ABSTRACT
Transgenic technology for mosquitoes is now more than two decades old, and a wide array of control sequences have been described for regulating gene expression in various life stages or specific tissues. Despite this, comparatively little attention has been paid to the development and validation of other transgene-regulating elements, especially 3ʹ-untranslated regions (3ʹUTRs). As a consequence, the same regulatory sequences are often used multiple times in a single transgene array, potentially leading to instability of transgenic effector genes. To increase the repertoire of characterized 3ʹUTRs available for genetics-based mosquito control, we generated fifteen synthetic sequences based on the base composition of the widely used SV40 3ʹUTR sequence, and tested their ability to contribute to the expression of reporter genes EGFP or luciferase. Transient transfection in mosquito cells identified nine candidate 3ʹUTRs that conferred moderate to strong gene expression. Two of these were engineered into the mosquito genome through CRISPR/Cas9-mediated site-specific insertion and compared to the original SV40 3ʹUTR. Both synthetic 3ʹUTRs were shown to successfully promote transgene expression in all mosquito life stages (larva, pupa and adults), similar to the SV40 3ʹUTR, albeit with differences in intensity. Thus, the synthetic 3ʹUTR elements described here are suitable for regulating transgene expression in Ae. aegypti, and provide valuable alternatives in the design of multi-gene cassettes. Additionally, the synthetic-scramble approach we validate here could be used to generate additional functional 3ʹUTR elements in this or other organisms.
KEYWORDS: Aedes aegypti, transgenic, 3ʹ-untranslated region, genome editing, synthetic biology
Introduction
Aedes mosquitoes are major vectors for several viruses of public health concern, including Zika virus (ZIKV), yellow fever virus (YFV), dengue virus (DENV) and chikungunya virus (CHIKV), with up to 3.9 billion people worldwide at risk [1]. Genetic approaches designed to control mosquito-borne diseases have been rigorously pursued and have led to advances in genome-editing and transgenesis technologies with the use of transposable elements (TEs) [2–5], homing endonucleases (HEs) [6,7], zinc finger nucleases (ZFNs) [8], and transcription activator-like effector nucleases (TALENs) [9,10]. The recent emergence of CRISPR technologies [11] has provided a pathway to new potential tools for the control of disease vector mosquito populations [12–18].
For efficient genetic control strategies, appropriate gene regulation is essential to achieve spatio-temporal activation of any potential effector molecules. Salivary gland or midgut-specific promoters controlling the expression of RNA interference (RNAi) effector sequences were able to suppress the infection cycle of DENV, reducing the risk of transmission to a vertebrate host [19–21]. For Anopheles mosquito vectors, fatbody-specific vitellogenin (Vg) or midgut-specific carboxypeptidase (Cp) promoters were able to drive expression of effector genes capable of killing malaria parasites [22]. Germline-specific expression of Cas9 controlled by the exu gene promoter significantly enhanced the rate of homology-directed repair events [23], contributing to a gene drive approach in Ae. aegypti [15]. The promoters of nanos [24,25] and oskar [26] were also shown in Ae. aegypti to activate gene expression in developing oocytes, and β2-tubulin [27] in the testis. Meanwhile, various promoter elements were characterized for general engineering purposes, such as the constitutive polyubiquitin (PUb) promoter [28], the eye-specific synthetic 3xP3 promoter [29], heat-activated Hsp70 promoters [30], and Pol III (U6)/7SK promoters for guide RNA transcriptional activity [31].
In eukaryotes, the 3ʹ-untranslated region (3ʹUTR) also functions as a significant regulatory cis-element responsible for mRNA stability, localization, or translational efficiency [32,33]. While numerous promoters are available for transgenic mosquitoes as mentioned above, the simian virus 40 3ʹUTR element (SV40 3ʹUTR) has been the primary element utilized for mRNA expression from transgenic marker genes. This often results in multiple uses of the same genetic element in a single transgenic construct, and it can cause an unwanted loss or conversion of the transgenic cargo genes from the transgenic mosquito genome [25]. For example, following the induction of a double-stranded DNA break, a eukaryotic DSB repair mechanism called single-strand annealing (SSA) can trigger the loss of gene regions flanked by repeated sequences [34]. Thus, there is value in developing a pool of 3ʹUTRs with similar activity but independent sequences. Here, we substantially expand the repertoire of 3ʹUTR variants for transgenesis-based strategies by shuffling the 3ʹUTR sequence of SV40. When positioned downstream of an EGFP or luciferase reporter sequence under the control of a strong promoter, several of these synthetic 3ʹUTRs exhibited moderate to substantial gene expression in mosquito cells. Two of these elements were selected for the generation of EGFP-expressing transgenic lines, and both were found to be functional in all developmental stages of Aedes aegypti when used in conjunction with the PUb promoter. Thus, we were able to increase the number of novel 3ʹUTRs currently available for genetic engineering in mosquitoes through synthetic biology, an approach that could be adapted to other emerging or established model organisms.
Results
Synthetic 3ʹUTR sequences contribute to reporter gene expression in mosquito cells
While the 3ʹUTR derived from the SV40 virus has been widely used to aid in the expression of transgenes in insects, including mosquitoes, little is known of the specific regulatory elements needed for this process. Since the SV40 3ʹUTR has been shown to broadly control gene expression in many exogenous insect species, we reasoned that this may be conferred primarily by base composition and the presence of AAUAAA motifs, suggesting synthetic alternatives could be developed through randomizing the existing sequence. Thus, we scrambled the SV40 3ʹUTR (~230 bp) to generate 16 synthetic sequences, each of which contained the same base composition and length as the starting sequence, along with two polyadenylation motifs (AATAAA) [35] (see Materials and Methods). Each synthetic 3ʹUTR, except for #9 (for which cloning attempts were unsuccessful), was placed downstream of the Ae. aegypti PUb promoter and an EGFP reporter (Fig. 1A), in place of the SV40 3ʹUTR, in a previously described expression cassette [28]. Following transfection into A20 or Aag2 mosquito cells with each plasmid construct, the intensity of EGFP fluorescence was determined by imaging cytometry, with normalization based on cell counts (Fig. 1 B and C). According to test results in A20 cells obtained from 8 biological replicates (Fig. 1B), fifteen synthetic 3ʹUTR sequences were classified into four groups: (a) #5 displaying strong activity (+++); (b) #2, 3, 12, 13, and 16 displaying intermediate activity (+); (c) #7, 8, and 15 displaying marginal, but significant activity (±); (d) #1, 4, 6, 10, 11, and 14 displaying no activity (‒). A similar pattern of differential gene expression was observed in Aag2 cell-based assays, while 3ʹUTR #7 did not show significant activity in this cell line (Fig. 1C). We also identified differential levels of 3ʹUTR-supported EGFP fluorescence by microscopic observation, while levels of internal control marker (DsRED) were equal throughout all transfected samples (Fig. 1D).
