ATP-driven conformational selection of Hsp90 conformations modulates the binding of clients and co-chaperones.
Abstract
Hsp90 is a molecular chaperone that interacts with a specific set of client proteins and assists their folding. The underlying molecular mechanisms, involving dynamic transitions between open and closed conformations, are still enigmatic. Combining nuclear magnetic resonance, small-angle x-ray scattering, and biochemical experiments, we have identified a key intermediate state of Hsp90 induced by adenosine triphosphate (ATP) binding, in which rotation of the Hsp90 N-terminal domain (NTD) yields a domain arrangement poised for closing. This ATP-stabilized NTD rotation is allosterically communicated across the full Hsp90 dimer, affecting distant client sites. By analyzing the interactions of four distinct clients, i.e., steroid hormone receptors (glucocorticoid receptor and mineralocorticoid receptor), p53, and Tau, we show that client-specific interactions with Hsp90 select and enhance the NTD-rotated state and promote closing of the full-length Hsp90 dimer. The p23 co-chaperone shifts the population of Hsp90 toward the closed state, thereby enhancing client interaction and processing.
INTRODUCTION
The molecular chaperone Hsp90 plays a crucial role in protein homeostasis by regulating the folding and activity of a set of proteins known as clients (1–3). Distinct from other chaperones, Hsp90 is selective for certain types of clients, including transcription factors, kinases, or even intrinsically disordered proteins (IDPs) (www.picard.ch/Hsp90Int/). Thus, Hsp90 has unique molecular properties to enable the recognition of a wide variety of proteins while keeping specificity and to provide a favorable environment for late stages of protein folding. Co-chaperones play an important role in these functions, by binding to Hsp90 and modulating its conformational states and activity (4) and by mediating client interaction [e.g., the kinase-specific Cdc37 (5, 6)].
Hsp90 is a dimeric protein composed of three domains. The N-terminal domain (NTD; residues 1 to 210 in yeast Hsp90) is responsible for adenosine triphosphate (ATP) binding and harbors several dynamic elements that respond to nucleotide binding (7, 8). A long, highly charged, and unstructured linker (residues 211 to 272) connects the NTD with the middle domain (MD; residues 273 to 529). The MD provides the main client binding site and is separated by a short disordered linker from the C-terminal domain (CTD; residues 534 to 674), which mediates dimerization. The C-terminal end of Hsp90 is a disordered, charged tail of 32 amino acids and comprises an EEVD motif for binding to tetratricopeptide repeat domain–containing co-chaperones.
The crystal structures of Hsp90 (7, 9, 10) represent snapshots of conformational states of Hsp90. Biophysical data indicate that apo-Hsp90 populates a dynamic ensemble of extended conformations in solution (11, 12). The presence of ATP and co-chaperones induces the formation of more compact structures, in which the NTD and MD are associated (13), as seen in a crystal structure of nucleotide-bound Hsp90. While the overall structure and topology of Hsp90 is highly conserved in eukaryotes, species-dependent differences exist for the relative population of Hsp90 conformational states across Hsp90 orthologs (14). In addition, clients have been proposed to modulate the conformational ensemble by binding preferentially to distinct Hsp90 domain arrangements (6, 11, 13, 15–17). However, the underlying structural mechanisms are poorly characterized.
The dynamic conformations and interactions of Hsp90 with clients render the structural analysis of these complexes challenging. Recently, cryo–electron microscopy (cryo-EM) structures have been reported for Hsp90-client complexes. The structure of the ternary complex between Hsp90, the client kinase Cdk4, and Cdc37 revealed how Cdk4 is maintained in an inactive state with the N-terminal and C-terminal lobe of the kinase separated by the internal cleft of Hsp90 dimer (6). Recent cryo-EM structures of two Hsp90-co-chaperone complexes with the ligand-binding domain of the glucocorticoid receptor (GR-LBD), a steroid hormone receptor (SHR), provide insight in client loading (GR-Hsp90-Hsp70-Hop) and maturation (GR-Hsp90-p23) (18, 19). In these structures, an N-terminal region and the helix α1 of the GR client are recognized in a similar fashion when bound to the Hsp90 dimer. Nuclear magnetic resonance (NMR) studies with subconstructs or full-length Hsp90 have been used to characterize complexes with the GR-LBD (16), the DNA binding domain (DBD) of p53 (20, 21), the IDP Tau (22), and the ternary complex of Hsp90, Tau, and the peptidyl-prolyl isomerase FKBP52 (23). However, the limited availability of chemical shift assignments for full-length Hsp90 has, so far, precluded a comprehensive description of client interactions and conformational dynamics of Hsp90. In spite of recent advances, the principles underlying the ATP-dependent dynamic conformational cycle of Hsp90 and its coupling to the recognition and processing of clients remain poorly understood. It is still unclear how nucleotide binding to the NTD can allosterically affect and modulate binding of clients at the Hsp90 MD and CTD (22, 24).
Here, we have analyzed the effects of ATP on the conformational ensemble of the full-length yeast Hsp90 dimer by combining methyl NMR spectroscopy and small-angle x-ray scattering (SAXS) experiments in solution. Together with biochemical and biophysical experiments, we systematically compare the binding of a representative set of clients to Hsp90, which include globular proteins, i.e., the LBDs of two SHRs [GR and mineralocorticoid receptor (MR)] with similar three-dimensional (3D) fold, the less stable p53-DBD (25), and the IDP Tau. Our analysis uncovers that ATP induces changes to the relative orientation of the NTD and MD without substantial closure of the overall protomer arrangement in the Hsp90 dimer. We find that client binding shifting the population of NTD-MD arrangements toward the NTD-rotated state is seen in the fully closed Hsp90 dimer. Differential effects are observed for these population shifts for the two SHRs, while p53-DBD and Tau bind independently of the Hsp90 conformational state. Our work demonstrates that client binding induces population shifts from open to closed states of the conformational ensemble that are allosterically communicated across all three domains of Hsp90. The p23 co-chaperone further stabilizes the closed conformations to promote processing of clients.
RESULTS
Extensive methyl labeling of full-length yeast Hsp90 and NMR assignment
In solution, Hsp90 exists as an ensemble of dynamically interconverting conformations, where binding of nucleotides to Hsp90 can induce more compact conformations (11, 12, 14). However, the structural details of the underlying transitions are poorly understood. Crystal structures of Hsp90 proteins are expected to represent static low-energy states (Fig. 1, A and B) that may exist within the dynamic conformational ensemble of the Hsp90 dimer in solution. For example, an intermediate open conformation is observed in the crystal structure of Escherichia coli Hsp90 [Protein Data Bank (PDB): 2IOQ (10)] (Fig. 1, A and C). In the closed conformation of the Hsp90 dimer in complex with p23 (7), the NTD is found to be rotated by about 90° and slightly tilted toward the internal side of the MD. This NTD-MD arrangement requires the ATP lid to fold over the binding pocket and adopt a “down” conformation. Secondary NTD dimerization involves the helix Nα1 and swapping of strand Nβ1 together with twisting of the MD (Fig. 1, B and D) (7, 8, 26). In this conformation, the loop of the MD containing R380 associates with the γ-phosphate of the nucleotide close to the catalytic E33 to yield the active state.
Fig. 1. Hsp90 structure, conformations, and methyl labeling strategy.
(A and B) Structures of different Hsp90 conformations with domains colored as indicated: intermediate open (A), homology model of Htpg (PDB: 2IOQ) (10), and closed (B), Hsp90-p23 complex in the presence of adenylyl-imidodiphosphate (AMP-PNP) with p23 removed (PDB: 2CG9) (7). Methyl groups of Ala, Ile, Leu, Met, and Val (AILVM) shown as spheres in the second protomer. Insets show zoomed views of ATP-induced conformational changes in the NTD leading to the closed state, with flexible elements colored in cyan; E33, R380, and ATP are shown as spheres. The open conformation is shown with the NTD-only structure (PDB: 1AM1), while the closed NTD conformation is seen in the closed full-length Hsp90 dimer. (C and D) Schematic representations of open and closed Hsp90 states. The secondary dimerization region of the NTD (presented in the fully closed dimer) is highlighted in red. The NTD rotation required for the secondary dimerization is indicated by arrows. (E) 1H-13C methyl transverse relaxation optimized spectroscopy (TROSY) spectrum of AILVM pro-R–labeled Hsp90 recorded at 950-MHz proton frequency. Signal positions in the spectra of individual domains are represented by open circles colored by domain.
To obtain a comprehensive view on conformational dynamics and client binding of the full-length Hsp90 dimer, we used NMR combined with extensive methyl labeling (27). This approach has been successfully applied to study chaperone-substrate complexes (28) and allosteric pathways in chaperones (29). On the basis of the amino acid distribution in Hsp90, the methyl groups of Ala, Ile, Leu, Val, and Met (AILVM) provide an excellent coverage for all three domains of full-length Hsp90 dimer (Fig. 1, A and B). Stereospecific 1H,13C labeling of pro-R groups of Leu-δ1 and Val-γ1 residues as well as of Ile-δ1, Ala-β, and Met-ε methyl groups was combined with complete deuteration (see Materials and Methods) (30). As seen in Fig. 1E, the 1H-13C methyl transverse relaxation-optimized spectroscopy (TROSY) spectrum of full-length AILpro-RVpro-RM Hsp90 superposes well with NMR spectra of the individual domains, indicating the absence of strong domain interactions. Assignments were obtained for 185 methyl groups corresponding to 87% of all AILVM methyl groups and more than 25% of Hsp90 sequence, covering all domains and disordered elements (fig. S1A).
ATP drives conformational changes of the NTD-MD orientation
The progression of the Hsp90 chaperone cycle depends on ATP-driven conformational changes. To investigate how nucleotide binding affects the Hsp90 conformation, we combined solution NMR and SAXS experiments. SAXS data of apo-Hsp90 are consistent with an ensemble of extended structures, indicated by a population-averaged Rg of 62.3 Å (Fig. 2A and table S1). Unexpectedly, ATP binding results only in a minor compaction of Hsp90 (Rg of 57.6 Å). Notably, the experimental pairwise distance distribution functions P(r) of ATP-bound Hsp90 show notable contributions of larger distances compared to the back-calculated P(r) of the closed conformation (Fig. 2A and fig. S2A). This suggests that, even in the presence of nucleotides, extended, not fully closed conformations of Hsp90 exist in solution.
Fig. 2. Effects of ATP binding to Hsp90 by NMR and SAXS.
(A) SAXS P(r) distribution functions of apo-Hsp90 (gray) and SAXS P(r) distribution bound to ATP (red) indicate minimal dimer closing. The distance distribution back-calculated for the closed conformation seen in the crystal structure (PDB: 2CG9) is shown as a black dashed line. (B) Chemical shift perturbations (CSPs) versus residue number upon ATP binding to Hsp90. The highest perturbations are highlighted with a rose background. Red negative bars denote resonances showing a minor state signal. (C) Zoomed views of the 1H-13C TROSY spectrum of methyl-labeled Hsp90 apo (gray) and bound to ATP (red; for clarity, only lower contour levels are shown). The presence of a second signal reflecting a minor state is marked by an asterisk and is connected to the major state signal by a dashed line. (D) Residues showing minor and major state signals and CSPs upon ATP binding (red and cyan spheres, respectively) are highlighted on the Hsp90 dimer structure. The client binding region is indicated in yellow.
