Abstract
Pe poke is a naturally fermented sticky soybean food of Myanmar. The present study was aimed to profile the whole microbial community structure and their predictive gene functionality of pe poke samples prepared in different fermentation periods viz. 3 day (3ds), 4 days (4ds), 5 days (5ds) and sun-dried sample (Sds). The pH of samples was 7.6 to 8.7, microbial load was 2.1–3.9 x 108 cfu/g with dynamic viscosity of 4.0±1.0 to 8.0±1.0cP. Metataxonomic profile of pe poke samples showed different domains viz. bacteria (99.08%), viruses (0.65%), eukaryota (0.08%), archaea (0.03%) and unclassified sequences (0.16%). Firmicutes (63.78%) was the most abundant phylum followed by Proteobacteria (29.54%) and Bacteroidetes (5.44%). Bacillus thermoamylovorans was significantly abundant in 3ds and 4ds (p<0.05); Ignatzschineria larvae was significantly abundant in 5ds (p<0.05), whereas, Bacillus subtilis was significantly abundant in Sds (p <0.05). A total of 172 species of Bacillus was detected. In minor abundance, the existence of bacteriophages, archaea, and eukaryotes were also detected. Alpha diversity analysis showed the highest Simpson’s diversity index in Sds comparable to other samples. Similarly, a non-parametric Shannon’s diversity index was also highest in Sds. Good’s coverage of 0.99 was observed in all samples. Beta diversity analysis using PCoA showed no significant clustering. Several species were shared between samples and many species were unique to each sample. In KEGG database, a total number of 33 super-pathways and 173 metabolic sub-pathways were annotated from the metagenomic Open Reading Frames. Predictive functional features of pe poke metagenome revealed the genes for the synthesis and metabolism of wide range of bioactive compounds including various essential amino acids, different vitamins, and enzymes. Spearman’s correlation was inferred between the abundant species and functional features.
Introduction
The community-specific ethnic fermented foods have been centre of interest for their unique gastronomy as well as colossal microbial diversity [1]. Myanmar has several ethnic fermented foods and beverages including fermented soybeans, which have been traditionally prepared and consumed by more than 135 different ethnic communities [2]. Among fermented foods, traditional methods of fermentation of locally grown soybean is an ancient practice mostly seen in North-western regions of Myanmar bordering with North East states of India and North Western parts of Myanmar bordering with Northern Thailand. Both mould-fermented and bacterial-fermented soybean are prepared and consumed widely in Myanmar [3]. Pe poke is an ethnic fermented soybean food of northern Myanmar. There is no historical documentation of origin of pe poke in Myanmar, however, it is believed that soybean has been introduced to Myanmar from Yunnan province of China [4]. During traditional method of preparation of pe poke, soybeans are soaked in water overnight, dewatered, boiled and wrapped in leaves, and are kept in a warm place for natural fermentation of 3–5 days (Fig 1a and 1b). Sometimes, freshly prepared pe poke is mashed with addition of salt and hot pepper, shaped as flat wafers, and are sun dried (Fig 1c). Some people prefer to eat pe poke immediately after fermentation and make into a typical Burmese-style cuisine as a side dish (Fig 1d) and fried fritters (Fig 1e) with boiled rice in main meal. This is mostly observed in the North-western regions of Myanmar bordering with India, where similar types of sticky fermented soybean foods are prepared such as hawaijar in Manipur, bekang in Mizoram, peruyaan and peron namsing in Arunachal Pradesh and axone or aakhone in Nagaland states of India [5]. Whereas, in the North-eastern regions of Myanmar bordering with Thailand, freshly prepared pe poke is made into flat wafers, and are sun dried, which is similar to thua nao of Northern Thailand [6]. Pe poke is one of the delicacies in the diets of ethnic people of Myanmar, however, the consumption of pe poke among younger generation is declining. Traditionally prepared pe poke is sold in local markets by marginal farmers in many regions of Myanmar. Pe poke is similar to other sticky fermented soybean foods of Asia such as kinema of India, Nepal and Bhutan, natto of Japan, thua nao of Thailand, cheonggukjang of Korea, douchi of Yunnan province of China and sieng of Laos [7, 8].
Fig 1. Traditional method of preparation of pe poke in Myanmar, (a) Boiling of soybeans; (b) freshly fermented pe poke; (c) sun-dried wafer-shaped pe poke; (d) pe poke curry; and (e) fried pe poke fritters.
Though pe poke is a popular ethnic food in Burmese gastronomy, but information on microbiology and nutritional aspects of pe poke is very rare, except few reports on Bacillus subtilis as the main fermenting bacterium in pe poke [9, 10]. It is necessary to understand the microbial community structure in pe poke, which is prepared by natural fermentation, moreover, such rare ethnic product has not been studied in details to profile its microbial community structure. We choose the shotgun metagenome sequence tool to profile the entire microbial community up to species, which is considered as one of the most reliable metataxonomic tools [11], that may sequence the genomes of untargeted cells in a microbial community to decode community structures including culturable and unculturable bacteria, yeasts, fungi, virus and archaea in food samples [12]. Hence, we aimed to study the metataxonomic of abundant domains in naturally fermented pe poke of Myanmar, prepared in different fermentation periods, by shotgun metagenomic sequencing method, supported by machine learning tools. Functional profiles of metagenomes were also predicted using the SqueezeMeta pipeline [13] and KEGG database [14]. We believe this is the first report of microbial community structures in naturally fermented pe poke by shotgun metagenome sequence tool.
Materials and methods
Sample collection and analysis of pH
Samples of pe poke, traditionally prepared in different fermentation periods viz. 3 days (3ds), 4 days (4ds), 5 days (5ds) and sun-dried sample (Sds) were collected from Pyinnolwin village in Mandalay state of Myanmar. Samples were collected in pre-sterile containers kept in ice-box carriers and transported to the Department of Industrial Chemistry, University of Mandalay and stored at 4°C. All samples were kept in ice-box carrier, by feeling with fresh ice in every 5–6 hours, till we reached to the Department of Microbiology, Sikkim University, Gangtok, India for immediate microbiological analysis. The pH of pe poke samples was determined by homogenizing 1 g of sample in physiological saline (0.85% sodium chloride, NaCl) and was measured using digital pH-meter (Orion 910003, Thermo Fisher Scientific, USA).
Total viable count
Samples were coarsely crushed by a sterile spatula, and ten grams of the sample were homogenized with 90 mL of 0.85% physiological saline in a stomacher lab blender 40 (Seward, United Kingdom) for 5 min. The homogenized samples were serially diluted in the same diluents, and 1 mL of appropriate diluents was plated in plate count agar (M091S, HiMedia, India) using pour plate method and incubated at 37°C for 24 h. The number of colonies was counted as colony forming unit (cfu/g).
Measurement of viscosity
The dynamic viscosity of pe poke samples was determined using the method described by Ratha and Jhon [15]. Thirty grams of samples were mixed with 30 mL of distilled water and subjected to vigorous shaking in a conical flask (250mL) for 30 min. The slimy part was collected and 30 mL of its aliquot (100 rpm at 20°C) was measured for dynamic viscosity in centipoise (cP) using a viscometer (DV1MRVTJ0, Brookfield AMETEK, MA, USA). The experiment was done in triplicate sets.
Genomic DNA extraction
Ten grams of coarsely crushed samples of pe poke were homogenised in Stomacher (400 Circulator, Seward, UK) with 90 mL of sterile 0.1 M phosphate buffer saline (pH 6.4) for 5 min. After homogenization, the homogenate was filtered and the filtrate was used for the extraction of genomic DNA using the Nucleospin® Food DNA kit (MACHEREY-NAGEL GmbH & Co. KG, Duran, Germany) as per the manufacturer’s protocol. Concentration of DNA was then quantified using spectrophotometer (Eppendorf, USA). The quality of DNA was checked in 0.8% agarose gel electrophoresis and visualized using Gel Doc EZ imager (BioRad, USA).
Metagenomics sequencing and library preparation
Pe poke metagenome library preparation for long reads sequencing was performed by following the method of Sevim et al. [16]. The 10 μg of DNA was used to create the ONT (Oxford Nanopore Technologies) library. The generated DNA fragments was sheared using Covaris g-tubes (Covaris Inc., Woburn, MA USA) and DNA was repaired using NEBNext FFPE (Formalin-Fixed, Paraffin-Embedded) Repair Mix (New England BioLabs, Ipswich, MA USA) according to the manufacturer’s instructions. AMPure XP beads (62 μl) were added to the FFPE-repair reaction and incubated at room temperature for 30 min on a Hula mixer, followed by two washes with 70% ethanol. Beads were then resuspended with 93 μl of nuclease free water and incubated for 30 min at room temperature on a Hula mixer; 90 μl of the eluate was then transferred to a clean 1.5 mL Eppendorf tube.
The fragmented and repaired DNA underwent end repair and A-tailing using the NEBNExt End Repair/dA-Tailing Module (New England BioLabs) following manufacturer’s protocol: The reaction volume was doubled to 120 μl, incubation was performed at 20°C for 20 min and at 65°C for 20 min. AMPure XP beads (120 μl) were added to the end-prep reaction and incubated for 30 min at room temperature on a Hula mixer, followed by two washes with 70% ethanol. Beads were then resuspended in 31 μl of nuclease free water and incubated for 30 min at room temperature on a Hula mixer; 61 μl of the eluate was then transferred to a clean 1.5 mL Eppendorf tube. The resulting DNA was quantified using the Qubit HS DNA kit.
