Abstract
We report the simultaneous isolation of one Aspergillus flavus strain from the aortic prosthesis of a heart surgery patient and another two isolates recovered from a dual-reservoir cooler-heater used in the operating room where this patient was operated on. Genetic typing of these three isolates by randomly amplified polymorphic DNA (RAPD) revealed identical genotypes. Eight unrelated control strains of A. flavus had eight different genotypes. These results clearly indicated the nosocomial origin of the A. flavus strain isolated from the patient. We suggest that the RAPD technique is a rapid and reliable tool to ascertain the epidemiology of infections caused by A. flavus.
Aspergillus spp. are ubiquitous, commonly occurring in soil, water, and decaying vegetation. Reservoirs in hospitals from which these fungi have been cultured include unfiltered air, ventilation systems, contaminated dust dislodged during hospital construction, carpeting, food, and ornamental plants (4). Members of the genus Aspergillus are opportunistic pathogens; most infections occur in severely immunocompromised patients. Aspergillus fumigatus is the most common agent of systemic infections, followed by Aspergillus flavus (2). Inhalation of fungal conidia is thought to be the primary means of acquiring aspergillosis. Another proposed route of acquisition of Aspergillus spp., specially A. flavus, is through ingestion of contaminated food (10).
Because Aspergillus spp. are ubiquitous in the environment, the simultaneous isolation of the same species from clinical and environmental specimens collected during epidemiological surveillance is not enough to demonstrate a common origin for the isolates. In these cases, only genotyping data provide conclusive evidence of nosocomiality. Although an ideal genotyping procedure applicable to a wide range of Aspergillus species remains to be determined, randomly amplified polymorphic DNA (RAPD) analysis has been shown to be a rapid and reliable method useful for A. fumigatus, A. flavus, and Aspergillus terreus (1, 7, 8, 9, 10).
Here we describe the isolation of one A. flavus strain causing an aortic prosthesis infection three months after aortic valve replacement and two A. flavus strains recovered from a cooling and heating unit used in the operating room. The RAPD patterns of these isolates were compared to check their genetic relatedness. A random selection of eight unrelated strains was used as a control population.
Case report.
A 54-year-old female received an aortic prosthesis in October 1998 because of aortic valve insufficiency. In January 1999, she had to be readmitted and operated on again because of an aortic aneurysm in the area next to the prosthesis. At this time, an A. flavus strain was isolated from the prosthesis. Liposomal amphotericin B (AmBisome; Nexstar Pharmaceuticals, Madrid, Spain) at 1 mg/kg of body weight/day was administered for 10 days. Because of the absence of clinical response, the dose of liposomal amphotericin B was increased to 3 mg/kg/day for another 5 days and then to 5 mg/kg/day. Despite this treatment, the patient died.
Environmental surveillance.
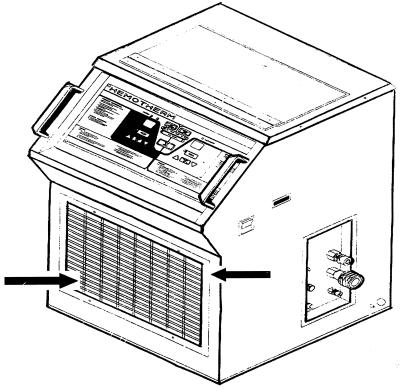
Air sampling is routinely performed in the room used for heart surgery in this hospital every 2 months by means of a volumetric sampler (1 m3/10 min). In addition, Sabouraud plates are systematically laid on the exits of the two HEPA filters installed in the room. As mentioned above, the prosthesis implantation was performed in October 1998. The previous and subsequent environmental controls performed in the operating room (September and November, respectively) were negative. This means that both the volumetric samples and the controls for the HEPA filters were negative. When A. flavus was isolated from the patient (January 1999), the environmental controls remained negative. Then epidemiological surveillance was conducted by taking samples with cotton-tipped swabs from the surfaces and equipment within the operating room. Two A. flavus strains were cultured from the grilles of the dual-reservoir cooler-heater (Hemotherm; CincinnatiSubzero, Cincinnati, Ohio) usually employed in heart surgery to maintain the extracorporal blood at the proper temperature. This electric device has internal coils filled with water to cool or heat the ducts with the circulating blood. The water is supplied from water pans placed inside the device. The water inside this reservoir was not cultured. Two A. flavus isolates were separately recovered from the cooling-heating unit, one from the upper part of the grille and the other from the lower part (Fig. 1).