To assess the effects of nucleotide binding at residue resolution, we analyzed the 1H,13C methyl TROSY NMR spectra in the absence and presence of ATP using an ATP-regenerating system (31). This is important, as nucleotide analogs show distinct effects on Hsp90 conformations differing from that observed for the authentic ligand ATP (fig. S2A). The presence of ATP results in large spectral changes (Fig. 2B and fig. S3A). As expected, chemical shift perturbations (CSPs) cluster in the NTD, which harbors the nucleotide binding site, and reflect large conformational changes of the ATP lid and proximal regions (7, 8, 10, 32, 33). Unexpectedly, in the presence of ATP, we observe a second set of NMR signals for numerous methyl signals in all three domains of Hsp90 (Fig. 2C and fig. S3A). The presence of these additional signals is indicative of a minor conformational state with a population of about ≈20% on the basis of the NMR signal integration that is in slow exchange (on the chemical shift time scale, i.e., kex < 1000 s−1) with the major species. Signal splitting is especially observed in the ATP lid, at the NTD-MD interface, and at the MD-CTD interface (Fig. 2D). Notably, the minor conformational state is observed in both the full-length Hsp90 dimer and a monomeric NTD-MD construct in the presence of adenylyl-imidodiphosphate (AMP-PNP), but not in the isolated NTD (fig. S3, B to D). This suggests that the minor conformation originates from changes in the NTD-MD arrangement depending on contacts between the two domains and excludes a potential trans-protomer effect in the full-length Hsp90 dimer.
ATP stabilizes an NTD-rotated conformation
Next, we characterized the NTD-MD arrangement by analyzing mutations that modulate the domain arrangements using NMR spectroscopy. Because we suspected that the minor conformation originates from changes in the ATP lid and the association of the two domains, we selected previously characterized mutants that either disrupt [R32A (34) and R380A (35)] or stabilize [A107N (26)] NTD-MD contacts. We first recorded 1H,13C methyl TROSY spectra to monitor the effect of nucleotide addition for wild-type and mutant monomeric NTD-MD constructs and the corresponding full-length Hsp90 dimer in the presence of a nucleotide. The R32A mutation decouples catalysis from conformational motions and prevents N-terminal closing, while R380A is catalytically inefficient. The NTD-MD mutants with impaired domain association show no signatures of the second conformation when bound to AMP-PNP (Fig. 3A, left). In contrast, A107N, which stabilizes the “lid-down” conformation and exhibits increased adenosine triphosphatase (ATPase) activity, shows a higher population of the second state both in the full-length and NTD-MD construct (Fig. 3A, right, and Fig. 3B). This confirms our hypothesis that the binding of ATP or analogs induces a conformational state, in which the NTD and MD are associated.
Fig. 3. NMR analysis of NTD-MD arrangements induced by nucleotide.
(A) Methionine region of the 1H-13C TROSY spectra of Hsp90 bound to AMP-PNP (gray). The position and NMR spectra of the mutants R32A and R380A (teal and maroon; left), which cannot induce the NTD rotation, and of the mutant A107N (orange; right), which stabilizes the NTD-rotated state, are indicated. Minor state signals are marked by asterisks and are connected to the major state signal by dashed lines. (B) Analysis of populations of the second state, derived from the integration of NMR signal intensities, for full-length (FL) and NTD-MD constructs of Hsp90 wild type (WT) and with the A107N mutation in the presence of AMP-PNP. Lower and upper whiskers indicate minimal and maximal values, respectively, and lower and upper boxes correspond to the first and third quartiles. Asterisks indicate statistically significant differences (P < 0.001) based on a t test with the null hypothesis assuming no difference. More information on statistical treatment of data is provided in Materials and Methods. (C and D) NMR observation of major and minor conformations in the wild-type Hsp90 dimer exemplified with the methyl group NMR signals of residues I350, which is located at the NTD-MD interface (C), and M105 near the ATP lid (D). (E) For the NTD-MD protein bound to AMP-PNP, spin labeled at D61C, the paramagnetic relaxation enhancement (PRE) effects for the major signal of I350 are consistent with the open conformation, i.e., the nonrotated NTD-MD arrangement. (F) For the spin label at Q314C in the A107N mutant of NTD-MD, strong PRE effects seen for the minor state signal of M105 are consistent with the NTD-rotated conformation. For complete analysis of the PRE data, see figs. S4 and S5. (G) Scheme illustrating how nucleotide binding shifts the population of open and NTD-rotated conformations, which are observed as major and minor state signals in the NMR spectra.
To characterize the structures of the two conformational states observed in the NMR spectra of full-length Hsp90 (Fig. 3, C and D), we measured paramagnetic relaxation enhancements (PREs) with spin labels attached to either domain in the NTD-MD construct in the presence of AMP-PNP. The distance dependence of the PRE effect allows to define the relative arrangement of domains in multidomain proteins (36, 37). The experimental PREs of the major state correlate well with the values back-calculated for the structure of the open dimer (Fig. 3E and fig. S4, A and B) and are inconsistent with the values back-calculated from the closed state, indicating that the major states correspond to the nonrotated NTD-MD arrangement. To characterize the domain arrangements for the minor state, we measured PREs with spin labels attached to the A107N NTD-MD protein, as with this mutation, the population of the “minor” state signals approaches 50% and enables more precise analysis of PRE effects (Fig. 3F and fig. S5, A and B). The PRE data for the A107N mutant show that the NTD-MD arrangement of the minor state corresponds to the NTD-rotated domain arrangement. Thus, in the absence of nucleotide, the major conformation represents the open, nonrotated NTD-MD arrangement, while nucleotide binding stabilizes the NTD-rotated conformation, which resembles the interdomain arrangement seen in the fully closed Hsp90 dimer. It is important to note, however, that some remaining differences between experimental PREs and those expected for the open and rotated states likely indicate a dynamic equilibrium where additional intermediate conformations are also sampled.
To derive a conformational ensemble of Hsp90 in the presence of ATP that reflects the NTD-MD rearrangements without undergoing substantial dimer closing, we used the ensemble optimization method (EOM) (38). The ensemble was based on the SAXS data of Hsp90-ATP, the structure of the CTD dimer, and different populations of the NTD-MD rotated states, which are indicated by the NMR minor state signals. As seen in fig. S2B, ensembles containing ≈20% of rotated NTD-MD arrangement describe the experimental scattering profile with highest agreement and are consistent with the NMR-derived populations. The EOM-derived ensemble also indicates the presence of open structures with interprotomer angles of the ATP-bound Hsp90 dimer between 75° and 250° (fig. S2C).
In summary, the combined analysis by NMR and SAXS indicates that ATP binding triggers coordinated motions involving ATP lid closing, NTD rotation, and MD association, without substantially affecting the overall closure of the Hsp90 dimer. This local NTD-MD rotation affects ≈20% of the Hsp90 molecules, resulting in allosteric changes that propagate across all domains including the client binding sites (Fig. 3G).
Hsp90 clients exhibit distinct conformational properties
For a comparative analysis of Hsp90 clients, we focused on four representative clients, including globular and disordered proteins (Fig. 4A and fig. S6A). As stringent Hsp90-dependent clients, we selected two related SHRs, the LBD of the GR (16, 39) and the LBD of the MR. Both SHRs bind Hsp90 through their LBDs, and they depend strictly on Hsp90 for activation (4, 40). However, GR and MR are affected differently by co-chaperones of Hsp90 in vivo (4). Considering their high sequence and structural similarity (41, 42), these differences suggest that there are distinct interactions with Hsp90. The DBD of the tumor suppressor protein p53 is a dynamic Hsp90 client with limited thermodynamic stability (25). The folding state of this domain when bound to Hsp90 has been controversial (20, 43, 44), although recent studies indicate that the Hsp90 interaction stabilizes a folded, DNA binding–competent state (45, 46). Last, the IDP Tau, linked to neurodegenerative diseases such as Alzheimer’s disease (47), is a client of Hsp90 that binds dynamically to multiple patches on the NTD and MD (48).
Fig. 4. Biophysical characterization of clients.
(A) X-ray structures of GR (81) and MR (82) (magenta and green; with the bound hormone shown as cyan spheres), NMR structure of p53 (blue) (27), and schematic representation of Tau (orange) including proline-rich regions (P), microtubule-binding repeats (R), and N-terminal inserts (I). The same color code is used in (B) to (D). (B) Secondary structure of clients monitored by CD. (C) Thermal stability of clients followed by CD. Melting temperatures are indicated on the plots. (D) Hydrophobicity of clients determined by ANS fluorescence. The normalized fluorescence of ANS-client conjugate with respect to ANS alone at 450 nm is plotted as a bar chart with σ from three measurements. AU, arbitrary units.
We assessed the general biophysical properties of the four selected clients with a view to determine similarities and differences. Circular dichroism (CD) and thermal unfolding experiments show that the GR-LBDs and MR-LBDs are highly similar in terms of folding and stability (Fig. 4, B and C). The p53-DBD exhibits a slightly lower melting temperature than the SHR-LBDs. In addition, its CD spectrum reveals a high content of random coil, indicating that the p53-DBD is in equilibrium with partially unfolded structures, indicative of a thermodynamically unstable protein (Fig. 4, B and C) (25). As expected for an IDP, Tau is devoid of stable secondary structures, and thermal unfolding is not cooperative. An important feature for chaperone recognition is the overall accessible hydrophobicity. On the basis of the binding of the hydrophobic dye 8-anilino-1-naphthalenesulfonic acid (ANS), the clients studied here show a range of exposed hydrophobic regions increasing from GR via MR and p53 to Tau (Fig. 4D). Together, the clients selected for our study are representative of different structures, stabilities, hydrophobicity, and folding cooperativity.
