Abstract
Dnmt2, a member of the DNA methyltransferase superfamily, catalyzes the formation of 5-methylcytosine at position 38 in the anticodon loop of tRNAs. Dnmt2 regulates many cellular biological processes, especially the production of tRNA-derived fragments and intergenerational transmission of paternal metabolic disorders to offspring. Moreover, Dnmt2 is closely related to human cancers. The tRNA substrates of mammalian Dnmt2s are mainly detected using bisulfite sequencing; however, we lack supporting biochemical data concerning their substrate specificity or recognition mechanism. Here, we deciphered the tRNA substrates of human DNMT2 (hDNMT2) as tRNAAsp(GUC), tRNAGly(GCC) and tRNAVal(AAC). Intriguingly, for tRNAAsp(GUC) and tRNAGly(GCC), G34 is the discriminator element; whereas for tRNAVal(AAC), the inosine modification at position 34 (I34), which is formed by the ADAT2/3 complex, is the prerequisite for hDNMT2 recognition. We showed that the C32U33(G/I)34N35 (C/U)36A37C38 motif in the anticodon loop, U11:A24 in the D stem, and the correct size of the variable loop are required for Dnmt2 recognition of substrate tRNAs. Furthermore, mammalian Dnmt2s possess a conserved tRNA recognition mechanism.
INTRODUCTION
5-Methylcytosine is one of the most abundant modifications in both DNA and RNA (5mC and m5C) (1,2). DNA:5mC and its derivatives play a prominent role in epigenetic gene regulation (3). In contrast to the well-established study of DNA:5mC, research into RNA:m5C is lagging behind. Recently, resulting from the development of high-throughput methods for RNA:m5C detection (4–6), such as RNA bisulfite sequencing and 5-azacytidine based sequencing, the RNA:m5C modification has been found to be widespread in different RNA species and organisms (7,8), and could influence mRNA export, ribosome assembly, RNA stability, and tRNA fragmentation, (9–12). Moreover, defects of RNA:m5C modification are closely related to human diseases, such as intellectual disability, cancer, male infertility, and metabolic disorders (13–16).
RNA:m5C is catalyzed by the NSun methyltransferase family and a member of DNA methyltransferase family (Dnmt), Dnmt2 (17). Intriguingly, Dnmt2 contains the conserved DNA:5mC catalytic motif (motif IV), but only displays a weak DNA:5mC methyltransferase activity (18,19). Indeed, Dnmt2 was identified to methylate cytosine 38 to form m5C in the anti-codon loop of tRNAAsp (GUC) in mouse, Arabidopsis thaliana and Drosophila melanogaster (20). The Dnmt2 and NSun family both use S-adenosyl methionine (SAM) as a methyl group donor, but possess quite different enzymatic mechanisms. Significantly, Dnmt2 utilizes the catalytic mechanism of Dnmts to methylate tRNA (21). In Dnmt2, the conserved Cys residue in motif IV functions as the nucleophile in RNA:m5C formation (21,22); while the NSun family contains both an RNA:m5U-like motif (motif VI) and a DNA:m5C-like motif (motif IV) (23), the Cys residue in motif VI is the nucleophile and the Cys residue in motif IV acts as a general base to initiate product release (23,24). To date, Dnmt2 is the only m5C methyltransferase identified to catalyze tRNA:m5C formation in a manner similar to that of DNA:5mC methylation, which distinguishes Dnmt2 from all other known RNA methyltransferases.
Modifications at the anti-codon loop of tRNA usually contribute to the efficiency and accuracy of decoding (25,26). m5C38, which is near the anti-codon loop, could increase the amino acid accepting capacity of mouse tRNAAsp(GUC) (27). In accordance with that, the charging level of tRNAAsp(GUC) was reduced in Dnmt2-deficient mouse cells (27). It was shown that Dnmt2 could ensure specific protein synthesis in the bone marrow (28), as well as some proteins with poly-Asp sequences (27). Furthermore, loss of mouse Dnmt2 and NSun2 (encoding a member of NSun family NSun2), affected tRNA stability and reduced the rates of overall mouse protein synthesis (29).
The Dnmt2-mediated m5C38 modification could also affect the tRNA stability and the production of tRNA-derived fragments (tsRNAs). In Drosophila melanogaster and mouse, the m5C38 modification protected tRNAs against ribonuclease cleavage under stress conditions (12,28). Consistently, under stress conditions, human DNMT2 re-localizes to stress granules and participates in RNA processing, which suggests that Dnmt2 plays an important role in tRNA stability (12,30,31). Recently, it has been demonstrated that Dnmt2 alters the sperm tsRNA expression profile, and mediates the intergenerational transmission of paternal metabolic disorders to offspring through the m5C modification on sperm tsRNAs (16,32). Notably, 5′ fragments of tRNAGly(GCC), a substrate of mouse Dnmt2, were extremely abundant in mouse mature sperm (33,34). These tRNAGly(GCC) fragments were upregulated by ∼2–3-fold in mouse sperm with a low protein diet, and suppressed the expression of the endogenous retroelement MERVL-related genes in mouse embryos (34). Intriguingly, 5′ tRNA fragments from many other tRNAs, such as tRNAGly(CCC) and tRNAGlu(CUC), which have not been identified as substrates of Dnmt2, were also affected by knockout of Dnmt2(34). Taken together, Dnmt2 has a vital influence in the biogenesis and functions of tRNA-derived fragments, although its mechanistic role remains to be determined.
Dnmt2 is not only related to the canonical functions of tRNA, but also is associated with physiological functions of living organisms and human diseases. Danio rerio Dnmt2 promotes proper organ differentiation, including the retina, liver, and brain (35). D. melanogaster Dnmt2 is required for efficient innate immune responses to control virus infection directly (36). Deficiency of Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts (37). In addition, mouse Dnmt2 correlated with hematopoiesis (28). While in human fibroblasts, knockdown of DNMT2 induced cellular senescence (38). Moreover, human DNMT2 is closely linked to virus infection and cancer development, suggesting the prominent role of Dnmt2 in higher eukaryotes (31,39).
Dnmt2 is highly conserved in most eukaryotes and a few bacteria, such as Geobacter sulfurreducens and Holophaga foetida (Supplementary Figure S1) (40). The RNA substrate of Dnmt2 is not limited to tRNAAsp(GUC), and several other tRNAs could also be modified by Dnmt2. Interestingly, the tRNA substrates for Dnmt2 in different species are not the same. RNA bisulfite sequencing demonstrated that tRNAGly(GCC) and tRNAVal(AAC) are also substrates of mouse and D. melanogaster Dnmt2 (12,28). In Schizosaccharomyces pombe, Dnmt2 preferably methylated tRNAAsp(GUC) and showed a weak enzymatic activity to tRNAGlu(UUC) in vivo (41). In addition, the m5C modification level was regulated by queuine incorporation at G34 of tRNAAsp(GUC) or S. pombe Dnmt2 overexpression (41). Dictyostelium discoideum Dnmt2 could catalyze m5C38 on tRNAAsp(GUC) in vivo and in vitro, but displayed weaker methylation activity on tRNAGlu(UUC), -(CUC) and tRNAGly(GCC) in vitro, but not in vivo (42). While in G. sulfurreducens, Dnmt2 could only methylate tRNAGlu(UUC) in vitro and in vivo (43). Above all, these studies have shown that the substrate specificity of Dnmt2 in different species are very divergent. To date, the tRNA substrates of higher eukaryotic Dnmt2s have been identified through RNA bisulfite sequencing (28,29,32); however, the exact recognition mechanism of mammalian Dnmt2 remains unknown, hindering our understanding of the working mechanism of Dnmt2.
In the present study, we took human DNMT2 as a representative, and uncovered the distinct substrate recognition mechanisms of Dnmt2s from different species. We successfully reconstituted the catalytic activity of hDNMT2 in vitro, mapped all the C38-containing tRNAs, and demonstrated that tRNAAsp(GUC), tRNAGly(GCC), and tRNAVal(AAC) are the authentic tRNA substrates of hDNMT2. Intriguingly, for tRNAAsp(GUC) and tRNAGly(GCC), we found that G34 is the discriminator element; whereas, for tRNAVal(AAC), the pre-existing inosine at position 34 (I34), which is formed by the adenosine deaminase tRNA specific (ADAT)2/3 complex, serves as the prerequisite for hDNMT2 recognition. We showed that the recognition mechanism for tRNAVal(AAC) by mammalian Dnmt2s is conserved, suggesting that the interdependent modifications between I34 and m5C38 exist widely in mammals. Besides these, the tRNA motifs: C32U33(G/I)34N35(C/U)36A37C38 in the anticodon loop, U11:A24 in the D stem, and the variable loop are also required for hDNMT2 recognition. Taken together, our findings demonstrated that the m5C38 modification is depended on the identity or modification on position 34, and further defined that mammalian Dnmt2 utilities a delicate network for tRNA recognition, which involved both the primary sequence and the tertiary structure of tRNA substrates.
MATERIALS AND METHODS
Materials
Tris–HCl buffer, tryptone, yeast extract, bovine serum albumin (BSA), sodium phosphate monobasic, sodium phosphate dibasic, Pfu DNA polymerase, adenosine triphosphate (ATP), cytidine triphosphate (CTP), guanosine triphosphate (GTP), uridine triphosphate (UTP), dithiothreitol (DTT), isopropyl-β-d-thiogalactopyranoside (IPTG), and ethylene diaminetetra acetic acid (EDTA) were purchased from Sangon Biotech (Shanghai, China). The DNA fragment rapid purification kit and the plasmid extraction kit were purchased from Vazyme company (Nanjing, China). Escherichia coli Rosetta (DE3) cells were purchased from Weidi biotechnology Co. (Shanghai, China). Oligonucleotide primers for polymerase chain reaction (PCR) were synthesized by TsingKe (Shanghai, China) and BioSune (Shanghai, China). The KOD-plus mutagenesis kit and KOD Plus Neo high-fidelity DNA polymerase were purchased from TOYOBO (Osaka, Japan). Pyrobest DNA polymerase and the dNTP mixture were purchased from Takara (Shiga, Japan). MgCl2, NaCl, KCl, guanosine monophosphate (GMP), sodium acetate (NaAc), benzonase, and phosphodiesterase I from Crotalus adamanteus venom were purchased from Sigma-Aldrich Co. LLC. (St Louis, MO, USA). [Methyl-3H] SAM (78.0 Ci/mmol) was purchased from Perkin Elmer Inc. (Waltham, MA, USA). Isopropyl β-D-thiogalactoside (IPTG) was purchased from AMRESCO (Solon, OH, USA). Dynabeads protein G, Lipofectamine 2000 transfection reagent, T4 DNA ligase, the ribonuclease inhibitor, Trizol, and all the restriction endonucleases were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Phusion high-fidelity DNA polymerase and SAM were obtained from New England BioLabs (Ipswich, MA, USA). The Superdex™ 200 column (10/300 GL; column volume, 23.562 ml) and 3 mm filter papers were from GE Healthcare (Fairfield, CT, USA). The anti-Dnmt2 antibody was purchased from Santa Cruz Biotechnology (sc-365001, Santa Cruz, CA, USA). Inorganic pyrophosphate was obtained from Roche Applied Science (Basel, Switzerland). Polyethyleneimine cellulose plates and Nitrocellulose membranes (0.22 μm) were purchased from Merck (Darmstadt, Germany). Nickel nitrilotriacetic (Ni2+-NTA) super flow resin was obtained from Qiagen, Inc. (Hilden, Germany).
