Abstract
The purpose of this research was to assess the effects of a microRNA (miRNA) cluster on platelet production. Human chromosome 19q13.41 harbors an evolutionarily conserved cluster of three miRNA genes (MIR99B, MIRLET7E, MIR125A) within 727 base-pairs. We now report that levels of miR-99b-5p, miR-let7e-5p and miR-125a-5p are strongly correlated in human platelets, and all are positively associated with platelet count, but not white blood count or hemoglobin level. Although the cluster regulates hematopoietic stem cell proliferation, the function of this genomic locus in megakaryocyte (MK) differentiation and platelet production is unknown. Furthermore, studies of individual miRNAs do not represent broader effects in the context of a cluster. To address this possibility, MK/platelet lineage-specific Mir-99b/let7e/125a knockout mice were generated. Compared to wild type littermates, cluster knockout mice had significantly lower platelet counts and reduced MK proplatelet formation, but no differences in MK numbers, ploidy, maturation or ultra-structural morphology, and no differences in platelet function. Compared to wild type littermates, knockout mice showed similar survival after pulmonary embolism. The major conclusions are that the effect of the Mir-99b/let7e/125a cluster is confined to a late stage of thrombopoiesis, and this effect on platelet number is uncoupled from platelet function.
Keywords: Mir-99b/let7e/125a cluster, megakaryocyte, platelets, conditional knockout mice
Introduction
Megakaryopoiesis (MKpoiesis) and thrombopoiesis are complex biological processes wherein hematopoietic stem cells differentiate to megakaryocyte (MK) progenitors, undergo endomitosis, mature, and releases platelets into the circulation. We and others have shown that MKs and platelets carry a rich repertoire of miRNAs, a class of small non-coding RNAs that regulate expression of protein-coding genes critical for normal functioning of MKs and platelets [1]. Platelet miR-125a-5p is positively associated with human platelet number and enhances human MK proplatelet formation [2]. The mir-125a precursor is a part of an evolutionary conserved miRNA cluster (MIR-99b/let7e/125a) within 727 base-pairs on human chromosome 19. This cluster is known to regulate primitive hematopoietic cells[3], but whether it regulates MKpoiesis and thrombopoiesis remains unknown. In vivo systemic inhibitors of miR-125a-5p lower mouse platelet counts. However, because miR-125a-5p has a broad tissue distribution, it is not known whether this miRNA effect is mediated by MKs. The goal of this study was to determine whether the entire cluster of three miRNAs might regulate platelet production in a MK-specific manner. We now report generation of MK/platelet lineage specific Mir-99b/let7e/125a cluster knockout mice strain Mir-99b/let7e/125afl/fl platelet factor 4 (PF4)-Cre, and investigate its contribution to platelet production and function. We show that loss of the Mir-99b/let7e/125a cluster in MKs reduces murine MK proplatelet formation and circulating platelet counts without altering MK differentiation or platelet function.
Material and Methods
Generation of Mir-99b/let7e/125afl/flPF4-Cre mouse strain
Conditional floxed Mir-99b/let7e/125a allele (Mir-99b/let7e/125afl/fl) mice were custom generated by Biocytogen (Worcester, MA)[4] using homologous recombination in C57BL6 embryonic stem cells. Initially PCR products of desired locus in the C57BL/6N mouse were sequenced and confirmed as the same as those from the NCBI and Ensembl database. The ideal homologous arms for the target vector forced us to include exon 1 of the Spaca6 gene within the loxP sites. Spaca6 encodes Sperm acrosome associated 6, which is not expressed in MKs or platelets. Cas9/sgRNA plasmids were generated and confirmed by DNA sequencing. Eight sets of sgRNAs were made and tested to select the most efficient set for CRISPR activity. Targeting vector with loxP flanking the three miRNA genes was constructed and verified by sequencing. RNA was prepared and mouse zygotes microinjected. Pup DNA was genotyped with PCR primers; seven pups had the targeted allele; pups were bred and seven confirmed by PCR as F1 heterozygotes. All animal studies were done under the approval by the Institutional Animal Care and Use Committee at the University of Utah.
TaqMan advanced miRNA assay
Leukocyte depleted platelet (LDP) were prepared as described previously [5]. Total RNA was isolated using TRIzol method (Thermo Fisher Scientific, Waltham, MA). Mature platelet miRNA expression of mmu-miR-99b-5p, mmu-let-7e-5p and mmu-miR-125a-5p was detected using TaqMan advanced miRNA assay as per manufactures’ protocol (Thermo Fisher Scientific, Waltham, MA).