Role of ATP for Hsp90-client interactions
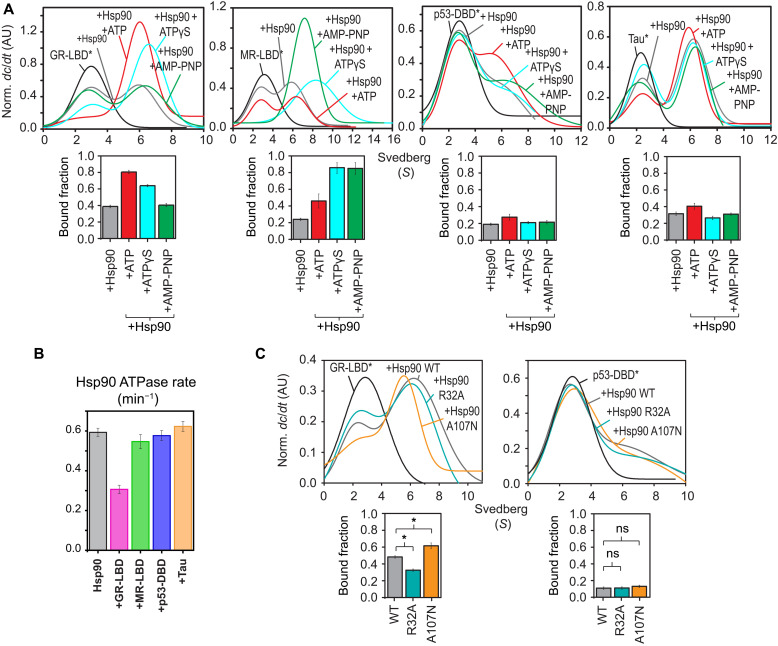
As nucleotide binding triggers conformational changes in Hsp90 that affect client interactions, we performed fluorescence anisotropy binding assays using labeled clients and unlabeled Hsp90. In the absence of nucleotides, all clients show similar affinities in the low micromolar range for Hsp90 (Table 1), consistent with previous studies (16, 20, 48). For the GR-LBD, the affinity significantly increases in the Hsp90-ATP state, while MR shows enhanced binding in the presence of ATP analogs [AMP-PNP and adenosine 5′-[γ-thio]triphosphate (ATPγS)]. In contrast, nucleotides have little effects on the interactions of Hsp90 with the p53-DBD and Tau. These trends were also seen in analytical ultracentrifugation (AUC) experiments with labeled clients and Hsp90 in different nucleotide states (Fig. 5A). Here, ATP favors complex formation in the case of the GR-LBD, while the Hsp90–MR-LBD complexes are enhanced in the presence of ATP analogs. In contrast, no effects of nucleotides were observed for the p53-DBD and Tau. It has been shown previously that the interaction of the GR-LBD with Hsp90 affects the kinetics of ATP hydrolysis (16). We tested this for all clients and found that only GR binding to Hsp90 causes a 50% reduction of the ATPase activity of Hsp90 (Fig. 5B), while the other clients tested do not affect the Hsp90 ATPase rate.
Table 1. Hsp90-client affinities determined by fluorescence anisotropy using labeled clients and unlabeled Hsp90 in different nucleotide states.
Values correspond to the average of three independent experiments and the error to the SD. The column at the right shows theoretical fractions of complexes on our standard NMR conditions (100 μM 2:2 stoichiometric mixture) based on the experimentally determined dissociation constant (KD) values.
| Client |
Hsp90
nucleotide state |
KD (μM) |
Theoretical %
complex |
| GR-LBD (16) | Apo | 2.8 ± 0.23 | 84 |
| ATP | 0.9 ± 0.11 | 91 | |
| AMP-PNP | 4.3 ± 0.28 | 81 | |
| ATPγS | 1.4 ± 0.11 | 88 | |
| MR-LBD | Apo | 5.7 ± 0.38 | 79 |
| ATP | 5.3 ± 0.58 | 80 | |
| AMP-PNP | 2.2 ± 0.25 | 86 | |
| ATPγS | 1.9 ± 0.06 | 87 | |
| P53-DBD | Apo | 5.3 ± 0.3 | 79 |
| ATP | 4.4 ± 0.21 | 81 | |
| AMP-PNP | 5.5 ± 0.25 | 79 | |
| ATPγS | 5.3 ± 0.25 | 79 | |
| Tau (isoform D) | Apo | 5.3 ± 0.65 | 81 |
| ATP | 5.0 ± 0.6 | 81 | |
| AMP-PNP | 5.4 ± 0.66 | 80 | |
| ATPγS | 5.5 ± 0.32 | 80 |
Fig. 5. Effect of nucleotides and conformation on Hsp90-client interactions.
(A) Top: AUC sedimentation velocity curves showing complex formation between labeled clients and Hsp90 in different nucleotide states. Bottom: Fraction of ATTO 488–labeled client* (GR-LBD*/MR-LBD*/p53-DBD*/Tau*) bound to Hsp90 in each case is calculated as described in Materials and Methods from the normalized sedimentation velocity curves and is shown as bar plots. Error bars correspond to the SD σ from three separate measurements. Labeled client sediments at 2.5 to 2.7S (black), while in the presence of Hsp90 (gray), and ATP (red), ATPγS (cyan), or AMP-PNP (green); it sediments at 6 to 6.2S, indicating complex formation. (B) Effects of clients on the ATPase activity of Hsp90. Error bars correspond to the SD from three independent measurements. (C) AUC traces monitoring complex formation between R32A and A107N mutants of Hsp90, GR-LBD*, and p53-DBD*. *P < 0.05. ns, not significant.
Overall, our experiments show that the GR-LBD preferentially binds to a specific conformation of Hsp90 during the ATPase cycle, which is consistent with the about sixfold higher affinity for the ATP-bound state and the effects on ATPase activity. The MR-LBD shows enhanced binding in the presence of ATP analogs and no effects on ATPase activity. Thus, the specific Hsp90 conformations with which they interact seem to differ. In contrast, p53-DBD and Tau do not show increased binding to nucleotide-bound forms of Hsp90, demonstrating that they bind in a conformation-independent manner. To provide further evidence for client specificity to Hsp90 conformations, we analyzed binding to the R32A and A107N mutants stabilizing opposite Hsp90 states. As seen in Fig. 5C, increased binding of GR-LBD to A107N and lower binding to R32A are consistent with a preferential binding to the ATP-induced rotated state, while the absence of effects for the p53-DBD reflect conformation-independent binding.
NMR-based mapping of common and client-specific binding sites in Hsp90
To understand the basis of client specificity for Hsp90 conformations, we mapped and compared the client binding sites of Hsp90. To this end, we combined NMR CSPs and line broadening in 1H,13C methyl TROSY spectra of free Hsp90 and in complex with clients with PRE experiments. All clients induce moderate spectral changes (Fig. 6A and fig. S6, B and C), reflecting the dynamic nature of Hsp90-client complexes. Notably, effects occur in all three Hsp90 domains, with stronger changes induced by the GR-LBD, smaller ones by the MR-LBD as well as the p53-DBD, and weak effects by Tau. To discriminate direct interaction effects from conformational changes on Hsp90 upon binding, we recorded intermolecular PRE experiments with spin-labeled clients (49), which provide an approximate measure of client proximity up to 30 Å (50, 51). To obtain representative PRE effects, we selected the spin labeling positions of clients on the basis of previously determined Hsp90 sites (fig. S6, A and D). These experiments reveal well-defined client contact regions, especially in the case of the globular folded clients (Fig. 6A). The weaker effects seen in complex with Tau likely reflect transient and dynamic interactions at multiple sites of Hsp90 for this intrinsically disordered client. In all cases, the broad spatial distribution of PRE effects likely reflects the dynamic binding with multiple low-affinity interactions that are reflected by the paramagnetic effects (52).
Fig. 6. Client binding regions of Hsp90 as determined by NMR spectroscopy.
(A) CSP (gray bars) upon client binding versus residue number. Black dashed lines correspond to the median + 1σ of the CSP. Intensity ratios between complexes of Hsp90 with spin-labeled clients in paramagnetic and diamagnetic states are shown as lines according to the following color code: GR-LBDs and MR-LBDs in magenta and green, respectively; p53-DBD in blue; and Tau in orange. Colored dashed lines correspond to the median − 1σ of the intensity ratios. Binding elements are highlighted and annotated at the top of each plot, together with Hsp90 domains. (B) Client binding sites determined by PRE, CSP, and intensity ratios mapped as described in the Supplementary Materials, mapped on the surface of Hsp90 in the open conformation, with the flexible NM linker removed. Helix Cα2 important for GR and MR interaction is colored in purple. (C) Analysis of surface conservation of Hsp90 determined by ConSurf (86) reveals high conservation of client binding sites. (D) Cylindrical cartoon representation of Hsp90 showing common binding sites for GR, MR, p53, and Tau in red (ATP lid and helices α3 and α7 of the MD) and helix Cα2 for GR and MR interaction (purple). Src loop and helix Mα2, important for the binding of globular clients, are also indicated. NTD is colored in wheat, MD in green, and CTD in light blue.
The client interactions identified by the NMR data are located in regions that are buried in closed conformations, indicating that initial interaction occurs with open states (fig. S7A). To extrapolate the binding surfaces from NMR effects observed for individual methyl probes, we performed a global analysis of PRE data, methyl CSPs, and intensity ratios as described in the Supplementary Materials (table S2 and fig. S7B). The integrated NMR effects mapped onto the open conformation define large interaction regions at the internal face of the open Hsp90 dimer that are highly conserved (Fig. 6, B and C). Folded clients (GR, MR, and p53) bind to large and continuous surfaces, being more extensive in the case of SHRs (table S2). The intrinsically disordered Tau shows the most divergent interaction pattern with discontinuous scattered patches, distinct from the globular clients.
The common sites shared by all clients define the primary client binding region of Hsp90, corresponding to the ATP lid in the NTD and the MD region near the MD-CTD interface (helices Mα3 and Mα7) (Fig. 6D). In the MD, globular proteins interact with the “Src loop” (35) and the N terminus of helix Mα2. However, differences between the SHRs and p53 indicate that distinct client-specific binding features exist. GR and MR both show notable effects with the CTD helix Cα2, which projects toward the MD (Fig. 6, B and D), while p53 shows the strongest effects to the NTD-MD interface and the linker region, consistent with previous studies (43, 53). In addition, electrostatic interactions with the disordered C-terminal extension contribute to p53 binding (20). The high conservation of the general binding surface of Hsp90 that shared the four clients suggests that common features of Hsp90 are used for the recognition of diverse clients. This also implies that client interactions of yeast Hsp90 represent well those of eukaryotic systems in general. This was further confirmed by an analysis of PRE and CSPs induced by the binding of spin-labeled MR to methyl-labeled human Hsp90β (fig. S8, A and B). The highly similar effects for the two orthologs confirm the conservation of eukaryotic client interactions.
CTD helix Cα2 is a discriminator for selective client binding to closed Hsp90 conformations
The spatial separation of nucleotide and client binding sites in the NTD, MD, and CTD regions raises the question of how these distinct functional interactions are communicated across full-length Hsp90. We therefore used NMR to monitor client binding to Hsp90 in the presence of ATP. As seen in fig. S9A, the GR-LBD causes large CSPs, which are almost one order of magnitude larger for ATP-bound Hsp90 compared to those seen in the absence of nucleotide. Smaller spectral changes are seen upon binding of MR-LBD, p53-DBD, and Tau. To demonstrate that the observed NMR spectral changes relate to bona fide clients, we performed NMR titrations with glutathione S-transferase (GST) as a nonclient protein. As expected, only negligible spectral changes are observed for GST (fig. S10, A to C). To compare the client binding surfaces in Hsp90 in the apo-bound and nucleotide-bound state, we performed intermolecular PRE experiments for complexes of Hsp90–AMP-PNP bound to spin-labeled clients. PRE effects observed were comparable to those seen for the apo state. This demonstrates that similar regions are involved in client binding in the presence of ATP, although the presence of nucleotide may shift the population of Hsp90 conformations that are recognized by clients, thereby enhancing the binding affinity (fig. S9B).