The resulting DNA ligation and clean-up were performed using the SQK-LSK108 kit (Oxford Nanopore Technologies, Oxford, United Kingdom) following manufacturer’s instructions. The ligation reaction was incubated at room temperature for 10 min and then overnight at 4°C. The ligated samples were purified using 40 μl of AMPure XP beads, incubated for 30 min at room temperature on a Hula mixer followed by two washes using the kit-provided wash buffer. The beads were resuspended in 15 μl of the kit-provided elution buffer and then incubated for 30 min at room temperature on a Hula mixer; 15 μl of the eluate was then transferred to a clean 1.5 mL tube and quantified using the Qubit HS DNA kit. The library was then sequenced on a MinION using the R9 flow cell sequencing chemistry and were processed using the MinKNOW software (v1.13.1).
Bioinformatics analysis
Metataxonomic
Raw data derived from MinION (TM) ONT (Oxford Nanopore Technologies) in fast5 format was converted into a fastq files using poretools v0.6.0 for the bioinformatics analysis of pe poke metagenome [17]. After conversion, the quality of the fastq files were then examined using NanoPlot [18] and generated the corrected-assembled data via canu-assembler [19]. A database derived from GenBank containing millions of protein sequences from bacteria, archaea, viruses, fungi, and other microbial eukaryotes was downloaded within Kaiju via kaiju-makedb -s nr_euk. Taxonomy assignment of the assembled quality sequences was performed using a taxonomical pipeline, Kaiju [20] in which a default “greedy algorithm” was used to map the sequences against the database [21]. A cut-off for a minimum required match length (m = 11, default), minimum match score of 80 (s = 80) and the E-value (E = 0.05) was set to filter the mismatches. Filtering of query sequences containing low-complexity regions was performed to avoid false positive taxon assignments that may cause by bogus matches or other sequencing noises [20]. Amino acid substitution model was performed with a total score for each match calculated as in amino acid sequence alignment and ranked a multiple match and taxon classification from the database. After translation of ORFs into a set of amino acid fragments, we ranked the fragments by their BLOSUM62 (BLOcks SUbstitution Matrix) score and start the database search with the highest scoring fragment [22]. The fragments were searched backwards via BWT (Burrows-Wheeler transform) algorithm against database [23] and the higher score of fragments in the search was used for classifying the reads and outputs the taxon identifier [20].
Predictive functional features
Predictive functional features of the metagenome was performed on Quality-filtered contigs using the SqueezeMeta pipeline version 1.3.0 [13]. After importing of data, the contigs of <500bp were removed using prinseq [24], followed by gene prediction of the assembled using Prodigal (v2.6.2) [25] and the predicted genes were searched for homologies against the functional databases using DIAMOND, computational tool for the alignment of sequencing reads against a protein reference database [26]. After running the DIAMOND, the method assigned as functions to each Open Reading Frames (ORF) was carried out using the fun3 method (fun3 method produced functional assignments to compare genes sequences against the functional database) for Clusters of Orthologous Groups/Non-supervised Orthologous Groups (COGs/NOGs) using evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) database [27] and Kyoto Encyclopedia of Genes and Genomes (KEGG) database [14]. In the process of analysis, the highest-scoring ORFs in the contig with an exceeding of 30% (default) were considered for annotation [13]. Best hits gene annotations were further processed for pathways prediction and enzyme classification [28]. Metabolic pathways assigned against the KEGG database was categorised in three level: high-level function (Level 1), lower-level function (Level 2) and the sub-pathways (Level-3) [29]. Enzymes involved in lysine biosynthesis, alanine, aspartate, glutamate, glycine, serine, threonine metabolism, pentose phosphate pathways, galactose metabolism and phosphotransferase system were mapped against the KEGG pathways database [30].
Statistical analysis
Inter species diversity
Significance among the abundant species (>1%) was calculated using Fisher exact test [31]. Inter species diversity of pe poke metagenome was performed among the samples (3ds, 4ds, 5ds and Sds) using Tukey’s test in IBM SPSS v20.0 [32]. Shared and unique species was calculated using InteractiVenn: a web-based tool for the analysis [33].
Alpha and beta diversity
Differences of species distribution among the samples was measured using diversity indices, Simpson and Shannon diversity index was calculated, and principal coordinate analysis (PCoA) was plotted based on Bray-Curtis dissimilarities using PASTv4 (Paleontological Statistics Software Package) [34]. Furthermore, UPGMA (Unweighted Pair Group Method with Arithmetic mean) hierarchical clustering was performed for similarity analysis based on microbial communities which was compared between the samples and support the result observed in beta diversity [35].
Predictive functional features
The predictive functional profiles of pe poke were tested using Tukey’s test to check the inter pathways distribution among the samples (3ds, 4ds, 5ds and Sds) using IBM SPSS v20.0 [32]. UPGMA hierarchical clustering was also performed to compare the functional distribution among the samples [35]. Heatmap visualization of functional profiles, level-1 and level-2 was carried out using a web tool: ClustVis [36]. Correlation between the major species and functional features was performed by a non-parametric Spearman’s rank correlation using IBM SPSS v20.0 (Statistical Package for the Social Sciences), and a network-based visualization was generated using MetScape v3.1.3 in Cytoscape v3.8.2.
Results
The pH of pe poke samples was 7.6 to 8.7 with the microbial load of 2.1–3.9 x 108 cfu/g. The dynamic viscosity of samples was 4.0±1.0 to 8.0±1.0 cP (centipoise).
Microbial community
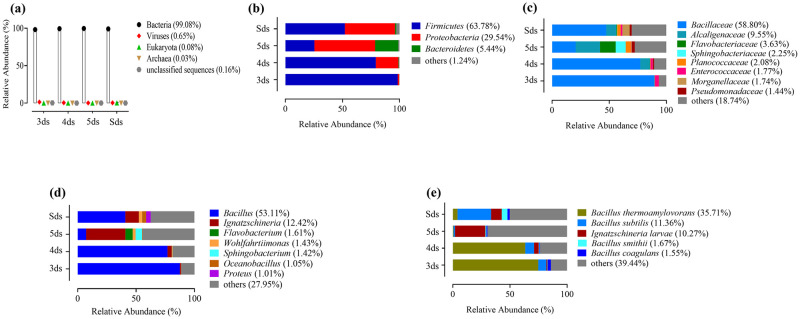
A total of 1085311 reads were obtained from all samples of pe poke with an average of 271327.7 reads per sample. Average length of the reads was found 901.5. Total number of bases recovered from the samples were 314,801,023 bases for 3ds, 176,709,193 bases for 4ds, 372,945,882 bases for 5ds and 41,455,352 bases for Sds, respectively. Shotgun metagenomic sequence analysis of pe poke metagenome showed different domains viz., bacteria, archaea, viruses and eukaryotes with 46 phyla, 328 families, 718 genera and 1475 species. Taxonomic classification at domain level revealed the abundance of bacteria (99.08%) followed by viruses (0.65%), eukaryota (0.08%), archaea (0.03%) and unclassified sequences (0.16%) (Fig 2a). At bacterial phylum level, Firmicutes was the most abundant phylum followed by Proteobacteria, Bacteroidetes and others (1.24%) (Fig 2b) including phyla with a relative abundance of <1% (S1 Table). Bacillaceae was the most abundant family followed by Alcaligenaceae, Flavobacteriaceae, Sphingobacteriaceae, Planococcaceae, Enterococcaceae, Morganellaceae, Pseudomonadaceae (Fig 2c) and others detected at <1% abundance (S2 Table). Taxonomic annotation revealed the abundance of Bacillus (53.11%) at genus level followed by Ignatzschineria, Flavobacterium, Wohlfahrtiimonas, Sphingobacterium, Oceanobacillus, Proteus (Fig 2d) and others detected at <1% abundance (S3 Table). At species level, Bacillus thermoamylovorans was the most abundant species in pe poke samples, followed by B. subtilis, Ignatzschineria larvae, B. smithii, B. coagulans (Fig 2e) and others detected at <1% abundance (S4 Table). No phylum, family, genus and species with a relative abundance of >1% were detected from other domains viz. archaea, viruses and eukaryote in pe poke metagenomes.
Fig 2. Relative abundance of microbial communities in pe poke (a) domain, (b) phyla, (c) family, (d) genera and (e) species.
