of glucose and rely on oxidative phosphorylation to generate the bulk of their ATP. In the glucose-rich environment of the mammal host, trypanosomes (BF) reduce mitochondrial function and up-regulate substrate-level phosphorylation (18). In addition, since T. brucei is a heme auxotroph (28), it requires addition of hemin to the culture media for growth of all stages of the parasites. To determine whether nutrient or growth condition differences between PF and BF might be responsible for the difference in Q, we tested if changes in glucose, hemin, or fetal bovine serum (FBS) in the media could affect Q levels. Cell growth was monitored for several days under these conditions (Figure 1B) and compared with growth in complete media. As expected, slower growth was observed in these conditions, highlighting the importance of these nutrients. We then isolated total RNA from cells growing in the above conditions and determined tRNATyr Q levels (Figure 1C). We found no significant change in Q levels in hemin- or glucose-lacking media when compared to complete media (Figure 1C). However, we observed a significant decrease in Q content in cells grown in serum-free media. The latter is not surprising and in agreement with previous reports showing that serum in media is the principal source of queuine (8,9), the free base of Q and the substrate used by the TGT enzyme for Q incorporation into tRNA. Taken together, these results show that neither lack of glucose nor hemin affect Q levels and the observed differences in the steady-state levels of Q in tRNA between PF and BF trypanosomes cannot be ascribed to these general nutrients.
Changes in the concentration of specific amino acids alter Q levels
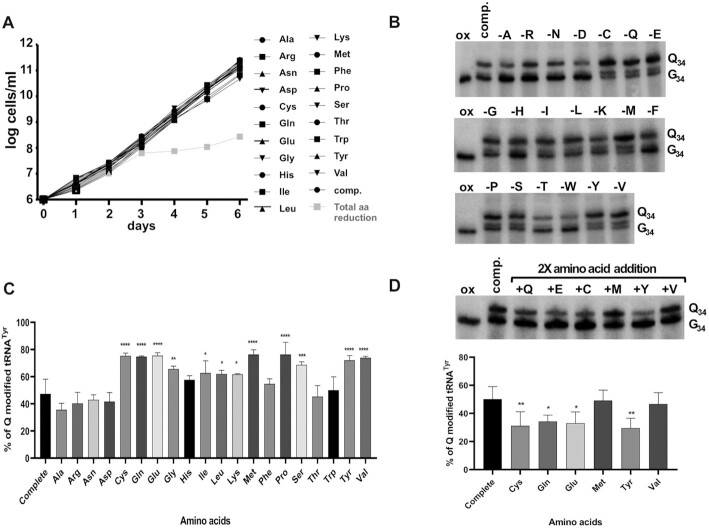
In mammals, there is an unusual connection between TGT and the ability for cells to convert phenylalanine to tyrosine. In mice a knockout of the TGT gene leads to reduced activity of phenylalanine hydroxylase (PAH) and accumulation of phenylalanine to toxic levels, and although the mechanism for this is not clearly understood, it somehow links queuosine formation to amino acid metabolism (29). In addition, the influence and importance of amino acid availability during the different stages of T. brucei development are well-documented. For example, the amino acid proline is the main carbon source for PF T. brucei (the insect form) (30). Likewise, various amino acids play critical roles in the metabolic reprogramming while transitioning from the insect to the mammal host (31). We therefore hypothesized a possible connection between amino acid availability and the micronutrient queuine as a way to monitor metabolic state during ‘normal’ growth of PF T. brucei. Reinforcing this hypothesis is the fact that amino acids such as tyrosine cannot be synthesized by these parasites as they lack the PAH enzyme. Therefore, trypanosomes are auxotrophs for some amino acids that are non-essential in the mammalian host (31). To test our hypothesis, we made a modified media in which individual amino acids were systematically left out of the synthetic portion of the media, effectively reducing, but not completely removing, the given amino acid, provided that serum is still amino acid rich. We monitored cell growth for 6 days in the reduced amino acid media and surprisingly found no major differences in growth rate when compared to complete media (Figure 2A). However, when all 20 amino acids were reduced, cells showed a marked growth defect after three days, indicative of important amino acid interconversions in the semi-complete media to maintain normal growth rates (Figure 2A). Total RNA was isolated from cells grown in each of the reduced amino acid conditions and Q-tRNA levels were compared to cells grown in complete media. We found that a decrease in the concentration of a majority of amino acids did not significantly change Q-tRNA levels (Figure 2B and C). However, reduction in several amino acids led to significant increases in the Q content of tRNATyr (>74%) as compared to tRNA isolated from cells grown in complete media (45% Q average).
Figure 2.
Q levels in tRNA change inversely proportional to changes in the concentration of specific amino acids. (A) growth curve of PF cells grown in media where the concentration of single amino acids was reduced. ‘Comp.’ refers to complete media, the symbols indicate the single amino acid that was lowered in the media. ‘total amino acid reduction’ refers to cells grown in media where all amino acids were omitted from the defined portion of the media. The graph plots cell growth as log of cells per ml based on cumulative cell counts plotted as a function of time in days. (B) APB-gel northern blots probing for tRNATyr as described in the Figure 1 legend. ‘Q34’ and ‘G34’ denote Q-modified and unmodified tRNA as above and ‘ox’ serves as negative control as before. (C) the percent Q-tRNA was quantified as before. The single amino acid whose concentration was lowered in the growth media is indicated by the single letter code. (D) the top panel represents a similar APB-gel northern experiment as in (A), but the total RNA used was derived from cells grown in media where the selected amino acids were increased two times by their normal concentration in complete media (comp.). The band intensities from the top panel were quantified and plotted as percent Q in tRNATyr as before. All graphs are representative of at least 3 different independent experiments. Data were compared by one-way analysis of variance (ANOVA) with significant differences indicated (*P< 0.05, **P< 0.01 and ***P< 0.001).