MK quantification and characterization
Mouse bone marrow was isolated and MKs quantified as previously described [6]. For ploidy analysis, day 5 ex vivo generated MKs were stained with propidium iodide (50 μg/ml, Thermo Fisher Scientific, Waltham, MA) for 1 hour at room temperature in the dark, and CD41a positive MKs stained for propidium iodide were analyzed for ploidy analysis using a Cytoflex flow cytometer. Transmission electron microscopy of day 5 ex vivo MKs was performed as described previously[2] at the University of Utah, Electron Microscopy Core Facility.
Platelet functional characterization
Platelet cell counts were performed on blood collected from the retro-orbital plexus into tubes containing EDTA (Sarstedt, Inc. Newton, NC) using a Hemavet 950FS blood cell counter (Drew Scientific, Miami Lakes, FL). 2x108 cells/ml washed murine platelets were activated with agonists, AYPGKF (50 μM, 100 μM and 150 μM) (GL Biochem (Shanghai) Ltd.) and collagen related peptide (CRP) (synthesized at Baylor College of Medicine and cross-linked with glutaraldehyde) (1 ng, 30 ng and 100 ng) or Tyrode’s buffer (resting), as done previously [7]. For platelet aggregation, 2x108 cells/ml washed platelets were stimulated with AYPGKF (30 μM, 150 μM) and CRP (30 ng, 100 ng), and change in the light transmission was measured by a lumi-aggregometer (Chrono-Log, Havertown, PA) at 37°C under stirring conditions.
For platelet spreading assays, washed platelets from knockout or littermates were plated on fibrinogen (100 μg/ml, Calbiochem-Novabiochem Corporation, San Diego, CA) or collagen (100 μg/ml, Chrono-Log, Havertown, PA) coated chamber slides for 45 minutes. Cells were fixed with 2% paraformaldehyde for 20 minutes on ice, and stained with Alexa Flour 488 Phalloidin (Thermo Fisher Scientific, Waltham, MA). Images were taken under 60x oil objective using a confocal microscope. Eight-week-old, gender matched knockout and littermates were used for platelet counts and functional assays, unless otherwise indicated.
Pulmonary embolism model
Collagen (0.2 mg/kg; Chronolog) and epinephrine (15 mg/kg; Sigma-Aldrich, St. Louis, MA) in 100 μL of PBS were administered through retro-orbital injection [8]. The time to death was defined as the time needed to the onset of respiratory arrest that lasted at least 2-10 minutes.
Statistics
Two-group comparisons of parametric data were done using a two-tailed Student t-test unless otherwise indicated. All statistical analyses were performed using Prism 9 software (GraphPad, La Jolla, CA). P value of 0.05 or less was considered as significant. All data are presented as mean ± standard error of the mean.
Results
miRNA-99b/let7e/125a cluster expression in differentiating human MKs and is positively correlated with platelet number
miR-125a-5p has been previously shown to correlate with human platelet number, so we initially tested whether miR-99b-5p and miR-let-7e-5p levels also correlated. Significant positive associations were observed between platelet count and each of the three miRNAs in 154 healthy donors, but not with white blood count or hemoglobin (Figure 1A), supporting the possibility that the whole cluster may impact plt numbers. Subsequent studies using real time PCR in human umbilical cord blood CD34 derived MK cultures showed the expression of miR-99b-5p, miR-let-7e-5p and miR-125a-5p increased from day 6 to 13 as MKs differentiate (Figure 1B). These results suggest the effect of this miRNA cluster may be at the level of platelet production by MKs.
Figure 1. The expression of miR-99b-5p, miR-let7e-5p and miR-125a-5p increases with human MK differentiation and associates positively with human platelet numbers.