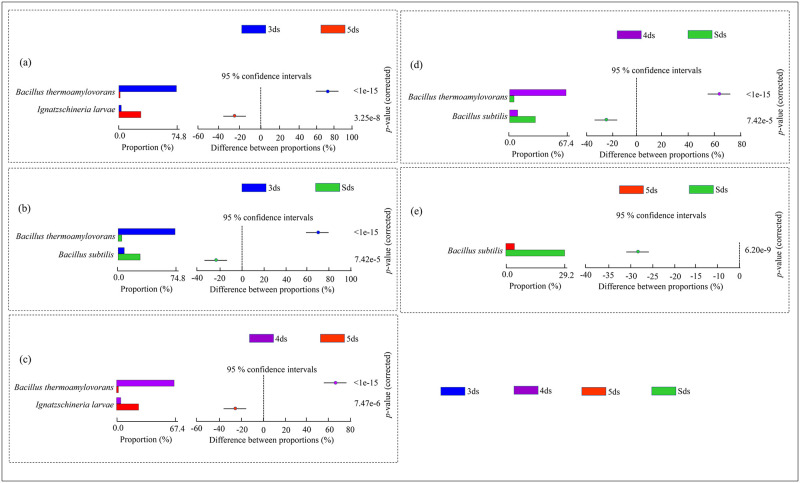
Firmicutes was found abundant phylum in samples of 3ds, 4ds and Sds, whereas Proteobacteria was observed abundant in 5ds. In comparison among 3ds, 4ds and Sds, Bacillus thermoamylovorans was significantly (p<0.05) abundant in 3ds (Fig 3a and 3b) and Ignatzschineria larvae was significantly (p<0.05) abundant in 5ds (Fig 3a); whereas, B. subtilis was significantly (p<0.05) abundant in Sds (Fig 3b). Similarly, when compared among 4ds, 5ds and Sds, B. thermoamylovorans was significantly (p<0.05) abundant in the 4ds (Fig 3c and 3d), Ignatzschineria larvae in 5ds (Fig 3c) and Bacillus subtilis was significantly (p<0.05) abundant in Sds (Fig 3d and 3e). In pe poke metagenomic analysis, a total of 172 species of Bacillus were detected, out of which the abundant species with a relative abundance of >1% were B. thermoamylovorans, B. subtilis, B. smithii and B. coagulans (S5 Table). Besides Bacillus, lactic acid bacteria (LAB) were also detected with a cumulative abundance of 1.78%, which included 94 species (S6 Table). Apart from bacterial domain, taxonomic annotation also revealed 16 species of archaea (S7 Table), 31 species of eukaryotes i.e., 3 species of yeasts, 15 species of filamentous moulds and 13 species of other eukaryotes (S8 Table), and 33 species of virus (bacteriophages), out of which 21 species were Bacillus phages (S9 Table).
Fig 3. Fisher exact test performed at species level to observe the significant differences and similarities of predominant species comparing between the samples.
Diversity indices
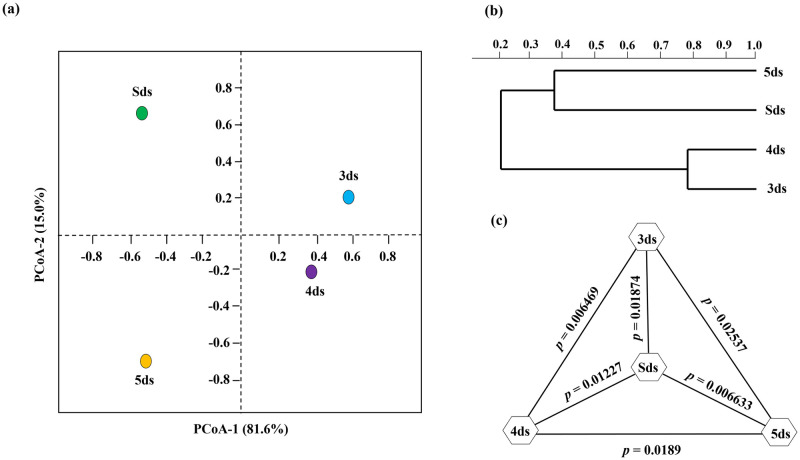
Alpha diversity analysis showed the highest Simpson’s diversity index in Sds comparable to other samples (Table 1). Similarly, a non-parametric Shannon’s diversity index was also highest in Sds (Table 1). Good’s coverage of 0.99 was observed in all the samples. Beta diversity analysis using PCoA (Fig 4a) and UPGMA (Fig 4b) showed no significant clustering. Statistically, in term of species abundance, the inter species diversity among the samples was calculated using Tukey’s test (Fig 4c) and a significant difference was observed between 3ds and 4ds (p = 0.006469), 5ds (p = 0.02537) and Sds (p = 0.01874). Similarly, 4ds was significantly different from 5ds (p = 0.0189) and Sds (p = 0.01227), and 5ds was significantly different from Sds (p = 0.006633).
Table 1. Indices of alpha diversity among the four samples of pe poke.
| Indices | Samples | |||
|---|---|---|---|---|
| 3ds | 4ds | 5ds | Sds | |
| Simpson (1-D) | 0.43 | 0.62 | 0.86 | 0.91 |
| Shannon | 2.25 | 3.14 | 5.46 | 5.99 |
| Goods Coverage | 0.99 | 0.99 | 0.99 | 0.99 |
Fig 4.
(a) PCoA of pe poke metagenomes based on microbial species. (b) UPGMA showed no clustering between samples, and (c) one-way ANOVA Tukey’s test revealed the significant differences among samples (p <0.05).
Shared and unique species
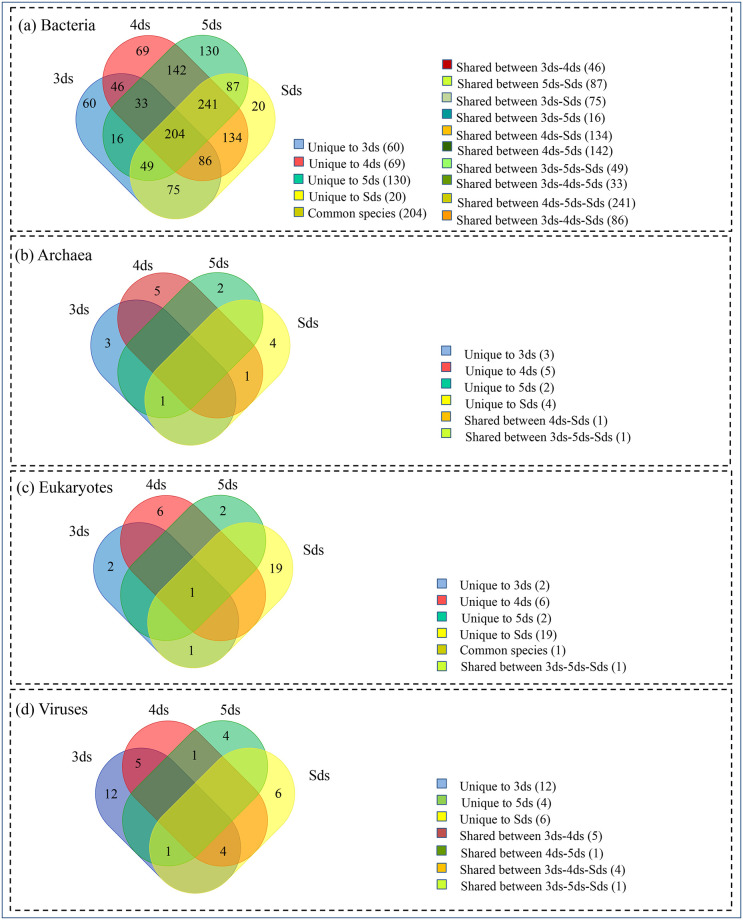
Metataxonomic annotation of pe poke metagenome revealed a huge diversity of microbial communities including shared and unique species (Fig 5a–5d). Based on different domains that have been classified via taxonomic classification, we observed about 204 bacterial core species were common in all the samples (Fig 1a). Additionally, the unique species were 60, 69, 130 and 20 in 3ds, 4ds, 5ds and Sds, respectively (S10 Table). Among archaeal species, no core species was found common to all samples. Methanobacterium formicicum was shared between 3ds, 5ds and Sds, and Halapricum salinum was shared between 4ds and Sds (Fig 1b). The unique species were 3, 5, 2 and 4 in 3ds, 4ds, 5ds and Sds, respectively (S11 Table). Among eukaryota, Mucor ambiguus was the core species commonly found in all samples. Batrachochytrium dendrobatidis shared between 3ds 5ds and Sds (Fig 1c) and the unique species were 2, 6, 2, 19 in the 3ds, 4ds, 5ds and Sds, respectively (S12 Table). In the category of viruses, no common species was observed. Aeribacillus phage AP45 was shared between 4ds and 5ds, and Geobacillus virus E3 was shared between 3ds, 5ds and Sds (Fig 5d). The unique species were 12, 4, 6 in 3ds, 5ds and Sds, respectively (S13 Table).
Fig 5. Shared, unique and common species in pe poke metagenome represented by InteractiVenn.