FIG. 1.
Dual-reservoir cooler-heater (Hemotherm) employed in heart surgery. The arrows indicate the upper and lower parts of the grille, where the two A. flavus strains were isolated.
Mycology.
The sources of the isolates discussed in this study are described in Table 1. Isolates 1, 2, and 3 were epidemiologically related strains: isolates 1 and 2 were isolated from the cooling-heating unit in the operating room. Isolate 3 was found on the cardiac prosthesis of the patient. The eight epidemiologically unrelated strains (no. 4, 5, 8, 10, 12, 13, 16, and 19) belonged to the mold collection at the Mycology Reference Laboratory (Instituto de Salud Carlos III, Madrid, Spain).
TABLE 1.
Characteristics and RAPD profiles of 11 A. flavus strains
| No. | Origin | Source | Date of isolation (mo/day/yr) | Genotype with RAPD primer:
|
Overall genotype | ||
|---|---|---|---|---|---|---|---|
| R-108 | R-151 | AP12h | |||||
| 1 | Baracaldo | Heat exchanger | 1/16/99 | A | A | A | I |
| 2 | Baracaldo | Heat exchanger | 1/16/99 | A | A | A | I |
| 3 | Baracaldo | Aortic prosthesis | 1/15/99 | A | A | A | I |
| 4 | Almería | Sputum | 3/9/98 | B | B | B | II |
| 5 | San Sebastian | Sputum | 3/16/98 | A | A | C | III |
| 8 | Almería | Oticum exudate | 5/29/98 | C | C | D | IV |
| 10 | Almería | Nail | 5/21/98 | D | D | E | V |
| 12 | Almería | Oticum exudate | 7/24/98 | E | E | F | VI |
| 13 | Toledo | Skin biopsy | 10/16/98 | F | F | G | VII |
| 16 | Madrid | Paranasal sinus | 1/23/96 | G | G | H | VIII |
| 19 | Zamora | Sputum | 3/10/97 | H | H | I | IX |
Molecular typing.
Conidia from each strain were inoculated into 3 ml of GYEP broth (2% glucose, 0.3% yeast extract, 1% peptone) and were grown overnight at 37°C. The mycelium mats were recovered and subjected to a DNA extraction protocol described previously (5). After RNase treatment, the samples were treated with proteinase K (Sigma-Aldrich, Alcobendas, Spain), and the DNA was purified again by extraction with phenol-chloroform and ethanol precipitation.
The primers used in this work were AP12h (5′ CGG CCC CTG T 3′) (11), R-108 (5′ GTA TTG CCC T 3′), and R-151 (5′ GCT GTA GTG T 3′) (1). The primers were 10 nucleotides in length, had G+C contents ranging from 50 to 80%, and did not contain palindromic sequences.
The RAPD reaction mixtures included 10 mM Tris HCl (pH 8)–50 mM KCl as a buffer; 200 μM (each) dATP, dTTP, dCTP, and dGTP; 2.5 mM MgCl2; 1 μM primer; 2.5 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer Cetus, Madrid, Spain); and 25 ng of template DNA in a 50-μl final volume.
The thermal cycler (Perkin-Elmer) was programmed with 40 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 36°C (27°C for primer R-108), and 2 min of primer extension at 72°C. A blank control with all the PCR reagents except DNA was always included. The resulting banding patterns were indexed by capital letters, and even a single band mismatch led to a different letter code.