Figure 1.
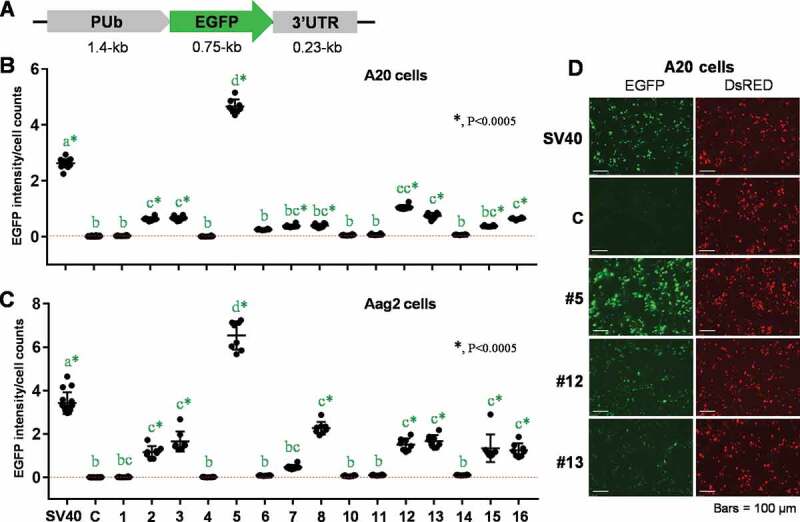
Effects of synthetic 3ʹUTRs on EGFP expression under the control of polyubiquitin gene (PUb) promoter sequence in mosquito cells. (A) The schematic representation of plasmid constructs, pSLfa-PUb-EGFP-3ʹUTRs. (B and C) Insect cell-based EGFP imaging analysis. Following transient transfections of each plasmid to A20 (B) and Aag2 (C) cells in 8 biological replicates, transformed cells were measured for EGFP fluorescent intensity by using UV-assisted imaging cytometry. The gene expression activity was determined by normalizing EGFP fluorescence intensities to total cell counts. The 3ʹUTRs with significant EGFP-expressing activity, compared to the negative control (c), were designated by green stars. Tukey’s multiple comparison test (One-way ANOVA): *, P < 0.0005. (D) The EGFP marker expression with synthetic or control 3ʹUTRs was examined under the fluorescent microscope at 48 hr post transfection. DsRED fluorescence was visualized along with EGFP as the internal control of transfection efficiency. Each entire experiment was repeated at least 3 times, representative results are shown
In addition to regulatory activity of 3ʹUTRs or 5ʹUTRs, gene expression is also dependent on codon composition of the target gene, which can provide regulatory codons associated with mRNA stability and charged tRNA availability [36,37]. To evaluate the versatility of synthetic 3ʹUTR sequences in combination with alternative promoters/reporters, we also placed six of them (#3, #5, #7, #12, #13, and #16) downstream of the firefly luciferase gene under the control of the IE1 promoter (Fig. 2A), which has been used previously in mosquito cells [25]. Each plasmid was co-transfected with an internal control plasmid encoding Renilla luciferase into A20 or Aag2 cells in two independent experiments, each of which included 8 biological replicates (Fig. 2 B and C). Compared to the control plasmid pGL3-basic (Luc-SV40), all tested plasmid containing synthetic 3ʹUTRs showed significant levels of gene expression in A20 cells, equivalent to the positive control, pGL3-IE1-Luc-SV40 (Fig. 2B). Meanwhile in Aag2 cells, 3ʹUTR #3 displayed the strongest activity, with #7 not significant for gene expression (Fig. 2C). Together, our results provide evidence that novel synthetic 3ʹUTR sequences randomized from the existing SV40 sequence can be functional with multiple reporter genes and 5ʹUTR sequences as expressed under the control of either endogenous or exogenous promoter elements in two independent mosquito cell lines.
Figure 2.