Predictive functional features
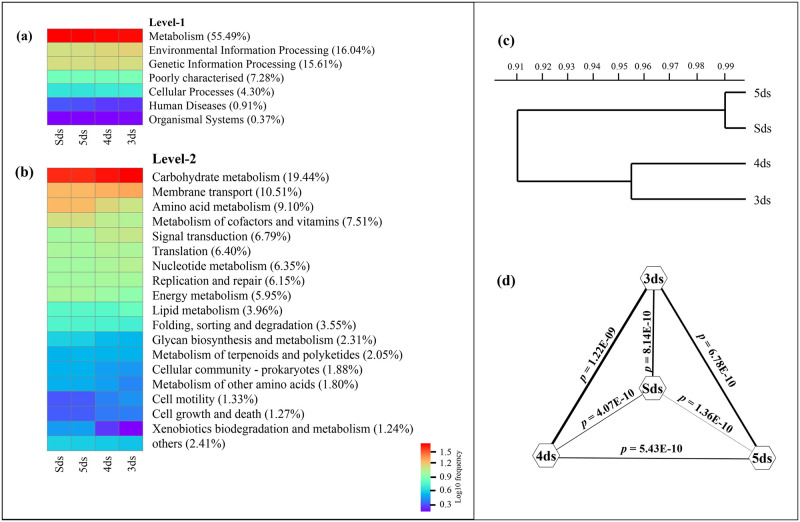
The mapping of metagenomic sequences against the databases of orthologous gene groups (COG and KO) revealed many enriched functional features. About 56% were assigned to COG functional genes and the remaining 44% ORFs were assigned to KEGG functional pathways. In COG annotation, general function prediction only was the abundant followed by DNA replication, recombination and repair, amino acid transport and metabolism, carbohydrate transport and metabolism, transcription, translation, ribosomal structure and biogenesis, inorganic ion transport and metabolism, energy production and conversion, cell envelope biogenesis, outer membrane (S14 Table). In KEGG database, a total number of 33 super-pathways and 173 metabolic sub-pathways were annotated from the metagenomic ORFs. At KO level-1, metabolism was the most abundant followed by environmental information processing, genetic information processing, cellular processes, human diseases, organismal systems and poorly characterised (Fig 6a). At KO level—2, the abundant functional prediction was carbohydrate metabolism followed by other metabolisms (Fig 6b) and super-pathways with relative abundance of <1% mapped against KEGG (S15 Table). Furthermore, at KO level-3, super-pathways with relative abundance of <1% mapped against KEGG showed genes related to ABC transporters was the most abundant followed by other predictive metabolic pathways (S16 Table). Based on the distribution of functional features, no clustering of samples was observed by performing the UPGMA analysis (Fig 6c). Tukey’s test was performed to check the significant differences of functional features between the samples (Fig 6d).
Fig 6.
The abundant functional features in pe poke metagenome (a) Level-1 and (b) Level-2 with a relative abundance more than 1%. (c) UPGMA showed no clustering between samples and (d) significant differences among samples observed using one-way ANOVA Tukey’s test.
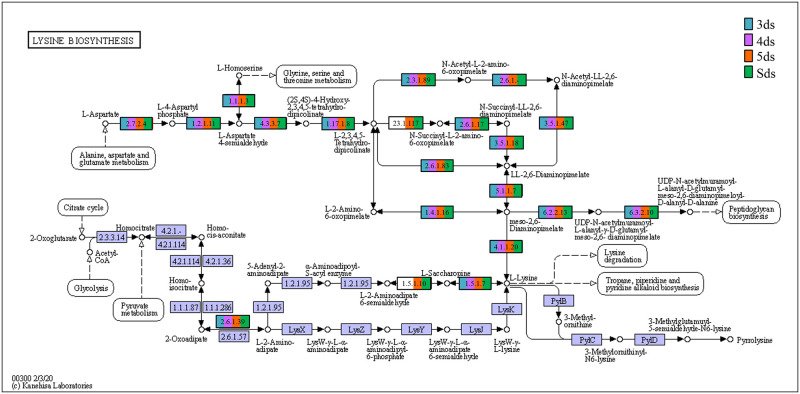
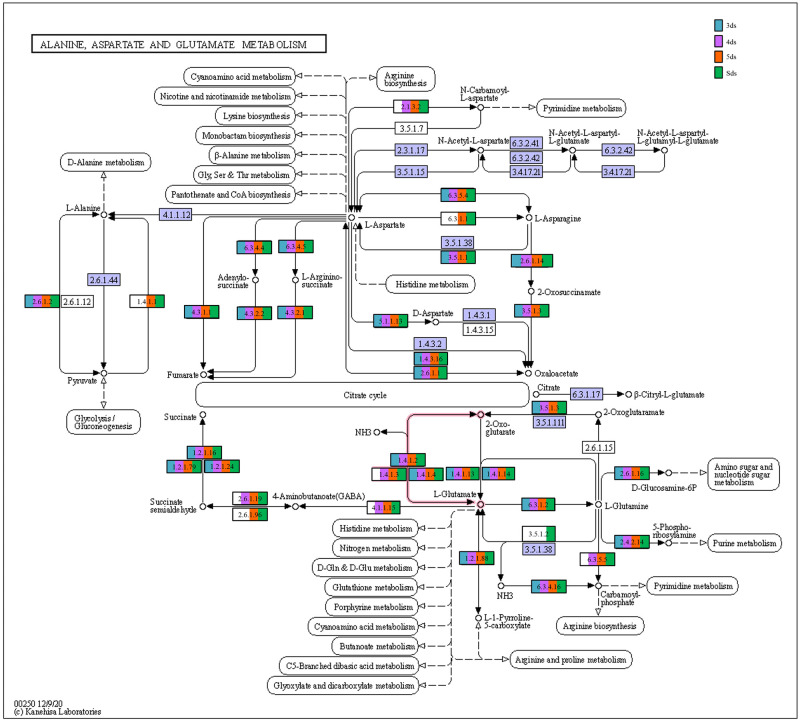
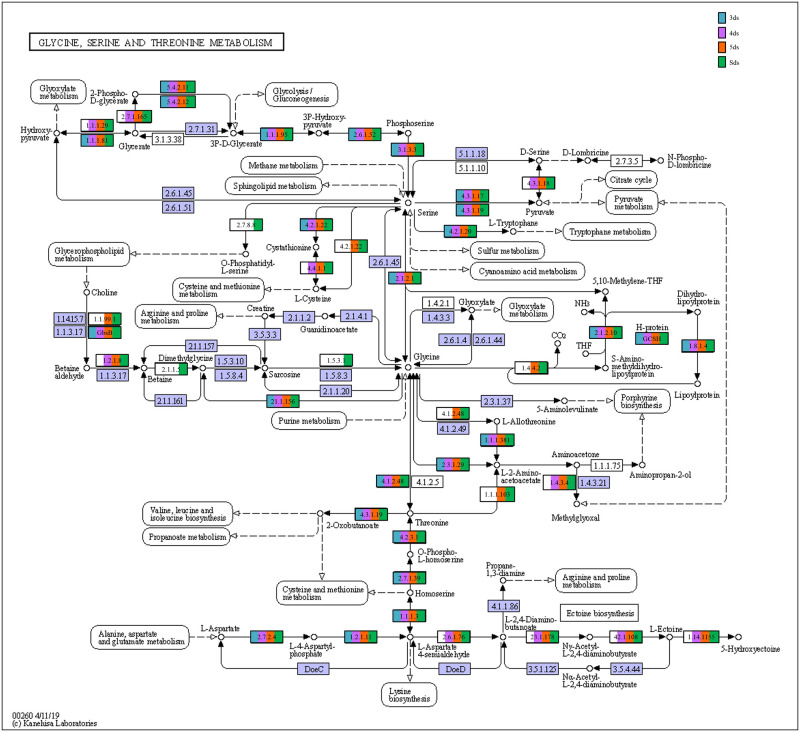
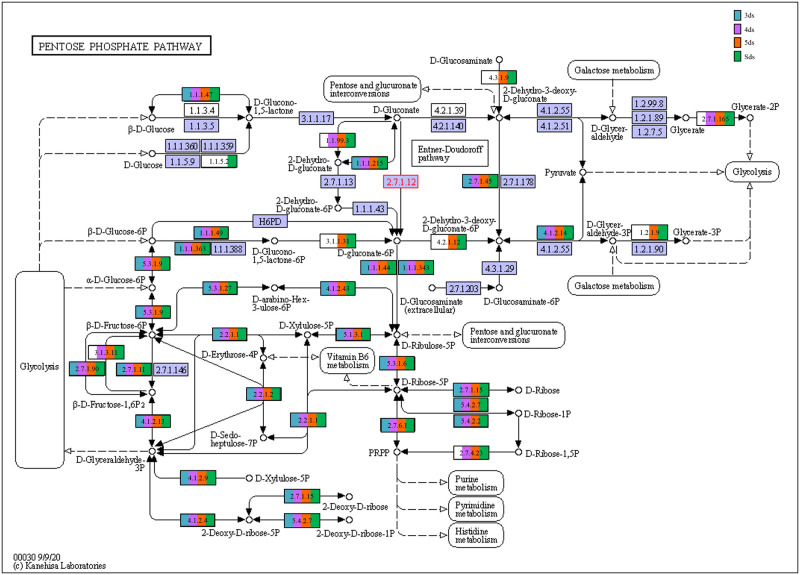
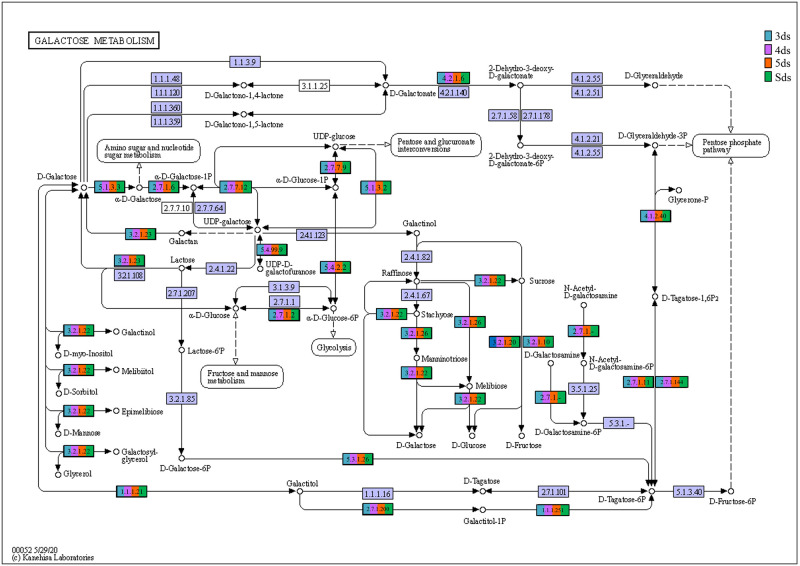
In enzyme classification, we detected genes encoding for enzymatic activity such as protease, serine protease, amylase, lipase, galactosidase, glucosidase, glutamate decarboxylase and functions involve in poly-γ-glutamic acid biosynthesis (S17 Table). Predictive metabolic pathways were mapped against the KEGG pathways database such as lysine biosynthesis (Fig 7), alanine, aspartate and glutamate metabolism (Fig 8), glycine, serine and threonine metabolism (Fig 9), pentose phosphate pathways (Fig 10) and galactose metabolism (Fig 11). The predictive enzymes involved in different pathways were observed such as lysine metabolism (S18a Table), alanine, aspartate and glutamate metabolism and 4-aminobutanoic acid (γ-aminobutyric acid, or GABA) (S18b Table), glycine, serine, threonine metabolism and ectoine biosynthesis (S18c Table), pentose phosphate pathways (S18d Table) and galactose metabolism (S18e Table).