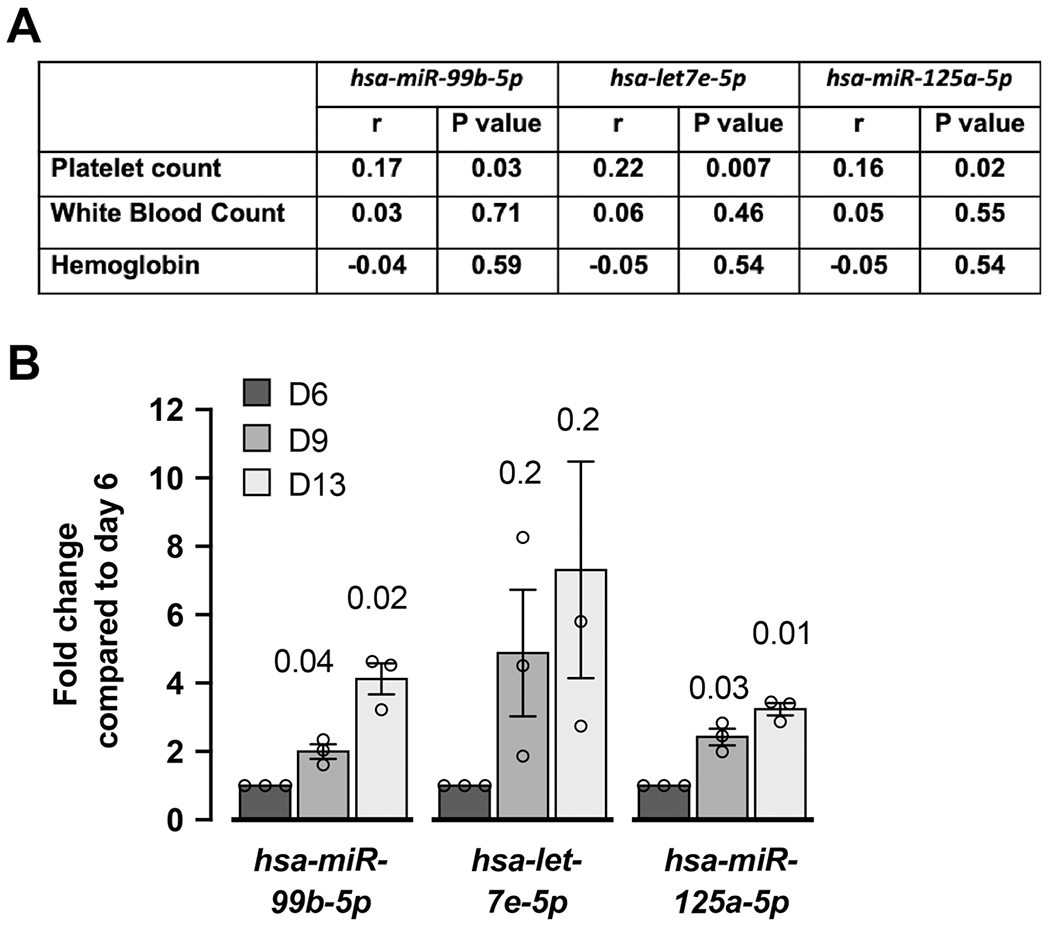
A. Correlations between miRNAs and blood platelet count, white blood count or hemoglobin level in 154 healthy donors from the Platelet RNA and Expression 1 data set. r, Pearson correlation coefficient. B. Human umbilical cord blood derived, CD61-purified MK cultures were assessed for the expression of mature miRNAs, miR-99b-5p, let-7e-5p and miR-125a-5p by real time PCR analysis at days 6, 9 and 13. Fold changes were calculated for their expression at days 9 and 13 compared to day 6. n=3 different cords (=independent experiments).
Mir-99b/let7e/125afl/flPF4-Cre mice are thrombocytopenic
Conditional floxed Mir-99b/let7e/125a allele (Mir-99b/let7e/125afl/fl) mice were custom generated by Biocytogen (Worcester, MA)[4] using homologous recombination in C57BL6 embryonic stem cells using the strategy shown in Figure 2A (see Methods for more details). Germ line transmission of genetic recombination in F1 animals was confirmed with long fragment PCR and sequencing analysis. Genotyping primers used in PCR analyses are shown in Supplemental Figure 1 (red arrows). No random insertions were observed by Southern blotting using an internal probe (Supplemental Figure 2). Mir-99b/let7e/125afl/fl mice were crossed with C57BL/6 mice expressing platelet factor 4 promoter-driven Cre-recombinase (PF4-Cre)[9] to generate mice lacking expression of Mir-99b/let-7e/125a in MKs and platelets (Mir-99b/let-7e/125afl/flPF4-Cre). Throughout the manuscript these knockout mice are referred as ‘Mir-99b/let7e/125a −/−’, and their littermate control mice are referred as ‘WT’.
Figure 2. Deletion of the Mir-99b/let7e/125a cluster lower blood platelet counts.