To further characterize client-specific binding and effects on the Hsp90 conformation, we compared the interaction of Hsp90 with the two SHR clients, GR-LBD and MR-LBD. These clients are structurally highly similar but, nevertheless, exhibit different effects on Hsp90. When we analyzed NMR spectral changes and the population of the NTD-rotated state in the presence of the GR-LBD, we observed perturbations exclusive for the rotated conformation, which are accompanied by a significant increase in its population (≈20 to ≈55%) (Fig. 7, A and B). Notably, CSPs are observed not only in the binding site but also at the NTD-MD interface (I320) and regions that are involved in additional dimer interfaces present in the fully closed Hsp90 dimer, for instance, M16 in helix Nα1 (7). This demonstrates that the GR-LBD binds preferentially to the NTD-rotated state and thereby further shifts the conformational equilibrium of the dimer toward closed conformations (Fig. 7, B and C). In contrast, the MR-LBD causes smaller spectral changes in distant regions and does not significantly change the population of the NTD-rotated conformation (Fig. 7, A and B). This reveals that, in spite of the high structural similarity, the two SHRs induce differential allosteric effects across Hsp90. The stronger effects caused by GR compared to MR (fig. S9A) suggest that a tighter binding to this conformational switch enables a stronger allosteric communication across the whole Hsp90 dimer. As expected, p53-DBD and Tau, whose binding are not affected by nucleotides, elicit overall small changes in the NMR spectra of Hsp90 and do not affect the population of the NTD-rotated state (Fig. 7B and fig. S9C).
Fig. 7. Client-induced allostery of Hsp90 in the presence of ATP.
(A) Residues that show allostery affects GR binding mapped to Hsp90 open and closed structures (yellow spheres). GR sites are colored in magenta and common sites for GR and MR in purple. Chemical shift changes on the 1H-13C TROSY spectrum for residues at the NTD dimerization, NTD-MD interface, and client site are shown at the right. Hsp90-ATP is shown in dark gray and GR complex in magenta. For residues showing two sets of signals, open and NTD-rotated conformations are indicated and connected by dashed lines. Specific shifts on the rotated state are indicated by an arrow. For MR complex (green), no important long-range effects are observed outside the binding site, indicating that MR binding does not trigger an allosteric response. Note that M16 reports on the NTD/NTD dimerization interface in the closed Hsp90 dimer. (B) Box plot showing populations derived from NMR signal integration of the rotated state, for Hsp90-ATP and client complexes. The data are presented as in Fig. 3B and asterisks indicate significant differences (P < 0.001). (C) Schematic representation of specific client binding to NTD-MD rotated conformation and induced dimer closing for GR. (D) P(r) distribution functions derived from the SAXS data of Hsp90-client complexes in the presence of ATP (color lines). Shorter distances with respect to Hsp90-ATP (solid black line) indicate shifts toward compact conformations in the complexes. The theoretical P(r) function of the closed conformation is shown as a dashed line.
SAXS analysis of Hsp90-client complexes in the presence of ATP was performed to determine how clients affect the overall conformation of the Hsp90 dimer (table S3). The SAXS-derived Rg and P(r) distributions show that the SHRs favor more compact conformations of Hsp90 (Fig. 7D). This effect is especially pronounced for the GR-LBD, which causes a reduction on the Rg from 61.3 Å for Hsp90-ATP to 58.8 Å. In contrast, SAXS data of the Hsp90 complexes with the p53-DBD and Tau exhibit only minor changes, in line with the absence of nucleotide effects on the interaction and binding independent of the overall Hsp90 conformation. Together, differential effects of clients on Hsp90 in the presence of ATP reveal specific allosteric changes upon binding to a conformational switch of Hsp90 that senses the rotation of the NTD. GR-LBD binding to the NTD-rotated state leads to a population shift of the NTD-MD arrangement toward the NTD-rotated state and thereby increases dimer closing involved in ATP hydrolysis. In contrast, the closely related MR-LBD elicits reduced allosteric effects, presumably due to weaker interactions with the conformational switch region around the Cα2 helix. Notably, both p53-DBD and Tau bind independently of the overall conformation of Hsp90 and show no allosteric effects on Hsp90.
The co-chaperone p23 stabilizes closed conformational states in Hsp90-client complexes
As our analysis shows that nucleotides and clients are able to modulate the conformational ensemble of Hsp90, we next studied the effect of the p23 co-chaperone. p23 is special as it affects the processing of all client proteins, for which a detailed analysis has been performed (4). It binds and stabilizes the closed conformation of Hsp90 in the presence of ATP (7), inhibits its ATPase activity (54, 55), and stabilizes bound clients (56). p23 consists of a globular CS (CHORD and Sgt1) domain and a largely disordered charged C-terminal tail. We have recently demonstrated that the p23 tail contains a helical segment that weakly interacts with the MD-CTD interface of Hsp90 and with GR, free or in complex with Hsp90 (57). Similar interactions are observed in a cryo-EM structure of the maturation complex of GR-Hsp90-p23 (18), and direct interactions have been previously reported for the p23 tail and p53 (24). However, mapping of the binding sites of globular clients onto the crystal structure of the Hsp90-p23 complex (7) shows that the p23 interaction substantially overlaps with client sites in Hsp90 (Fig. 8A). Therefore, it is not clear how p23 interactions with Hsp90 can stabilize client binding.
Fig. 8. p23 forms ternary complexes and stabilizes closed Hsp90 conformations in client complexes.
(A) Binding sites of globular clients (GR, MR, and p53; pink) mapped onto the surface of Hsp90 in complex with p23 (cyan). A helical segment in the p23 C-terminal tail is indicated as a cyan tube. (B) Zoomed views of the 1H-13C TROSY spectrum of Hsp90-ATP (black) showing representative peak shifts upon p23 binding (cyan) for residues at the NTD-MD interface (I320) and client site (M589), which report on allosteric effects on NTD-MD arrangement associated with dimer closing and client binding, respectively (see also fig. S12). Signals corresponding to the open and rotated states are annotated and connected by dashed lines. In the spectra with p23, only the lowest contour level of NMR spectra for the Hsp90-ATP complex is shown for comparison. (C) Box plot showing the populations of the NTD-rotated states for Hsp90-ATP in the absence and the presence of p23. The data are presented as in Fig. 3B. (D and E) Spectral changes following the assembly of the ternary Hsp90-ATP-client-p23 complexes by successive addition of GR (magenta) (D) or MR (green) (E) to Hsp90-ATP (black) followed by p23 (cyan). Shifts on the rotated states are indicated by an arrow, and the first contour level of the previous assembly state is included. (F) AUC sedimentation velocity curves of ATTO 488–labeled client* (black) in the presence of Hsp90-ATP (red) and Hsp90-ATP-p23 (cyan) indicate the formation of stable ternary complexes in all cases. The fraction of bound complex as calculated from the sedimentation curves for each client is shown in the insets.
To determine how p23 can affect and modulate Hsp90 conformation to support client interactions, we first studied the effects of p23 binding to Hsp90 in the presence of ATP. Upon addition of p23, the 1H,13C heteronuclear multiple-quantum coherence (HMQC) spectrum of methyl-labeled Hsp90 shows chemical shift changes only for the NTD-rotated state signals induced by the nucleotide. The p23 interactions directly affect residues in the ATP lid and the N-terminal region of helix Mα2 (fig. S11A) and extend to the secondary dimerization region, the NTD-MD interface, and the Cα2 at the client binding site (residues I320 and M589, respectively; Fig. 8B and fig. S12B). These interactions stabilize the closed state, as is reflected by a significant increase of the population of the NTD-rotated state from ≈20% in Hsp90-ATP to ≈35% in the presence of p23, as determined by the relative NMR signal intensities (Fig. 8C).
Next, we performed p23 titrations in the presence of the two SHRs, as p23 is known to support the activation of GR and MR. Addition of p23 to preformed Hsp90-ATP–client complexes yields similar direct NMR spectral changes indicating that the CS domain binds and displaces client interactions at this site (fig. S11, B and C). However, distinct chemical shift changes upon p23 binding are seen for residues in the Hsp90 MD (for example, for the methyl group of I320), secondary dimerization region, and within the core client binding site (residue M589; Fig. 8, D and E, and fig. S12, C and D). In the case of the GR-LBD, where client binding already strongly promotes dimer closing (Fig. 7), p23 binding does not induce further spectral changes and thus merely binds to the conformation that is already selected by the client (Fig. 8D). In contrast, addition of MR-LBD to Hsp90 in the absence of p23 only perturbs signals at the binding site (M589) and does not affect residues at the NTD-MD interface, i.e., I320 (Fig. 8E). The addition of p23 causes additional chemical shift changes for I320 that resemble those observed for GR/p23. This indicates that p23 stabilizes and increases the population of closed conformations of Hsp90 bound to MR. Notably, the chemical shift changes seen for M589 (Fig. 8E), which is an important residue in the Cα2 switch helix of the CTD, indicate that p23 reinforces allosteric effects that stabilize the closed Hsp90 conformations also in the presence of a bound client.
To investigate the stability of the Hsp90-client interactions, we evaluated the ability of p23 to form complexes with fluorescently labeled clients and Hsp90-ATP by AUC. Hsp90-ATP readily forms ternary complexes with p23 and the clients studied. The ternary complexes sediment at 8 to 8.2S, while Hsp90-client complexes sediment at 6.2S (Fig. 8F). The comparison of the fraction of client bound to Hsp90 in the presence and absence of p23 (Fig. 8F, insets) shows that, in the case of the GR-LBD, p23 does not cause an additional increase. This likely reflects that the closed conformation of Hsp90 is already stabilized by the GR-LBD. In the case of the MR-LBD, the addition of p23 increases the population of the complex, which is consistent with the stabilization of the closed conformation seen by NMR. Last, for p53-DBD and Tau, the addition of p23 has very little effect on client binding, confirming the independence of their binding from the Hsp90 conformation.
In summary, these results indicate that p23 stabilizes the closed state and promotes allosteric changes to the client binding sites of Hsp90. Although the p23 interaction overlaps with client sites, only the CS domain interaction directly competes with clients. The transient and weak interactions of the p23 C-terminal region can be displaced by clients. The subsequent binding of the C-terminal tail of p23 to the bound client (57) results in a stabilization of the ternary complex.