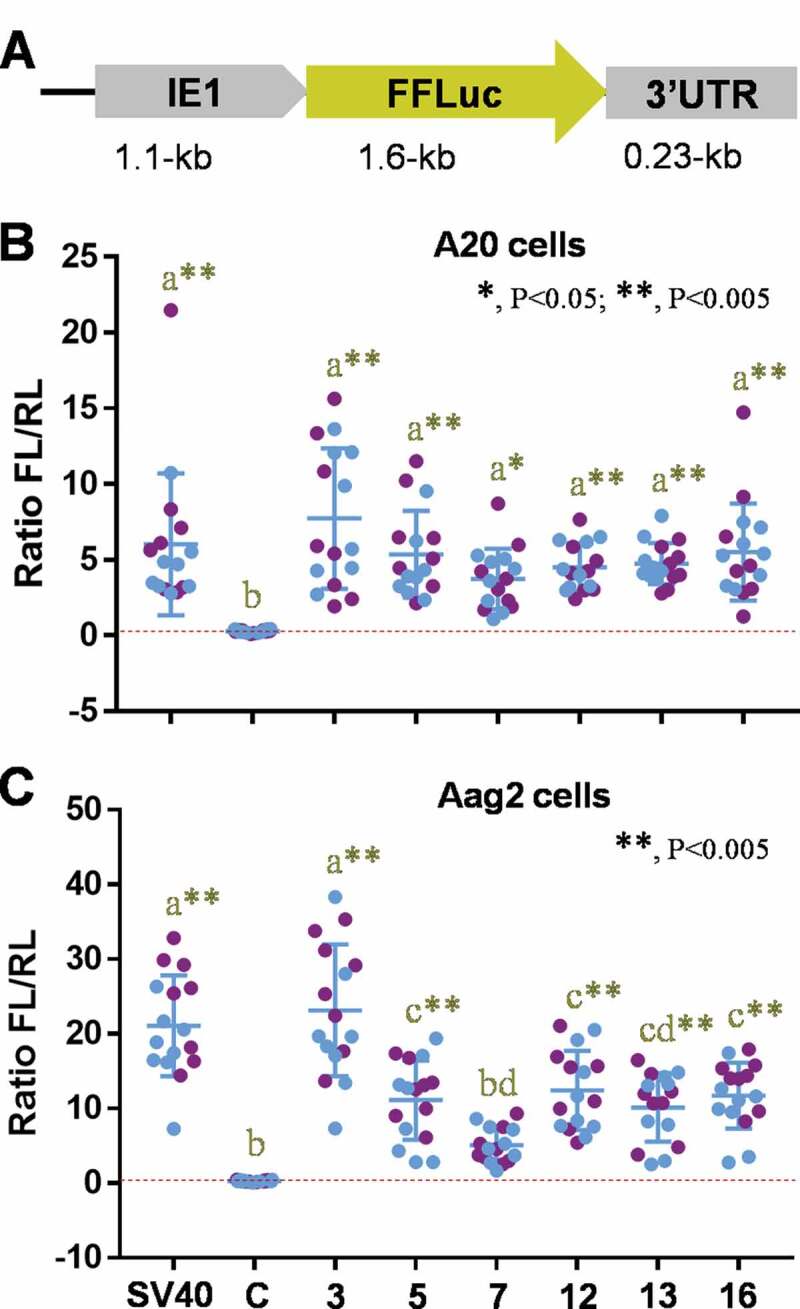
Effects of synthetic 3ʹUTRs on firefly luciferase expression under the control of baculovirus Immediate-Early gene (IE1) promoter sequence in mosquito cells. (A) The schematic representation of plasmid constructs, pSLfa-IE1-Luc-3ʹUTRs. (B and C) Dual-luciferase assay using A20 (B) or Aag2 (C) cells. The individual plasmid was co-transfected to cells with the internal control plasmid expressing Renilla luciferase in two independent assays of 8 biological replicates (blue and purple dots). The 3ʹUTRs with significant luciferase activity, compared to the negative control (c), were designated by yellow stars. Tukey’s multiple comparison test (One-way ANOVA): *, P < 0.05; **, P < 0.005
The activity of synthetic 3ʹUTR sequence did not correlate with in silico predicted features
As we observed substantial variation in EGFP or firefly luciferase expression levels between the synthetic 3ʹUTR sequences, we sought to identify any common sequence elements that might be associated with strong gene expression. We analysed each synthetic sequence using the RegRNA2.0 webserver [38] to predict RNA motifs (Fig. 3). While polyadenylation sites (PS) were identified in most of the sequences, no known motifs appeared to be associated with gene expression levels. We also sought to determine if any novel motifs were enriched in the active synthetic sequences using the MEME motif enrichment tool [39]. Once again, we were not able to identify any combinations of motifs that were present in the effective 3ʹUTR group (SV40, #2, #3, #5, #12, #13, #16) but absent in the non-effective group (#1, #4, #6, #10, #11, #14, #15) (Table S1), potentially due to the small sample size. Similarly, no correlations could be found between the predicted secondary structures (Figure S1) and minimum free energies (MFE) (Figure S2) of the 3ʹUTR RNA sequences and the gene expression levels associated with synthetic 3ʹUTRs. Depending on the positions of polyadenylation signal (PAS), mature 3ʹUTR sequences can be predicted to be different lengths, which may in turn influence RNA stability or translation efficiency. However, no such relationship appeared from our tests (Fig. 3), as the 3ʹUTR #5 sequence showed the highest activity but is anticipated to be of similar length to #2, #8, and #15, all of which showed much lower activity. Likewise, the starting SV40 sequence is amongst the longest, but was much more active than the structurally similar #1, #10, #14, all of which had little to no activity.
Figure 3.