Construction of expression vectors
The coding sequences of human DNMT2 and mouse Dnmt2 were amplified from cDNAs, which were obtained by RT-PCR from total RNA extracted from Hela and NIH/3T3 cells, respectively. The Danio rerio and D. melanogaster Dnmt2 coding sequences were chemically synthesized by TsingKe. These four coding sequences were separately inserted between the BamHI and XhoI sites of pET28a vector with the DNA sequence encoding an N-terminal His6 tag. The coding sequences of human ADAT2 and ADAT3 were initially amplified from the Hela cells cDNA, then inserted between the EcoRI /HindIII and NdeI/XhoI sites of the pRSFDuet-1 expression vector (Novagen) respectively, which results in ADAT2 with the DNA sequence encoding an His6 tag at its N-terminus and ADAT3 with the DNA sequence encoding an S-tag at its C-terminus and were translated to ADAT2 and ADAT3, separately. In eukaryotes, the heterodimeric enzyme ADAT, including ADAT2 and ADAT3, catalyzes inosine modification at position 34 (I 34) in seven or eight different eukaryotic tRNAs (44,45). All the primers used for the construction of recombined vectors containing DNMT2/Dnmt2, ADAT2 and ADAT3 are listed in supplementary Table S1.
Expression and purification of Dnmt2 and ADAT2/3
The recombinant pET28a plasmids containing cDNAs coding the various Dnmt2 proteins were transformed separately into E. coli Rosetta (DE3). Single clones were cultivated in 100 ml of LB liquid medium containing 100 μg/ml of kanamycin at 37°C for 4 h, and then sub-cultured in 1 l of the same medium at 37°C. When the optional density reached A600 ∼0.6, protein overproduction was induced by adding 0.5 mM IPTG into the growth medium, followed by cultivation at 18°C for 16 h. The cultured cells were harvested by centrifugation (5000 × g at 22°C for 10 min).
The Dnmt2 proteins from different species were purified using the same method as described below. E. coli cells were suspended in lysis buffer (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 2 mM DTT, 10% glycerol, 10 mM imidazole) and sonicated in an ice bath. The supernatant was collected by centrifugation of the crude extracts at 16000 × g at 4°C for 1 h, and then incubated with Ni-NTA Superflow resin for 30 min. Subsequently, the resin was washed with 100 ml of 20 mM imidazole in lysis buffer to remove the nonspecific binding proteins, and eluted stepwise using 250 mM imidazole in lysis buffer. The eluted fraction was concentrated using Dialysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM DTT), followed by gel filtration chromatography using a Superdex™ 200 column. The eluted fractions were pooled, and then concentrated. The protein concentrations were determined by UV absorbance at 280 nm, and the molar absorption coefficient was calculated according to the sequence of each protein (46).
Co-expression of ADAT2 and ADAT3 from the recombinant pRSFduet–ADAT2–ADAT3 and purification of the heterodimer of ADAT2 and ADAT3 (ADAT2–ADAT3) were similar to that of Dnmt2. The ADAT2–ADAT3 complex was purified on a Ni-NTA Superflow resin, followed by gel filtration chromatography with a Superdex™ 200 column. The purified ADAT2–ADAT3 complex was stored in 50 mM Tris–HCl, pH 8.0, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 2 mM DTT with 50% glycerol.
tRNA transcription and mutagenesis
The DNA sequence of the T7 promoter and genes encoding 13 human cytoplasmic C38-containing tRNAs investigated in this study were ligated between the EcoRI and BamHI sites of pTrc99b to construct pTrc99b-T7-tRNAs. The sequences of these tRNAs are listed in Supplementary Table S2. Mutants of the hctRNAGly(GCC), hctRNAAsp(GUC) and hctRNAVal(AAC) were obtained using the KOD-plus mutagenesis kit. All tRNAs were produced by in vitro transcription using T7 RNA polymerase, as described previously (47). Transcribed tRNAs were purified by urea denaturing 12% polyacrylamide gel electrophoresis (PAGE). Then, tRNAs were refolded by fast heating and slow cooling down at room temperature in the presence of 5 mM MgCl2. Finally, tRNAs were stored at –20°C. The tRNA concentrations were determined by UV absorbance at 260 nm, and the molar absorption coefficient was calculated according to the sequence of each tRNA (48).
Methyltransferase activity assay
The methyltransferase activity of hDNMT2 for a wide range of tRNAs and their mutants was measured in a reaction mixture containing 50 mM Tris–HCl, pH 7.5, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 5 μM human cytoplasmic tRNA and 20 μM [3H]-SAM at 37°C. The reaction was initiated by adding hDNMT2 (0.5 μM). At various time intervals (3, 6, 9 and 12 min), aliquots were quenched by spotting on filters and washed with 5% trichloroacetic acid. The amount of radioactive [3H]-methyl-tRNA was measured in a Beckman Las6500 scintillation counting apparatus (Beckman Coulter, Indianapolis, IN, USA).
In vitro A34 to I34 deamination of tRNAVal(AAC)
The A34 to I34 deamination of tRNAVal(AAC) was performed as follows: 5 μg of in vitro transcribed tRNAVal(AAC) was incubated with 1.3 μM ADAT2–ADAT3 complex in a reaction buffer comprising 50 mM Tris–HCl, pH 8.0, 100 mM KCl, 2 mM DTT, and 200 μM SAM for 1 h at 37°C. tRNAVal(AAC) that had been incubated in the same buffer without adding the ADAT2–ADAT3 complex was the negative control. The tRNAVal(AAC) incubated with ADAT2–ADAT3 was purified by the standard phenol extraction procedure and precipitated using a three-fold volume of ethanol. Then, the tRNA was dissolved in DEPC-treated water and quantified using a NanoDrop ND-2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The I34 formation of tRNAVal(AAC) was checked using mass spectrometry analysis.
Mass spectrometry analysis of RNA modifications
tRNA (400 ng) was hydrolyzed by 0.5 μl Benzonase, 0.5 μl phosphodiesterase I, and 0.5 μl bacterial alkaline phosphatase overnight at 37°C in a 20 μl reaction buffer containing 4 mM NH4OAc. The solution was then diluted with 200 μl H2O and 10 μl was subjected to ultra-performance liquid chromatography-mass spectrometry/mass spectrometry (UPLC-MS/MS). The nucleosides were separated using UPLC on a C18 column (Agilent Zorbax Eclipse Plus C18, 2.1 × 50 mm, 1.8-Micron; Agilent, Santa Clara, CA, USA) or a Hilic column (Atlantis Silica HILIC Column, 3 μm, 2.1 mm × 150 mm, USA). For the separation on the C18 column, the elution solvents consisted of H2O plus 0.1% formic acid (solvent A) and methanol plus 0.1% formic acid (solvent B); for the separation on the Hilic column, the elution solvents consisted of 50% acetonitrile plus 0.1% formic acid (solvent A) and 90% acetonitrile plus 0.1% formic acid (solvent B). Subsequently, the detection was performed on a triple-quadruple mass spectrometer (Agilent 6400 QQQ or AB Sciex Q-TRAP 6500+) in the positive ion multiple reaction-monitoring (MRM) mode. The nucleosides were quantified using the nucleoside-to-base ion mass transitions of 268.1–136.2 (A), 258.1–126.1 (m5C) and 269.1–137.1 (I), which were monitored and recorded.
Isolation of endogenous specific tRNA by biotinylated DNA probe
Total RNA was extracted using Trizol according to the manufacturer's instructions. Individual endogenous tRNAs were isolated from total RNA of HEK293T cell with their own 5′ biotinylated DNA oligonucleotides, and purified by Streptavidin Agarose Resin as described previously (49,50). The probes for tRNAAsp(GUC) and tRNAGly(GCC) selection used in this study are listed in Supplementary Table 3. Five micrograms of biotinylated DNA probes were incubated with 15 μl of the high-capacity streptavidin-conjugated agarose beads in 100 mM Tris–HCl, pH 7.5 at room temperature for 90 min. After incubation, the oligonucleotide-coated beads were washed three times in 10 mM Tris–HCl, pH 7.5 and equilibrated in 6 × NTE solution (20 × NTE solution is 4 M NaCl, 0.1 M Tris–HCl pH 7.5, 50 mM EDTA, 5 mM β-mercaptoethanol). Then, total RNA was added into the above 6 × NTE solution that contains oligonucleotide-coated beads and heated for 5 min at 70°C. Subsequently, the mixture was naturally cooled to room temperature, and washed with 3 × NTE for three times and with 1 × NTE twice. The specific tRNA retained on the beads was eluted with 0.1 × NTE at 70°C and precipitated using 75% ethanol.