DISCUSSION
Our study provides a comprehensive analysis of client recognition by Hsp90, highlighting the effect of ATP binding and conformational dynamics in controlling and modulating Hsp90 conformations. To investigate the effects of nucleotides on Hsp90 conformation and interactions, we used an ATP-regenerating system, as ATP analogs have different effects depending on the client type (Fig. 5). Our solution NMR and SAXS data show that, both in the apo- and ATP-bound states, the Hsp90 dimer adopts an ensemble of open conformations with a wide range of opening angles between the two protomers (10). Notably, we found that ATP binding induces a distinct minor conformation of Hsp90, where the NTD is rotated relatively to the MD, resembling the NTD-MD arrangement seen in the closed Hsp90 dimer (Fig. 9) (7). In spite of this local domain arrangement, the Hsp90 dimer, overall, still samples a range of open states when bound to ATP, as indicated by SAXS data and consistent with previously reported fluorescence resonance energy transfer experiments (11). This rotated NTD-MD arrangement requires conformational changes of the ATP lid and enables contacts with the MD required for ATP hydrolysis. Ultimately, these changes lead to the N-terminal dimerization and full activation of the Hsp90 dimer in the closed conformation. Thus, our data provide direct experimental evidence for the allosteric communication across the three domains of Hsp90, which has been suggested so far on the basis of computational studies (58, 59). The NMR spectral changes that we observed provide direct evidence that the local NTD rotation induces effects that are transmitted to the MD and CTD domains, where client binding occurs. This allosteric communication forms the basis for the modulation and selection of Hsp90 conformations by clients and co-chaperones.
Fig. 9. Client binding and conformational selection in Hsp90 cycle.
Conformational cycle of Hsp90 showing population shifts promoted by binding of nucleotides, clients, and the co-chaperone p23. In the presence of ATP, Hsp90 is in equilibrium of extended conformations differing in the local rotation of the NTD-MD domains. Conformational changes associated to the NTD-MD rotation involve ATP lid closing and engagement between E33-R380 of the NTD and MD (top inset). These changes transmit allosterically to helix Cα2 (colored cylinders; bottom inset). (Allosteric signal is indicated by red dots.) Binding of clients to specific conformation(s) causes their stabilization, as shown in the insets. This is especially the case for the GR-LBD, which binds preferentially to the helix Cα2 in the conformation of the NTD-MD rotated state (highlighted in red). Stabilization of the rotated state leads to the largest allosteric effects across the full protein due to population shift toward the closed conformation. The highly related MR-LBD shows lower preference for closed conformations, while p53-DBD and Tau bind highly independent of the Hsp90 conformation. In the late stages of the cycle, the co-chaperone p23 forms ternary complexes by binding to the MD and NTD of Hsp90, thereby stabilizing the client by direct contacts with its C-terminal tail. Upon ATP hydrolysis, the ternary complex dissociates releasing p23 and the active client.
By comparing the binding surfaces of clients with distinct structural features and stabilities with the effects on Hsp90 conformation, we have identified general and client-specific regions and their involvement on functional allostery. Generally used, conserved binding sites are located at the internal MD-CTD interface of the open Hsp90 dimer and are shared by all clients studied, with additional contributions from the ATP lid. These sites are extended and harbor short amphipathic binding motifs, i.e., 423KNIKLG428, 505IDEY508, 554LKEIL558, and 589MERIM593, which, in part, have been implicated in client binding in vivo (60–62).
Specific differences between the binding sites of globular clients revealed a major switch point of Hsp90 sensitive to clients. For the GR-LBDs and MR-LBDs, the Cα2 helix in the Hsp90 CTD is an important unique binding site. This is consistent with the fact that mutations in this element or its surroundings have been shown to impair GR activation in vitro and in vivo (2, 60, 62). It has been demonstrated that K594 in this element is an important switch point that allosterically regulates the ATPase activity, dimer closing kinetics, co-chaperone modulation, and client maturation of Hsp90 (59). In addition, clients bind to the MD in close proximity of W300, another allosteric regulation point sensitive to client binding that modulates long-range conformational transitions involved in client maturation (63). This region also harbors the conserved Y293, a phosphorylation site affecting Hsp90’s structural dynamics and involved in the acceleration effect of Aha1 on Hsp90 closing (64). In exquisite agreement, our NMR data reveal allosteric effects and the presence of two conformational states in these regions of Hsp90 in the presence of ATP, which are further influenced by client binding. We provide evidence that the second conformational state is coupled to the rotation of the NTD, as seen for the NMR signals of residue M589 in the Cα2 helix (Fig. 2A). Thus, GR-LBD interactions with the NTD-rotated conformation of this helix, indicated by specific NMR spectral changes, result in the stabilization of this state and stalling of the fully closed, N-terminally dimerized state required for ATP hydrolysis (Fig. 7) (4, 16). This explains the higher affinity for the ATP state and the inhibition of the ATPase activity observed for the GR-LBD. In addition, the relevance of NTD rotation for GR-LBD interaction provides a rationale for the strong negative influence of shortened or rigidified variants of the NTD-MD linker with impaired interdomain mobility on GR activation in vivo (65).
The MR-LBD, in contrast, causes smaller spectral changes in the Cα2 region. This may indicate that different amino acids are involved in the binding interfaces involving MR and GR. The smaller spectral changes might thus partially reflect the different chemical environment of the interactions. However, the reduced spectral changes may also indicate weaker binding to this switch region and thus explain the attenuated allosteric closing of the Hsp90 dimer. Given the high sequence similarity and conservation of structural features in the two SHRs (fig. S13, A to C), it is unlikely that the minor differences explain the distinct effects on Cα2 alone. It is possible that differential conformational dynamics of the two SHR clients rather than their structures contributes to their different interactions with Hsp90 (66). A recent NMR study reveals coupled motions involving the hormone binding pocket, the activation region, and the helix α1 in GR (67). These results are also consistent with single-molecule force spectroscopy measurements showing coupled detachment of the helix α1 in GR and hormone binding (68). Moreover, our ANS binding experiments reveal higher exposure of hydrophobic sites for the GR-LBD compared to the MR-LBD, despite similar surface properties seen in their crystal structures. We speculate that differences in internal motion of the LBDs may affect the propensity of the helix α1 and the N-terminal region of the client to detach and interact with Hsp90, which is required for client loading and maturation in recently reported cryo-EM structures (fig. S13D) (18, 19).
In contrast, p53-DBD and Tau, which do not interact with the Cα2 helix, do not induce substantial allosteric changes. The p53-DBD binds to the NTD-MD interface, involving sites of the NTD that are regulated by the charged linker connecting the two domains (53). A discriminating feature of the disordered protein Tau is the scattered contacts compared to globular clients, consistent with highly dynamic and transient interactions. These clients do not affect the Hsp90 ATPase activity and bind with similar affinities independent of the nucleotide state of the chaperone (Fig. 9). The p53-DBD and Tau are characterized by low thermodynamic stability and/or aggregation propensity. This suggests that Hsp90 has a holdase function for these proteins by binding and protecting them from aggregation (69). This function of Hsp90 might also be important for stabilizing clients and displacing them from unproductive interactions with Hsp70, thus favoring spontaneous folding trajectories (70). Therefore, we propose that the interaction with Cα2 is an important feature to discriminate between the chaperone and holdase functions of Hsp90.
The principle of conformational selection also extends to the co-chaperone p23, which selects the rotated conformation of the NTD in the presence of clients, thus promoting dimer closure and allosteric changes on the client binding site. The effects of p23 are stronger for clients that induce less Hsp90 dimer closing alone, suggesting a role of p23 especially in assisting conformational changes in Hsp90-client complexes that do not readily achieve closed conformations of Hsp90. We hypothesize that the p23-induced structural changes of clients are crucial for their activation, including direct contacts with the C-terminal helix of p23 for additional stabilization of the bound client. The substantial overlap between the co-chaperone and client binding sites at the NTD of Hsp90 suggests that p23 carries additional functions by blocking client interactions with the ATP lid. In the open conformation of Hsp90, this element points toward the client binding sites in the MD and the CTD and forms a continuous binding surface. In the closed state, these interactions with the ATP lid are sterically disfavored. Therefore, we propose that the ATP lid binds to clients in a conformation-sensitive manner, interacting with them only in the open conformation, while being displaced upon dimer closing or by p23. The interactions of the C-terminal tail of p23 with the client sites at the MD-CTD interface are transient and nonspecific, covering a broad region in the main client sites (57). Our data show that clients are readily able to displace the p23 C-terminal tail, which then bind and stabilize to the bound client. This dual role of the p23 tail is consistent with a function in proofreading of clients by blocking unwanted interactions of nonclients to the Hsp90 MD-CTD interface.
Our studies define Hsp90 as a molecular chaperone that uses extensive “modular” binding sites for client interaction, where their availability and accessibility is modulated by conformational dynamics, nucleotides, and co-chaperones. Reshaping of dynamic client interactions upon Hsp90 conformational changes is critical for promoting structural transitions of the bound clients (71, 72). The presence of multiple client binding motifs in Hsp90 seems required to handle the large number of clients that are processed by Hsp90 and suggests that conformational dynamics of both partners plays a major role in molecular recognition. This also agrees with the notion that Hsp90 may interact with partially folded but conformationally dynamic clients, including IDPs. Overall, the picture emerging is that, although many regions are shared in the binding of clients, specific features exist that dictate preferential binding to certain regions of Hsp90. We hypothesize that multiple binding sites of Hsp90 are also important for the different client loading pathways. In this respect, a client may initially access different regions on the Hsp90 surface depending on which route it adopts for loading onto Hsp90. Thus, loading through Hsp70 and Hop (Hsp70/90 organizing protein), direct Hsp70-Hsp90 interactions (73), or additional co-chaperone–supported binding (6) may result in different initial interactions with Hsp90 but, nevertheless, leads to the formation of productive complexes with closed Hsp90. ATP-induced allostery enables communication between different domains of Hsp90, which can be fine-tuned and further modulated by co-chaperones such as p23, to adapt the Hsp90 conformational cycle in a client-specific manner.
MATERIALS AND METHODS
Protein expression and purification
Expression and purification of full-length AILpro-RVpro-RM yeast Hsp90 (Hsp82) was carried out by adapting previous protocols (74). Briefly, His-tagged Hsp90 containing Tobacco Etch Protease (TEV) digestion site was expressed in M9 minimal medium with 99.8% D2O (Sigma-Aldrich) supplemented with 2 g/liter of glucose-D7 (Cambridge Isotopes, Tewksbury, USA). Cells were adapted to deuterated medium in three steps of 0, 50, and 99.9% D2O. Precursors for stereospecific AILpro-RVpro-RM labeling were added according to the manufacturer’s protocol (NMR-Bio, Grenoble, France) before induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Expression was done overnight at 30°C. Cells were lysed by French press and purified by Ni2+ affinity chromatography. After TEV cleavage, protein was further purified by anion exchange and gel filtration chromatography steps.