We then tested what would happen to Q-tRNA levels when those same amino acids, which showed a significant increase in Q upon reduction, were provided in excess by increasing their concentration to 2× of that found in complete media. Again, total RNA from the different cultures was separated using APB gel electrophoresis followed by northern blotting and probing for tRNATyr (Figure 2D). We found a trend towards lower Q levels with reductions in tyrosine, cysteine and glutamate concentrations (down to 23–25% from 45% in complete media), we also saw a slight decrease with reduced glutamine (down to 32%) (Figure 2D). In the case of methionine and valine, Q levels remained similar to the complete media. In conclusion, changes in the concentration of certain amino acids lead to changes in Q modification levels and in the case of tyrosine, cysteine and glutamate the amount of amino acid in the media was inversely correlated to Q modification levels.
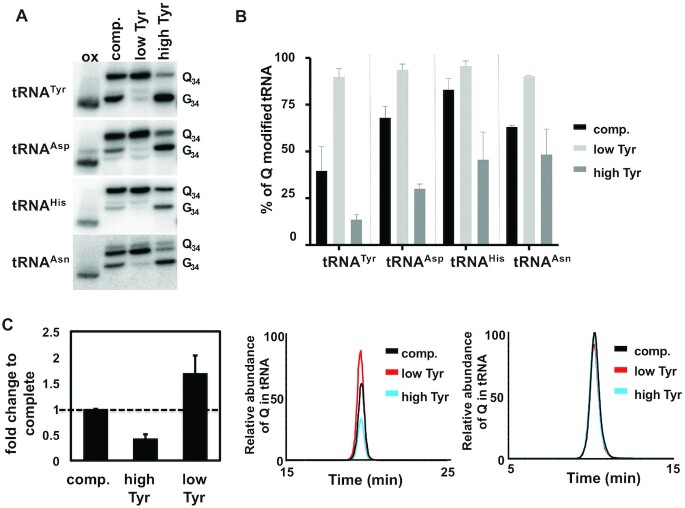
We also tested further increasing the amino acids to 4× their concentration in complete media; all with the exception of tyrosine led to growth reduction perhaps indicative of cytotoxicity (not shown). Therefore, for further studies we focused solely on tyrosine, which at 4× its normal concentration led to a further decrease in Q-tRNATyr to less than 10% (Figure 3A and B) and no apparent defects in cell growth (Supplementary Figure S1). Under 4× tyrosine, we also probed for the other Q-containing tRNAs (tRNAAsp, tRNAHis and tRNAAsn); in all cases their Q content responded to tyrosine concentration changes in the media and followed the same inverse relationship as seen for tRNATyr (low tyrosine/high Q and high tyrosine/low Q) (Figure 3A). Notably, the steady-state levels of the Q content of different tRNAs from cells grown in complete media varied, tyrosine showing the lowest level at ∼45%. These results were confirmed by liquid chromatography–mass spectrometry (LC–MS), where the levels of Q in total tRNA were calculated from the extracted ion chromatogram (XIC) normalized to that of guanosine (Figure 3C). Despite these measurements being taken over the total Q-containing tRNA population, which may have a smoothing effect on the data, we still observed the same trend with the levels of Q in tRNA remaining inversely proportional to the tyrosine concentration added to the growth media. These results further confirm the relationship between certain amino acids and Q-tRNA content observed by APB-gel/northern blots with tRNA-specific probes.
Figure 3.
Q-levels in all Q-containing tRNAs change in response to alterations in tyrosine concentration in the media. (A) APB gel-northern blots to determine the levels of Q in the four TGT tRNA substrates in response to changes in tyrosine concentration in the media using radioactive oligonucleotide probes specific for each tRNA as indicated. ‘Q34’, ‘G34’ and ‘ox’ refer to Q-containing, unmodified and the oxidized control respectively. (B) band intensities in (A) were quantified as before and plotted as percent Q in each tRNA. ‘comp.’ refers to total RNA from cells grown in complete media and ‘low’ and ‘high’ refer to cells grown in reduced tyrosine media or media containing four times the amount of tyrosine found in complete media respectively. (C) The graph on the left shows the average fold change in Q from the extracted-ion chromatograph (XIC) in the middle panel, when the levels for all four tRNAs are added together and normalized against the unmodified canonical nucleotides (relative abundance of Q in tRNA in the y-axis), as a control an XIC of changes in G (relative abundance of G in tRNA in the y-axis) under the different conditions is provided (right panel). The graphs are representative of at least three independent experiments.