Construction of a human DNMT2 knock out HEK293T cell line
The human DNMT2 gene was knocked out using the CRISPR-Cas9 mediated gene targeting technology. Briefly, we designed two single guide RNAs (sgRNAs) targeting exon 1 of DNMT2 (http://crispr.mit.edu/). Sense and antisense oligonucleotides for the sgRNAs were cloned into vector pX330-mCherry (plasmid #98750; Addgene, Watertown, MA, USA) (51). HEK293T cells (1.0 × 106 cells per well) were transfected with the pX330-mCherry-sgRNA plasmids (1 μg) using lipofectamine 2000 according to the manufacturer's instructions. Twelve hours after transfection, the 293T cells expressing the red fluorescent protein were sorted using flow cytometry (FACS Aria SORP, Becton Dickinson, Franklin Lakes, NJ, USA) and seeded into 96-well plates. Two weeks after the transfection, colonies were isolated and genomic DNAs were extracted. DNMT2 KO cell lines were selected by confirming the frameshift mutations in the target region. The genotyping of HEK293T stable cell lines were analyzed by DNA sequencing of PCR products using the following primers:
hDNMT2-identify-F: GGAGAGGCTGGTCTAATTTC
hDNMT2-identify-R: CAGGATGAAGGACCGAGTCT
Western blotting
The knockout efficiency of DNMT2 was measured using western blotting. The cells were washed using ice-cold phosphate-buffered saline (PBS) twice and lysed using ice-cold lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 20 mM NaF, 1% NP-40) supplemented with a ProteinSafe™ Protease Inhibitor Cocktail (TransGen Biotech, Beijing, China). The supernatant was collected by centrifugation at 12000 × g for 10 min. Then, the cell lysates were separated using 10% SDS-PAGE together with pre-stained molecular protein standards, and transferred to a polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 5% (w/v) non-fat dried milk and incubated with the corresponding primary antibodies overnight at 4°C. The membranes were washed three times using PBS buffer plus 0.05% Tween-20 (PBST) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 and 0.5 ‰ Tween-20), and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at room temperature for 1 h. The membranes were treated with the chemiluminescent substrate after washing three times with PBST buffer. Imaging of the target protein was performed using the Amersham imager 680 system (GE).
RESULTS
Reconstitution the tRNA:m5C38 catalytic activity of hDNMT2 in vitro
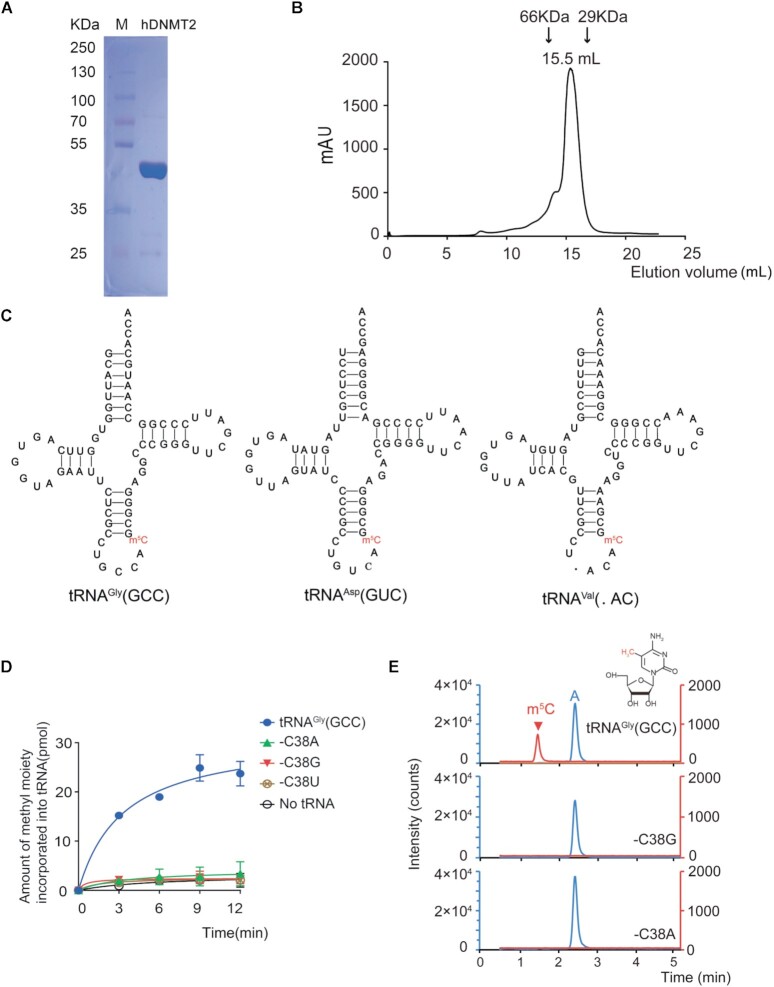
To reconstitute the enzymatic activity of hDNMT2, hDNMT2 was purified using Ni-NTA and gel filtration after isolation from E. coli transformants and analyzed using SDS-PAGE (Figure 1A). The hDNMT2 consists of 391 amino acid residues and a C-terminal His6 tag. Recombinant hDNMT2 was eluted at 15.5 ml by gel filtration, corresponding to a calculated molecular mass of 51.4 kDa, as compared with the theoretical molecular mass of purified hDNMT2 of 49.3 kDa, indicating that hDNMT2 exists as a monomer in solution (Figure 1B). This purified hDNMT2 was used for the subsequent methyl transfer assays. In Hela cells, RNA bisulfite sequencing demonstrated that human cytoplasmic tRNAGly(GCC), tRNAAsp(GUC) and tRNAVal(.AC) (‘.’ represents the unknown nucleoside) contain the m5C38 modification, suggesting that these three tRNAs are potential substrates of hDNMT2 (Figure 1C) (8). Indeed, we found that tRNAGly(GCC) could be efficiently methylated by hDNMT2 in vitro (Figure 1D). To confirm that C38 was the methylation site of hDNMT2 in vitro, we created C38A, C38G, and C38U substitutions in tRNAGly(GCC) to form three mutants: tRNAGly(GCC)-C38A, -C38G and -C38U. None of these mutants could be methylated by hDNMT2 (Figure 1D). To determine if m5C is the final product of the modification reaction by hDNMT2 in vitro, the methylated tRNAGly(GCC) was digested to nucleosides and subjected to UPLC-MS/MS analysis, and the modification was proved to be m5C (Figure 1E). However, the m5C modification was not detected in the tRNAGly(GCC) mutants incubated with hDNMT2 (Figure 1E). Thus, the reconstituted assay system for the tRNA:m5C38 catalytic activity of hDNMT2 was accurate and efficient in vitro.
Figure 1.
HDNMT2 catalyzes m5C38 modification on tRNAGly(GCC) in vitro. (A) SDS-PAGE analysis of the purified recombinant hDNMT2. Standard molecular weights are shown on the left. (B) The purified hDNMT2 was analyzed by gel filtration chromatography on a Superdex™ 200 column. HDNMT2 was eluted at 15.5 ml. The evolution volume of the standard proteins was marked above the graph. (C) The secondary structures of tRNAGly(GCC), tRNAAsp(GUC), and tRNAVal(.AC). (D) The capacity of tRNAGly(GCC) and the three mutants which were generated by in vitro transcription: -C38A, -C38G, -C38U to be methylated by hDNMT2. (E) tRNAGly(GCC) and the -C38G, and -C38A mutants incubated with hDNMT2 analyzed by UPLC-MS/MS analysis after digestion. Chromatogram of m5C (Q1/Q3 = 258.1/126.1) and A (Q1/Q3 = 268.1/136.2) are described, respectively. Error bars represent the standard errors of three independent experiments in Figures 1–8.
hDNMT2 methylates only tRNAGly(GCC) and tRNAAsp(GUC) among all the thirteen human cytoplasmic tRNA transcripts containing C38
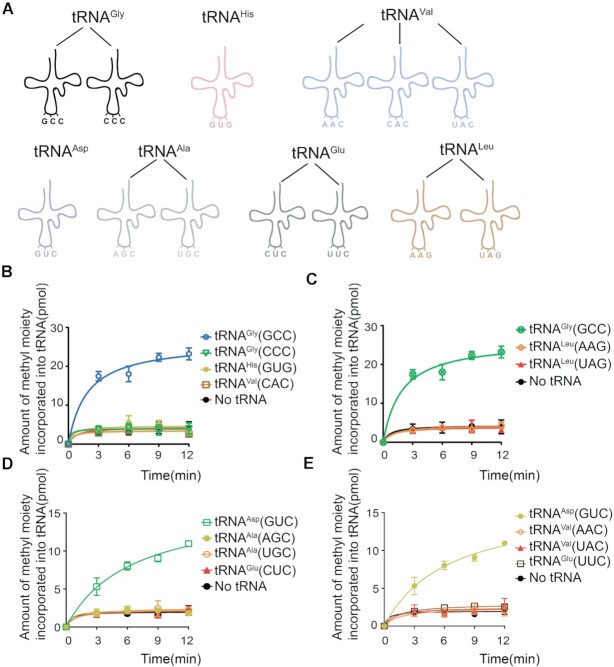
Mammalian Dnmt2 could modulate the production of several different kinds of tRNA fragments, which raised the possibility that hDNMT2 might have wider range of substrates than those previously identified by RNA bisulfite sequencing (8). Human cells contain 49 kinds of cytoplasmic tRNA molecular species (52), thirteen of which contain C38 (Figure 2A). To uncover the potential substrates of hDNMT2, we obtained all human cytoplasmic tRNAs transcripts containing C38 (tRNAAla(AGC) and -(UGC), tRNAAsp(GUC), tRNAGlu(CUC) and -(UUC), tRNAGly(GCC) and -(CCC), tRNAHis(GUG), tRNALeu(AAG) and -(UAG), tRNAVal(AAC), -(CAC) and -(UAC)) and determined whether they could be methylated by hDNMT2. Remarkably, only tRNAGly(GCC) and tRNAAsp(GUC), could be methylated by hDNMT2 in vitro (Figure 2B-E). The modification in tRNAAsp(GUC) when methylated by hDNMT2 was confirmed to be m5C (Supplementary Figure S2).
Figure 2.
HDNMT2 methylates only tRNAGly(GCC) and tRNAAsp(GUC) among all the thirteen human cytoplasmic tRNA transcripts containing C38. (A) Schematic diagrams of all the thirteen human cytoplasmic tRNAs containing C38: two tRNAGly isoacceptors, tRNAHis(GUG), three tRNAVal isoacceptors, tRNAAsp(GUC), two tRNAAla isoacceptors, two tRNAGlu isoacceptors, and two tRNALeu isoacceptors. (B–E) The capacity of the thirteen human cytoplasmic C38-containing tRNAs to be methylated by hDNMT2.