The stabilized variant of human GR-LBD (amino acids 527 to 777; F602S/A605V/V702A/E705G/M752T), rat MR-LBD (amino acids 709 to 981; C805S), and human p53-DBD (amino acids 94 to 312) were expressed and purified as described before (4, 16, 46). Human Tau isoform D was expressed at an optical density (OD) of 0.7 to 0.8 with 1 mM IPTG for 3 hours at 37°C. Cell pellet was resuspended in Ni-NTA (nitrilotriacetic) buffer A [20 mM tris, 500 mM NaCl, 10 mM imidazole, and 1 mM dithiothreitol (DTT) (pH 8)] containing protease inhibitor G (Serva) and 1 mM phenylmethylsulfonyl fluoride. The cells were lysed by sonication, and the cellular extract was boiled for 20 min and cooled on ice after which a white precipitate is formed. The boiling and cooling step was followed by centrifugation at 20,000 rpm for 45 min, and the clear supernatant was loaded on a Ni-NTA column (5 ml of HisTrap HP, GE Healthcare) equilibrated in Ni-NTA buffer A. Tau was eluted in 60% Ni-NTA buffer B [20 mM tris, 500 mM NaCl, 500 mM imidazole, and 1 mM DTT (pH 8)] and was also present in the wash fractions (100 ml, 5% buffer B). The fractions containing Tau protein [as checked by SDS–polyacrylamide gel electrophoresis (SDS-PAGE)] were pooled and diluted (about equal to four times) in cold double-distilled water. TEV protease was added to the diluted protein, and it was dialyzed overnight at 4°C in Ni-NTA buffer A (except imidazole). TEV cleavage was followed by a reverse Ni-NTA step to separate the cleaved from the uncleaved protein. The flow-through fractions containing the pure cleaved protein were checked by SDS-PAGE, pooled, and concentrated to 5 ml. The concentrated protein was loaded on the gel filtration column (Superdex 200, 26/60 pg, GE Healthcare) equilibrated in 40 mM Hepes, 150 mM KCl, 5 mM MgCl2, and 1 mM Tris(2-carboxyethyl)phosphine (TCEP) (pH 7.5) buffer. Fractions containing pure protein were pooled and concentrated using 3-kDa molecular weight cutoff Amicon concentrators before flash-freezing in liquid nitrogen and storing at −80°C. The protein concentration was checked with a BCA kit. Previous studies on the isoform F of Tau showed transient and dynamic binding to multiple patches on the NTD and MD of human Hsp90 involving the microtubule binding repeats (48). Isoform D of Tau, lacking the N-terminal negatively charged insert comprised between residues 45 and 102 of isoform F, contains the regions involved in Hsp90 interaction, and its binding to yeast Hsp90 was confirmed by NMR experiments of 15N-labeled Tau.
The cysteine mutants of the client proteins used for spin labeling for NMR-PRE experiments, MR-LBD (C805S/A841C), p53-DBD (C124S/C229S/C277S) containing one exposed cysteine C182, and Tau isoform D (C264S) were purified in the same way as their wild-type counterparts as described above, which harbors C233. For GR-LBD, the naturally exposed single-cysteine C638 was used. The mutations did not affect the function of proteins as checked by the hormone binding for GR-LBDs and MR-LBDs, respectively, and DNA binding of p53-DBD. Binding to Hsp90 was also not altered for any of the client protein mutants as investigated by AUC. Spin labeling reactions were carried out by adding 10× excess of 100 mM 3-(2-Iodoacetamido)-PROXYL free radical (IPSL) in dimethyl sulfoxide (Sigma-Aldrich) to the proteins in 1 M tris-HCl (pH 8) and 200 mM NaCl buffer, followed by overnight incubation at 4°C in the dark. Afterward, samples were buffer-exchanged to NMR buffer and checked by mass spectrometry. In the case of MR-LBD A841C, mass spectrometry analysis revealed an increase of mass corresponding to double spin labeling, which is due to the additional reaction with the partially exposed C907.
NMR spectroscopy
NMR experiments of full-length Hsp90 and its complexes were carried out at 30°C in a Bruker AV950 MHz spectrometer (Bruker, Karlsruhe, Germany) equipped with a cryogenically cooled probe. NMR samples were in buffer containing 20 mM tris-D11, 100 mM NaCl, 5 mM MgCl2, 2 mM DTT-D10, 0.02% NaN3 (pH 7) (pH meter reading) in 99.9% D2O. Dextran sulfate sodium (5 μM) was used as an internal standard. For p53, the concentration of tris, NaCl, and MgCl2 was reduced to the half, and 5 mM DTT-D10 was used. Because of the unstable nature of GR, experiments that involved GR binding were supplemented with 50 μM dexamethasone. Nucleotides were added to a final concentration of 5 mM. The ATP-regenerating system was prepared as described (31).
2D spectra were processed by Bruker TopSpin 3.5 (Bruker, Karlsruhe, Germany); 3D spectra were recorded using nonuniform sampling and processed by NMRPipe (75). All spectra were analyzed by CcpNmr software (76). CSP were calculated according to the following formula
where σH/C is 1 SD of the methyl 1H and 13C chemical shifts, respectively.
PRE effects were derived from 1H-13C methyl TROSY experiments (77) recorded for the oxidized and reduced samples using a recycle delay of 4 to 5 s. Paramagnetic broadening was estimated from the intensity ratios between the oxidized and reduced forms (37, 49, 50). To estimate theoretical intensity ratios from distances derived from structures, our previous protocol (37) was adapted to methyl-based experiments. Briefly, four copies of the spin label were used as an ensemble representing the conformational flexibility. Paramagnetic relaxation contributions (R2,para) were extracted on the basis of the Solomon-Bloembergen equation (78)
where k is a constant (1.23 × 1016 Å6 s−2), r is the distance (in angstroms), τC is the tumbling correlation time estimated from the molecular weight assuming a globular protein (3.3 × 10−8 s), and ω1H is the 1H Larmor frequency. Theoretical intensity ratios were calculated as (37, 49)
The time t corresponds to the transfer time during the 1H,13C methyl TROSY HMQC pulse sequence and was set to 7.6 ms, assuming that the main contribution is transverse proton relaxation during the insensitive nuclei enhancement by polarization transfer (INEPT) delays. R2,dia was obtained from the 1H linewidth and set to 64 Hz; however, residue-specific variations ±15 Hz do not significantly affect the PRE analysis.
Populations of NTD-MD open and rotated states were determined the from the signal intensities of the major (Iopen) and minor state signals (Irot). The following formula was used to extract the percentage of rotated state population for each residue
Minimal, maximal, and median values as well as first and third quartiles are then plotted in the box-and-whisker plots. Data points above 1.5 times the interquartile range were excluded from the plots and from further analysis. To determine statistically significant differences between conformational populations upon mutation, client or p23 binding, ratios derived from individual residues were compared using a two-tailed t test.
Chemical shift assignment of full-length Hsp90
For the assignment of full-length AILpro-RVpro-RM Hsp90 from yeast, 15N- and ILVM-labeled individual domains were expressed in an otherwise deuterated background. Methyl resonances were partially assigned by a combination of 15N- and 13C-edited nuclear Overhauser effect spectroscopy (NOESY) experiments and further extended by 3D CCH NOESY experiments. Ala resonances were partially assigned from the Cβ chemical shifts from HNCACB/HNcoCACB experiments and a combination of 15N- and 13C-edited NOESY experiments recorded with 13C, 15N-labeled samples (79, 80). To minimize the overlap in Leu and Val signals in the full-length spectrum, potential overlaps between the two diastereomeric methyl groups were estimated by superimposing the spectra of the three individual domains. Stereospecific assignments were predicted by considering the downfield shift of pro-S groups in the 1H dimension (81). To assign the 1H-13C methyl TROSY spectrum of the full-length protein, assignments were transferred from the individual domains and complemented by 3D CCH NOESY experiments on full-length Hsp90. Assignments were supported by comparison with the NTD-MD construct and complemented by mutagenesis of several residues in the full-length protein lacking NOE networks, namely, M16L, M105L, M116L, M382L, M389L, M589I, M593I, M603I, and M607I in the absence and presence of AMP-PNP. In addition, L15I, I96L, and L331I mutants were produced on the individual domains in the absence and presence of ATP for the NTD. Assignments are deposited in the Biological Magnetic Resonance Data Bank under accession number 50472. For the assignment of Ile, Met resonances of human Hsp90, a similar approach was used, on the basis of standard backbone assignment of individual domains (80), followed by methyl resonance assignment transferred by matching 13Cα and 13Cβ frequencies from backbone experiments and CCH total correlation spectroscopy experiments in methyl-labeled, 13C, 2H uniformly labeled samples. The individual domain assignment was further validated on the basis of the structure of the domains and their 3D CCH NOESY spectra. Methyl groups in full-length Hsp90 were assigned by transferring the assignment from the isolated domain and were validated using 3D CCH NOESY experiments. A list of assigned resonances is included in table S4.
Small-angle x-ray scattering
All SAXS experiments of Hsp90-client complexes were carried out on a Rigaku BioSAXS-1000 instrument with a HF007 microfocus copper target (40 kV, 30 mA), OptiSAXS optics, and a HyPix-3000 detector. For q calibration, a silver behenate sample (Alfa Aesar) was used. Transmissions were measured with a photodiode beam stop. Samples were measured in eight 900-s frames checked for beam damage, circular averaged, solvent-subtracted, and normalized to concentration by the SAXSLab software (version 3.1.1). Scattering data of Hsp90 bound to nucleotides were recorded by in-line gel filtration chromatography coupled to SAXS as described previously (34). For client complexes, samples containing 70 μM Hsp90 in 25 mM Hepes (pH 7.5), 150 mM KCl, and 5 mM MgCl2 buffer containing 5 mM ATP were used, and different stoichiometric amounts of clients were added as summarized in table S3. Storage buffer of the clients was removed by successive steps of concentration and dilution before the SAXS measurements. Subtracted curves were analyzed with PRIMUS and GNOM programs implemented in ATSAS 3.0.2 package (82).
For the EOM (38, 83) of Hsp90 bound to ATP, the scattering curve of Hsp90 bound to ATP comprising q values from 0.00683 to 0.5 Å−1 was used as input. Structures of NTD-MD and dimeric CTD constructs were used to generate the initial pool of conformations, defining a five-residue randomized linker (residues 528 to 532) to consider interdomain flexibility. To account for the different NTD-MD orientations, NTD-MD constructs derived from the 2CG9 [rotated (7)] and 2IOQ [open (10)] structures were used separately to generate two pools of conformations. Each pool consisted of 500 structures, from which 50% were of C2 symmetry. The EOM was run using 25 curves per ensemble, fixing different population of rotated NTD-MD structures. The genetic algorithm was run for 200 cycles.
Biophysical characterization of clients
The client proteins were structurally characterized by CD spectroscopy and ANS fluorescence. CD measurements were performed on a Chirascan Q100 spectrometer attached with a Peltier unit (Applied Photophysics) in 25 mM NaH2PO4, 100 mM NaCl, and 1 mM TCEP (pH 7.9). Apo–GR-LBD was prepared by extensive dialysis of GR-LBD in 20 mM Hepes, 150 mM NaCl, and 0.04% CHAPS (pH 7.5). Protein concentrations used were 3 μM for each protein except p53-DBD, which was used at 5 μM. Far ultraviolet (UV) CD spectra (260 to 200 nm) were recorded at 20°C in 1-mm quartz cuvette. Thermal transitions were recorded at 220 nm from 20° to 80°C at a rate of 0.5°C/min. To monitor the surface hydrophobicity of clients, ANS fluorescence spectra were recorded on a Jasco fluorescence spectrometer FP-8500 (Jasco, Groß-Umstadt, Germany). Measurements were performed in 20 mM Hepes, 20 mM KCl, 5 mM MgCl2, and 1 mM TCEP (pH 7.5). GR-LBD, MR-LBD, p53-DBD, or Tau at a concentration of 1 μM was incubated with 50 μM ANS dye at room temperature (RT) for 15 min before recording their corresponding fluorescence spectra at 25°C from 400 to 600 nm after excitation at 370 nm. Excitation and emission slit widths of 5 nm each were used. Fluorescence intensity at 450 nm of protein-ANS conjugate was normalized with respect to the fluorescence of ANS-alone sample.