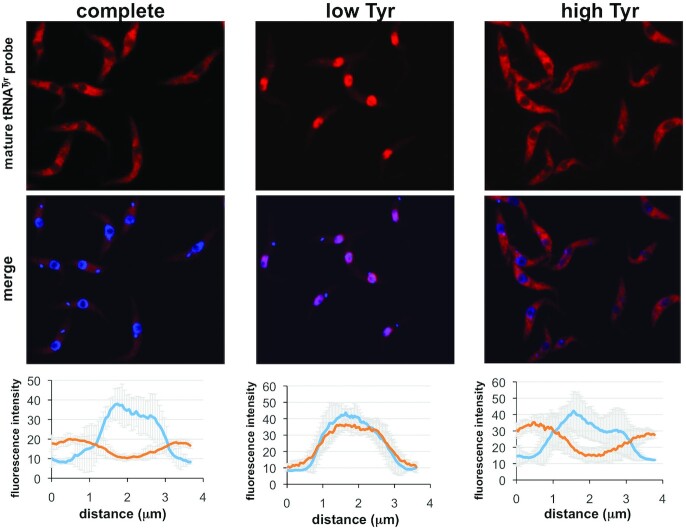
Reduced tyrosine in media leads to nuclear tRNATyr accumulation
In a previous study, we showed that down-regulation of expression of the nuclear transporters TbMex67/TbMtr2 caused longer nuclear retention of certain tRNAs in T. brucei (32). More specifically TbMex67 down-regulation led to selective retention of the four TGT substrates (tRNAHis, –Asn, –Asp and –Tyr). Increased dwell time in the nucleus when TbMex67 was down-regulated also led to increases in the levels of Q in these tRNAs. This suggested a competition between Q formation and nuclear export, whereby longer dwell time in the nucleus led to increased Q content, given that TbTGT is a nuclear enzyme (20). It is also well established that tRNA retrograde transport combined with slower secondary export (re-export) to the cytoplasm leads to longer nuclear tRNA retention in other systems in response to changes in nutrient levels (33,34). We thus explored the fate of tRNATyr during conditions of high and low tyrosine in the media as compared to complete media (as above). FISH (fluorescent in situ hybridization) was performed with a fluorescent oligonucleotide probe specific for mature tRNATyr (as described). We found that growth in low tyrosine media leads to longer retention of tRNATyr in the nucleus (Figure 4), thus the increased level of Q under those conditions can be explained in part by a nuclear retention mechanism. When grown in the high tyrosine media, the tRNA distribution resembles the situation with complete media, where the bulk of the tRNA is in the cytoplasm without hint of nuclear accumulation (Figure 4). We also probed for the other Q-containing tRNAs (tRNAHis, –Asp and –Asn), but only observed sporadic perinuclear localization, which was not as pronounced as what was observed with tRNATyr. In addition, probing for tRNAGlu, a non-Q tRNA did not show any traces of increased dwell time when tyrosine levels were low in the media (Supplementary Figure S3).
Figure 4.
Low tyrosine in the media leads to nuclear accumulation of tRNATyr. The experiment shows fluorescent in situ hybridization (FISH) with a fluorescently labeled oligonucleotide probe specific for mature (spliced) tRNATyr (red). Co-localization was performed with DAPI (blue) staining the nuclear and mitochondrial DNA ‘complete’, ‘low’ and ‘high’ refer to cells grown in complete, low tyrosine or high tyrosine media as indicated. Each graph shows the intensity profile of the individual fluorophores of eight randomly selected cells, where the values represent the relative intensity average ± SD. Distance refers to 3.5 μm line centered on the nucleus and with overhangs spanning the cytoplasm.
Q content changes dynamically with changes in tyrosine concentrations
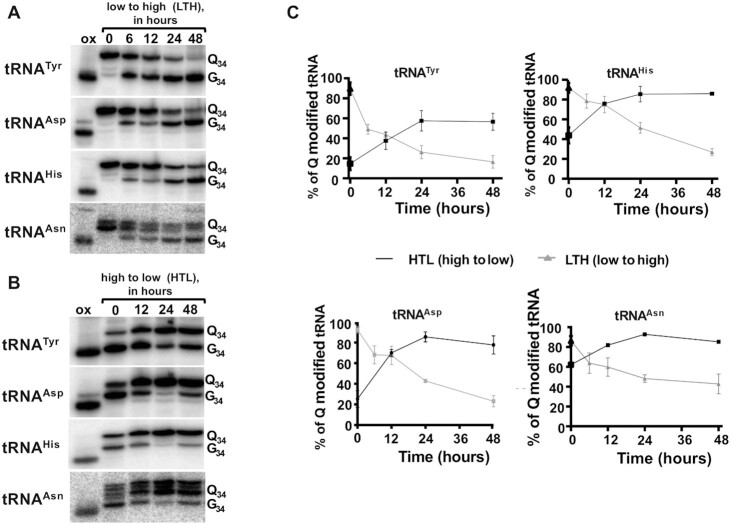
Next, we tested the kinetics of Q-tRNA changes in response to variations in tyrosine concentration in the media. For these experiments, cells were grown in complete media, then washed with phosphate buffer saline and cultured in either low tyrosine, high tyrosine or complete media. Cells were collected at 0, 12, 24 and 48 h and total RNA was isolated and analyzed by APB gel-northern blots. In the case of low tyrosine media, the initial level of Q-tRNATyr was 40% showing an increase up to 45% within 12 h (Figure 5A and B), further increase after 24 h, finally reaching a maximum in 48 hours. Comparable maximum levels of Q were also seen with the other three tRNAs with tRNAAsp and tRNAHis reaching a level of 95%, but tRNAHis showed the highest basal Q levels of 80% at time 0, but it still increased to 95% Q in the next 48 h (Figure 5B). Similarly, we determined the Q levels after switching cells from complete to high tyrosine media (Figure 6A and B). In this case all of the Q-containing tRNAs showed a downward trend in Q levels. Again, tRNATyr showed the lowest levels of Q under high tyrosine conditions, down to less than 10% Q content. Next, we grew cells for 48 h in low tyrosine media and then switched to the high tyrosine media, and vice versa. We monitored Q levels going from low to high tyrosine (low to high, LTH) (Figure 7A), or high to low tyrosine (high to low, HTL) (Figure 7B). Cells were collected within 48 h after the switch and the levels of Q in the tRNA determined (Figure 7C). In all cases, but particularly in the switch from ‘low’ to ‘high’ tyrosine media, changes in the levels of Q in tRNAs were observed as early as 6 hours after the switch. Again, this was most pronounced with tRNATyr, which went down from ∼95% to ∼50% Q content within 6-hours (Figure 7C. top left graph). T. brucei has a doubling time of ∼8–10 h in culture and reaches a culture saturation density after 48 h of growth. Therefore, the observed drops in Q levels cannot simply be ascribed to dilution as the cells are passaged.