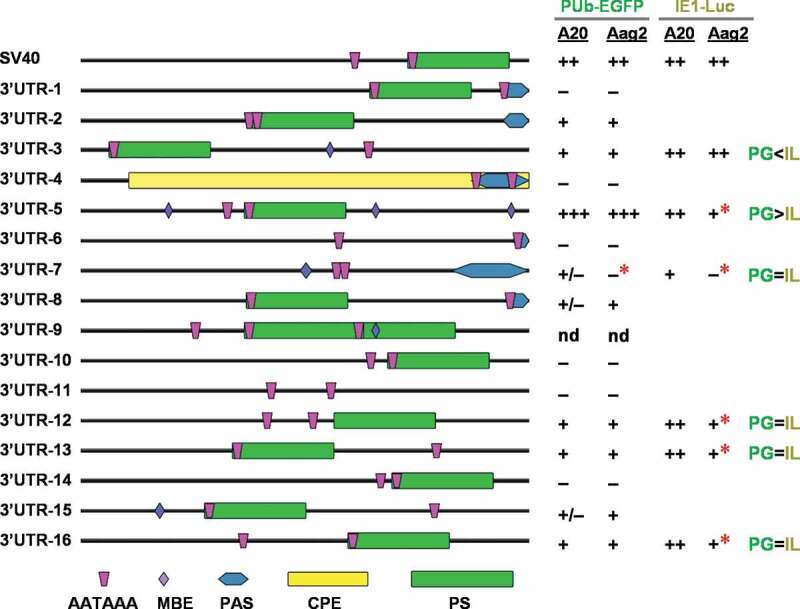
Predicted features and summarized gene expression activity of SV40 and synthetic 3ʹUTR fragments. Features were predicted using the RegRNA2.0 webserver [http://regrna2.mbc.nctu.edu.tw/detection.html] [38], and included polyadenylation site, UTRsite motifs, AU-rich elements, ERPIN and Rfam predictions, long-stem RNA structural patterns, fRNAdb, and miRNA target sites (Ae. aegypti) using the default parameters. All features identified in one or more sequences are shown; MBE, Musashi Binding Element; PAS, Polyadenylation signal; CPE, cytoplasmic polyadenylation signal; PS, polyadenylation site; AATAAA, manually inserted polyadenylation motif. Cell-based test results from Figs. 1 and Figs. 2 are presented; red stars (*), cell type-dependent activity change; nd, not done; PG, PUb-EGFP-3ʹUTR; IL, IE1-Luc-3ʹUTR
While the underlying mechanism remains unclear regarding how these small regulatory elements (~230 bp) assist in transcript expression, our results demonstrate that their gene expression levels can be varied through potential interactions with cis-regulatory elements in 5ʹUTR or reporter gene in a cell type-dependent manner (Fig. 3). While 3ʹUTR #7, 12, 13, and 16 displayed similar expression levels regardless of reporter gene (Fig. 3, PG = IL), #3 was more active with IE1-Luc than PUb-EGFP (Fig. 3, PG<IL), and #5 was the opposite (Fig. 3, PG>IL). Meanwhile, 3ʹUTR #7 showed no activity in the embryo-derived Aag2 cell line (Fig. 3, red stars), unlike a marginal level of gene expression in the larva-derived somatic A20 cell line. With the IE1-Luc elements, all the tested 3ʹUTR sequences, except for #3, were less able to support gene expression in Aag2 cells, compared to A20 (Fig. 3, red stars). Albeit not attempted for IE1-Luc, 3ʹUTR #2, 8, and 15 may also have potential for broad utilization. Thus, we conclude that some of the active synthetic fragments (n = 9) are suitable candidates for assisting in the control of transgene expression in the mosquito.
Synthetic 3ʹUTRs support EGFP expression in Ae. aegypti transgenic mosquitoes
To determine if the synthetic 3ʹUTRs could be utilized in mosquito transgene expression, we generated two site-specific insertions at the kynurenine 3-monooxygenase (kmo) gene locus, each of which contained an EGFP gene, PUb promoter, as well as a 3ʹUTR sequence with either full (+++, 3ʹUTR#5) or reduced level (+, 3ʹUTR#13) activity (Fig. 4A). A positive control was also developed that contained the EGFP gene under the control of the PUb promoter and SV40 3ʹUTR. For simplicity, we refer to these transgenic lines as kmoEGFP−3ʹUTR#5, kmoEGFP−3ʹUTR#13, and kmoEGFP-SV40 (Table 1). For all site-specific insertions, the genomic integration of the donor DNA constructs was confirmed by PCR analysis using primer pairs that were designed to amplify the sequences between the transgene and flanking genomic sequences (Fig. 4B). The donor plasmid for kmoEGFP-SV40 contains a partial fragment of the DsRED gene between the homology arm 1 and the PUb promoter, which increased the size of the PCR amplicon compared to others (Fig. 4B).
Figure 4.
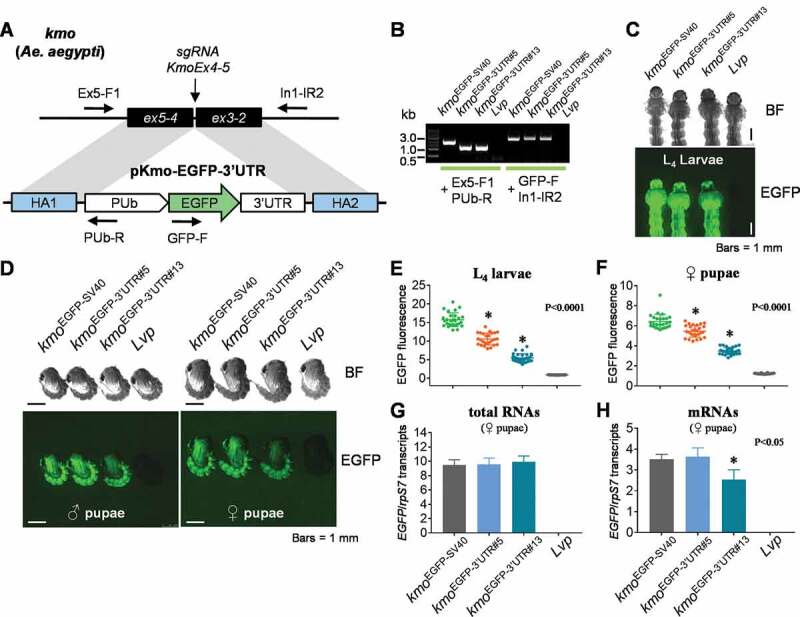
EGFP expression patterns and levels influenced by synthetic 3ʹUTRs in Ae. aegypti transgenic lines. (A) The schematic representation of the plasmid constructs of DNA donors and Aedes aegypti kmo gene locus. The vertical arrow indicates the DNA double strand break site induced by the CRISPR/Cas9-sgRNA complex. The horizontal arrows indicate PCR primers (Table S2) used for identifying chromosomal integration of the transgene. (B) PCR-based genotyping analysis for donor plasmid constructs in transgenic lines. Two pairs of PCR primers were utilized to recognize the junction areas between cargo genes and chromosomal sequences outside of HAs (Figure 4A). (C and D) Synthetic 3ʹUTR-controlled transgene levels and patterns in mosquito growth stages of L4 instars (C) and pupae (D). BF, bright field; EGFP, UV-induced EGFP fluorescence. (E and F) The ImageJ-based quantification of EGFP fluorescence. About 25 microscopic images of L4 larvae (E) or female pupae (F) were taken, and the EGFP intensity of individual specimen was measured by ImageJ. Tukey’s multiple comparison test (One-way ANOVA): *, P < 0.0001. (G and H) RT-qPCR analysis for 3ʹUTR-regulated transcript levels. Total RNA was purified from 10 female pupae of each transgenic mosquito line. cDNAs were synthesized by using random hexamers for the generation of total RNA levels (G) or using oligo-dT for mRNA levels (H). Quantitative PCR was performed in triplicate to evaluate EGFP transcripts normalized by ribosomal protein S7 transcripts (rpS7). Tukey’s multiple comparison test (One-way ANOVA): *, P < 0.05