To verify the tRNA substrates of hDNMT2 in vivo, we knocked out DNMT2 in HEK293T cells using the CRISPR-Cas9 system and obtained the knockout (KO) cell lines in which both alleles contained frameshift mutations (Supplementary Figure S3A). The knockout efficiency of DNMT2 is shown as Supplementary Figure S3B. To validate whether tRNAGly(GCC) and tRNAAsp(GUC) were the substrates of hDNMT2 in vivo, we isolated these two tRNAs from the wild-type (WT) and the DNMT2 KO cell line, and subjected them to UPLC-MS/MS to analyze their respective m5C levels. For tRNAGly(GCC) and tRNAAsp(GUC), compared to that of WT cells, the level of m5C in both decreased significantly in DNMT2 KO cells (Supplementary Figure S3C, D). Taken together, our results showed that tRNAGly(GCC) and tRNAAsp(GUC) are the tRNA substrates of hDNMT2 in vitro and in vivo.
The tRNAAsp(GUC), tRNAGly(GCC) and tRNAVal(AAC) had been previously identified as substrates of mouse Dnmt2 by substrate identification in Dnmt2–/– mice and RNA bisulfite sequencing (20,28). Additionally, in HeLa cells, RNA bisulfite sequencing revealed that human cytoplasmic tRNAGly(GCC), tRNAAsp(GUC), and tRNAVal(.AC) possess m5C38 modification (8). However, based on our results, only tRNAAsp(GUC) and tRNAGly(GCC), but not tRNAVal(AAC), -(CAC) and -(UAC), could be methylated by hDNMT2 in vitro (Figure 2B–E). Notably, the 34th nucleotide of tRNAVal(.AC) was read as G after reverse transcription followed by sequencing (8). However, tRNAVal has only -(AAC), -(CAC) and -(UAC) isoacceptors, and there is no -(G34AC) isoacceptor, suggesting that some prior modifications in cellular endogenous tRNAVal might serve the essential recognition elements for methylation by hDNMT2.
The formation of m5C38 on tRNAVal(AAC) depends on the pre-existing A-to-I modification at position 34
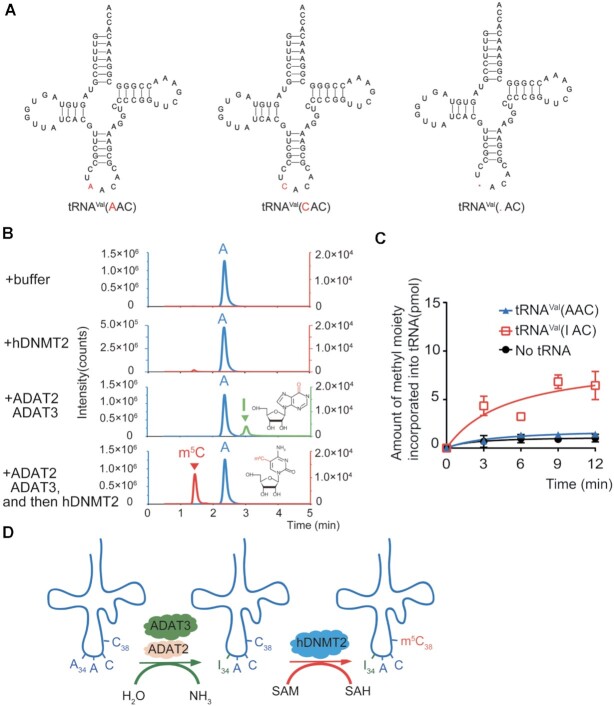
As mentioned above, tRNAVal(.AC) possesses the m5C38 modification according to the RNA bisulfite sequencing (8). In addition, the 34th nucleotide of tRNAVal(.AC) was read as G after reverse transcription followed by sequencing. The sequence alignment shows that tRNAVal(.AC) has the same sequence as tRNAVal(AAC) and -(CAC), except at position 34 (Figure 3A). Therefore, it is impossible to isolate the separate tRNAVal(CAC) or -(AAC) from cells using the biotinylated DNA probes, which hinders the detection of m5C38 modification level in every separate tRNAVal by UPLC-MS/MS.
Figure 3.
I34 is the prerequisite for hDNMT2-mediated m5C38 modification in tRNAVal(AAC). (A) The secondary structures of tRNAVal(AAC), -(CAC) and -(.AC). (B) UPLC-MS/MS analysis of m5C (Q1/Q3 = 258.1/126.1) of tRNAVal(AAC) after incubation with or without hDNMT2. UPLC-MS/MS analysis of I (Q1/Q3 = 269.1 to 137.1) of tRNAVal(AAC) after incubation with the ADAT2-ADAT3 complex. The products of tRNAVal(AAC) incubated with ADAT2-ADAT3 and then reacted with hDNMT2 were also digested and detected by UPLC-MS/MS analysis. (C) The capacity of tRNAVal(AAC) and tRNAVal(IAC) to be methylated by hDNMT2. (D) Schematic diagram showing that in tRNAVal(AAC), hDNMT2-mediated m5C38 modification depends on the A-to-I modification at position 34 formed by the ADAT2-ADAT3 complex.
However, tRNAVal has only -(AAC), -(CAC) and -(UAC) isoacceptors, but there is no -(GAC) isoacceptor in the human genome, raising the question that whether tRNAVal(.AC) is result from tRNAVal(AAC) or tRNAVal(CAC) with modification at position 34. Considering that tRNAVal(AAC) contains an adenosine to inosine (A-to-I) modification at position 34 (52), and inosine will be read as G during reverse transcription followed by sequencing, we speculated that tRNAVal(.AC), which possesses the m5C38 modification, might derive from tRNAVal(AAC) with an A-to-I modification at position 34. Thus, we tested whether the A-to-I modification at position 34 has an impact on the formation of m5C38.
The A-to-I modification at position 34 is catalyzed by human the ADAT2-ADAT3 protein complex (53,54); therefore, we co-expressed and purified the human ADAT2-ADAT3 complex, and generated the tRNAVal(AAC) carrying I34 (tRNAVal(IAC)) (Figure 3B). The formation of I34 in tRNAVal(AAC) via the human ADAT2–ADAT3 complex was verified by UPLC-MS/MS analysis (Figure 3B). Critically, hDNMT2 could methylate tRNAVal(IAC) but not tRNAVal(AAC) (Figure 3C). We further showed that after incubation with hDNMT2, the m5C modification could indeed be detected in tRNAVal(IAC) by UPLC-MS/MS analysis (Figure 3B). Thus, our results showed that m5C38 on tRNAVal(AAC) catalyzed by hDNMT2 is dependent on the pre-existing A-to-I modification at position 34 (Figure 3D).
G34 or I34 function as the determinant for hDNMT2 recognition
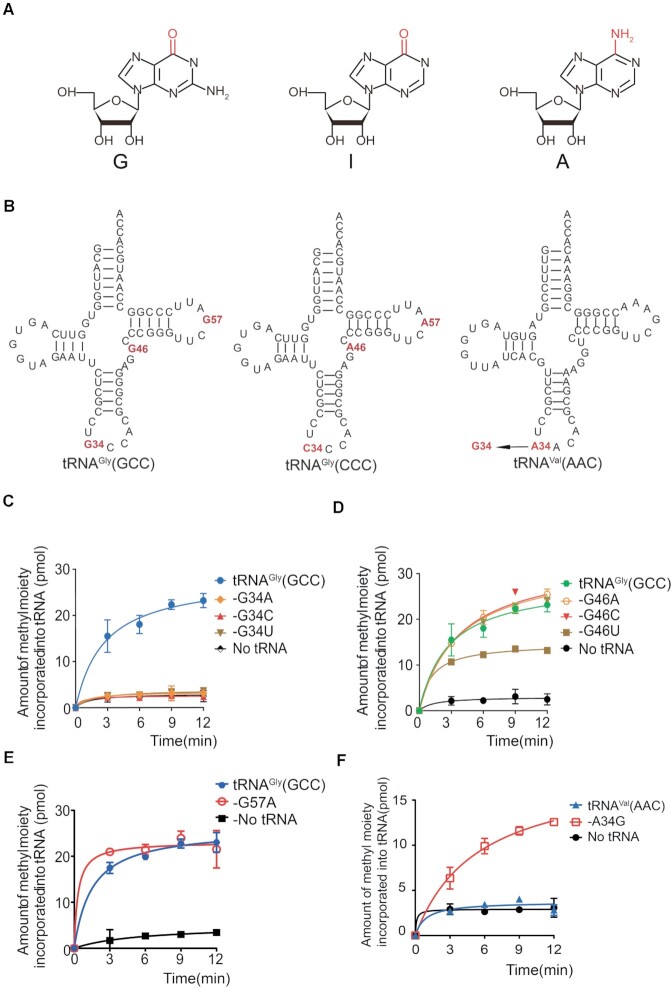
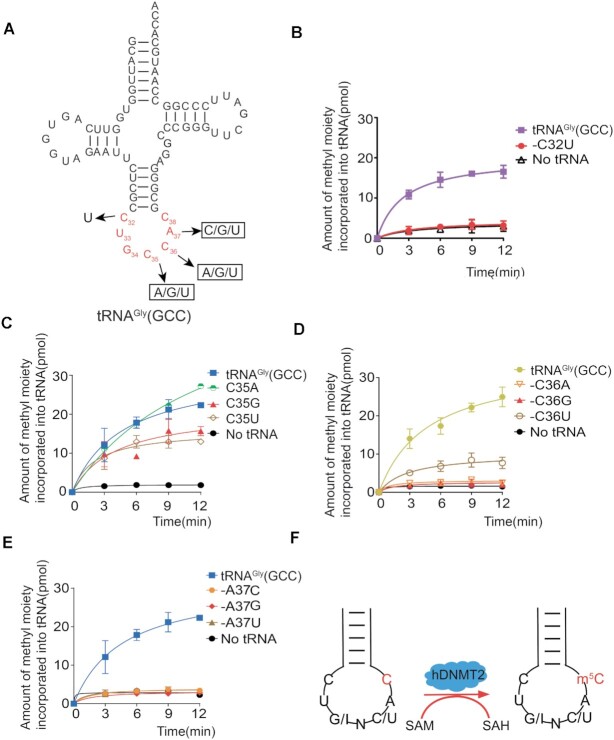
To determine the substrate specificity of hDNMT2, we next characterized the elements of tRNA recognized by hDNMT2. Based on our results, hDNMT2 could catalyze m5C38 on tRNAGly(GCC), tRNAAsp(GUC), and tRNAVal(IAC). Notably, inosine is similar to guanine in that both have a carbonyl oxygen at position 6 (Figure 4A), suggesting that the 34th nucleoside might play a role in hDNMT2 substrate recognition. The sequence alignment of the substrate tRNAGly(GCC) and the non-substrate tRNAGly(CCC) showed that there are only three different nucleotides between them, which are located at position 34, 46 and 57 (Figure 4B). To determine whether these three nucleosides are essential for hDNMT2 catalysis, we mutated the nucleotides at these positions, separately, and the methyl transferase activity of hDNMT2 for the tRNA mutants was detected.