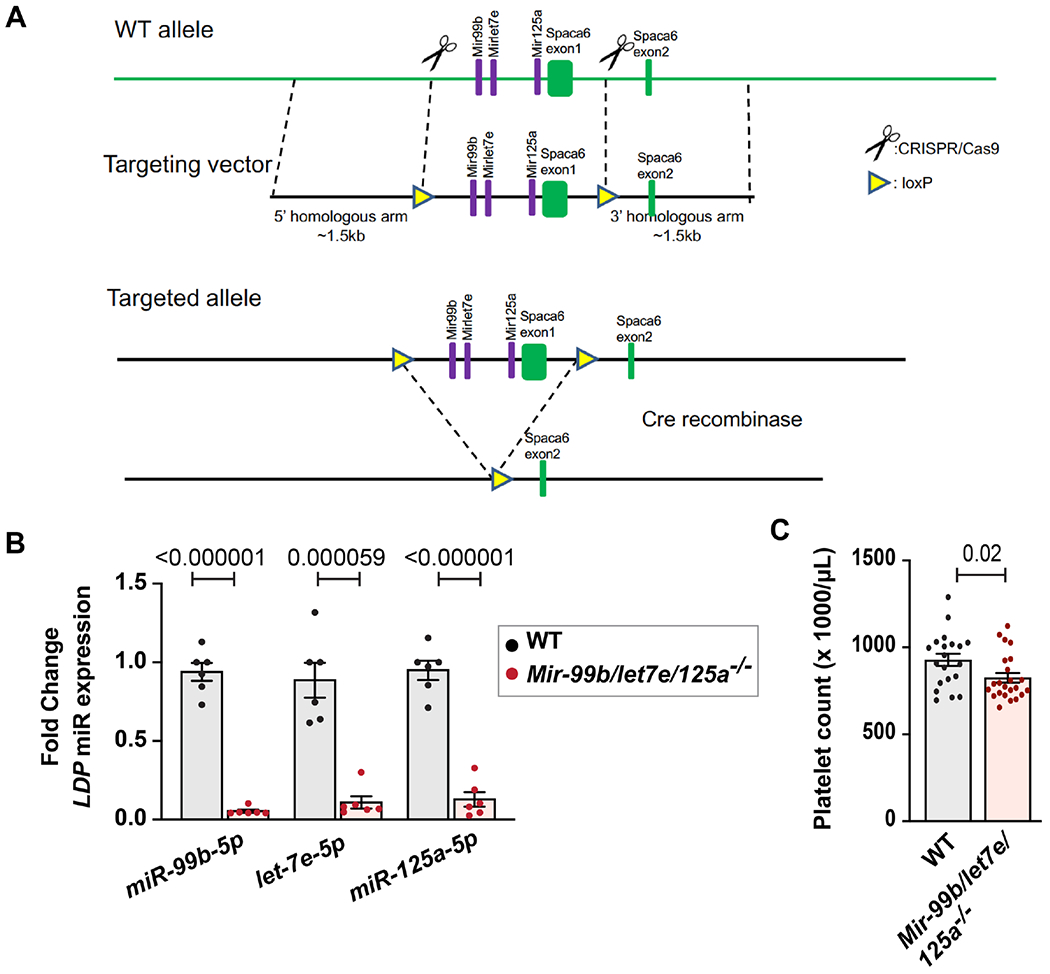
A. Schema showing experimental design. Mice were generated with the targeted allele containing loxP sites flanking the Mir-99b/let7e/125a cluster. These mice were bred with PFA-Cre mice resulting in mice with MKs lacking the Mir-99b/let7e/125a cluster. B. Fold change of mmu-miR-99b-5p, mmu-let-7e-5p and mmu-miR-125a-5p in leukocyte depleted platelets (LDP) isolated from Mir-99b/let-7e/125a −/− mice (n=6) compared to their littermates (n=6), as assessed by TaqMan advance miRNA assay. miR-320a-5p (expression did not change in knockout vs littermates, data not shown) was used as a housekeeping gene for fold change calculation. C. Peripheral blood platelet count measured by Hemavet for Mir-99b/let7e/125a−/− mice (n=23) or littermates (n=20) (8 weeks old, and gender matched).
Mir-99b/let7e/125a−/− mice did not display any overt physical abnormalities, were fertile, and bred normally. The expression of miR-99b-5p, miR-let-7e-5p and miR-125a-5p was abolished in Mir-99b/let7e/125a −/− purified platelets, confirming successful deletion of the cluster (Figure 2B). A modest but significant (p=0.02) reduction in peripheral blood platelet count was observed in Mir-99b/let-7e/125a −/− compared to wild type littermates. (Figure 2C).
Mir-99b/let7e/125afl/flPF4-Cre mice have defective thrombopoiesis
Since this cluster is known to regulate HSC proliferation, we assessed whether MK-specific deletion would affect early stages of MKpoiesis, but observed no differences between knockout mice and wild type littermates in the number of mature MKs (Figure 3A), the levels of MK polyploidy (Figure 3B) or MK ultra-structural morphology (Figure 3C). However, compared to MKs from wild-type littermates, MKs from Mir-99b/let7e/125a −/− mice displayed a 71.4% reduction (p=0.01) in MK proplatelet formation (Figure 3D). These results suggested the effect of the miRNA cluster on platelet number may be at the late stages of thrombopoiesis.
Figure 3. The Mir-99b/let7e/125a cluster is required for normal MK proplatelet formation, but not for differentiation, ploidy or cytoplasmic maturation.