Fig 7. Enzymes involve in lysine biosynthesis detected in pe poke metagenome.
Fig 8. Enzymes involved in alanine, aspartate and glutamate metabolism and 4-aminobutanoic acid (γ-aminobutyric acid, or GABA) detected in pe poke metagenome.
Fig 9. Enzymes involved in glycine, serine and threonine metabolism and ectoine biosynthesis detected in pe poke metagenome.
Fig 10. Enzymes involved in pentose phosphate pathways.
Fig 11. Enzymes involved in galactose metabolism.
Correlation between predominant species and predictive functions
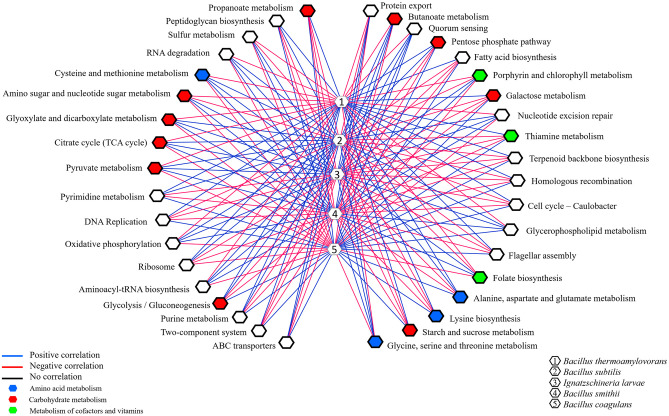
Spearman’s correlation was inferred between the abundant species and functional features (Fig 12). Bacillus thermoamylovorans, B. subtilis, B. smithii and B. coagulans were positively correlated with alanine, aspartate and glutamate metabolism, pentose phosphate pathway, and glycolysis/gluconeogenesis. Lysine biosynthesis and galactose metabolism were positively correlated with B. thermoamylovorans and B. coagulans, whereas negatively correlated with B. subtilis and B. smithii. Glycine, serine and threonine metabolism and cysteine and methionine metabolism was positively correlated with B. subtilis and B. smithii, and negatively correlated with B. thermoamylovorans and B. coagulans. Among metabolism of cofactors and vitamins, B. thermoamylovorans and B. coagulans were positively correlated with thiamine metabolism; B. smithii with porphyrin and chlorophyll metabolism and B. subtilis with folate biosynthesis (Fig 12).
Fig 12. Spearman-s correlation was performed between the predominant species and functional features that has a relative abundance >1% using IBM SPSS (Statistical Package for the Social Sciences) Statistics v.20 and represented via correlation-based network.
Discussion
Microbial community
Pe poke is an alkaline (pH 7.8–8.7) naturally fermented soybean food of Myanmar, which is prepared traditionally by ethnic Burmese people. Though pe poke is considered as a sticky fermented soybean food, however, the dynamic viscosity of samples was 8.0±1.0cP as compared with that of natto, a highly sticky Japanese fermented soybean with dynamic viscosity of >23cP [37]. The microbiological population of pe poke, as determined by cultural method, showed a viable load of 108 cfu/g, indicating its richness in microbial diversity. Since fermentation periods during natural fermentation of pe poke vary from 3 to 5 days, we collected the samples fermented for 3 days, 4 days and 5 days, and also the sun-dried samples for profiling the microbial community using the shotgun metagenome sequence tool to know the abundant microbial domains with their predictive functional features. Bacteria were detected as the most abundant domain, and the least abundant domains were archaea, eukaryotes and viruses, which reflects the comprehensive general picture of the microbial communities of pe poke. The higher abundance of Firmicutes and the presence of Proteobacteria, Bacteroidetes and Actinobacteria in the minority groups were previously reported in other fermented soybean foods such as kinema of India, Nepal and Bhutan [8], douchi of China [38] and da-jiang of Korea [39]. Bacillaceae and Bacillus were reported in pe poke as the abundant family and genus, respectively. A colossal interspecies diversity of Bacillus with more than 172 species was detected in pe poke metagenomes by shotgun sequence tool. By cultural method, only B. subtilis was reported in pe poke [9, 10]. At species level, we observed the abundance of B. thermoamylovorans in 3ds and 4ds, Ignatzschineria larvae in 5ds, and B. subtilis in Sds sample, respectively. B. thermoamylovorans is a heat resistant [40] and amylolytic bacterium [41], which is reported in cheonggukjang [42], kinema [8] and douchi [43], and it may also involve in producing thermo-stable enzymes during fermentation at high temperatures [44]. B. subtilis, the second abundant species in pe poke, is one of the major bacterial species in many Asian fermented soybean foods [8, 45–47]. We also observed B. coagulans, which is resistance to high temperatures, and produces various enzymes applicable to food industry [48]. The abundance of B. smithii in pe poke metagenome was also previously reported in fermented soybean foods such as tungrymbai of Meghalaya state and bekang of Mizoram state of North-East India [46]. Abundance of Bacillus species indicates high proteolytic activity, amylase activity and lipase activity [49–52]. Ignatzschineria larvae was also found abundant in 5 days-pe poke, probably contaminated from flies [53], during prolonged fermentation under unhygienic condition. Some LAB were also detected in samples of pe poke, which may have beneficial antimicrobial activity against pathogenic bacteria [54].
Myoviridae, Podoviridae and Siphoviridae were the abundant families of viruses belonging to the order Caudovirales in pe poke. In fermented soybean food, bacteriophages have been reported to cause food spoilage [55] and the abnormal effect on products that may cause reduction of viscous poly-γ-glutamic acid in fermented soybean foods [56]. Bacteriophages may kill the beneficial starter, hamper the bacterial growth, delay fermentation process, yield low-quality, and lower down the bioactivities of the food product [57]. However, some suggested an alternative hypothesis that the presence of bacteriophages is considered to be a very useful therapy in reducing pathogenic bacteria in food products [58]. The presence of archaeal and eukaryotic species were in low abundances in pe poke metagenomes. Archaea contributes to development of taste, aroma flavour, dietary supplements, acetate production during fermentation, and even protect food from spoilage by yeasts [59]. Domain Eukarya consisted of yeasts, filamentous moulds, different species of algae, protozoa and parasites was detected in low abundances in pe poke. Filamentous moulds are known to contribute flavour in fermented soybean product [60] and possess high proteolytic activity [61].
Diversity index, which considers both number of species as well as relative abundance of each species for evaluating diversity [62], showed highest value for the Sds of pe poke, probably due to the duration of fermentation that may cause the changes in species abundance [63]. A goods’ coverage observed in our study indicates a maximum microbial diversity [64] in the samples. In beta diversity, we observed a discrete association among metagenome samples corroborated by PCoA plot, based on their taxonomic features, which may be due to the changes with fermentation time and environmental factors [39, 65]. Several unique and shared species were observed in different samples, probably due to abiotic factors or unusual associations among species from different domains [66].
We found that natural fermentation days of 3–4 days may be suitable for consumption of pe poke due to abundance of B. thermoamylovorans and B. subtilis, which are considered as safe fermenting bacteria in fermented foods [8, 67] comparable to 5 days pe poke with abundance of Proteobacteria, which contains several pathogenic bacteria [68].