Figure 4.
G34 or I34 function as the determinants for hDNMT2 recognition. (A) The formulas of guanosine (G), inosine (I) and adenosine (A). (B) The secondary structures of tRNAGly(GCC), -(CCC) and tRNAVal(AAC). The capacity of tRNAGly(GCC) with various mutations at position 34 (C), 46 (D) and 57 (E) to be methylated by hDNMT2. (F) The capacity of tRNAVal(AAC) and -A34G to be methylated by hDNMT2.
We constructed G34A, G34C, and G34U substitutions in tRNAGly(GCC) to form three mutants: tRNAGly(GCC)-G34A, -G34C and -G34U. Surprisingly, none of these mutants could be methylated by hDNMT2 (Figure 4C). However, hDNMT2 could methylate all three mutants tRNAGly(GCC)-G46A, -G46C and -G46U at position 46, and tRNAGly(GCC)-G57A at position 57 (Figure 4D-E). Our results showed that G34 is a critical element for tRNA recognition by hDNMT2.
We observed that the human cytoplasmic tRNAVal(AAC) transcript could not be methylated by hDNMT2, but when A was mutated to G at position 34, it could be efficiently methylated by hDNMT2 (Figure 4F). Combined with the observation that I34 is the prerequisite for m5C38 modification in tRNAVal (AAC), our results showed that G34 or I34 serve as the determinant for hDNMT2 recognition.
C32U33(G/I)34N35(C/U)36A37C38 in the anticodon loop is essential for hDNMT2 recognition
Besides the nucleotides at the 34th and 38th positions, the nucleotide residues at other positions in the anticodon loop region involved in RNA recognition by hDNMT2 were then identified (Figure 5A). Given that the 32nd nucleotide of human cytoplasmic tRNAs is semi-conserved and exist as C or U, we mutated the C32 to U32 to generate the tRNAGly(GCC)-C32U mutant. The result showed that hDNMT2 could not methylate the tRNAGly(GCC)-C32U mutant, indicating that C32 is crucial for hDNMT2 recognition (Figure 5B). According to the tRNA database (52,55), U33 is conserved in human cytoplasmic tRNAs. Thus, we speculated that U33 might not serve as a discriminative element for hDNMT2 recognition. As expected, hDNMT2 could methylate the tRNAGly(GCC)-U33 mutants replaced with the other three nucleotides, respectively, indicating that the 33rd nucleotide is dispensable for hDNMT2 recognition (Supplementary Figure S4).
Figure 5.
The hDNMT2 recognition elements in the anti-codon loop of tRNA. (A) The secondary structure of tRNAGly(GCC), summarizing the mutations in the anti-codon loop. The capacity of wild-type tRNAGly(GCC) and mutants with various mutations at position 32 (B), 35 (C), 36 (D) and 37 (E) to be methylated by hDNMT2. (F) Schema showing hDNMT2′s tRNA recognition elements in the anticodon loop.
In the anticodon loop, the 35th or 36th nucleotides were substituted with the other three nucleotides, separately, to obtain tRNAGly(GCC)-C35A, -C35G, -C35U, -C36A, -C36G and -C36U mutants. Remarkably, the tRNAGly(GCC)-C35A, -C35G, -C35U mutants could all be methylated by hDNMT2 (Figure 5C), while tRNAGly(GCC)-C36G or -C36A could not, and the efficiency of hDNMT2-mediated methylation of tRNAGly(GCC)-C36U was lower than that for tRNAGly(GCC) (Figure 5D). Our results showed that the 35th nucleotide is not essential; however, the 36th must be pyrimidine C or U for substrate recognition by hDNMT2.
Based on the tRNA database (52,55), the 37th nucleotide of human cytoplasmic tRNA is A or G. It is noticeable that among the three identified tRNA substrates of hDNMT2, this nucleotide always is A37. To determine whether hDNMT2 recognized the 37th nucleotide, tRNAGly(GCC)-A37G was constructed and the methylation by hDNMT2 was assessed. The results showed that tRNAGly(GCC)-A37G could not be methylated by hDNMT2 completely (Figure 5E). Furthermore, tRNAGly(GCC)-A37C and -A37U mutants were not the substrates of hDNMT2 (Figure 5E), indicating that A37 is a critical element for hDNMT2 recognition.
Collectively, we found the essential elements for hDNMT2 recognition: C32, U33, the determinant element G34 or I34, N35 (N = A, G, C, U), C or U36, A37 in the anticodon loop. Thus, the recognition motif in the anticodon loop of tRNA for hDNMT2 recognition is C32U33(G/I)34N35(C/U)36A37C38 (Figure 5F).
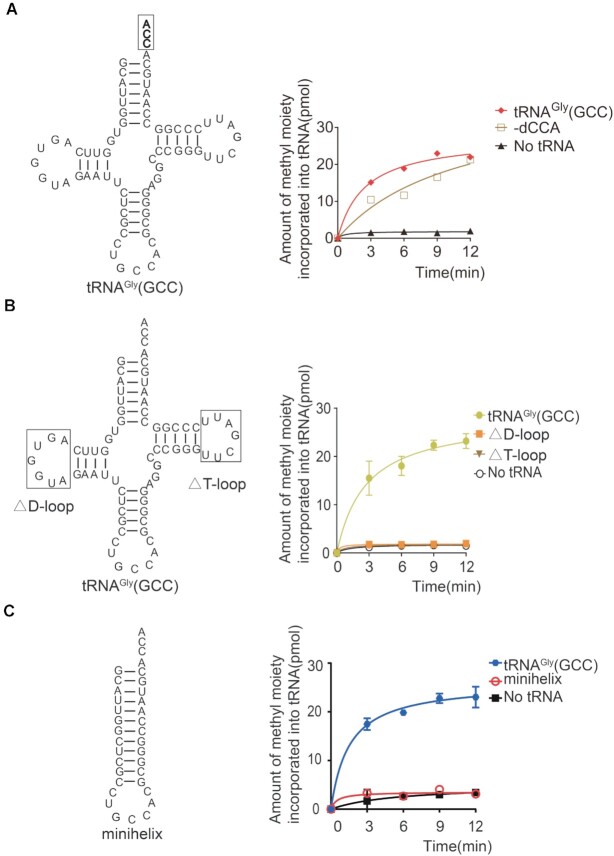
HDNMT2 recognizes a well-folded tRNA substrate
Considering that all tRNAs contain the 3′ end CCA, we determined whether the 3′ end CCA is essential for hDNMT2 recognition. Our results showed that hDNMT2 could methylate the tRNAGly(GCC) mutant (Figure 6A), implying that the common 3′ end CCA of the tRNAs was not essential for hDNMT2 recognition.
Figure 6.
HDNMT2 recognizes well-folded tRNA substrates. (A) The capacity of a tRNAGly(GCC) mutant lacking the CCA terminus to be methylated by hDNMT2. (B) Schematic diagram showing the truncations of the D-loop and T-loop of tRNAGly(GCC), respectively. The capacity of the two truncated tRNAGly(GCC) to be methylated by hDNMT2. (C) Schematic diagram showing the tRNAGly(GCC)-minihelix, which is formed by the entire anticodon stem loop fused with the acceptor stem. The capacity of the tRNAGly(GCC)-minihelix to be methylated by hDNMT2.
Based on the sensitivity to the tertiary structure of the tRNA substrates, tRNA methyltransferases can be categorized into two groups (56). The first group only utilizes well-folded tRNA molecules as substrates, while the second group can efficiently methylate truncated tRNA fragments independent of the tRNA tertiary structure (56). We wondered whether the L-shaped structure of tRNA or the anticodon stem loop alone was sufficient for hDNMT2 recognition. The methyl transferase activity of hDNMT2 for the truncation mutants of the full D-loop or T-loop of tRNAGly(GCC) showed that neither of the two mutants could be methylated (Figure 6B), indicating that the tertiary structure of tRNA is required for hDNMT2 recognition. Based on the identified recognition element in the anticodon loop for hDNMT2 recognition, we constructed the tRNA minihelix derived from tRNAGly(GCC). This minihelix only retained the acceptor stem, anticodon stem, and loop domains, without the D stem and loop, the variable loop, and TΨC-regions (Figure 6C). The results showed that hDNMT2 could not methylate the minihelix of tRNAGly(GCC) (Figure 6C), suggesting that hDNMT2 recognizes a well-folded tRNA substrate.
Elements within the D-stem and variable loop are involved in recognition by hDNMT2
As mentioned above, hDNMT2 was unable to methylate the tRNA minihelix that contained the acceptor stem and the anticodon stem loop region of tRNAGly(GCC), indicating that additional elements of the tRNA besides the anticodon loop are important for hDNMT2 recognition. According to the tertiary structure of tRNA, the D stem and the variable loop regions are relatively close to the anticodon loop in space. To investigate whether there are other recognition elements in the D stem region of tRNA, we compared the secondary structure of tRNAGly(GCC), tRNAAsp(GUC) and tRNAVal(IAC), and presumed that the two base pairs 11:24 and 12:23, which are spatially close to m5C38, might have an impact on the recognition of hDNMT2.
Based on the tRNA database, both the 11st and 24th nucleotides are semi-conserved and exist separately as pyrimidine and purine, which are Watson-Crick U11:A24 or C11:G24 base pairs. However the 11:24 base pair of tRNAGly(GCC), tRNAAsp(GUC), and tRNAVal(IAC) are all U11:A24 (Figure 1C). To investigate the impact of the base pair between the 11st and 24th on hDNMT2 recognition, we substituted the U11:A24 base pair to C11:G24 in tRNAGly(GCC). Remarkably, the tRNAGly(GCC)-C11:G24 mutant was not methylated by hDNMT2 (Figure 7A), implying that U11:A24 base pair is important for hDNMT2 recognition.