Transgenic mosquito lines were examined for EGFP expression levels and patterns of expression in life stages prior to mature adults (Fig. 4 C and D). In both L4 larvae and pupae, kmoEGFP−3ʹUTR#5 was associated with robust expression of EGFP, but the EGFP level of kmoEGFP−3ʹUTR#13 was lower than that observed in the other lines. ImageJ analysis (n > 25 images) showed variations in EGFP fluorescence intensity of L4 larvae and female pupae: kmoEGFP-SV40 > kmoEGFP−3ʹUTR#5 > kmoEGFP−3ʹUTR#13 (Fig. 4 E and F). Despite the relatively weaker activity of 3ʹUTR #13 in cell-based assays, this element was still sufficient to support a significant level of transgenic marker gene expression in the mosquito. This suggests that many of the other synthetic 3ʹUTR sequences we validated in vitro could also be good candidates for mosquito genome engineering.
To begin to understand how the examined 3ʹUTR sequences contribute to the differential regulation of gene expression, we measured total or mature RNA (polyA+) transcripts at the pupal stage in each female transgenic mosquito strain (Fig. 4 G and H; Table S3). While the level of EGFP fluorescence varied between strains (Fig. 4F), levels of total EGFP RNA were equivalent in all transgenic lines (Fig. 4G). This is not surprising as the marker genes were under the control of the same PUb promoter, and suggests that the synthetic 3ʹUTRs did not adversely affect transcriptional activity. However, the levels of mature EGFP mRNA in the kmoEGFP−3ʹUTR#13 strain were found to be significantly lower than that of the other two strains (Fig. 4H). This pattern is consistent with the variation in EGFP fluorescence intensity observed in between kmoEGFP−3ʹUTR#5 and kmoEGFP−3ʹUTR#13 strains, potentially suggesting their differential roles in RNA maturation/stability. Meanwhile, we observed that EGFP mRNA levels between kmoEGFP-SV40 and kmoEGFP−3ʹUTR#5 were identical (Fig. 4H), but their EGFP fluorescence intensities were significantly different (Fig. 4F). This suggests that the synthetic 3ʹUTR#5 may be less efficient in promoting translation.
In either male or female adult mosquitoes, both kmoEGFP−3ʹUTR#5 and kmoEGFP−3ʹUTR#13 showed EGFP expression in the whole body, but the EGFP level was reduced compared to that of kmoEGFP-SV40 (Fig. 5 A and B). As the PUb promoter is known to be active in organs such as the midgut, as well as in somatic ovarian cells [28], we sought to determine if the synthetic 3ʹUTR fragments would alter the expression patterns of genes expressed from this regulatory element. We dissected ovaries from blood-fed female mosquitoes and examined EGFP fluorescence (Fig. 5 C and D). An identical pattern of EGFP fluorescence was detected in the somatic tissue of the developing ovaries in kmoEGFP−3ʹUTR#5 and kmoEGFP−3ʹUTR#13 mosquitoes at 24 hr and 48 hr post bloodmeal, respectively, again with stronger intensity of expression in kmoEGFP−3ʹUTR#5. The kmoEGFP−3ʹUTR#5 and kmoEGFP−3ʹUTR#13 strains also displayed robust EGFP expression in the foregut, similar to kmoEGFP-SV40. While both strains displayed EGFP expression in the midgut (Fig. 5E), levels were lower in lines containing the synthetic 3ʹUTRs in comparison to those observed in the line with the SV40 control sequence. Nevertheless, the synthetic 3ʹUTRs described here did not substantially alter patterns of expression from the PUb promoter in any significant way, facilitating the expression of a marker gene at levels similar (or reduced) to those observed with the control SV40 sequence. Thus, the two synthetic sequences, 3ʹUTR#5 with high activity (+++) and #13 with low activity (+) as determined by our cell-based assays following in silico nucleotide randomization, were able to facilitate EGFP expression from the PUb promoter in broad life stages and tissues of Ae. aegypti and will be useful for the identification and tracking of transgenic Ae. aegypti strains.
Figure 5.