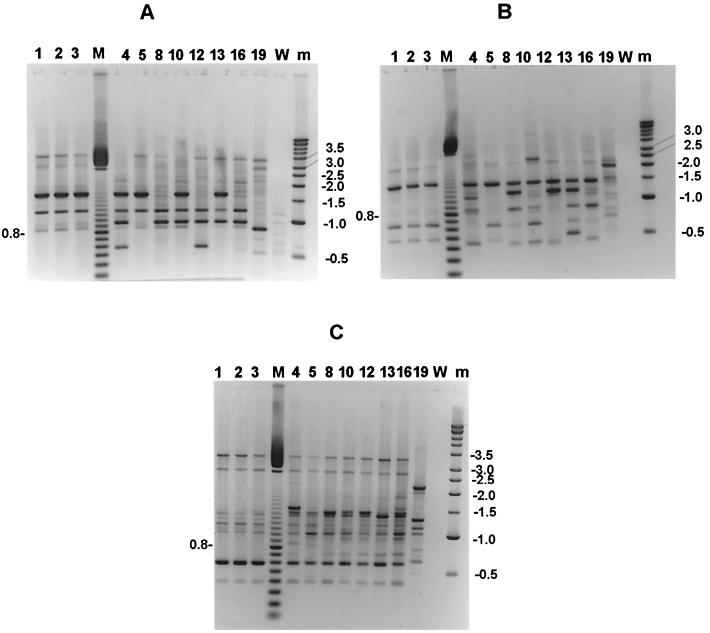
The genotypes of the 11 A. flavus isolates obtained by RAPD analysis are summarized in Table 1. The patterns obtained with each of the primers were easily analyzed because of the number, spreading, and intensity of the bands (Fig. 2). Some unspecific bands appeared in lane W (control without DNA) of Fig. 2A which correspond to the patterns obtained with primer R-108; they were due to the low annealing temperature used with this primer (27°C).
FIG. 2.
DNA band patterns obtained with three different arbitrary primers. (A) Primer R-108; (B) primer R-151; (C) primer AP12h. Lane M, 100 bp ladder (Pharmacia); the size in kilobase pairs of the most intense fragment is shown at the left of each panel. Lane m, kilobase pair ladder (Pharmacia), with sizes noted at the right of each panel. The disposition of the strains is the same in the three panels. Lanes 1 and 2, A. flavus strains isolated from the heat exchanger in the operating room; lane 3, A. flavus strain from the aortic prosthesis of the case patient; lanes 4, 5, 8, 10, 12, 13, 16, and 19, unrelated isolates used as controls; lane W, blank control for each PCR procedure.
The discriminatory power of each of the three different primers used was satisfactory: each of the unrelated strains harbored a distinct pattern with any of the primers tested. However, the isolate from the patient and the two strains obtained from the electric device in the operating room harbored a unique DNA pattern, as shown by any of the three primers tested. To test the reliability of these results, the DNAs from the three test isolates were extracted again, quantified, and then amplified. The bands scored for each strain were exactly the same in the first and second runs.
Strain 5 had the same genotype as strains 1, 2, and 3 when analyzed with primers R-108 and R-151. However, it had a unique genotype when analyzed with primer AP12h. There was no known epidemiological relationship among these strains, as strain 5 was obtained from a patient in a hospital distant from the one where the case patient was operated on.
In spite of the increasing reports of infections caused by A. fumigatus, the isolation of A. flavus has been less frequently described. We report the isolation of A. flavus strains with the same molecular pattern from the aortic prosthesis of one patient and from environmental samples from the room used for heart surgery where the patient was operated on. That RAPD analysis is a truly rapid and reliable tool for epidemiological investigations of infections due to Aspergillus spp. has been previously stated (7). However, when this method is used to analyze the genetic relationship among isolates from the same hospital, it is necessary to consider two factors. First, a collection of unrelated isolates analyzed by the same method must be included in order to compare the clustering of strains in related and unrelated groups. Second, the combination of data generated by at least two primers is recommended to increase the discriminatory power of the technique. One advantage of this technique is that the primers are universal and they can be used for genomic analysis of a wide variety of species. In fact, R-108 and R-151 have been used successfully to amplify DNA segments from A. fumigatus (1).