Figure 7.
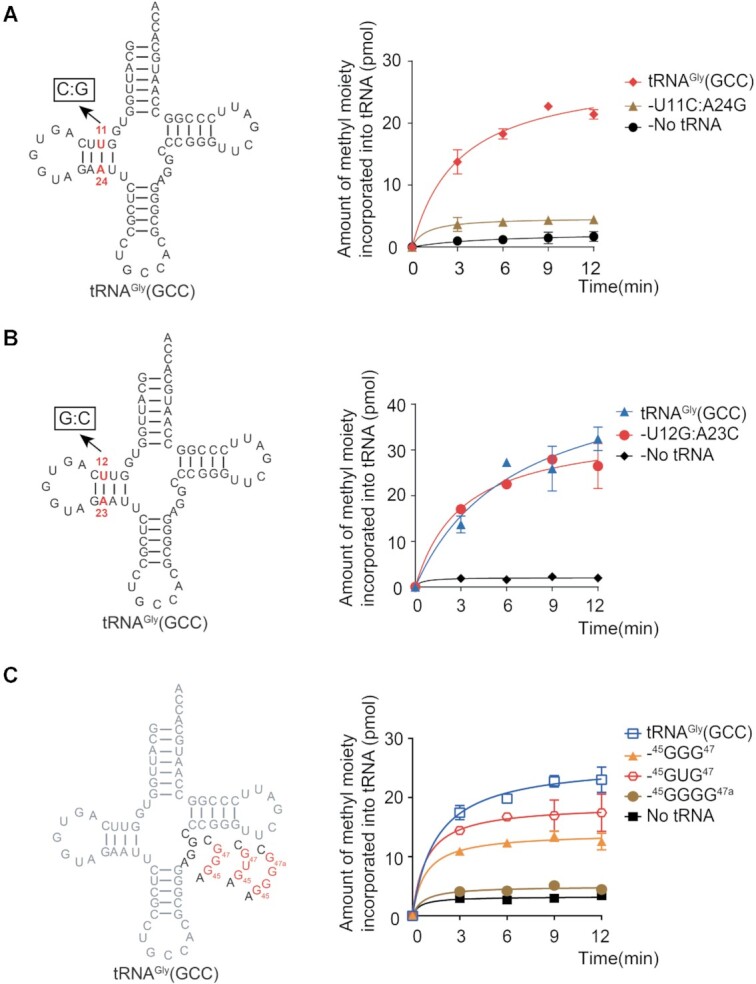
The D-stem and variable loop are recognized by hDNMT2. (A, B) Mutants of the D-stem region. The capacity of tRNAGly(GCC)-U11C:A24G (A) and -U12G:A23C (B) to be methylated by hDNMT2. (C) Mutants of the variable loop. The capacity of tRNAGly(GCC) with several mutations in variable loop to be methylated by hDNMT2.
To verify whether the 12:23 base pair is recognized by hDNMT2, the base pair U12:A23 was replaced with the more rigid base pair G12:C23. The mutant tRNAGly(GCC)-U12G:A23C was still efficiently methylated by hDNMT2 (Figure 7B), indicating that the base pair between the 12nd and the 23rd nucleotides is not involved in tRNA recognition by hDNMT2. Indeed, the 12nd and the 23rd nucleotides are not conserved in the three tRNA substrates of hDNMT2, they are U12:A23, A12:U23 and G12:C23 in tRNAGly(GCC), tRNAAsp(GUC) and tRNAVal(IAC), respectively.
To further confirm the D-stem region is recognized by hDNMT2, we constructed the tRNAGly(GCC)-U25G and -A24U&U25G mutants to destroy the hydrogen bonds of the G10:U25 and U11:A24 base pairs, respectively. The results showed that the methylation of these two mutants by hDNMT2 was completely lost (Supplementary Figure S5), suggesting that D-stem of tRNA is involved in the substrate recognition of hDNMT2.
A previous study showed that for tRNAAsp(GUC) recognition by hDNMT2, the variable loop is also important, and in particular, the GG dinucleotide in the variable loop might serve as the anti-determinant (43). Intriguingly, the GG dinucleotide in the variable loop is observed in tRNAGly(GCC) and tRNAVal(IAC) (Figure 1C). Moreover, five nucleotides were present in the variable loop of tRNAVal(IAC), while the other two substrates only contain four nucleotides, suggesting that the recognition mechanism in the variable loop might be complicated. To study whether the number of nucleotides in the variable loop affects hDNMT2 recognition, we first added G or U between G45 and G46 in tRNAGly(GCC) to form tRNAGly(GCC)-45GGG47 or -45GUG47 mutants with a variable loop of five nucleotides (Figure 7C). Compared with that of the wild-type tRNAGly(GCC), the methylation activity of hDNMT2 toward the two mutants was decreased by varying amounts (Figure 7C). The tRNAGly(GCC)-45GGGG47a mutant with the variable loop of six nucleotides, in which was GG was inserted between G45 and G46 of tRNAGly(GCC), was completely incapable of being methylated by hDNMT2 (Figure 7C). Above all, these results indicated that the nucleotide composition and size of the variable loop are also important for hDNMT2 recognition.
The stringent requirement for G/I34 on tRNA is only associated with mammalian Dnmt2 substrate specificity
The above results showed that G/I34 of tRNAs function as the determinant for hDNMT2 recognition. Furthermore, C32U33(G/I)34N35(C/U)36A37C38 in the anticodon loop, base pair U11:A24 in the D stem, and the number of nucleotides in the variable loop are also involved in hDNMT2 recognition. Many studies have shown that the substrate specificity of Dnmt2 in different species is extremely divergent, as shown in Figure 8D. This phenomenon indicated that Dnmt2s from different species might possess distinct mechanisms for substrate recognition. To test the hypothesis, we first verified whether the substrate recognition element of hDNMT2, especially the determinant G/I34, is conserved in M. musculus, D. rerio and D. melanogaster Dnmt2 proteins, respectively.
Figure 8.
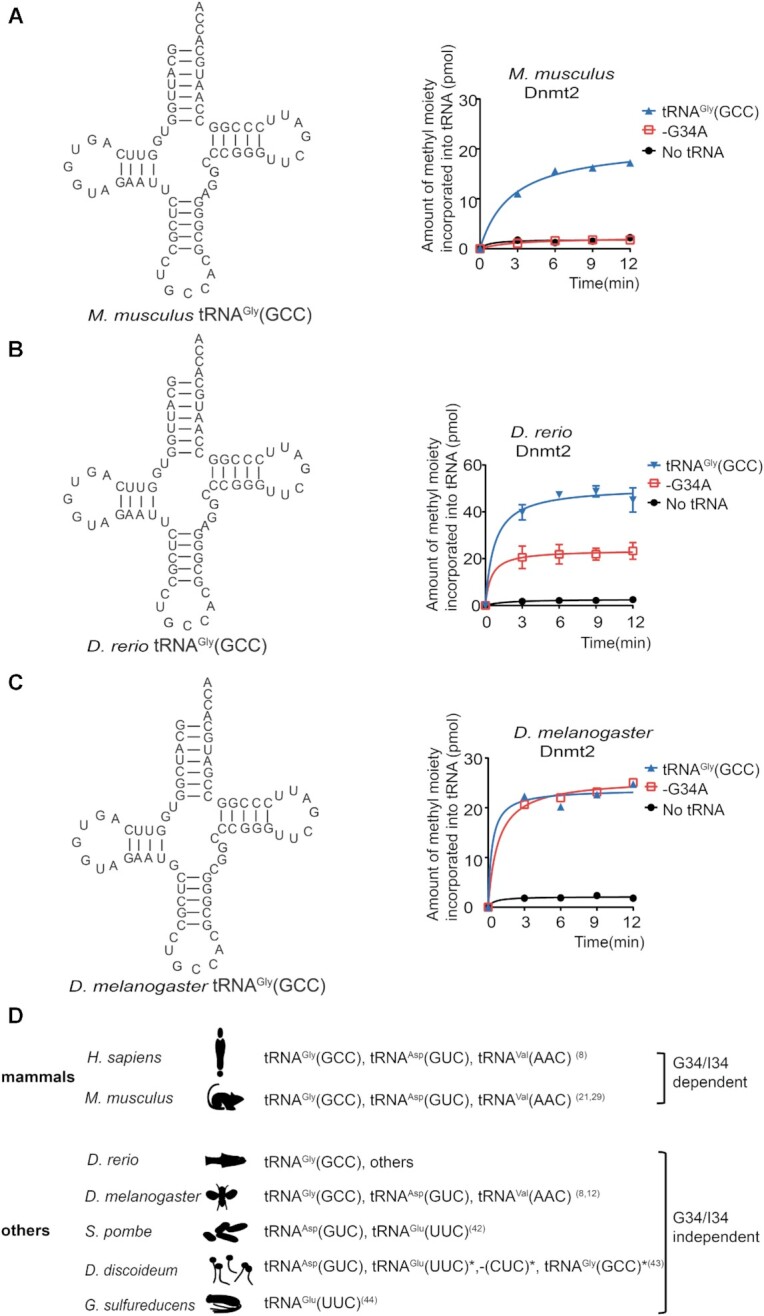
G/I34 is the prerequisite for m5C38 formation by mammalian Dnmt2s. The capacity of orthologs of tRNAGly(GCC) from different species and their -G34A mutants to be methylated by their cognate Dnmt2 from M. musculus (A), D. rerio (B) and D. melanogaster (C), respectively. (D) Summary of the known tRNA substrates of Dnmt2s from different species. Our results, together with those of previous studies, showed that G/I34 in tRNA substrates is required for mammalian Dnmt2 recognition; while in other lower eukaryotes and prokaryotes, G/I34 is dispensable for Dnmt2 recognition. The tRNAs labeled with * can only be methylated by the cognate Dnmt2 in vitro, but not in vivo.