Figure 5.
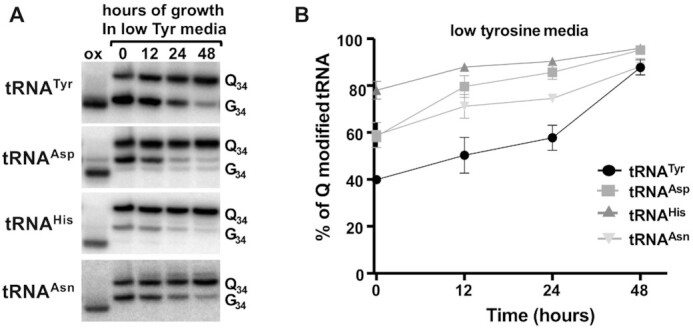
Q levels in tRNA increase in response to low tyrosine concentration in the growth media. The figure shows the results of growing cells in media containing ‘low tyrosine’, where total RNA was isolated at various time points in the growth curve as indicated. (A) shows a representative APB-gel northern blot using radioactive oligonucleotide probes specific for the four Q-containing tRNAs. ‘Q34’, ‘G34’ and ‘ox’ refer to Q-modified, unmodified and the oxidized control as in previous figures. (B) quantification of the results from (A), where percent Q modified tRNA was calculated as described before. The graph is the result of three independent experiments.
Figure 6.
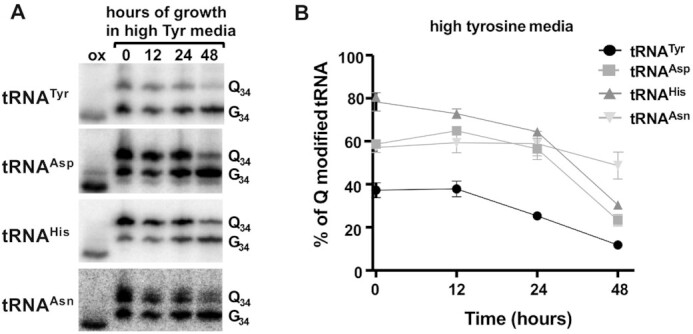
Q levels in tRNA decrease in response to high tyrosine concentration in the growth media. The figure shows the results of growing cells in media containing ‘high tyrosine’, where total RNA was isolated at various time points in the growth curve as indicated. (A) shows a representative APB-gel northern blot using radioactive oligonucleotide probes specific for the four Q-containing tRNAs. ‘Q34’, ‘G34’ and ‘ox’ refer to Q-modified, unmodified and the oxidized control respectively as in previous figures. (B) quantification of the results from (A), where percent Q modified tRNA was calculated as described before. The graph is the result of three independent experiments.
Figure 7.
Q levels in tRNA change dynamically with variations in tyrosine concentration. The levels of Q in tRNA as the cells are switch from growth in low to high tyrosine media (LTH) and from high to low tyrosine media (HTL) is shown. RNA samples were prepared from cells at different times in the growth curve as before. (A) Q levels when switching from low to high tyrosine. (B) similar experiment as in (A) but instead cells where switched from high to low tyrosine. (C) quantification of Q levels from three independent experiments as in (A) and (B).
Taken together these results show that Q levels respond to the external tyrosine concentration of the media. In low tyrosine media, all four Q containing tRNAs reached a Q content higher than 90% within 48 h. Whereas, in high tyrosine media all the four Q-containing tRNAs showed a substantial decrease in Q levels within 48 hours. Importantly, the decrease in Q levels cannot be ascribed to limiting availability of queuine in the media since in all cases the concentration of serum (the source of all forms of Q) was kept constant.
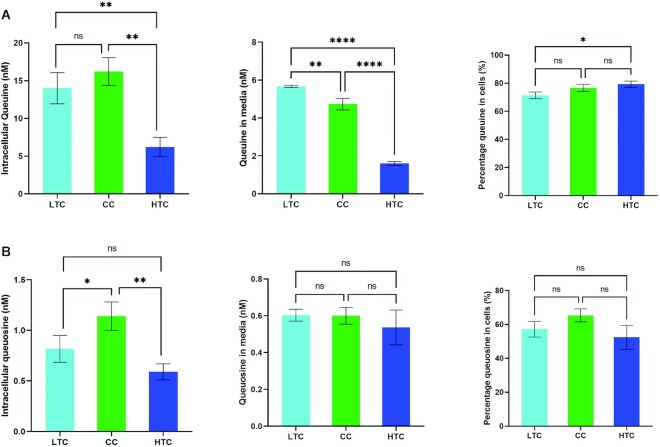
We further tested whether the observed reduction in Q levels in tRNA were due to impeded membrane transport. We hypothesize that it is possible that tyrosine at high concentrations may act as a competitive inhibitor of queuine/queuosine transport reducing the intracellular levels of these substrates and thus impacting the ability of TGT to modify its cognate tRNAs. To test, this we determined the intracellular levels of queuine and queuosine and compared these against total levels (intracellular plus growth media) when cells were grown in low tyrosine, high tyrosine or complete media (Figure 8). Although we observed different total levels of each under the three conditions (Figure 8A, B) for reasons we cannot explain, the percentage of each component inside cells compared to the total inside and outside was within a similar range regardless of growth conditions (Figure 8). This leads us conclude that the observed reduction in Q content in tRNAs when cells are grown in high tyrosine media are not the result of impaired Q/q transport.