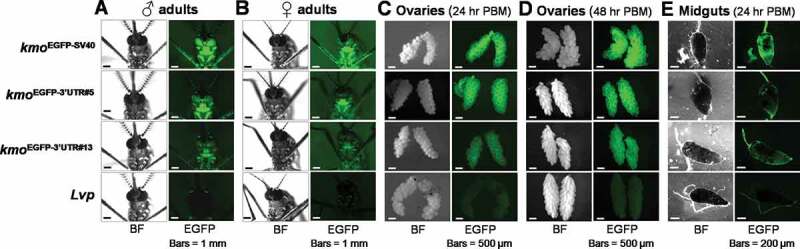
EGFP expression patterns influenced by synthetic 3ʹUTRs in the whole body, ovaries, and midguts of Ae. aegypti transgenic adult mosquitoes. (A and B) Synthetic 3ʹUTR-controlled transgene levels and patterns in male (A) and female (B) adults. (C and D) Transgenic mosquito ovaries dissected at 24 hr (C) and 48 hr (D) post bloodmeal (PBM), respectively. (E) Midgut tissues obtained at 24 hr post bloodmeal. BF, bright field; EGFP, UV-induced EGFP fluorescence
Discussion
In eukaryotic genes, 3ʹUTRs are significant regulatory elements whose presence and activity help determine the potential of mRNA to be converted into functional protein through post-transcriptional mechanisms [32,33]. It is common for the activity of a 3ʹUTR to depend on one or more effectors recruited by RNA-binding proteins (RBPs), which bind to the 3ʹUTR in a sequence- or secondary structure-specific manner [32]. For mRNA localization in polarized cells, 3ʹUTR-interacting RBPs bind motor proteins, allowing mRNAs to move along filaments of the cytoskeleton. In Drosophila oocytes, oskar mRNAs are transported along microtubules to the posterior pole of the oocyte [40–42], where Oskar protects nanos mRNA stability by precluding deadenylating effectors that bind to the 3ʹUTR sequence [43], which subsequently determines a morphogenic gradient important for embryo pattern formation [44,45]. In our results, randomizing the sequence composition of the established SV40 3ʹUTRs allowed synthetic 3ʹUTRs to control reporter expression levels, although no specific cis-element motif was yet identified as being critical to the stability of the mRNA or translational efficiency. Future experiments may also address how the 3ʹUTRs described here differ from each other in terms of mRNA localization, export from the nucleus, and recruitment to ribosomes. Likewise, scaling up the cell-based reporter assays we developed here could enable screening of hundreds of additional 3ʹUTR elements, which may aid in the discovery of specific critical motifs. Ultimately, while our goal in this study was the development of novel useful tools, detailed mutagenesis of both the SV40 and synthetic derivatives will likely be necessary to identify the critical features of these elements to provide a mechanistic basis for our observed results.
In this study, we demonstrated that synthetic 3ʹUTRs, generated by randomizing the established element SV40 sequence, were able to support the expression of a transgene under the control of the Ae. aegypti PUb promoter. Based upon the expression of EGFP and luciferase marker genes in mosquito cells, 9 (60%) out of the 15 synthetic sequences will be potentially available for various bioengineering purposes with diverse genetic elements and cell types. Transgenesis-based site-specific insertion experiments allowed us to compare the activity of these synthetic 3ʹUTRs at an identical chromosomal position, controlling for the influence of local chromatin structure on gene expression levels. Thus, synthetic 3ʹUTRs can be applied in the expression of various marker genes for purposes related to the screening of transgenic mosquitoes. This new repertoire of novel 3ʹUTRs will allow researchers to avoid using repetitive sequences in transgenic cargos where multiple expression cassettes are present and thereby to prevent unwanted transgene deletion due to identical sequence repeats [25], ultimately preserving transgene integrity in the target host genome. These synthetic biology techniques enlarge the genome engineering toolbox available for the design of genetic approaches to the control of disease-vector mosquitoes.
Materials and Methods
Constructing novel 3ʹUTR repertoire
A 226 nt sequence corresponding to the SV40 3ʹUTR commonly used in conjunction with transgene expression was scrambled using the Shuffle DNA function as part of the Sequence Manipulation Suite webserver [https://www.bioinformatics.org/sms2/shuffle_dna.html] [35]. From a pool of random sequences, 16 synthetic fragments were selected based on the presence of at least one AATAAA polyadenylation motif. As the SV40 starting sequence contains two such motifs, each selected sequence was manually adjusted to incorporate a second such site, which was selected based upon similarity to AATAAA in order to minimize sequence alterations. Compensatory changes in each sequence were made in order to keep the base composition identical to the starting SV40 sequence.
Subcloning
The shuffled 3ʹUTR sequences were synthesized to include two flanking restriction enzyme sites, NotI at the 5ʹ-terminus and EcoRI or XhoI at the 3ʹ-terminus (Epoch Life Sci.) (Appendix 1); these fragments were individually subcloned into the corresponding sites of pSLfa-PUb-EGFP-SV40 [28], with each synthetic fragment replacing SV40, and confirmed by sequencing. These pSLfa-based plasmid constructs were used to evaluate the regulatory activity of the synthetic 3ʹUTRs on EGFP expression in mosquito cells. For the plasmid constructs for luciferase gene expression, the synthetic 3ʹUTR sequences were individually subcloned into pGL3-IE1-Luc, replacing SV40 (Epoch Life Sci.). For the development of the kmoEGFP−3ʹUTR#5 or kmoEGFP−3ʹUTR#13 transgenic strains, the donor plasmid constructs, pKmo-PUb-EGFP-3ʹUTR#5 (Appendix 2) or pKmo-PUb-EGFP-3ʹUTR#13 (Appendix 3), were constructed by using Golden Gate Assembly (NEB) of three plasmid constructs: pGSP1-KmoHA1; pGSP2-PUb-EGFP-3ʹUTR#5 or pGSP2-PUb-EGFP-3ʹUTR#13; pGSP3-KmoHA2. For pGSP2-PUb-EGFP-3ʹUTRs, the MluI-XhoI fragment of PUb-EGFP-3ʹUTR was ligated into the pGSP2-mcs plasmid backbone. The pGSP3-KmoHA2 was obtained from pGSP3.8 C-PUb-EGFP-KmoHA2 by removing the Bsu36I-PmeI fragment of PUb-GFP-SV40. For the kmoEGFP-SV40 strain, the donor plasmid construct, pBR-KmoEx4 (Appendix 4) was constructed by using Golden Gate Assembly (NEB) of three plasmid constructs: pGSP1-KmoHA1; pGSP2-REDh-SV40; pGSP3.8 C-PUb-EGFP-KmoHA2.