We found that M. musculus, D. rerio and D. melanogaster Dnmt2s, which were successfully purified from E. coli transformants could all methylate the cognate tRNAGly(GCC) (Figure 8A–C). The data are consistent with the previous identified tRNA substrates of Dnmt2s from these species. Significantly, the methylation of tRNAGly(GCC)-G34A by M. musculus Dnmt2 was completely lost (Figure 8A), indicating that G34 still serves as the determinant for M. musculus Dnmt2 recognition. Considering that tRNAVal(AAC) containing the I34 modification is a substrate of M. musculus Dnmt2, as indicated by RNA bisulfite sequencing (28,52), and the mammalian Dnmt2s share high sequence similarity with each other (Supplementary Figure S1), we presumed that G/I34 functions as a common determinant for the substrate specificity of mammalian Dnmt2.
We found that D. rerio and D. melanogaster Dnmt2 could both methylate the cognate tRNAGly(GCC)-G34A (Figure 8B, C), indicating that Dnmt2 from these two species could methylate the tRNAs containing G34 or A34. Indeed, a previous study has shown that tRNAAsp(GUC) and tRNAVal(AAC) are also substrates of D. melanogaster Dnmt2 (12). Furthermore, we summarized the identified tRNA substrates of Dnmt2 from different species. S. pombe Dnmt2 could catalyze the methylation on the cognate tRNAAsp(GUC) and tRNAGlu(UUC) in vivo and in vitro, although the methylation of tRNAGlu(UUC) was weaker than that of tRNAAsp(GUC) (41). In D. discoideum, Dnmt2 could methylate tRNAAsp(GUC) in vivo and in vitro and also showed weaker methylation activity on tRNAGlu(UUC), -(CUC) and tRNAGly(GCC) in vitro, but not in vivo (42). In addition, only tRNAGlu(UUC) is methylated to form m5C38 by G. sulfurreducens Dnmt2 in vitro and in vivo (43). Collectively, these findings suggest that G/I34 is independent of the substrate recognition of Dnmt2 from lower species (Figure 8D).
In conclusion, the specific recognition of mammalian Dnmt2 of its substrate is stringent and requires G/I34 as the determinant. By contrast, Dnmt2 from lower eukaryotes and prokaryotes has looser requirements and independent of G/I34 (Figure 8D).
DISCUSSION
The substrate specificity of Dnmt2 during evolution
In this study, we have screened all human cytoplasmic tRNAs containing C38 and identified the accurate tRNA substrates of hDNMT2, i.e., tRNAGly(GCC), tRNAAsp(GUC), and tRNAVal(AAC), by assaying the enzyme activity of hDNMT2 in vitro. The substrates recognized by hDNMT2 are based on the following elements: (i) the determinant G/I34; (ii) the recognition motif C32U33(G/I)34N35(C/U)36A37C38 in the anticodon loop; (iii) the tertiary structure of the tRNA substrates; (iv) the U11:A24 in the D stem; and (v) the nucleotides and size of the variable loop (Figure 9). In accordance with our study, RNA bisulfite sequencing of Hela cells revealed that only tRNAGly(GCC), tRNAAsp(GUC) and tRNAVal(.AC) contain the m5C38 modification among the tRNA species (8). Additionally, tRNAVal(.AC) might be derived from tRNAVal(AAC) containing I34, considering that I was read as G after reverse transcription followed by sequencing (53,54). Thus, our results demonstrated the stringent and delicate tRNA substrate requirements of hDNMT2, especially for the 34th nucleotide.
Figure 9.
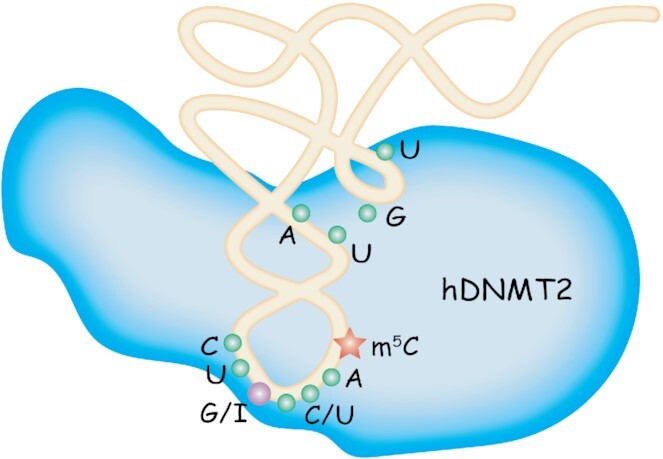
Model of tRNA substrates recognized by hDNMT2.
During the submission of our work, another in vitro biochemical study of hDNMT2 was published, demonstrating new tRNA substrates besides tRNAGly(GCC), tRNAAsp(GUC) and tRNAVal(AAC) (57), which is not consistent with the RNA bisulfite sequencing from Hela cells (8). However, in that study, the enzyme concentration used was extremely high, and the time taken to measure the enzymatic activity was quite long, at 70 min (57). What is more, there was no confirmation of whether the modification is indeed m5C after that long incubation in such an high concentration of the enzyme, especially for the new tRNA substrates (57). Based on these observations, we prefer to believe that our enzymatic assay system of hDNMT2 is closer to the in vivo system, and our result of substrate specificity of hDNMT2 is consistent with the high-throughput sequencing data (8).
In M. musculus, tRNAGly(GCC), tRNAAsp(GUC) and tRNAVal(AAC) are the only tRNA substrates of Dnmt2, as indicated by sequencing (20,28). We found that the formation of the m5C38 modification by M. musculus Dnmt2 is also dependent on the G/I34, suggesting that the dependence on G/I34 of m5C38 formation is a conserved substrate recognition mechanism of mammalian Dnmt2s (8,20,28). In lower eukaryotes, such as D. rerio, D. melanogaster, S. pombe and D. discoideum, G/I34 is dispensable for the substrate recognition of Dnmt2s (12,41,42). Furthermore, in a few prokaryotes, such as the G. sulfurreducens, the substrate recognition of Dnmt2 does not require G/I34 as a prerequisite (43). The underlying mechanism of the substrate recognition of Dnmt2 during evolution still awaits further studies, especially its relevance to the diverse biological functions of Dnmt2s.
The modulation by modifications at position 34 on m5C38 formation
In eukaryotes, such as mammals, S. pombe and D. discoideum, queuosine (Q) occurs at G34 of tRNAAsp(GUC) (58), the most common tRNA substrate of Dnmt2s (20). Queuosine is formed from the precursor queuine, which is salvaged from environmental sources, diet, and/or gut microbiota in eukaryotes (58). In S. pombe, and D. discoideum, the enzymatic activities of Dnmt2s are strongly stimulated by the pre-existing Q34 modification in the substrate tRNAAsp(GUC) (41,59). Furthermore, the mechanism of Q34 stimulation of m5C38 formation is evolutionarily conserved in mammals (60). Therefore, the m5C38 modification in tRNAAsp(GUC) modulated by the nutritionally determined Q34 formation is widespread in eukaryotes.
Based on the result of the present study, we showed that for mammalian tRNAVal(AAC), only when A34 is deaminized to form I34, could the m5C38 modification be formed by Dnmt2s, suggesting that I34 is a prerequisite for the formation of m5C38. In mammalian tRNAs, eight tRNAs are I34-modified (52). It is noteworthy that among the thirteen C38-containing tRNAs, tRNAVal(AAC), tRNALeu(AAG) and tRNAAla(AGC) are I34-modified (52). However, U32 instead of C32 is present on both tRNALeu(AAG) and tRNAAla(AGC), which exclude them from being recognized by hDNMT2.
Previous studies have shown that I34 could potentiate the wobble-pairing flexibility of the anti-codon, because I is able to pair with A, C or U (61). Considering that tRNAVal(AAC) needs to decipher both GUU and GUC codons of mRNA in mammals (52,55), further investigation is to determine whether the intricate I34 and m5C38 modifications play a role in decoding the non-cognate GUC codon is warranted.
Our results, together with those of previous studies, showed that the intricate network of modifications on site 34 and m5C38 exists at the ASL (anticodon stem and loop) region. First, I34 of tRNAVal(AAC) functions as the prerequisite for m5C38 formation by Dnmt2s in mammals. Second, Q34 in tRNAAsp(GUC) promotes the formation of the m5C38 modification in mammals (60), as well as in S. pombe, and D. discoideum (41,59). Third, in light of the formation process of Q34, the environmental conditions or nutrients could modulate the level of m5C38 in tRNAAsp(GUC) (62). These results indicated that the modulation of the 34th nucleotide during m5C38 formation is intricate and varies for different tRNA substrates. Intriguingly, the interplay among tRNA modifications are diverse and usually observed in the ASL region, especially between modifications at position 34 and 37 (63,64). The exact mechanism of how the 34th nucleotide modulates m5C38 formation and the associated biological effect require further study.
Dnmt2-mediated m5C modification on tsRNA
The Dnmt2-mediated m5C modification has a prominent role in the production of tsRNA (12,30). Dnmt2 could alter the sperm tsRNA expression profile, and mediate the transgenerational inheritance of paternal metabolic disorders to offspring through the m5C modification on sperm tsRNAs (16,32). Moreover, 5′ fragments of tRNAGly(GCC), which contain the Dnmt2-mediated m5C modification, were extremely abundant in mouse mature sperm (33,34). According to our results, the m5C38 modification catalyzed by Dnmt2 depends on the tertiary structure of tRNA. Therefore, we speculated that these m5C containing sperm tsRNAs might be generated from intact tRNA that carry the m5C38 modification. However, some sperm abundant tsRNAs, such as those derived from tRNAGly(CCC) and tRNAGlu(CUC), which are not the substrates of mammalian Dnmt2s, are also affected by knockout of Dnmt2 (33,34). Accordingly, it remains possible that Dnmt2 might regulate the production of m5C on tsRNA through other mechanisms, independent of its tRNA modification activity, which requires further study.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the Multi-Omics Facility in School of Life Science and Technology of ShanghaiTech University in Shanghai, and the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with the UPLC-MS/MS work.
Contributor Information
Zhi-Xuan Huang, State Key Laboratory of Molecular Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, 320 Yue Yang Road, Shanghai 200031, China; University of Chinese Academy of Sciences, Beijing 100039, China; School of Life Science and Technology, ShanghaiTech University, 393 Middle Hua Xia Road, Shanghai 201210, China.
Jing Li, School of Life Science and Technology, ShanghaiTech University, 393 Middle Hua Xia Road, Shanghai 201210, China.
Qing-Ping Xiong, State Key Laboratory of Molecular Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, 320 Yue Yang Road, Shanghai 200031, China; University of Chinese Academy of Sciences, Beijing 100039, China.