Figure 8.
Intracellular concentrations of free q and Q are not altered by tyrosine concentration. The figure shows accurately quantified levels of free q and Q as determined by a specific LC–MS/MS method employing a 7-point calibration curve. LTC, HTC and CC refer to cells grown in low tyrosine, high tyrosine or complete media as indicated. (A) shows the levels of free q intracellularly and in media along with the cellular q expressed as a % of total q, (B) shows corresponding data for free Q. Data were compared by One-way analysis of variance (ANOVA) with significant differences indicated (*P< 0.05, **P< 0.01, ***P< 0.001 and ****P< 0.0001).
Queuosine in tRNA is important for codon choice
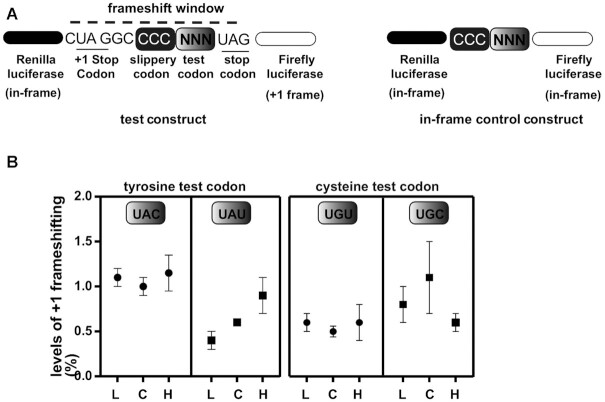
Q-containing tRNAs in eukaryotes are responsible for reading the codons for tyrosine, aspartate, histidine, and asparagine; all belong to two-codon boxes formed by C- or U- ending codons (NAC/NAU, where N = G, C, U and A). Recently, using a dual-luciferase reporter assay, we showed that in T. brucei Q-containing tRNAs ensure efficient decoding of U-ending codons (9), similar in nature to Xenopus (5). In light of these results, we decided to explore how changes in the levels of Q effected by variations in tyrosine concentration affect translational efficiency in T. brucei. We used a dual-luciferase reporter system, where Renilla luciferase (R-luc) and Firefly luciferase (F-luc) are encoded as part of a construct to detect ribosomal + 1 frameshifting (Figure 9A). In this assay +1 frameshifting is used as a proxy for translational efficiency of the test codon; if ribosomes slow down at the test codon it would lead to increased frameshifting and increased activity of the downstream reporter (F-luc). In this dual reporter system, the sequences are placed in such a way that the R-luc and F-luc protein-coding regions are separated by a frame-shifting box. R-luc is in the 0 frame, while F-luc is in the +1 frame (Figure 9A). Therefore, F-luc translation requires ribosomal +1 frameshifting. The sequence between the R-luc and F-luc coding sequences also contains a slippery proline codon (CCC), followed by a test codon (NNN) and a stop codon (UAG) in the 0 frame. A stop codon is also present in the +1 frame just upstream of the slippery codon so that accidental ribosome slippage will not lead to F-luc production (Figure 9A). This construct provides an extremely short window for +1 frameshifting. As control, we also use a construct in which R-luc and F-luc genes are in the same frame, thus not sensitive to frameshifting (Figure 9A). To get the final +1 frameshifting values, the ratio of F-luc/R-luc (frame-shift construct) activity is divided by the ratio F-luc/R-luc (in-frame control) activity. An increase in the ratios in F-luc activity compared to R-luc, normalized to the in-frame construct, implies increased frameshifting due to slower translation of a given test codon.
Figure 9.
Q changes in tRNA driven by fluctuations in tyrosine concentrations affect decoding of the U-ending codons for tyrosine. A dual-luciferase reporter assay where a translational frameshifting window is used to assess the efficiency of decoding the U-ending or C-ending codons for tyrosine in response to changes in media tyrosine concentration. (A) a schematic of the assay where the test codon was either UAA or UAU for tyrosine. Non-Q requiring cysteine codons were used as a control to highlight the Q dependence. The ‘in-frame’ constructs, which lack a frameshifting window, were used for normalization. (B) the results of 3 independent experiments. L, C and H refer to cells expressing the dual-luciferase reports and grown in either ‘low’, ‘complete’ or ‘high’ tyrosine media.
Given that tRNATyr showed the greatest alterations in Q content in response to tyrosine changes in the growth media, we used the two tyrosine codons as (UAC and UAU) test codons (Figure 9B). As controls, we also tested the two cysteine codons, which also belong to a two-codon box, both end in C or U, but do not use a Q-containing tRNA for decoding. Clonal cell lines constitutively expressing the various cassettes above were grown for 48 hours in normal, low, and high tyrosine media and assayed for dual luciferase expression. Our results show no difference in +1 frameshifting for the C-ending tyrosine codons (UAC) in any of the three conditions tested. However, +1 frameshifting increased with the U-ending tyrosine (UAU) test codon in cells grown in high-tyrosine media, a situation that was corrected in the low tyrosine media. No changes in +1 frameshifting were observed with the cysteine codons in any of the three media tested. These results taken together show that changing the levels of Q, in response to altering the availability of tyrosine in the media, leads to inefficient reading of the U-ending codon for tyrosine. As corollary, Q in T. brucei is important for reading the U-ending codons and to respond to changes in the availability of specific amino acids. None of the conditions tested had any major impact on growth rate or the levels of translating ribosomes (Supplementary Figure S1). This suggest that fluctuations of Q levels in tRNA lead to codon-biased translation and may affect the expression of certain genes depending on their content of the U-ending codons that depend on Q-containing tRNAs for decoding.