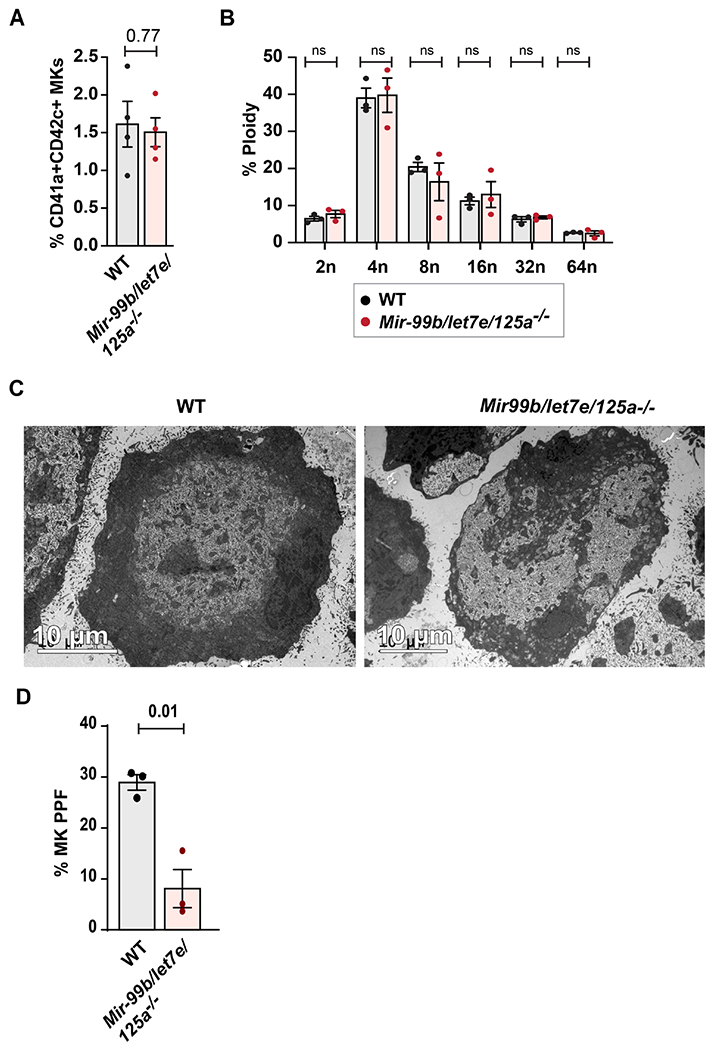
A. The percentage of CD41a+ CD42c+ bone marrow MKs were measured by flow cytometric analysis in Mir-99b/let-7e/125a −/− or littermates (n=4 each group). B. Ploidy analysis of ex vivo cultured bone marrow MKs at day 5, detected by propidium iodide staining. Total percentage of CD41a+ propidium iodide positive MKs representing 2n, 4n, 8n, 16n, 32n and 64n ploidy were plotted for Mir-99b/let-7e/125a −/− or littermate mice (n=3 each). Sub-2n and apoptotic cells were gated out in this analysis. C. Representative transmission electron microscopic images showing ex vivo cultured bone marrow derived MK for Mir-99b/let-7e/125a −/− or littermate mice (n=3). D. Proplatelet formation assay for ex vivo cultured MKs isolated from femurs of Mir-99b/let7e/125a −/− or littermates (n=3 each). Quantification was done blinded to the treatment. Schematic representation shows experimental design for generating Mir-99b/let7e/125a cluster mice that were flanked with loxP sites.
Mir-99b/let7e/125a −/− mice has no detectable platelet function defects
In additional to the quantitative effect on platelet numbers, it was of interest to assess whether MK/platelet-specific expression of Mir-99b/let7e/125a alters platelet function. Compared to platelets from wild type littermates, platelets from Mir-99b/let-7e/125a −/− mice showed no differences in measures of platelet agonist-induced αIIbβ3 integrin activation (Figure 4A), alpha-granule release (Figure 4B) or platelet aggregation (Figure 4C). In addition, platelets from the knockout mice showed no differences in ultra-structural morphology (Figure 4D) or spreading on fibrinogen or collagen (Figure 4E–F). There was also no difference in mean platelet volume (data not shown). Finally, compared to platelets from wild type littermates, platelets from Mir-99b/let-7e/125a −/− mice showed no significant difference in survival in a pulmonary thromboembolism model (Figure 4G). Thus, Mir-99b/let7e/125a cluster is dispensable for murine platelet function.
Figure 4. MK/platelet specific loss of Mir-99b/let7e/125a does not alter platelet function or in vivo thrombosis.