ATPase assay
ATPase measurements of yeast Hsp90 were performed in a 150-μl reaction volume in Varian Cary 50 UV-visible spectrometer at 30°C. Hsp90 (3 μM) was incubated with or without 10 μM of each client protein at RT for 15 min in ATPase assay buffer [40 mM Hepes, 20 mM KCl, and 5 mM MgCl2 (pH 7.5)] containing ATP-regenerating system of NADH [reduced form of NAD+ (nicotinamide adenine dinucleotide)], pyruvate kinase, lactate dehydrogenase, and phosphoenolpyruvate. The measurement was started by the addition of 2 mM ATP to the reaction mix, and decrease in the absorption kinetics of NADH-to-NAD+ conversion at 340 nm was recorded. Background ATPase activity due to any contaminations was subtracted from the total activity by the addition of 50 μM Hsp90 inhibitor radicicol to each of the reaction to get Hsp90-specific activity. ATPase activity was calculated from the OD versus time slopes using the molar extinction coefficient of NADH (6200 M−1 cm−1) as described previously (84).
Fluorescence labeling of proteins
For anisotropy and AUC measurements, GR-LBD and MR-LBD were randomly labeled on cysteine residues with ATTO 488 maleimide (ATTO-TEC) according to the manufacturer’s protocol with a few modifications. Briefly, the LBDs were diluted in 20 mM Hepes, 150 mM NaCl, and 0.04% CHAPS (pH 7.5) buffer such that their final concentration was always less than or equal to 50 μM. Labeling was performed with threefold excess of the fluorescent dye at 25°C for 1.5 to 2 hours. The free dye was removed with extensive dialysis in cold conditions. Tau was labeled specifically at the N terminus using ATTO 488 N-hydroxysuccinimide ester in 50 mM K2HPO4, 50 mM KCl, and 1 mM TCEP (pH 6.8) at RT for 2 hours with threefold excess of dye. The free dye was removed from the labeled protein using a PD-10 column (GE Healthcare). The degree of labeling was determined according to the formula given on the manufacturer’s site. p53-DBD was labeled as previously described (46). Labeling does not change the activity of the proteins as reported before (4, 16, 46).
Analytical ultracentrifugation
AUC measurements were conducted with a ProteomLab Beckman XL-A centrifuge (Beckman Coulter, Brea, California) equipped with an AVIV fluorescence detection system (Aviv Inc., Lakewood, USA). For Hsp90 interaction studies, 500 nM labeled GR-LBD*, MR-LBD*, or Tau* (asterisk indicates labeled protein) were incubated with 10 μM unlabeled yeast Hsp90. p53-DBD* was used at a concentration of 1 μM. Wherever mentioned, nucleotides (ATP/ATPγS/AMP-PNP) were added at 2 mM concentration. For binding study with p23, 6 μM unlabeled yeast p23 was added to Hsp90 and client reaction mix in the presence of ATP. All the reactions were incubated for 2 hours at 25°C in 20 mM Hepes, 20 mM KCl, 5 mM MgCl2, and 1 mM TCEP (pH 7.5) before measuring in AUC at 20°C. The reactions were prepared in a total of 320 μl of reaction volume. Data analysis was performed using SEDVIEW and OriginPro 9.1 (85). The fraction of labeled protein bound (f) was calculated from sedimentation curves as follows
where means the normalized dc/dt value of free labeled protein in the absence of any addition; represents the normalized dc/dt value of free labeled protein left after complex formation in each case. The maximum value of dc/dt was used for each fitted curve.
Fluorescence anisotropy
Fluorescence anisotropy measurements were performed on a Jasco fluorescence spectrometer FP-8500 (Jasco, Groß-Umstadt, Germany) equipped with polarizers to measure the affinity of the clients for Hsp90 under different nucleotide conditions. Five hundred nanomolars of labeled GR-LBD*, MR-LBD*, Tau*, or p53-DBD* was incubated with increasing concentrations of unlabeled Hsp90 (0.3 to 30 μM) at 25°C for 2 hours before recording the final steady-state anisotropy values of the labeled proteins. Nucleotides (ATP, ATPγS, and AMP-PNP) were added at 2 mM final concentration. The final anisotropy values at each Hsp90 concentration were averaged over a period of 50 s with a time interval of 5 s and plotted against respective Hsp90 concentration. The plots were fitted to one site binding model in OriginPro 9.1 to determine the dissociation constant of client binding to Hsp90. The experiments were performed at 25°C in 20 mM Hepes, 20 mM KCl, 5 mM MgCl2, and 5 mM DTT (pH 7.5) in a 150-μl reaction volume. The excitation and emission wavelengths of 490 and 520 nm were used, respectively.
Acknowledgments
We thank M. Biebl for providing p23 samples, B. Simon for advice with scripts for PRE data analysis, and T. Madl for discussions. We acknowledge G. Gemmecker for help with NMR experiments and members of the Sattler und Buchner groups for discussions.
Funding: This work was funded by German Research Foundation DFG grant CRC1035, project 201302640 (to J.B. and M.S.); German Research Foundation DFG grant EXC214, Center for Integrated Protein Science Munich (CIPSM) (to J.B. and M.S.); Helmholtz Association Initiative and Networking Fund under project number ZT-I-0003 (to M.S.); National Institute on Aging of the National Institutes of Health, grant RF1AG055088 (to L.B.); the Alexander Von Humboldt Foundation (to V.D.); EMBO Long-term Fellowship ALTF 243-2018 (to F.D.); European Commission FP7-PEOPLE-20112011-IIF 301193, Hsp90NMR (to L.F.); and EMBO Long-term Fellowship ALTF 1255-2011 (to L.F.).
Author contributions: A.L. produced isotope-labeled proteins and performed NMR experiments, assignments, and NMR analyses. L.F. performed preliminary NMR experiments. S.A. designed NMR experiments. L.B. provided Tau constructs and critical assessment of the manuscript. V.D. generated p53 and MR constructs, purified all client proteins used in the study, and performed and analyzed the biochemical experiments. D.R. purified proteins, and both V.D. and D.R. performed AUC. R.S. and A.L. performed SAXS experiments and analyzed scattering data. Data from human Hsp90 was provided by F.D. M.S. and J.B. designed the study and supervised the experiments. A.L., V.D., J.B., and M.S. wrote the manuscript. All authors contributed and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. NMR chemical shift assignments are available in the Biological Magnetic Resonance Data Bank (BMRB) repository under the accession code 50472 and BMRB code 50474. Other data generated and analyzed in this study (NMR CSPs, PRE, and SAXS data) are available at https://doi.org/10.5281/zenodo.5534905.
Supplementary Materials
This PDF file includes:
Tables S1 to S4
Figs. S1 to S13
References
REFERENCES AND NOTES
- 1.McClellan A. J., Xia Y., Deutschbauer A. M., Davis R. W., Gerstein M., Frydman J., Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131, 121–135 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Nathan D. F., Vos M. H., Lindquist S., In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. U.S.A. 94, 12949–12956 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D., Karras G. I., Lindquist S., Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahasrabudhe P., Rohrberg J., Biebl M. M., Rutz D. A., Buchner J., The plasticity of the Hsp90 Co-chaperone system. Mol. Cell 67, 947–961.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Keramisanou D., Aboalroub A., Zhang Z., Liu W., Marshall D., Diviney A., Larsen R. W., Landgraf R., Gelis I., Molecular mechanism of protein kinase recognition and sorting by the Hsp90 kinome-specific cochaperone Cdc37. Mol. Cell 62, 260–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verba K. A., Wang R. Y., Arakawa A., Liu Y., Shirouzu M., Yokoyama S., Agard D. A., Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352, 1542–1547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H., Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440, 1013–1017 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prodromou C., Roe S. M., Piper P. W., Pearl L. H., A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nat. Struct. Biol. 4, 477–482 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Dollins D. E., Warren J. J., Immormino R. M., Gewirth D. T., Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol. Cell 28, 41–56 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiau A. K., Harris S. F., Southworth D. R., Agard D. A., Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127, 329–340 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Hellenkamp B., Wortmann P., Kandzia F., Zacharias M., Hugel T., Multidomain structure and correlated dynamics determined by self-consistent FRET networks. Nat. Methods 14, 174–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krukenberg K. A., Forster F., Rice L. M., Sali A., Agard D. A., Multiple conformations of E. coli Hsp90 in solution: Insights into the conformational dynamics of Hsp90. Structure 16, 755–765 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessling M., Richter K., Buchner J., Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 16, 287–293 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Southworth D. R., Agard D. A., Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol. Cell 32, 631–640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krukenberg K. A., Street T. O., Lavery L. A., Agard D. A., Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 44, 229–255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz O. R., Freiburger L., Rutz D. A., Krause M., Zierer B. K., Alvira S., Cuellar J., Valpuesta J. M., Madl T., Sattler M., Buchner J., Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol. Cell 53, 941–953 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Mickler M., Hessling M., Ratzke C., Buchner J., Hugel T., The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat. Struct. Mol. Biol. 16, 281–286 (2009). [DOI] [PubMed] [Google Scholar]
- 18.C. M. Noddings, R. Y.-R. Wang, D. A. Agard, GR chaperone cycle mechanism revealed by cryo-EM: Reactivation of GR by the GR:Hsp90:p23 client-maturation complex. bioRxiv 2020.09.12.294975 [Preprint]. 13 September 2020; 10.1101/2020.09.12.294975. [DOI]
- 19.R. Y.-R. Wang, C. M. Noddings, E. Kirschke, A. G. Myasnikov, J. L. Johnson, D. A. Agard, GR chaperone cycle mechanism revealed by cryo-EM: Inactivation of GR by GR:Hsp90:Hsp70:Hop client-loading complex. bioRxiv 2020.11.05.370247 [Preprint]. 5 November 2020; 10.1101/2020.11.05.370247. [DOI]