To our knowledge, few publications have described the attempt to type both clinical A. flavus from patients with invasive aspergillosis and environmental isolates, searching for a common source in the hospital setting (3, 7, 8). The authors of these studies stated that it is not an easy task to find strong evidence of the nosocomial origin of invasive aspergillosis.
Our work demonstrates that, in selected rooms where air sampling is performed routinely, other surveillance methods should not be discarded. It must be remembered that a volumetric sampler analyzes air contamination for a very limited period of time. However, profound changes in the airborne conidium concentration inside the hospital have been detected even between consecutive measurements (8). Furthermore, routine air sampling can produce a false sense of security and a decline in other maintenance and cleansing procedures. The combination of these factors may be the explanation for what happened in this case. First, the air-sampling procedure was not able to detect conidia, either when the patient was operated on for the first time or when the second operation was performed. Second, A. flavus conidia, whose source could not be elucidated, had probably settled on the grilles of the cooling-heating unit. When this electric device was manipulated during the surgical procedure, they were aerosolized, producing a transient burst. The grilles, cooling coils, water pans, and drain pans of ventilation systems are known to be likely locations for fungal growth, especially when there is standing water. It seems probable that these items of the cooling and heating system used in this hospital were not properly disassembled and cleaned with disinfectants.
This epidemiological surveillance confirmed that a nosocomial origin of Aspergillus infections can be demonstrated even after several months of delay between the acquisition of the fungus and the development of the infection. For this reason, other procedures must be added to routine air surveillance, such as taking samples with a cotton-coated swab from the surfaces where spores may settle and remain for a long time (6). In consequence, when a case of aspergillosis is detected in an operating room, a very thorough search for the fungus should be encouraged. These data also indicate that more emphasis must be placed on the important role that fomites can play as potential reservoirs for fungi.
Acknowledgments
This work at the Servicio de Micología was supported in part by grant 1078/99 from Instituto de Salud Carlos III.
REFERENCES
- 1.Aufauvre-Brown A, Cohen J, Holden D W. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J Clin Microbiol. 1992;30:2991–2993. doi: 10.1128/jcm.30.11.2991-2993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodey G P, Vartivarian S. Aspergillosis. Eur J Microbiol Infect Dis. 1989;8:413–437. doi: 10.1007/BF01964057. [DOI] [PubMed] [Google Scholar]
- 3.Buffington J, Reporter R, Lasker B A, McNeil M M, Lanson J M, Ross L A, Mascola L, Jarvis W R. Investigation of an epidemic of invasive aspergillosis: utility of molecular typing with the use of random amplified polymorphic DNA probes. Pediatr Infect Dis J. 1994;13:386–393. [PubMed] [Google Scholar]
- 4.Fridkin S K, Jarvis W R. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holden D W. DNA miniprep method for Aspergillus fumigatus (and other filamentous fungi) In: Maresca B, Kobayasi G S, editors. Molecular biology of pathogenic fungi. New York, N.Y: Telos Press; 1994. pp. 3–4. [Google Scholar]
- 6.Latgé J P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leenders A, van Belkum A, Janssen S, de Marie S, Kluytmans J, Wielenga J, Löwenberg B, Verbrugh H. Molecular epidemiology of an apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–351. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rath P M, Ansorg R. Value of environmental sampling and molecular typing of aspergilli to assess nosocomial sources of aspergillosis. J Hosp Infect. 1997;37:47–53. doi: 10.1016/s0195-6701(97)90072-4. [DOI] [PubMed] [Google Scholar]
- 9.Rath P M, Kamphoff S, Ansorg R. Value of different methods for the characterisation of Aspergillus terreus strains. J Med Microbiol. 1999;48:161–166. doi: 10.1099/00222615-48-2-161. [DOI] [PubMed] [Google Scholar]
- 10.Rhame F S. Prevention of nosocomial aspergillosis. J Hosp Infect. 1991;18(Suppl. A):466–472. doi: 10.1016/0195-6701(91)90058-g. [DOI] [PubMed] [Google Scholar]
- 11.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]