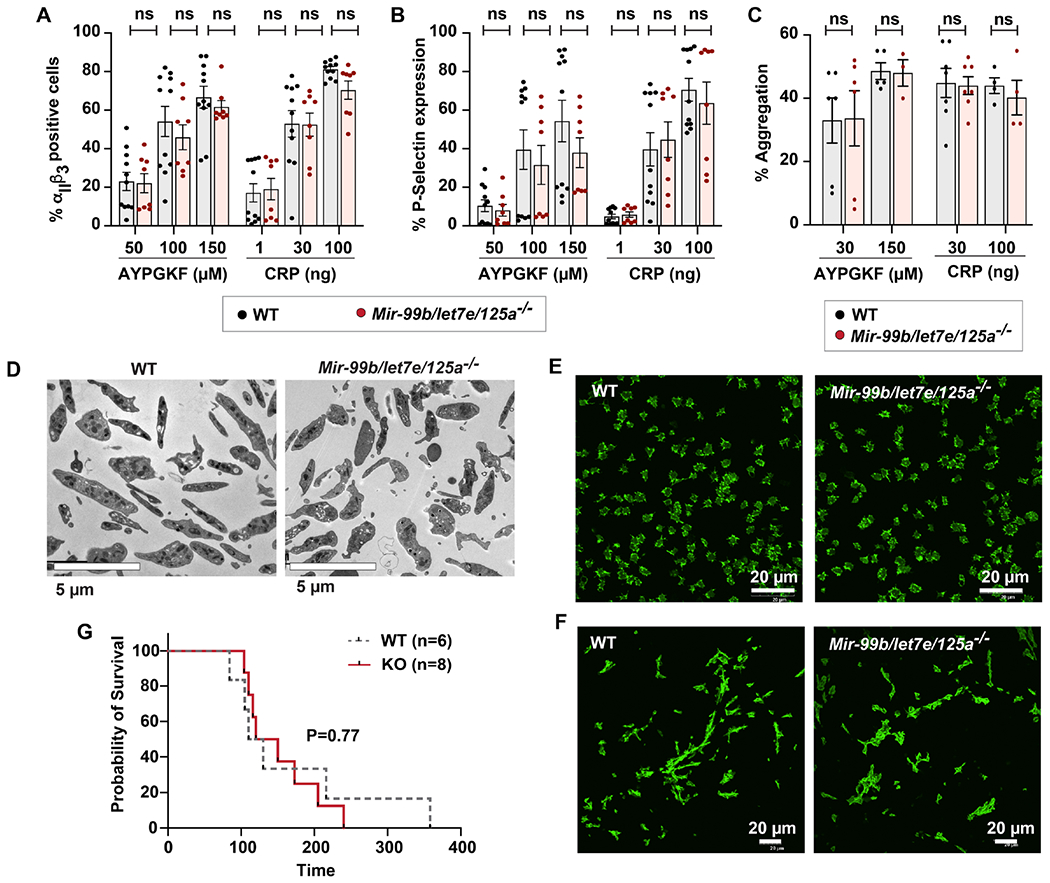
A-B. Washed platelets from Mir-99b/let7e/125a −/− (n=8) or littermates (n=11) were activated with 50, 100 or 150 μM AYPGKF and 1, 30 or 100 ng/ml collagen receptor peptide (CRP). A. JON/A binding and B. P-selectin was measured by flow cytometry. C. Washed platelets from Mir-99b/let7e/125a −/− (n=8) or littermates (n=11) were activated with AYPGKF (30 and 100 μM) or collagen (30 and 100 ng). Aggregation was measured using Chronolog aggregometer and plotted (n=3 to 7). D-E. Phalloidin staining of washed platelets isolated from Mir-99b/let7e/125a −/− or littermates, that were adhered on D. immobilized fibrinogen or E. collagen (n=3 each). F. Pulmonary thromboembolism was induced by injecting Mir-99b/let7e/125a −/− mice or littermates with collagen and epinephrine (n=10 each). Time to death was monitored and plotted.
Discussion
Prior work has shown diverse roles of miRNAs in MK and platelet physiology [10–18]. miR-125a-5p regulates MK proplatelet formation via targeting actin bundling protein, L-plastin [2]. In humans, about 40% of miRNA genes localize as clusters in the genome [19]. miRNA clusters are regulated by genetic and epigenetic modifications, and such clusters regulate many cellular processes, including proliferation, differentiation, immunity, apoptosis, DNA repair and self-renewal [20]. Aberrant expression of miRNA clusters can impact the pathogenesis of many diseases, including carcinogenesis [21]. The Mir-99b/let7e/miR125a cluster has been linked to oncogenesis[22], tumor metastasis [23], inflammation [24–26] and mechanisms leading to cystic fibrosis pathogenesis [27]. This is the first report of the generation of a MK/platelet specific Mir-99b/let-7e/125−/− mice strain and its characterization in MK-platelet function. The major findings in this report are that expression of this cluster in murine MKs is required for normal MK proplatelet formations and normal platelet production. Prior studies overexpressing the miR-99b/let7e/125a cluster have suggested that miR-125a-5p may account for the majority of effects on murine hematopoiesis [3], consistent with our MK-specific findings in this report and prior reports in cultured human MKs [2].