DISCUSSION
Soon after the discovery of tRNA, the connection between tRNA modifications and environmental cues was established. It was reported that when cells are starved of particular amino acids; for example, methionine, tRNAs became undermethylated (35). This observation was later expanded to include several other modifications, which changed not only in response to amino acid starvation but also in response to several other types of stressors (12–15). These early observations eventually led to the discovery of codon-biased translation; a way to efficiently control the expression of stress-response genes via coupling their codon bias to modification levels (16).
The idea of carefully adjusting translational rates to the metabolic status of cells via modifications has also been explored by several laboratories. For example, the recently established connection between the levels of sulfur-containing amino acids (cysteine and methionine), sulfur-containing tRNAs (tRNAGlu, –Gln and –Lys), translational efficiency and the synthesis of protein transporters for those same amino acids (36). Later it was shown that even phosphate levels can be sensed via changes in tRNA modification enacting a switch in sugar metabolism to maintain homeostasis (37).
We were among the first to raise the possibility that fluctuations in the modification content of tRNAs not only occur during situations of environmental extremes, but also during normal growth when cells have sufficient nutrients and are growing exponentially. Although seemingly counterintuitive, the latter idea suggests the existence of dynamic modifications in tRNA, which can be actively monitored and altered by cells accordingly, as translational and metabolic demands change. To test our hypothesis, we turned here to the Q modification, a reaction that involves the replacement of G34 for Q34 in tRNAAsp, tRNAAsn, tRNAHis and tRNATyr. Queuosine is unique among eukaryotic modifications in that cells must rely on transport of some form of Q (either the queuine base or Q nucleotides or nucleosides) from the media for it to be ultimately incorporated into tRNA via a reaction catalyzed by TGT (4). The finding that the steady-state levels of Q differ between the two developmental stages of T. brucei, implicate this modification in playing important roles in codon selection in these different stages of the parasites. Based on this, we hypothesized that Q modification may be an excellent candidate for a rapidly tunable modification. A connection between amino acid biosynthesis and TGT has been established in other systems (29), but the issue of possible tunability was never directly addressed. We found that whereas changes in more general nutrients such as glucose or hemin had little effect, decreases in the levels of individual amino acids in the growth media led to an increase in the steady-state levels of Q in tRNA. However, only glutamine, glutamate, cysteine and tyrosine showed changes in Q level that are inversely proportional to the concentration of the particular amino acid in the media. Tyrosine was unique in that it did not invoke toxicity or cause growth defects at 4x times its normal concentration. Tyrosine also showed a great dynamic range in the changes on Q it effected, going from as high as >90% Q in low tyrosine media to as low as 10% Q at four times its concentration. Although, most prominent with tRNATyr, analogous fluctuations were also observed for the other three Q-containing tRNA in response to differences in tyrosine concentration in the media.
Considering that tRNATyr undergoes interesting intracellular transport dynamics, led us to investigate how intracellular transport and processing may affect Q content. Beyond this, we focused on tRNATyr for two practical reasons: (i) it is the only intron-containing tRNA in T. brucei and splicing is a cytoplasmic event (19), (ii) it gets modified with Q but only in its spliced form and TGT is a nuclear enzyme; this tRNA undergoes retrograde transport into the nucleus following cytoplasmic splicing (20). In the latter case, Q content serves as a marker for nuclear re-import via retrograde transport. Using this set of observations, we suggest that the increase in Q-content in tRNATyr upon reduction of tyrosine in the media is partly due to increased nuclear dwell time by the spliced tRNA as shown by FISH. This is in line with our previous observations on tRNA nuclear export (32). Significantly, upon switching cells from ‘low’ to ‘high’ tyrosine the tRNA redistributed in a manner analogous to when the cells are grown in complete media and becomes mostly cytoplasmic. We emphasize that despite the reduction in individual amino acids for the experiments described, cells maintain a growth rate comparable to that seen with complete media and in fact we did not observe any difference in the amounts of translating ribosomes when analyzed by polysome fractionation (Supplementary Figure S1).
A more puzzling question is that of the observed reduction in Q content when cells are switched from ‘low’ to ‘high’ tyrosine media. As previously shown, the absence of Q has no effect on either the levels of tRNATyr nor its amino acylation efficiency. It is possible that Q is actively removed by TGT when tyrosine is ‘high’, given that the ability of TGT to efficiently perform the reverse reaction (Q to G exchange) is well documented (38). Additionally, it is possible that the levels of the enzyme themselves are changing as tyrosine concentrations change. Finally, tyrosine may also act as some sort of allosteric regulator of the enzyme (if acting directly), whereby high tyrosine could inhibit the enzyme. However, we deem the latter unlikely, since TGT is nuclear, and our results show that during ‘high’ tyrosine the bulk of the spliced tRNA is localized to the cytoplasm. Admittedly, we cannot formally refute some of these possibilities and addressing such questions will require future additional experimentation.
Beyond TGT regulation, it is also possible that tyrosine acts as a competitive inhibitor for Q transport, whereby under high tyrosine the levels of intracellular substrates for TGT drop and since tRNA transcription is constitutive, this leads to accumulation of unmodified tRNA. Our measurements of the levels of both intracellular and extracellular q/Q showing that there are not drastic differences in the cellular uptake of q/Q argue against this. However, we cannot formally exclude the possibility that once inside the cells, high levels of tyrosine impair Q transport to the nucleus where TGT resides. Lastly, there could exist a cytoplasmic activity that actively removes Q under high tyrosine conditions. In this scenario such an activity would be reminiscent of the demethylases that are part of the mechanism that resets mRNA function following m6A modification, the so-called ‘erasers’. Whether or not such an activity exists for Q will remain an open question.