Mosquito cell-based assay for EGFP fluorescence intensities
Aedes aegypti larval A20 [46] and embryonic Aag2 [47] cells were grown in Leibovitz’s (1X) L-15 Medium + L-Glutamine (Gibco) with the addition of 10% Foetal Bovine Serum (Atlanta Biologicals), 2% Tryptose Phosphate Broth (Gibco), and 1% Penicillin Streptomycin solution (Corning Inc.) in a 28°C incubator (Thermo Scientific Forma Series 3). Transfection was performed at 80% cell confluency in 96-well plates. The transfection mix per well included 250 ng/μl of each plasmid (pSLfa-PUb-EGFP-3ʹUTR), 250 ng/μl of an internal control plasmid (pMos-PUb-DsRED-SV40), 0.5 μl of TransIT-Insect Reagent (Mirus), and an amount of 1x OptiMEM (Life Technologies) to reach a total reaction volume of 10 μl. Plasmid pSLfa-PUb-EGFP-SV40 [28] and pSLfa-PUb-MCS (Addgene #52,908) were used as positive and negative control, respectively. Following 30 minute-incubation at room temperature, each transfection mix was gently mixed with cell growth media up to 100 μl and applied to adherent cells in 96-well plates, which were subsequently allowed to grow at 28°C for 6 hours. Transfected cells in each well were transferred to a new 96-well plate with an optically clear bottom for EGFP fluorescence measurement 24 h later using a SpectraMax MiniMax 300 Imaging Cytometer (Molecular Devices).
Dual-luciferase assay with mosquito cells
Each plasmid pGL3-IE1-Luc-3ʹUTR (100 ng/μl) was co-transfected with pMos-PUb-Renilla-SV40 (20 ng/μl), the internal control plasmid expressing Renilla luciferase, to the A20 or Aag2 cells at 80% confluency (96-well plates) in 8 biological replicates. Plasmid pGL3-IE1-Luc-SV40 and pGL3-basic (Promega) were used as positive and negative control, respectively. After 48 hr incubation at 28°C, the synthetic 3ʹUTR-dependent luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega) and the SpectraMax MiniMax 300 Imaging Cytometer (Molecular Devices).
Development of transgenic mosquito strains
For microinjection into Aedes aegypti embryos, the injection mixture consisted of 250 ng/μl of plasmid donor, 400 ng/μl of Cas9 protein (PNAbio) and 100 ng/μl of sgRNA-KmoEx4 (Table S2). This mixture was incubated at 37°C for 20 minutes to allow for Cas9-sgRNA complexing. The mixture was loaded into a tip-bevelled injection needle made of a laser-cut capillary tube (Sutter Instrument Co. Model P-2000) and injected into the posterior end of pre-blastoderm embryos with a constant pressure supported from a FemtoJet (Eppendorf FemtoJet 4i), as previously described [48–51]. G0 adults were outcrossed with Lvp wild-type mosquitoes at a 1-to-5 ratio of male-to-female. The G1 transgenic mosquitoes were identified by screening EGFP fluorescence in the whole body under a fluorescent microscope (Leica M165 FC).
Analysis of EGFP expression patterns in transgenic mosquitoes
The transgenic mosquitoes at various life stages (L4 Larvae, Pupae, and Adults) were examined for EGFP expression with the use of a fluorescent microscope (Leica M165 FC). Photos were taken by a microscope camera system (Leica DFC3000 G/Leica 10,450,028). The image capturing settings for larval photos were 10x magnification with 130.6 ms exposure time and 3.5 gain. Pupal photos were taken at 7.3x magnification with 254.34 ms exposure time and 3.5 gain. For quantitative evaluation of EGFP expression levels in L4 larvae or female pupae, fluorescent intensities of around 25 images were individually measured by ImageJ. Both male and female adult photos were taken at 25x and 12.5x magnification, respectively, with 180 ms exposure time and 3.5 gain. For EGFP expression patterns in ovaries and the midgut dissected at 24 or 48 hours post bloodmeal, the image capturing settings for photos were taken at 25x magnification with 180 ms exposure time and 3.5 gain.
RT-qPCR
Total RNA was purified from groups of 10 female pupae using Trizol (Invitrogen), and first-strand cDNA was synthesized from 1 µg of total RNA using SuperScript IV VILO Reverse Transcription Kit (Life Technologies). Quantitative PCR (qPCR) was performed using SsoAdvanced universal SYBR Green supermix (BioRad) in 35 cycles of denaturation at 95°C for 30 sec and annealing/extension at 57°C for 30 sec. Gene-specific primer sets were used for amplification of EGFP and ribosomal protein S7 transcripts as the PCR control, respectively (Table S2). Amplification efficiency (E) of primer pairs, Ct values obtained from three biological replicates, and calculations for ratios of EGFP to rpS7 transcripts are provided in Table S3.
Supplementary Material
Acknowledgments
We thank Chanell Dawson and Bryan Contreras in the Adelman laboratory at Texas A&M University for excellent technical assistance.