miRNAs in genomic clusters are often transcribed into a common precursor pri-miRNA,, e.g. the miR-17/92 cluster and its paralogues[28], and there is RT-PCR evidence for a single transcript in HeLa cells that contains each of the three miRNAs in the miR-99b/let7e/125a cluster.[27] The strong correlations we observed in human platelets support a similar mechanism of transcriptional regulation of a polycistronic RNA containing all three genes.
Conclusions
This is the first report of generating MK/platelet specific miRNA cluster knockout mice strain and its characterization in MK-platelet physiology. The MK-specific expression of Mir-99b/let7e/125a positively regulates murine platelet numbers without any deleterious effects on MK differentiation or platelet activation/function. This discovery has potential implications for several translational applications including hematologic malignancies, in vitro platelet production and transfusion medicine. Lastly, numerous studies have linked each of the miRNAs of the cluster individually to a different condition. But since each miRNA can have numerous targets, over-expressing or silencing these miRNAs one at a time shows their individual roles in a specific physiology or pathophysiology. Deleting a cluster of miRNAs asks a different question altogether - that of “synergy.” This is only the second example we are aware of in which an entire cluster of miRNAs has been deleted, and the only MK-specific miRNA cluster deletion. As such, our flowed mice may prove instructive for assessing function in other cell types.
Supplementary Material
Highlights:
The Mir-99b/let7e/125a cluster regulates late thrombopoiesis and platelet count
First generation of megakaryocyte-platelet-specific miRNA knockout
Funding:
This work was supported by grant from the National Institutes of Health National Heart, Lung and Blood Institute (HL14124) and funding from the Division of Hematology and Hematologic Malignancies, University of Utah. The authors thank Ms. Diana Lim for assistance with her analysis of figures and software consultation, and Ms. Linda Nikolova for technical assistance in the Electron Microscopy Core Laboratory.
Abbreviations:
- MK
megakaryocyte
- Mkpoiesis
megakaryopoiesis
- PF4
platelet factor 4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- [1].Rowley JW, Chappaz S, Corduan A, Chong MM, Campbell R, Khoury A, Manne BK, Wurtzel JG, Michael JV, Goldfinger LE, Mumaw MM, Nieman MT, Kile BT, Provost P, Weyrich AS, Dicer1-mediated miRNA processing shapes the mRNA profile and function of murine platelets, Blood, 127 (2016) 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bhatlekar S, Manne BK, Basak I, Edelstein LC, Tugolukova E, Stoller ML, Cody MJ, Morley SC, Nagalla S, Weyrich AS, Rowley JW, O’Connell RM, Rondina MT, Campbell RA, Bray PF, miR-125a-5p regulates megakaryocyte proplatelet formation via the actin-bundling protein L-plastin, Blood, 136 (2020) 1760–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E, van Os R, Bystrykh LV, de Haan G, Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells, Blood, 119 (2012) 377–387. [DOI] [PubMed] [Google Scholar]
- [4].Huffaker TB, Ekiz HA, Barba C, Lee SH, Runtsch MC, Nelson MC, Bauer KM, Tang WW, Mosbruger TL, Cox JE, Round JL, Voth WP, O’Connell RM, A Stat1 bound enhancer promotes Nampt expression and function within tumor associated macrophages, Nat Commun, 12 (2021) 2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS, Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes, Blood, 118 (2011) e101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shi DS, Smith MC, Campbell RA, Zimmerman PW, Franks ZB, Kraemer BF, Machlus KR, Ling J, Kamba P, Schwertz H, Rowley JW, Miles RR, Liu ZJ, Sola-Visner M, Italiano JE Jr., Christensen H, Kahr WH, Li DY, Weyrich AS, Proteasome function is required for platelet production, J Clin Invest, 124 (2014) 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Manne BK, Badolia R, Dangelmaier C, Eble JA, Ellmeier W, Kahn M, Kunapuli SP, Distinct pathways regulate Syk protein activation downstream of immune tyrosine activation motif (ITAM) and hemITAM receptors in platelets, J Biol Chem, 290 (2015) 11557–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Manne BK, Munzer P, Badolia R, Walker-Allgaier B, Campbell RA, Middleton E, Weyrich AS, Kunapuli SP, Borst O, Rondina MT, PDK1 governs thromboxane generation and thrombosis in platelets by regulating activation of Raf1 in the MAPK pathway, J Thromb Haemost, 16 (2018) 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tiedt R, Schomber T, Hao-Shen H, Skoda RC, Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo, Blood, 109 (2007) 1503–1506. [DOI] [PubMed] [Google Scholar]