In more general terms, it is not clear what exact mechanism(s), beyond intracellular transport, leads to the Q fluctuations observed with the tyrosine titration experiments. It is possible that the transporters themselves, change in response to alterations of amino acid concentration in the media. However, changes in the levels of the transporters could have a more general effect on tRNA export from the nucleus, but the export of tRNAGlu (a non-Q containing tRNA) was unaffected thus partly ruling out the possibility that the observed increase in Q-tRNAs are due to variations in transporter levels. We also performed FISH with the other Q-containing tRNAs (Asp, His and Asn) and we only observed a sporadic perinuclear localization, but their partitioning was never as pronounced as that observed with tRNATyr; yet Q levels also increase with those tRNAs. We suggest that their primary export from the nucleus is slowed down sufficiently for TGT to win the race, when tyrosine levels are low. It is clear that because of its unusual transport dynamics, tRNATyr stays in the nucleus longer implying that nuclear dwell time is indeed influenced by both the rate of primary export, the rate of retrograde transport and the rate of re-export. In terms of Q levels then, all those rates at some point have to be considered in relation to the intrinsic rate of Q catalysis.
Given that the variations in Q levels did not significantly affect growth rate or the levels of translating ribosomes, it was important to assess more specifically what the effects such fluctuations have on tyrosine codon choice. The dual luciferase assays confirmed that in the controlled background of such an assay, an increased in translational frameshifting was observed when cells were grown in high tyrosine media (low Q), this effect disappeared when cells were grown in complete or low tyrosine media. Notably in this assay, frameshifting acts as a proxy for translation efficiency by virtue of a translational slow down at the A-site of the ribosome leads to pausing and increased frameshifting. Taken together, we conclude that in this specific case the presence of Q aids in the efficient translation of the U-ending codons for tyrosine. Clearly, at a more global level, preference for C-ending or U-ending codons as a result of Q fluctuations will likely be influenced by codon context on a given mRNA and are best assessed by a more global translatome-wide analysis such as ribosome profiling. However, one thing is clear, the observed dynamic changes in Q levels correlate well with steady-state levels of Q observed in tRNAs from the two different developmental stages of this parasite. This raises the possibility that indeed Q formation may play an important role during the developmental program in enforcing codon-biased translation; a situation reminiscent of the Drosophila system (6). Due to the complexity of developmental plans, likely no single factor will be solely responsible for controlling development and tRNA Q content may be one driver in such a multivariate situation.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Alfonzo, Green, Limbach and Paris laboratories for their comments and suggestions.
Contributor Information
Sameer Dixit, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Alan C Kessler, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Jeremy Henderson, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Xiaobei Pan, School of Biological Sciences, Institute for Global Food Security, Queen's University Belfast, Belfast, UK.
Ruoxia Zhao, Rieveschl Laboratories for Mass Spectrometry, Department of Chemistry, University of Cincinnati, Cincinnati, OH, USA.
Gabriel Silveira D’Almeida, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Sneha Kulkarni, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
Mary Anne T Rubio, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
Eva Hegedűsová, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
Robert L Ross, Rieveschl Laboratories for Mass Spectrometry, Department of Chemistry, University of Cincinnati, Cincinnati, OH, USA.
Patrick A Limbach, Rieveschl Laboratories for Mass Spectrometry, Department of Chemistry, University of Cincinnati, Cincinnati, OH, USA.
Brian D Green, School of Biological Sciences, Institute for Global Food Security, Queen's University Belfast, Belfast, UK.
Zdeněk Paris, Institute of Parasitology, Biology Centre, Czech Academy of Sciences, České Budějovice, Czech Republic; Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic.
Juan D Alfonzo, Department of Microbiology and The Center for RNA Biology, The Ohio State University, Columbus, OH, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM132254, GM084065-11 to J.D.A.] NIH [GM058843 to P.A.L.] (in part); Czech Science Foundation [20-11585S to Z.P.]; ERDF/ESF project Centre for Research of Pathogenicity and Virulence of Parasites [CZ.02.1.01/0.0/16 019/0000759 to Z.P.]; University of South Bohemia [036/2017/P to S.K.]; Medical Research Council [MC_PC_18038 to B.D.G.]; Health and Social Care in Northern Ireland (HSCNI) [STL/5460/18 to BDG]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackman J.E., Alfonzo J.D.. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA. 2013; 4:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kersten H., Kersten W.. Chapter 2: Biosynthesis and function of queuine and queuosine tRNAs. J. Chromatogr. Libr. 1990; 45:B69–B108. [Google Scholar]
- 3. Gregson J.M., Crain P.F., Edmonds C.G., Gupta R., Hashizume T., Phillipson D.W., Mccloskey J.A.. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-ß-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (Archaeosine))*. J. Biol. Chem. 1993; 268:10076–10086. [PubMed] [Google Scholar]