Hao Li, School of Life Science and Technology, ShanghaiTech University, 393 Middle Hua Xia Road, Shanghai 201210, China.
En-Duo Wang, State Key Laboratory of Molecular Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, 320 Yue Yang Road, Shanghai 200031, China; University of Chinese Academy of Sciences, Beijing 100039, China; School of Life Science and Technology, ShanghaiTech University, 393 Middle Hua Xia Road, Shanghai 201210, China.
Ru-Juan Liu, School of Life Science and Technology, ShanghaiTech University, 393 Middle Hua Xia Road, Shanghai 201210, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2017YFA0504000, 2020YFA0803401]; National Natural Science Foundation of China [91940302, 32022040, 31870811, 31770842, 32000919]; China Postdoctoral Science Foundation Grants [2020M671253]. Funding for open access charge: National Key Research and Development Program of China [2017YFA0504000, 2020YFA0803401]; National Natural Science Foundation of China [91940302, 31870811, 32022040, 31770842, 32000919]; China Postdoctoral Science Foundation Grants [2020M671253].
Conflict of interest statement. None declared.
REFERENCES
- 1. Trixl L., Lusser A.. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA. 2019; 10:e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeng Y., Chen T.. DNA methylation reprogramming during mammalian development. Genes (Basel). 2019; 10:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breiling A., Lyko F.. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenet. Chromatin. 2015; 8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schaefer M., Pollex T., Hanna K., Lyko F.. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009; 37:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helm M., Motorin Y.. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017; 18:275–291. [DOI] [PubMed] [Google Scholar]
- 6. Khoddami V., Cairns B.R.. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013; 31:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edelheit S., Schwartz S., Mumbach M.R., Wurtzel O., Sorek R.. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013; 9:e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T.. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012; 40:5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heissenberger C., Liendl L., Nagelreiter F., Gonskikh Y., Yang G., Stelzer E.M., Krammer T.L., Micutkova L., Vogt S., Kreil D.P.et al.. Loss of the ribosomal RNA methyltransferase NSun5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019; 47:11807–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W.et al.. 5-methylcytosine promotes mRNA export-NSun2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017; 27:606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J., Li H., Long T., Dong H., Wang E.D., Liu R.J.. Archaeal NSun6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019; 47:2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., Lyko F.. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010; 24:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khan M.A., Rafiq M.A., Noor A., Hussain S., Flores J.V., Rupp V., Vincent A.K., Malli R., Ali G., Khan F.S.et al.. Mutation in NSun2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012; 90:856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chellamuthu A., Gray S.G.. The RNA methyltransferase NSun2 and its potential roles in cancer. Cells. 2020; 9:1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris T., Marquez B., Suarez S., Schimenti J.. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol. Reprod. 2007; 77:376–382. [DOI] [PubMed] [Google Scholar]
- 16. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y.et al.. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016; 351:397–400. [DOI] [PubMed] [Google Scholar]
- 17. Bohnsack K.E., Höbartner C., Bohnsack M.T.. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019; 10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hermann A., Schmitt S., Jeltsch A.. The human DNMT2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 2003; 278:31717–31721. [DOI] [PubMed] [Google Scholar]
- 19. Kunert N., Marhold J., Stanke J., Stach D., Lyko F.. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003; 130:5083–5090. [DOI] [PubMed] [Google Scholar]
- 20. Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H.. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006; 311:395–398. [DOI] [PubMed] [Google Scholar]
- 21. Jurkowski T.P., Meusburger M., Phalke S., Helm M., Nellen W., Reuter G., Jeltsch A.. Human Dnmt2 methylates tRNAAsp molecules using a DNA methyltransferase-like catalytic mechanism. RNA (New York, N.Y.). 2008; 14:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeltsch A., Nellen W., Lyko F.. Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006; 31:306–308. [DOI] [PubMed] [Google Scholar]
- 23. Bujnicki J.M., Feder M., Ayres C.L., Redman K.L.. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004; 32:2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu R.J., Long T., Li J., Li H., Wang E.D.. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017; 45:6684–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Willbanks A., Wood S., Cheng J.X.. RNA Epigenetics: fine-tuning chromatin plasticity and transcriptional regulation, and the implications in human diseases. Genes (Basel). 2021; 12:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garber K. Epigenetics comes to RNA. Science. 2019; 365:16–17. [DOI] [PubMed] [Google Scholar]
- 27. Shanmugam R., Fierer J., Kaiser S., Helm M., Jurkowski T.P., Jeltsch A.. Cytosine methylation of tRNAAsp by Dnmt2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015; 1:15010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tuorto F., Herbst F., Alerasool N., Bender S., Popp O., Federico G., Reitter S., Liebers R., Stoecklin G., Gröne H.J.et al.. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015; 34:2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F.. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012; 19:900–905. [DOI] [PubMed] [Google Scholar]
- 30. Thiagarajan D., Dev R.R., Khosla S.. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011; 6:103–113. [DOI] [PubMed] [Google Scholar]
- 31. Dev R.R., Ganji R., Singh S.P., Mahalingam S., Banerjee S., Khosla S.. Cytosine methylation by Dnmt2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem. J. 2017; 474:2009–2026. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y.F., Zhang X.D., Shi J.C., Tuorto F., Li X., Liu Y.S., Liebers R., Zhang L.W., Qu Y.C., Qian J.J.et al.. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 2018; 20:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng H., Shi J., Zhang Y., Zhang H., Liao S., Li W., Lei L., Han C., Ning L., Cao Y.et al.. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012; 22:1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F.et al.. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016; 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rai K., Chidester S., Zavala C.V., Manos E.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R.. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Gene Dev. 2007; 21:261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Durdevic Z., Hanna K., Gold B., Pollex T., Cherry S., Lyko F., Schaefer M.. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013; 14:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewinska A., Adamczyk-Grochala J., Kwasniewicz E., Wnuk M.. Downregulation of methyltransferase Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J. Cell Physiol. 2017; 232:3714–3726. [DOI] [PubMed] [Google Scholar]
- 38. Lewinska A., Adamczyk-Grochala J., Kwasniewicz E., Deregowska A., Semik E., Zabek T., Wnuk M.. Reduced levels of methyltransferase Dnmt2 sensitize human fibroblasts to oxidative stress and DNA damage that is accompanied by changes in proliferation-related miRNA expression. Redox Biol. 2018; 14:20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elhardt W., Shanmugam R., Jurkowski T.P., Jeltsch A.. Somatic cancer mutations in the Dnmt2 tRNA methyltransferase alter its catalytic properties. Biochimie. 2015; 112:66–72. [DOI] [PubMed] [Google Scholar]
- 40. Jurkowski T.P., Jeltsch A.. On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2. PLoS One. 2011; 6:e28104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Becker M., Müller S., Nellen W., Jurkowski T.P., Jeltsch A., Ehrenhofer-Murray A.E.. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012; 40:11648–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Müller S., Windhof I.M., Maximov V., Jurkowski T., Jeltsch A., Förstner K.U., Sharma C.M., Gräf R., Nellen W.. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res. 2013; 41:8615–8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shanmugam R., Aklujkar M., Schäfer M., Reinhardt R., Nickel O., Reuter G., Lovley D.R., Ehrenhofer-Murray A., Nellen W., Ankri S.et al.. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res. 2014; 42:6487–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Torres A.G., Rodriguez-Escriba M., Marcet-Houben M., Santos Vieira H.G., Camacho N., Catena H., Murillo Recio M., Rafels-Ybern A., Reina O., Torres F.M.et al.. Human tRNAs with inosine 34 are essential to efficiently translate eukarya-specific low-complexity proteins. Nucleic Acids Res. 2021; 49:7011–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gerber A.P., Keller W.. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999; 286:1146–1149. [DOI] [PubMed] [Google Scholar]
- 46. Gill S.C., Hippel P.H.. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989; 182:319–326. [DOI] [PubMed] [Google Scholar]
- 47. Li Y., Chen J.F., Wang E.D., Wang Y.L.. T7 RNA polymerase transcription of Escherichia coli isoacceptors tRNA(Leu). Sci. China Life Sci. 1999; 42:185–190. [DOI] [PubMed] [Google Scholar]
- 48. Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007; 35:W43–W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang Q., Yao P., Eriani G., Wang E.D.. In vivo identification of essential nucleotides in tRNALeu to its functions by using a constructed yeast tRNALeu knockout strain. Nucleic Acids Res. 2012; 40:10463–10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J., Wang Y.N., Xu B.S., Liu Y.P., Zhou M., Long T., Li H., Dong H., Nie Y., Chen P.R.et al.. Intellectual disability-associated gene ftsj1 is responsible for 2′-O-methylation of specific tRNAs. EMBO Rep. 2020; 21:e50095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013; 13:659–662. [DOI] [PubMed] [Google Scholar]
- 52. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A.et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gerber A.P., Keller W.. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999; 286:1146–1149. [DOI] [PubMed] [Google Scholar]
- 54. Torres A.G., Piñeyro D., Rodríguez-Escribà M., Camacho N., Reina O., Saint-Léger A., Filonava L., Batlle E., Ribas de Pouplana L.. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015; 43:5145–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grosjean H., Edqvist J., Straby K.B., Giege R.. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol. 1996; 255:67–85. [DOI] [PubMed] [Google Scholar]
- 57. Li H., Zhu D., Wu J., Ma Y., Cai C., Chen Y., Qin M., Dai H.. New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1. RNA Biol. 2021; 18:2531–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fergus C., Barnes D., Alqasem M.A., Kelly V.P.. The queuine micronutrient: charting a course from microbe to man. Nutrients. 2015; 7:2897–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Muller M., Hartmann M., Schuster I., Bender S., Thuring K.L., Helm M., Katze J.R., Nellen W., Lyko F., Ehrenhofer-Murray A.E.. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015; 43:10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tuorto F., Legrand C., Cirzi C., Federico G., Liebers R., Muller M., Ehrenhofer-Murray A.E., Dittmar G., Grone H.J., Lyko F.. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018; 37:e99777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Crick F.H. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966; 19:548–555. [DOI] [PubMed] [Google Scholar]
- 62. Ehrenhofer-Murray A.E. Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules. 2017; 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Han L., Phizicky E.M.. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018; 24:1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li J., Zhu W.Y., Yang W.Q., Li C.T., Liu R.J.. The occurrence order and cross-talk of different tRNA modifications. Sci. China Life Sci. 2021; 64:1423–1436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.