Predictive functional features
The predictive functional analysis of pe poke metagenome, mapped against KEGG database, suggested the abundance of metabolism including pathways for carbohydrate metabolism and amino acid metabolism. Abundance of genes related to carbohydrate metabolism (pentose phosphate pathway, and glycolysis) is important for microbial metabolism [69]. The genes for predictive enzymes such as α-glucosidase, α-galactosidase, and β-galactosidase were detected in galactose metabolism pathway in pe poke, essential for degradation of starch and oligosaccharides into simpler forms during fermentation [70]. Genes involved in the processing of lignocellulose were also detected in pe poke metagenome, which suggested that plants-derived carbohydrate act as the source of energy for aerobic (via tricarboxylic acid, TCA cycle) or anaerobic (fermentation) microbes [71]. It was also reported that β-glucosidase could be involved in the hydrolysis of cello-oligosaccharides [72], and biosynthesis of isoflavone glycosides [73], and also involved in digestion and hydrolysis of macromolecules present in soybean seeds during fermentation [74]. Genes related to glycine, serine and threonine metabolism detected in pe poke, may enhance the nutritional value of the product [8, 75]. The abundance of genes related to alanine, aspartate, glutamate metabolism in pe poke metagenome may contribute to the enhancement of taste and flavour of the product [76]. Folate biosynthesis, the key pathway of new therapies against infectious diseases caused by various microorganisms [77] was detected in pe poke. The positive correlation between Bacillus subtilis and folate biosynthesis was observed in pe poke, the key pathway of new therapies against infectious diseases [78] and also confers the protection against inflammation, cancer, anaemia, cardiovascular diseases [79].
Abundance of genes related to ABC transporters specific for peptides were detected in pe poke metagenomes, which may felicitate the uptake of di-/tripeptides [80]. The active role of the microbial population in the transformation of polysaccharide and short-chain carbohydrate in pe poke has been supported by the phosphotransferase system (PTS), the source of transport and phosphorylation of various sugar which forms mono/disaccharides, amino sugars, polyols, and other sugar derivatives [73].
In enzyme classification, we observed the presence of predictive enzymes involvement in the biosynthesis of lysine, alanine, aspartate, glutamate, glycine, serine and threonine, which enhance the nutritional value of the product [54]. Additionally, we also detected enzymes typically encoded by ectABCD gene cluster of bacteria [81] that have an excellent function-preserving property [82]. Genes related to serine protease such as fibrinolytic enzymes were detected in pe poke, which may play as antithrombotic agents [83, 84]. Gene related to signal transduction system that regulates poly-γ-glutamic acid (PGA) synthesis [85, 86].
A positive correlation observed between Bacillus species and predictive amino acids metabolism indicates the ability to accumulate most of amino acids such as alanine, aspartate, glutamate, glycine, serine, and threonine that enhance the nutritional values in the product [87], and also contributes to taste perception and flavour enhancement [88]. A positive correlation between lysine biosynthesis and B. thermoamylovorans was detected in pe poke, which was also reported in douchi [89]. Lysine has several health promoting benefits to consumers [90]. B. coagulans showed positive correlation with biosynthesis of thiamine (vitamin B1), one of the major growth factors that promotes the growth of B. coagulans [91], similarly B. coagulans showed a positive correlation with galactose metabolism, where α- and β-galactosidases (detected in galactose metabolism pathway) can hydrolyse a non-digestible galactoside present in the food matrix [92]. B. subtilis showed positive correlation with predictive folate (vitamin B9) biosynthesis; B. subtilis is reported to harbour pathways component for folate (vitamin B9) production [79]. Though prediction of some pathways related to human disease were also observed, but their abundance were too low to make any significant impact.
Conclusion
Pe poke is a popular traditional fermented soybean cuisine in the Burmese food culture, however, its microbiology and functional properties have not been studied in details, except few reports on Bacillus sp. Hence, we profiled the microbial community in samples of pe poke, which were naturally fermented for 3 days, 4 days, 5 days, respectively and also sundried pe poke, by shotgun metagenomic analysis. Colossal diversity of microbial communities in pe poke was observed. We found that natural fermentation days of 3–4 days may be suitable for consumption of pe poke due to abundance of Bacillus thermoamylovorans and B. subtilis. Several predictive biosynthesis of amino acids, vitamins and other bioactive compounds have been inferred indicting the functional properties of this unique Burmese fermented soybean food, and moreover, the information obtained from this study may help to sensitise the commercial producers and consumers aware on microbial community, the health benefits, hygiene and general safety in pe poke.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
Jyoti Prakash Tamang is grateful to International Centre for Integrated Mountain Development (ICIMOD) Chair for financial support to visit Myanmar and collection of samples.
Data Availability
The sequences of pe poke metagenomes were submitted to National Center for Biotechnology Information (NCBI) under Bio project ID PRJNA694857 with Sequence Read Archive (SRA) Number: SRR13574418 (3ds), SRR13574417 (4ds), SRR13574416 (5ds) and SRR13574415 (Sds). The data has been released publicly on request under the following URL. Bio-project ID PRJNA694857 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA694857) SRR13574418 (3ds): https://www.ncbi.nlm.nih.gov/sra/SRR13574418 SRR13574417 (4ds): https://www.ncbi.nlm.nih.gov/sra/SRR13574417 SRR13574416 (5ds): https://www.ncbi.nlm.nih.gov/sra/SRR13574416 SRR13574415 (Sds): https://www.ncbi.nlm.nih.gov/sra/SRR13574415.
Funding Statement
Jyoti Prakash Tamang is grateful to the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India for financial support through DBTDAICENTER project, sanction number: BT/B1/14/042/2017.
References
- 1.Tamang JP, Cotter P, Endo A, Han NS, Kort R, Liu SQ, et al. Fermented foods in a global age: East meets West. Compre Rev Food Sci Food Safe. 2020; 19(1): 184–217. doi: 10.1111/1541-4337.12520 [DOI] [PubMed] [Google Scholar]
- 2.Gravers, M. (2007). Exploring ethnic diversity in Burma. NIAS Studies in Asian Topics Series, Nordic Institute of Asian Studies, Copenhagen, Denmark.
- 3.Pepok Tanaka T. In: Kiuchi K, Nagai T, Kimura K, editors. Advanced science on natto. Tokyo: Kenpakusha; 2008. Pp. 218–221. [Google Scholar]
- 4.Shurtleff W, Aoyagi A. 2010. History of soybeans and soyfoods in South Asia/Indian subcontinent (1656–2010): Extensively annotated bibliography and sourcebook. Lafayette, CA, USA: Soyinfo Center; 2010. [Google Scholar]
- 5.Tamang JP. Naturally fermented ethnic soybean foods of India. J Ethnic Foods. 2015; 2(1): 8–17. doi: 10.1016/j.jef.2015.02.003 [DOI] [Google Scholar]
- 6.Chukeatirote E, Dajanta K, Apichartsrangkoon A. 2010. Thua nao, Indigenous Thai fermented soybean: a review. J Biol Sci. 2010; 10: 581–583. [Google Scholar]
- 7.Tamang JP, Watanabe K, Holzapfel WH. Diversity of microorganisms in global fermented foods and beverages. Front Microbiol. 2016; 7, 377. doi: 10.3389/fmicb.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharnaior P, Tamang JP. Bacterial and fungal communities and their predictive functional profiles in kinema, a naturally fermented soybean food of India, Nepal and Bhutan. Food Res Int. 2021; 140: 110055. doi: 10.1016/j.foodres.2020.110055 [DOI] [PubMed] [Google Scholar]
- 9.Mitsuboshi S, Tanaka T, Murahashi A, Muramatsu K, Kiuchi K. Characteristics of Burmese fermented soy food, pe pok, and development of itohiki-natto employing its bacterial isolates. J Japanese Soc Food Sci Technol. 2007; 54: 528–538. [Google Scholar]