Funding Statement
This study was supported by the National Institute of Allergies and Infectious Diseases of the National Institutes of Health [AI148787], and by the USDA National Institute of Food and Agriculture, Hatch project 1018401. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the USDA
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
KC and ZNA conceived the research and experimental design and wrote the manuscript. KMM revised the manuscript. KC conducted the experiments. CV performed microscopic imaging analysis of transgenic strains and tissue dissection. CV and EJ participated in the EGFP expression test in mosquito cells. All authors read and approved the final manuscript.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Norrby R. Outlook for a dengue vaccine. Clin Microbiol Infect. 2014;20:92–94. [DOI] [PubMed] [Google Scholar]
- [2].Bourque G, Burns KH, Gehring M, et al. Ten things you should know about transposable elements 06 Biological Sciences 0604 Genetics. Genome Biol. 2018;19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kokoza VA, Martin D, Mienaltowski MJ, et al. Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene. 2001;274(1–2):47–65. [DOI] [PubMed] [Google Scholar]
- [4].Coates CJ, Jasinskiene N, Miyashiro L, et al. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95(7):3748–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jasinskiene N, Coates CJ, Benedict MQ, et al. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci U S A. 1998;95(7):3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Traver BE, Anderson MAE, Adelman ZN. Homing endonucleases catalyze double-stranded DNA breaks and somatic transgene excision in Aedes aegypti. Insect Mol Biol. 2009;18(5):623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc R Soc B Biol Sci. 2003;270(1518):921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29(2):135–136. [DOI] [PubMed] [Google Scholar]
- [10].Aryan A, Anderson MAE, Myles KM, et al. TALEN-Based Gene Disruption in the Dengue Vector Aedes aegypti. PLoS One. 2013;8(3):e60082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gantz VM, Jasinskiene N, Tatarenkova O, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112(49):E6736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hammond A, Galizi R, Kyrou K, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34(1):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kyrou K, Hammond AM, Galizi R, et al. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol. 2018;36(11):1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li M, Yang T, Kandul NP, et al. Development of a confinable gene drive system in the human disease vector Aedes aegypti. Elife. 2020;9:e51701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simoni A, Hammond AM, Beaghton AK, et al. A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat Biotechnol. 2020;38(9):1054–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Carballar-Lejarazú R, Ogaugwu C, Tushar T, et al. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 2020;117(37):22805–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Galizi R, Hammond A, Kyrou K, et al. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci Rep. 2016;6(1):31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mathur G, Sanchez-Vargas I, Alvarez D, et al. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2010;19:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Franz AWE, Sanchez-Vargas I, Adelman ZN, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci U S A. 2006;103(11):4198–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yen PS, James A, Li JC, et al. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito Aedes aegypti. Commun Biol. 2018;1(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dong Y, Simões ML, Dimopoulos G. Versatile transgenic multistage effector-gene combinations for Plasmodium falciparum suppression in Anopheles. Sci Adv. 2020;6(20). eaay5898. DOI: 10.1126/sciadv.aay5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li M, Bui M, Yang T, et al. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc Natl Acad Sci U S A. 2017;114(49):E10540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Calvo E, Walter M, Adelman ZN, et al. Nanos (nos) genes of the vector mosquitoes, Anopheles gambiae, Anopheles stephensi and Aedes aegypti. Insect Biochem Mol Biol. 2005;35(7):789–798. [DOI] [PubMed] [Google Scholar]
- [25].Aryan A, Anderson MAE, Myles KM, et al. Germline excision of transgenes in Aedes aegypti by homing endonucleases. Sci Rep. 2013;3(1):1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Juhn J, James AA. oskar gene expression in the vector mosquitoes, Anopheles gambiae and Aedes aegypti. Insect Mol Biol. 2006;15(3):363–372. [DOI] [PubMed] [Google Scholar]
- [27].Smith RC, Walter MF, Hice RH, et al. Testis-specific expression of the β2 tubulin promoter of Aedes aegypti and its application as a genetic sex-separation marker. Insect Mol Biol. 2007;16(1):61–71. [DOI] [PubMed] [Google Scholar]
- [28].Anderson MAE, Gross TL, Myles KM, et al. Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect Mol Biol. 2010;19:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402(6760):370–371. [DOI] [PubMed] [Google Scholar]
- [30].Carpenetti TLG, Aryan A, Myles KM, et al. Robust heat-inducible gene expression by two endogenous hsp70-derived promoters in transgenic Aedes aegypti. Insect Mol Biol. 2012;21(1):97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Anderson MAE, Purcell J, Verkuijl SAN, et al. Expanding the CRISPR Toolbox in Culicine Mosquitoes: in Vitro Validation of Pol III Promoters. ACS Synth Biol. 2020;9(3):678–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mayr C. Regulation by 3′-untranslated regions. Annu Rev Genet. 2017;51(1):171–194. [DOI] [PubMed] [Google Scholar]
- [33].Mayr C. What are 3′ UTRs doing? Cold Spring Harb Perspect Biol. 2019;11(10). DOI: 10.1101/cshperspect.a034728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ivanov EL, Sugawara N, Fishman-Lobell J, et al. Genetic Requirements for the Single-Strand Annealing Pathway of Double-Strand Break Repair in Saccharomyces cerevisiae. Genetics. 1996;142(3):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stothard P. The sequence manipulation suite: javaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28(6). 10.2144/00286ir01 [DOI] [PubMed] [Google Scholar]
- [36].Bazzini AA, Viso F, Moreno‐Mateos MA, et al. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016;35(19):2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu Q, Medina SG, Kushawah G, et al. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife. 2019;8:e45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chang TH, Huang HY, Hsu JBK, et al. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(S2):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Machanick P. Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gáspár I, Sysoev V, Komissarov A, et al. An RNA-binding atypical tropomyosin recruits kinesin-1 dynamically to oskar mRNPs. EMBO J. 2017;36(3):319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zimyanin VL, Belaya K, Pecreaux J, et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134(5):843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim-Ha J, Webster PJ, Smith JL, et al. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119(1):169–178. [DOI] [PubMed] [Google Scholar]
- [43].Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133(22):4573–4583. [DOI] [PubMed] [Google Scholar]
- [44].Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176(1):36–50. [DOI] [PubMed] [Google Scholar]
- [46].Pudney M, Varma MGR, Leake CJ. Establishment of cell lines from larvae of culicine (Aedes species) and anopheline mosquitoes. Tissue Cult Assoc Man. 1979;5(1):997–1002. [Google Scholar]
- [47].Peleg J. Growth of arboviruses in monolayers from subcultured mosquito embryo cells. Virology. 1968;35(4):617–619. [DOI] [PubMed] [Google Scholar]
- [48].Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Aryan A, Myles KM, Adelman ZN. Targeted genome editing in Aedes aegypti using TALENs. Methods. 2014;69(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Basu S, Aryan A, Haac ME, et al. Methods for TALEN evaluation, use, and mutation detection in the mosquito Aedes aegypti. Methods Mol Biol. 2016;1338:157–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lorenz R, Bernhart SH, Höner zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011. 6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.