- [10].Georgantas RW 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI, CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control, Proc Natl Acad Sci U S A, 104 (2007) 2750–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D, Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder, J Exp Med, 205 (2008) 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Romania P, Lulli V, Pelosi E, Biffoni M, Peschle C, Marziali G, MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors, Br J Haematol, 143 (2008) 570–580. [DOI] [PubMed] [Google Scholar]
- [13].Chapnik E, Rivkin N, Mildner A, Beck G, Pasvolsky R, Metzl-Raz E, Birger Y, Amir G, Tirosh I, Porat Z, Israel LL, Lellouche E, Michaeli S, Lellouche JP, Izraeli S, Jung S, Hornstein E, miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis, Elife, 3 (2014) e01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR, MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors, Dev Cell, 14 (2008) 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Navarro F, Gutman D, Meire E, Caceres M, Rigoutsos I, Bentwich Z, Lieberman J, miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53, Blood, 114 (2009) 2181–2192. [DOI] [PubMed] [Google Scholar]
- [16].Kondkar AA, Bray MS, Leal SM, Nagalla S, Liu DJ, Jin Y, Dong JF, Ren Q, Whiteheart SW, Shaw C, Bray PF, VAMP8/endobrevin is overexpressed in hyperreactive human platelets: suggested role for platelet microRNA, J Thromb Haemost, 8 (2010) 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, Chen J, McKnight GS, Lopez JA, Yang L, Jin Y, Bray MS, Leal SM, Dong JF, Bray PF, Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity, Blood, 117 (2011) 5189–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Basak I, Bhatlekar S, Manne BK, Stoller M, Hugo S, Kong X, Ma L, Rondina MT, Weyrich AS, Edelstein LC, Bray PF, miR-15a-5p regulates expression of multiple proteins in the megakaryocyte GPVI signaling pathway, J Thromb Haemost, 17 (2019) 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H, Clustering and conservation patterns of human microRNAs, Nucleic Acids Res, 33 (2005) 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kabekkodu SP, Shukla V, Varghese VK, J DS, Chakrabarty S, Satyamoorthy K, Clustered miRNAs and their role in biological functions and diseases, Biol Rev Camb Philos Soc, 93 (2018) 1955–1986. [DOI] [PubMed] [Google Scholar]
- [21].Kabekkodu SP, Shukla V, Varghese VK, Adiga D, Vethil Jishnu P, Chakrabarty S, Satyamoorthy K, Cluster miRNAs and cancer: Diagnostic, prognostic and therapeutic opportunities, Wiley Interdiscip Rev RNA, 11 (2020) e1563. [DOI] [PubMed] [Google Scholar]
- [22].Zhang Y, Gao JS, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, Ramratnam B, MicroRNA 125a and its regulation of the p53 tumor suppressor gene, FEBS Lett, 583 (2009) 3725–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma J, Zhan Y, Xu Z, Li Y, Luo A, Ding F, Cao X, Chen H, Liu Z, ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma, Cancer Lett, 398 (2017) 37–45. [DOI] [PubMed] [Google Scholar]
- [24].Hildebrand D, Eberle ME, Wolfle SM, Egler F, Sahin D, Sahr A, Bode KA, Heeg K, Hsa-miR-99b/let-7e/miR-125a Cluster Regulates Pathogen Recognition Receptor-Stimulated Suppressive Antigen-Presenting Cells, Front Immunol, 9 (2018) 1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pelosi A, Alicata C, Tumino N, Ingegnere T, Loiacono F, Mingari MC, Moretta L, Vacca P, An Anti-inflammatory microRNA Signature Distinguishes Group 3 Innate Lymphoid Cells From Natural Killer Cells in Human Decidua, Front Immunol, 11 (2020) 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Urena-Peralta JR, Perez-Moraga R, Garcia-Garcia F, Guerri C, Lack of TLR4 modifies the miRNAs profile and attenuates inflammatory signaling pathways, PLoS One, 15 (2020) e0237066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Endale Ahanda ML, Bienvenu T, Sermet-Gaudelus I, Mazzolini L, Edelman A, Zoorob R, Davezac N, The hsa-miR-125a/hsa-let-7e/hsa-miR-99b cluster is potentially implicated in Cystic Fibrosis pathogenesis, J Cyst Fibros, 14 (2015) 571–579. [DOI] [PubMed] [Google Scholar]
- [28].Mogilyansky E, Rigoutsos I, The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease, Cell Death Differ, 20 (2013) 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