- 10.Kamada M, Hase S, Fujii K, Miyake M, Sato K, Kimura K, et al. Whole-genome sequencing and comparative genome analysis of Bacillus subtilis strains isolated from non-salted fermented soybean foods. PLoS One. 2015; 10(10):e0141369. doi: 10.1371/journal.pone.0141369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021; 11, 3030. doi: 10.1038/s41598-021-82726-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leech J, Cabrera-Rubio R, Walsh AM, Macori G, Walsh CJ, Barton W, et al. Fermented-food metagenomics reveals substrate-associated differences in taxonomy and health-associated and antibiotic resistance determinants. mSystems. 2020; 5 (6), e00522–20. doi: 10.1128/mSystems.00522-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamames J, Puente-Sánchez F. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front Microbiol. 2019; 9: 3349. doi: 10.3389/fmicb.2018.03349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28(1): 27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ratha P, Jhon DY. Factors increasing poly-γ-glutamic acid content of cheongguk-jang fermented by Bacillus subtilis 168. Food Sci Biotechnol. 2019; 28(1): 103–110. doi: 10.1007/s10068-018-0424-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevim V, Lee J, Egan R, Clum A, Hundley H, Lee J, et al. Shotgun metagenome data of a defined mock community using Oxford Nanopore, PacBio and Illumina technologies. Sci Data. 2019; 6(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 2014; 30(23): 3399–3401. doi: 10.1093/bioinformatics/btu555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 2018; 34(15): 2666–2669. doi: 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017; 27(5): 722–736. http://www.genome.org/cgi/doi/10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016; 7(1): 1–9. doi: 10.1038/ncomms11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput. Biol. 2000; 7(1–2): 203–214. doi: 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- 22.Lazar IM, Karcini A, Ahuja S, Estrada-Palma C. Proteogenomic analysis of protein sequence alterations in breast cancer cells. Sci Rep. 2019; 9(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pokrzywa R, Polanski A. BWtrs: a tool for searching for tandem repeats in DNA sequences based on the Burrows–Wheeler transform. Genomics. 2010; 96(5): 316–321. doi: 10.1016/j.ygeno.2010.08.001 [DOI] [PubMed] [Google Scholar]
- 24.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011; 27(6): 863–864. doi: 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11(1): 1–11. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021; 18: 366–368. doi: 10.1038/s41592-021-01101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016; 44(D1): D286–D293. doi: 10.1093/nar/gkv1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Y, Doak TG. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLOS Comput. Biol. 2009; 5(8): e1000465. doi: 10.1371/journal.pcbi.1000465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scala G, Serra A, Marwah VS, Saarimäki LA, Greco D. FunMappOne: a tool to hierarchically organize and visually navigate functional gene annotations in multiple experiments. BMC Bioinformatics. 2019; 20(1): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016; 44(D1): D457–D462. doi: 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H.Y. (2017). Statistical notes for clinical researchers: Chi-squared test and Fisher’s exact test. Restorative Dentistry & Endodontics 42(2): 152–155. doi: 10.5395/rde.2017.42.2.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haynes W. (2013) Tukey’s Test. In: Dubitzky W, Wolkenhauer O, Cho KH, Yokota H, editors. Encyclopedia of systems biology. New York: Springer; 2013. [Google Scholar]
- 33.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015; 16(1): 1–7. doi: 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammer Ø, Harper DA, Ryan PD. (PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electr. 2001; 4(1): 1–9. [Google Scholar]
- 35.Parks DH, Beiko RG. Measuring community similarity with phylogenetic networks. Mol Biol Evo. 2012; 29(12): 3947–3958. doi: 10.1093/molbev/mss200 [DOI] [PubMed] [Google Scholar]
- 36.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015; 43(W1): W566–W570. doi: 10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Q, Wolf-Hall C, Chang K. Natto characteristics as affected by steaming time, bacillus strain, and fermentation time. J Food Sci. 2001; 66: 167–173. doi: 10.1111/j.1365-2621.2001.tb15601.x [DOI] [Google Scholar]
- 38.Yang L., Yang H.L., Tu Z.C., and Wang X.L. (2016). High-throughput sequencing of microbial community diversity and dynamics during douchi fermentation. PLOS One 11(12): e0168166. doi: 10.1371/journal.pone.0168166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie M, An F, Zhao Y, Wu R, Wu J. Metagenomic analysis of bacterial community structure and functions during the fermentation of da-jiang, a Chinese traditional fermented food. LWT-Food Sci Technol. 2020; 129: 109450. doi: 10.1016/j.lwt.2020.109450 [DOI] [Google Scholar]
- 40.Berendsen EM, Krawczyk AO, Klaus V, de Jong A, Boekhorst J, Eijlander RT, et al. Bacillus thermoamylovorans spores with very-high-level heat resistance germinate poorly in rich medium despite the presence of ger clusters but efficiently upon exposure to calcium-dipicolinic acid. Appl Environ Microbiol. 2015; 22: 7791–7801. doi: 10.1128/AEM.01993-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choonut A, Prasertsan P, Klomklao S, Sangkharak K. Study on mcl-PHA Production by Novel Thermotolerant Gram-Positive Isolate. J Polymer Environ. 2020;28: 2410–2421 [Google Scholar]
- 42.Lee JY, Shim JM, Yao Z, Liu X, Lee KW, Kim HJ, et al. Antimicrobial activity of Bacillus amyloliquefaciens EMD17 isolated from Cheonggukjang and potential use as a starter for fermented soy foods. Food Sci Biotechnol. 2016; 25(2): 525–532. doi: 10.1007/s10068-016-0073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T, Xiong S, Jiang S, Wang M, Wu Q, Wei H. Molecular identification of microbial community in Chinese douchi during post-fermentation process. Food Sci Biotechnol. 2011; 20(6): 1633–1638. doi: 10.1007/s10068-011-0225-0 [DOI] [Google Scholar]
- 44.Nam YD, Yi SH, Lim SI. Bacterial diversity of cheonggukjang, a traditional Korean fermented food, analyzed by barcoded pyrosequencing. Food Contr. 2012; 28(1): 135–142. doi: 10.1016/j.foodcont.2012.04.028 [DOI] [Google Scholar]
- 45.Kubo Y, Rooney AP, Tsukakoshi Y, Nakagawa R, Hasegawa H, Kimura K. Phylogenetic analysis of Bacillus subtilis strains applicable to natto (fermented soybean) production. Appl Environ Microbiol. 2011; 77(18): 6463–6469. doi: 10.1128/AEM.00448-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chettri R, Tamang JP. Bacillus species isolated from tungrymbai and bekang, naturally fermented soybean foods of India. Int J Food Microbiol. 2015; 197: 72–76. doi: 10.1016/j.ijfoodmicro.2014.12.021 [DOI] [PubMed] [Google Scholar]
- 47.Ju S, Cao Z, Wong C, Liu Y, Foda MF, Zhang Z, et al. Isolation and optimal fermentation condition of the Bacillus subtilis subsp. natto strain wtc016 for nattokinase production. Ferment. 2019; 5(4): 92. doi: 10.3390/fermentation5040092 [DOI] [Google Scholar]
- 48.Konuray G, Erginkaya Z. Potential use of Bacillus coagulans in the food industry. Foods. 2018; 7(6): 92. doi: 10.3390/foods7060092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamang JP, Nikkuni S. Effect of temperatures during pure culture fermentation of Kinema. World J Microbiol Biotechnol. 1998; 14(6): 847–850. doi: 10.1023/A:1008867511369 [DOI] [Google Scholar]
- 50.Rai AK, Sanjukta S, Chourasia R, Bhat I, Bhardwaj PK, Sahoo D. Production of bioactive hydrolysate using protease, β-glucosidase and α-amylase of Bacillus spp. isolated from kinema. Biores Technol. 2017; 235: 358–365. doi: 10.1016/j.biortech.2017.03.139 [DOI] [PubMed] [Google Scholar]
- 51.Yan S, Wu G. Bottleneck in secretion of α-amylase in Bacillus subtilis. Microb Cell Fac. 2017; 16(1): 1–8. doi: 10.1186/s12934-017-0738-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma RJ, Wang YH, Liu L, Bai LL, Ban R. Production enhancement of the extracellular lipase LipA in Bacillus subtilis: Effects of expression system and Sec pathway components. Protein Exp Purif. 2018; 142: 81–87. doi: 10.1016/j.pep.2017.09.011 [DOI] [PubMed] [Google Scholar]
- 53.Toth EM, Borsodi AK, Euzeby JP, Tindall BJ, Marialigeti K. Proposal to replace the illegitimate genus name Schineria Toth et al. 2001 with the genus name Ignatzschineria gen. nov. and to replace the illegitimate combination Schineria larvae Toth et al. 2001 with Ignatzschineria larvae comb. nov. Int J Syst Evol Microbiol. 2007; 57:179–180. doi: 10.1099/ijs.0.64686-0 [DOI] [PubMed] [Google Scholar]
- 54.Tamang JP, Shin DH, Jung SJ, Chae SW. Functional properties of microorganisms in fermented foods. Front Microbiol. 2016; 7: 578. doi: 10.3389/fmicb.2016.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagai T, Yamasaki F. Bacillus subtilis (natto) bacteriophages isolated in Japan. Food Sci Technol Res. 2009; 15(3): 293–298. doi: 10.3136/fstr.15.293 [DOI] [Google Scholar]
- 56.Chukeatirote E, Phongtang W, Kim J, Jo A, Jung LS, Ahn J. Significance of bacteriophages in fermented soybeans: A review. Biomol Concept. 2018; 9(1): 131–142. doi: 10.1515/bmc-2018-0012 [DOI] [PubMed] [Google Scholar]
- 57.Ghosh K, Kang HS, Hyun WB, Kim KP. High prevalence of Bacillus subtilis-infecting bacteriophages in soybean-based fermented foods and its detrimental effects on the process and quality of Cheonggukjang. Food Microbiol. 2018; 76: 196–203. doi: 10.1016/j.fm.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 58.Krasowska A, Biegalska A, Augustyniak D, Łoś M, Richert M, Łukaszewicz M. Isolation and characterization of phages infecting Bacillus subtilis. BioMed Res Int. 2015; 2015: 1–10. doi: 10.1155/2015/179597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim MK, Seo WT, Lee YB, Cho KM. Analyses of archaeal communities in Doenjang and Ganjang using a culture-independent manner based on 16S rRNA sequences. Food Sci. Biotechnol. 2013; 22(2): 449–454. doi: 10.1007/s10068-013-0100-2 [DOI] [Google Scholar]