- 4. Fergus C., Barnes D., Alqasem M.A., Kelly V.P.. The queuine micronutrient: charting a course from microbe to man. Nutrients. 2015; 7:2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meier F., Suter B., Grosjean H., Keith G., Kubli E.. Queuosine modification of the wobble base in tRNA-His influences ‘ in vivo ’ decoding properties. EMBO J. 1985; 4:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zaborske J.M., Bauer DuMont V.L., Wallace E.W.J., Pan T., Aquadro C.F., Drummond D.A.. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014; 12:e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Müller M., Legrand C., Tuorto F., Kelly V.P., Atlasi Y., Lyko F., Ehrenhofer-Murray A.E.. Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res. 2019; 47:3711–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tuorto F., Legrand C., Cirzi C., Federico G., Liebers R., Müller M., Ehrenhofer-Murray A.E., Dittmar G., Gröne H.-J., Lyko F.. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018; 37:e99777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kulkarni S., Rubio M.A.T., Hegedűsová E., Ross R.L., Limbach P.A., Alfonzo J.D., Paris Z.. Preferential import of queuosine-modified tRNAs into Trypanosoma brucei mitochondrion is critical for organellar protein synthesis. Nucleic Acids Res. 2021; 49:8247–8260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zallot R., Brochier-Armanet C., Gaston K.W., Forouhar F., Limbach P.A., Hunt J.F., de Crécy-Lagard V.. Plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family. ACS Chem. Biol. 2014; 9:1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Müller M., Hartmann M., Schuster I., Bender S., Thüring K.L., Helm M., Katze J.R., Nellen W., Lyko F., Ehrenhofer-Murray A.E.. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015; 43:10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan C.T.Y., Pang Y.L.J., Deng W., Babu I.R., Dyavaiah M., Begley T.J., Dedon P.C.. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012; 3:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Endres L., Dedon P.C., Begley T.J.. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015; 12:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber S., Leonardi A., Dedon P., Begley T., Huber S.M., Leonardi A., Dedon P.C., Begley T.J.. The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics. 2019; 7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan C., Pham P., Dedon P.C., Begley T.J.. Lifestyle modifications: coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018; 19:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dedon P.C., Begley T.J.. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem. Res. Toxicol. 2014; 27:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coustou V., Biran M., Breton M., Guegan F., Rivière L., Plazolles N., Nolan D., Barrett M.P., Franconi J.M., Bringaud F.. Glucose-induced remodeling of intermediary and energy metabolism in procyclic Trypanosoma brucei. J. Biol. Chem. 2008; 283:16343–16354. [DOI] [PubMed] [Google Scholar]
- 18. Bringaud F., Rivière L., Coustou V.. Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol. Biochem. Parasitol. 2006; 149:1–9. [DOI] [PubMed] [Google Scholar]
- 19. Lopes R.R.S., Silveira G., de O., Eitler R., Vidal R.S., Kessler A., Hinger S., Paris Z., Alfonzo J.D., Polycarpo C.. The essential function of the Trypanosoma brucei Trl1 homolog in procyclic cells is maturation of the intron-containing tRNA Tyr. RNA. 2016; 22:1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kessler A.C., Kulkarni S.S., Paulines M.J., Rubio M.A.T., Limbach P.A., Paris Z., Alfonzo J.D.. Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. brucei. RNA Biol. 2018; 15:528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wirtz E., Leal S., Ochatt C., Cross G.A.M.. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 22. Hirumi H., Hirumi K.. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989; 75:985–989. [PubMed] [Google Scholar]
- 23. Chomczynski P. Single-step method of RNA isolation by acid guanidinium extraction. Anal. Biochem. 1987; 162:156–159. [DOI] [PubMed] [Google Scholar]
- 24. Hannah R., Sherf B.A., Navarro S.L., Hannah R.R., Wood K. V. Dual-luciferase TM reporter assay: an advanced co-reporter technology integrating firefly and renilla luciferase assays. Promega Notes Magazine. 1996; 57:2. [Google Scholar]
- 25. Xu H., Esberg A., Byström A.S.. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015; 43:9489–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shaheen H.H., Hopper A.K.. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:11290–11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Igloi G.L., Kössel H.. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res. 1985; 13:6881–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kořený L., Oborník M., Lukeš J.. Make it, take it, or leave it: heme metabolism of arasites. PLoS Pathog. 2013; 9:e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rakovich T., Boland C., Bernstein I., Chikwana V.M., Iwata-Reuyl D., Kelly V.P.. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J. Biol. Chem. 2011; 286:19354–19363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamour N., Rivière L., Coustou V., Coombs G.H., Barrett M.P., Bringaud F.. Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 2005; 280:11902–11910. [DOI] [PubMed] [Google Scholar]
- 31. Marchese L., Nascimento J.D.F., Damasceno F.S., Bringaud F., Michels P.A.M., Silber A.M.. The uptake and metabolism of amino acids, and their unique role in the biology of pathogenic trypanosomatids. Pathogens. 2018; 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hegedűsová E., Kulkarni S., Burgman B., Alfonzo J.D., Paris Z.. The general mRNA exporters Mex67 and Mtr2 play distinct roles in nuclear export of tRNAs in Trypanosoma brucei. Nucleic Acids Res. 2019; 47:8620–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whitney M.L., Hurto R.L., Shaheen H.H., Hopper A.K.. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol. Biol. Cell. 2007; 18:2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang H.-Y., Hopper A.. Multiple layers of stress-induced regulation in tRNA biology. Life. 2016; 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernhardt D., Darnell J.E.. tRNA synthesis in hela cells: a precursor to tRNA and the effects of methionine starvation on tRNA synthesis. J. Mol. Biol. 1969; 42:43–56. [DOI] [PubMed] [Google Scholar]
- 36. Damon J.R., Pincus D., Ploegh H.L.. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol. Biol. Cell. 2015; 26:270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta R., Walvekar A.S., Liang S., Rashida Z., Shah P., Laxman S.. A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife. 2019; 8:e44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y.C., Kelly V.P., Stachura S. V, Garcia G.A. Characterization of the human tRNA-guanine transglycosylase: confirmation of the heterodimeric subunit structure. RNA. 2010; 16:958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.