- 60.Ge J, Wang J, Chen L, Song G, Ping W. (2019). The Dynamics analysis of fungal community diversity during the fermentation process of Chinese traditional soybean paste. Waste Bio Valor. 2019; 11: 4789–4797. doi: 10.1007/s12649-019-00800-z [DOI] [Google Scholar]
- 61.Wang S, Giller K, Kreuzer M, Ulbrich SE, Braun U, Schwarm A. Contribution of ruminal fungi, archaea, protozoa, and bacteria to the methane suppression caused by oilseed supplemented diets. Front Microbiol. 2017; 8: 1864. doi: 10.3389/fmicb.2017.01864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lucas R, Groeneveld J, Harms H, Johst K, Frank K, Kleinsteuber S. A critical evaluation of ecological indices for the comparative analysis of microbial communities based on molecular datasets. FEMS Microbiol Eco. 2017; 93, 1. doi: 10.1093/femsec/fiw209 [DOI] [PubMed] [Google Scholar]
- 63.Zhang P, Wu R, Zhang P, Liu Y, Tao D, Yue X, et al. Structure and diversity of bacterial communities in the fermentation of da-jiang. Annl Microbiol. 2018; 68(8): 505–512. doi: 10.1007/s13213-018-1355-x [DOI] [Google Scholar]
- 64.Mardis ER. Next-generation DNA sequencing methods. Ann Rev Genom Hum Genetics. 2008; 9: 387–402. doi: 10.1146/annurev.genom.9.081307.164359 [DOI] [PubMed] [Google Scholar]
- 65.Lee S, Lee S, Singh D, Oh JY, Jeon EJ, Ryu HS, et al. Comparative evaluation of microbial diversity and metabolite profiles in doenjang, a fermented soybean paste, during the two different industrial manufacturing processes. Food Chem. 2017; 221: 1578–1586. doi: 10.1016/j.foodchem.2016.10.135 [DOI] [PubMed] [Google Scholar]
- 66.Wolfe BE, Dutton RJ. Fermented foods as experimentally tractable microbial ecosystems. Cell. 2015; 161(1): 49–55. doi: 10.1016/j.cell.2015.02.034 [DOI] [PubMed] [Google Scholar]
- 67.Lee NK, Kim WS, Paik HD. Bacillus strains as human probiotics: characterization, safety, microbiome, and probiotic carrier. Food Sci Biotechnol. 2019; 28(5): 1297–1305. doi: 10.1007/s10068-019-00691-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. BioMed Res Int. 2017; ID 9351507, 7: 2017. doi: 10.1155/2017/9351507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sulaiman J, Gan HM, Yin WF, Chan KG. Microbial succession and the functional potential during the fermentation of Chinese soy sauce brine. Front Microbiol. 2014; 5: 556. doi: 10.3389/fmicb.2014.00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mhuantong W, Charoensawan V, Kanokratana P, Tangphatsornruang S, Champreda V. Comparative analysis of sugarcane bagasse metagenome reveals unique and conserved biomass-degrading enzymes among lignocellulolytic microbial communities. Biotechnol Biofuels. 2015; 8(1): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roth JCG, Hoeltz M, Benitez LB. Current approaches and trends in the production of microbial cellulases using residual lignocellulosic biomass: a bibliometric analysis of the last 10 years. Arch Microbiol. 2020; 202, 935–951. doi: 10.1007/s00203-019-01796-9 [DOI] [PubMed] [Google Scholar]
- 72.Wilkens C, Busk PK, Pilgaard B, Zhang WJ, Nielsen KL, Nielsen PH, et al. Diversity of microbial carbohydrate-active enzymes in Danish anaerobic digesters fed with wastewater treatment sludge. Biotechnol Biofuel. 2017; 10(1): 1–14. doi: 10.1186/s13068-017-0840-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choe M, Park H, Lee CR, Kim YR, Seok YJ. The general PTS component HPr determines the preference for glucose over mannitol. Sci Rep. 2017; 7: 4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Švejstil R, Musilová Š, Rada V. Raffinose-series oligosaccharides in soybean products. Sci Agricul Bohem. 2015; 46(2): 73–77. doi: 10.1515/sab-2015-0019 [DOI] [Google Scholar]
- 75.Dajanta K, Apichartsrangkoon A, Chukeatirote E, Frazier RA. Free-amino acid profiles of thua nao, a Thai fermented soybean. Food Chem. 2011; 125(2): 342–347. doi: 10.1016/j.foodchem.2010.09.002 [DOI] [Google Scholar]
- 76.Parthasarathy A, Cross PJ, Dobson RC, Adams LE, Savka MA, Hudson AO. A three-ring circus: metabolism of the three proteogenic aromatic amino acids and their role in the health of plants and animals. Front Mol Biosci. 2018; 5: 29. doi: 10.3389/fmolb.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bertacine Dias MV, Santos JC, Libreros-Zúñiga GA, Ribeiro JA, Chavez-Pacheco SM. Folate biosynthesis pathway: mechanisms and insights into drug design for infectious diseases. Future Med Chem. 2018; 10(8): 935–959. doi: 10.4155/fmc-2017-0168 [DOI] [PubMed] [Google Scholar]
- 78.Revuelta JL, Serrano-Amatriain C, Ledesma-Amaro R, Jiménez A. Formation of folates by microorganisms: towards the biotechnological production of this vitamin. Appl Microbiol Biotechnol. 2018; 102(20): 8613–8620. doi: 10.1007/s00253-018-9266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khatri I, Sharma G, Subramanian S. Composite genome sequence of Bacillus clausii, a probiotic commercially available as Enterogermina®, and insights into its probiotic properties. BMC Microbiol. 2019; 19(1): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El-Awady R, Saleh E, Hashim A, Soliman N, Dallah A, Elrasheed A et al. The role of eukaryotic and prokaryotic ABC transporter family in failure of chemotherapy. Front Pharmacol. 2017; 7:535. doi: 10.3389/fphar.2016.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, et al. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3 (2) in response to salt and heat stresses. Appl Environmen Microbiol. 2008; 74(23): 7286–7296. doi: 10.1128/AEM.00768-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Czech L, Hermann L, Stöveken N, Richter AA, Höppner A, Smits SH, et al. Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: genetics, phylogenomics, biochemistry, and structural analysis. Genes. 2018; 9(4): 177. doi: 10.3390/genes9040177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh TA, Devi KR, Ahmed G, Jeyaram K. Microbial and endogenous origin of fibrinolytic activity in traditional fermented foods of Northeast India. Food Res Int. 2014; 55: 356–362. doi: 10.1016/j.foodres.2013.11.028 [DOI] [Google Scholar]
- 84.Hu Y, Yu D, Wang Z, Hou J, Tyagi R, Liang Y, et al. (2019). Purification and characterization of a novel, highly potent fibrinolytic enzyme from Bacillus subtilis DC27 screened from Douchi, a traditional Chinese fermented soybean food. Sci Rep. 9(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo Z, Guo Y, Liu J, Qi H, Zhao M, Zou W, et al. Microbial synthesis of poly-γ-glutamic acid: current progress, challenges, and future perspectives. Biotechnol Biof. 2016; 9(1): 1–12. doi: 10.1186/s13068-016-0537-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hsueh YH, Huang KY, Kunene SC, Lee TY. Poly-γ-glutamic acid synthesis, gene regulation, phylogenetic relationships, and role in fermentation. Int J Mol Sci. 2017; 18(12): 2644. doi: 10.3390/ijms18122644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hyeon H, Min CW, Moon K, Cha J, Gupta R, Park SU, et al. Metabolic profiling-based evaluation of the fermentative behavior of Aspergillus oryzae and Bacillus subtilis for soybean residues treated at different temperatures. Foods. 2020; 9(2): 117. doi: 10.3390/foods9020117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jang M, Jeong DW, Heo G, Kong H, Kim CT, Lee JH. Genetic background behind the amino acid profiles of fermented soybeans produced by four Bacillus spp. J Microbiol Biotechnol. 2021; 31(3): 447–455. doi: 10.4014/jmb.2012.12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen C, Xiang JY, Hu W, Xie YB, Wang TJ, Cui JW, et al. Identification of key microorganisms involved in Douchi fermentation by statistical analysis and their use in an experimental fermentation. J Appl Microbiol. 2015; 119(5): 1324–1334. doi: 10.1111/jam.12917 [DOI] [PubMed] [Google Scholar]
- 90.Gillner DM, Becker DP, Holz RC. Lysine biosynthesis in bacteria: a metallodesuccinylase as a potential antimicrobial target. J Biol Inorg Chem. 2013. Feb;18(2):155–63. doi: 10.1007/s00775-012-0965-1 Epub 2012 Dec 8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cao J, Yu Z, Liu W, Zhao J, Zhang H, Zhai Q, et al. Probiotic characteristics of Bacillus coagulans and associated implications for human health and diseases. J Function Foods. 2020; 64: 103643. doi: 10.1016/j.jff.2019.103643 [DOI] [Google Scholar]
- 92.Aulitto M, Strazzulli A, Sansone F, Cozzolino F, Monti M, Moracci M, et al. Prebiotic properties of Bacillus coagulans MA-13: production of galactoside hydrolyzing enzymes and characterization of the transglycosylation properties of a GH42 β-galactosidase. Microbial Cell Fact. 2021; 20(1): 1–17. doi: 10.1186/s12934-021-01553-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The sequences of pe poke metagenomes were submitted to National Center for Biotechnology Information (NCBI) under Bio project ID PRJNA694857 with Sequence Read Archive (SRA) Number: SRR13574418 (3ds), SRR13574417 (4ds), SRR13574416 (5ds) and SRR13574415 (Sds). The data has been released publicly on request under the following URL. Bio-project ID PRJNA694857 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA694857) SRR13574418 (3ds): https://www.ncbi.nlm.nih.gov/sra/SRR13574418 SRR13574417 (4ds): https://www.ncbi.nlm.nih.gov/sra/SRR13574417 SRR13574416 (5ds): https://www.ncbi.nlm.nih.gov/sra/SRR13574416 SRR13574415 (Sds): https://www.ncbi.nlm.nih.gov/sra/SRR13574415.