- 20.Hagn F., Lagleder S., Retzlaff M., Rohrberg J., Demmer O., Richter K., Buchner J., Kessler H., Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nat. Struct. Mol. Biol. 18, 1086–1093 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Park S. J., Kostic M., Dyson H. J., Dynamic interaction of Hsp90 with its client protein p53. J. Mol. Biol. 411, 158–173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oroz J., Chang B. J., Wysoczanski P., Lee C. T., Perez-Lara A., Chakraborty P., Hofele R. V., Baker J. D., Blair L. J., Biernat J., Urlaub H., Mandelkow E., Dickey C. A., Zweckstetter M., Structure and pro-toxic mechanism of the human Hsp90/PPIase/Tau complex. Nat. Commun. 9, 4532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colombo G., Morra G., Meli M., Verkhivker G., Understanding ligand-based modulation of the Hsp90 molecular chaperone dynamics at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 105, 7976–7981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehn A., Moroni E., Zierer B. K., Tippel F., Morra G., John C., Richter K., Colombo G., Buchner J., Allosteric regulation points control the conformational dynamics of the molecular chaperone Hsp90. J. Mol. Biol. 428, 4559–4571 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Canadillas J. M., Tidow H., Freund S. M., Rutherford T. J., Ang H. C., Fersht A. R., Solution structure of p53 core domain: Structural basis for its instability. Proc. Natl. Acad. Sci. U.S.A. 103, 2109–2114 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prodromou C., Panaretou B., Chohan S., Siligardi G., O’Brien R., Ladbury J. E., Roe S. M., Piper P. W., Pearl L. H., The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 19, 4383–4392 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosen M. K., Gardner K. H., Willis R. C., Parris W. E., Pawson T., Kay L. E., Selective methyl group protonation of perdeuterated proteins. J. Mol. Biol. 263, 627–636 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Huang C., Rossi P., Saio T., Kalodimos C. G., Structural basis for the antifolding activity of a molecular chaperone. Nature 537, 202–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuravleva A., Clerico E. M., Gierasch L. M., An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gans P., Hamelin O., Sounier R., Ayala I., Dura M. A., Amero C. D., Noirclerc-Savoye M., Franzetti B., Plevin M. J., Boisbouvier J., Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew. Chem. Int. Ed. Eng. 49, 1958–1962 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Karagoz G. E., Duarte A. M., Ippel H., Uetrecht C., Sinnige T., van Rosmalen M., Hausmann J., Heck A. J., Boelens R., Rudiger S. G., N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc. Natl. Acad. Sci. U.S.A. 108, 580–585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehner A., Furrer J., Richter K., Schuster I., Buchner J., Kessler H., NMR chemical shift perturbation study of the N-terminal domain of Hsp90 upon binding of ADP, AMP-PNP, geldanamycin, and radicicol. Chembiochem 4, 870–877 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Dollins D. E., Immormino R. M., Gewirth D. T., Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. J. Biol. Chem. 280, 30438–30447 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Mader S. L., Lopez A., Lawatscheck J., Luo Q., Rutz D. A., Gamiz-Hernandez A. P., Sattler M., Buchner J., Kaila V. R. I., Conformational dynamics modulate the catalytic activity of the molecular chaperone Hsp90. Nat. Commun. 11, 1410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H., Structural and functional analysis of the middle segment of hsp90: Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11, 647–658 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Mackereth C. D., Madl T., Bonnal S., Simon B., Zanier K., Gasch A., Rybin V., Valcarcel J., Sattler M., Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475, 408–411 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Simon B., Madl T., Mackereth C. D., Nilges M., Sattler M., An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew. Chem. Int. Ed. Eng. 49, 1967–1970 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Bernado P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I., Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Seitz T., Thoma R., Schoch G. A., Stihle M., Benz J., D’Arcy B., Wiget A., Ruf A., Hennig M., Sterner R., Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening. J. Mol. Biol. 403, 562–577 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Faresse N., Ruffieux-Daidie D., Salamin M., Gomez-Sanchez C. E., Staub O., Mineralocorticoid receptor degradation is promoted by Hsp90 inhibition and the ubiquitin-protein ligase CHIP. Am. J. Physiol. Ren. Physiol. 299, F1462–F1472 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Bamberger C. M., Wald M., Bamberger A. M., Schulte H. M., Inhibition of mineralocorticoid and glucocorticoid receptor function by the heat shock protein 90-binding agent geldanamycin. Mol. Cell. Endocrinol. 131, 233–240 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Caamano C. A., Morano M. I., Patel P. D., Watson S. J., Akil H., A bacterially expressed mineralocorticoid receptor is associated in vitro with the 90-kilodalton heat shock protein and shows typical hormone- and DNA-binding characteristics. Biochemistry 32, 8589–8595 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Park S. J., Borin B. N., Martinez-Yamout M. A., Dyson H. J., The client protein p53 adopts a molten globule-like state in the presence of Hsp90. Nat. Struct. Mol. Biol. 18, 537–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudiger S., Freund S. M., Veprintsev D. B., Fersht A. R., CRINEPT-TROSY NMR reveals p53 core domain bound in an unfolded form to the chaperone Hsp90. Proc. Natl. Acad. Sci. U.S.A. 99, 11085–11090 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boysen M., Kityk R., Mayer M. P., Hsp70- and Hsp90-mediated regulation of the conformation of p53 DNA binding domain and p53 cancer variants. Mol. Cell 74, 831–843.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Dahiya V., Agam G., Lawatscheck J., Rutz D. A., Lamb D. C., Buchner J., Coordinated conformational processing of the tumor suppressor protein p53 by the Hsp70 and Hsp90 chaperone machineries. Mol. Cell 74, 816–830.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Ittner L. M., Ke Y. D., Delerue F., Bi M., Gladbach A., van Eersel J., Wolfing H., Chieng B. C., Christie M. J., Napier I. A., Eckert A., Staufenbiel M., Hardeman E., Gotz J., Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 142, 387–397 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Karagoz G. E., Duarte A. M., Akoury E., Ippel H., Biernat J., Moran Luengo T., Radli M., Didenko T., Nordhues B. A., Veprintsev D. B., Dickey C. A., Mandelkow E., Zweckstetter M., Boelens R., Madl T., Rudiger S. G., Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 156, 963–974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Battiste J. L., Wagner G., Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39, 5355–5365 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T., The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature 502, 519–523 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Vahidi S., Ripstein Z. A., Bonomi M., Yuwen T., Mabanglo M. F., Juravsky J. B., Rizzolo K., Velyvis A., Houry W. A., Vendruscolo M., Rubinstein J. L., Kay L. E., Reversible inhibition of the ClpP protease via an N-terminal conformational switch. Proc. Natl. Acad. Sci. U.S.A. 115, E6447–E6456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang C., Iwahara J., Clore G. M., Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Lopez A., Elimelech A. R., Klimm K., Sattler M., The charged linker modulates the conformations and molecular interactions of Hsp90. Chembiochem 22, 1084–1092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richter K., Walter S., Buchner J., The co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J. Mol. Biol. 342, 1403–1413 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Siligardi G., Hu B., Panaretou B., Piper P. W., Pearl L. H., Prodromou C., Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 279, 51989–51998 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Morishima Y., Kanelakis K. C., Murphy P. J. M., Lowe E. R., Jenkins G. J., Osawa Y., Sunahara R. K., Pratt W. B., The Hsp90 cochaperone p23 is the limiting component of the multiprotein Hsp90/Hsp70-based chaperone system in vivo where it acts to stabilize the client protein·Hsp90 complex. J. Biol. Chem. 278, 48754–48763 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Biebl M., Lopez A., Rehn A., Freiburger L., Lawatscheck J., Blanck B., Sattler M., Buchner J., Elements in the flexible tail of the co-chaperone p23 coordinate client binding and progression of the Hsp90 chaperone cycle 90-binding agent geldanamycin. Nat. Commun. 12, 828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Astl L., Stetz G., Verkhivker G. M., Allosteric mechanism of the Hsp90 chaperone interactions with cochaperones and client proteins by modulating communication spines of coupled regulatory switches: Integrative atomistic modeling of Hsp90 signaling in dynamic interaction networks. J. Chem. Inf. Model. 60, 3616–3631 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Rehn A., Lawatscheck J., Jokisch M. L., Mader S. L., Luo Q., Tippel F., Blank B., Richter K., Lang K., Kaila V. R. I., Buchner J., A methylated lysine is a switch point for conformational communication in the chaperone Hsp90. Nat. Commun. 11, 1219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bohen S. P., Yamamoto K. R., Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc. Natl. Acad. Sci. U.S.A. 90, 11424–11428 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang L., Ricketson D., Getubig L., Darimont B., Unliganded and hormone-bound glucocorticoid receptors interact with distinct hydrophobic sites in the Hsp90 C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 103, 18487–18492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genest O., Reidy M., Street T. O., Hoskins J. R., Camberg J. L., Agard D. A., Masison D. C., Wickner S., Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol. Cell 49, 464–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rutz D. A., Luo Q., Freiburger L., Madl T., Kaila V. R. I., Sattler M., Buchner J., A switch point in the molecular chaperone Hsp90 responding to client interaction. Nat. Commun. 9, 1472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu W., Beebe K., Chavez J. D., Boysen M., Lu Y., Zuehlke A. D., Keramisanou D., Trepel J. B., Prodromou C., Mayer M. P., Bruce J. E., Gelis I., Neckers L., Hsp90 middle domain phosphorylation initiates a complex conformational program to recruit the ATPase-stimulating cochaperone Aha1. Nat. Commun. 10, 2574 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daturpalli S., Kniess R. A., Lee C. T., Mayer M. P., Large rotation of the N-terminal domain of Hsp90 is important for interaction with some but not all client proteins. J. Mol. Biol. 429, 1406–1423 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Boczek E. E., Reefschlager L. G., Dehling M., Struller T. J., Hausler E., Seidl A., Kaila V. R., Buchner J., Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc. Natl. Acad. Sci. U.S.A. 112, E3189–E3198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kohler C., Carlstrom G., Gunnarsson A., Weininger U., Tangefjord S., Ullah V., Lepisto M., Karlsson U., Papavoine T., Edman K., Akke M., Dynamic allosteric communication pathway directing differential activation of the glucocorticoid receptor. Sci. Adv. 6, eabb5277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suren T., Rutz D., Mossmer P., Merkel U., Buchner J., Rief M., Single-molecule force spectroscopy reveals folding steps associated with hormone binding and activation of the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 115, 11688–11693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burmann B. M., Gerez J. A., Matecko-Burmann I., Campioni S., Kumari P., Ghosh D., Mazur A., Aspholm E. E., Sulskis D., Wawrzyniuk M., Bock T., Schmidt A., Rudiger S. G. D., Riek R., Hiller S., Regulation of α-synuclein by chaperones in mammalian cells. Nature 577, 127–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moran Luengo T., Kityk R., Mayer M. P., Rudiger S. G. D., Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Dezwaan D. C., Freeman B. C., HSP90: The Rosetta stone for cellular protein dynamics? Cell Cycle 7, 1006–1012 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Hiller S., Chaperone-bound clients: The importance of being dynamic. Trends Biochem. Sci. 44, 517–527 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Bhattacharya K., Weidenauer L., Luengo T. M., Pieters E. C., Echeverria P. C., Bernasconi L., Wider D., Sadian Y., Koopman M. B., Villemin M., Bauer C., Rudiger S. G. D., Quadroni M., Picard D., The Hsp70-Hsp90 co-chaperone Hop/Stip1 shifts the proteostatic balance from folding towards degradation. Nat. Commun. 11, 5975 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buchner J., Weikl T., Bügl H., Pirkl F., Bose S., Purification of Hsp90 partner proteins Hop/p60, p23, and FKBP52. Methods Enzymol. 290, 418–429 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 76.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Tugarinov V., Hwang P. M., Ollerenshaw J. E., Kay L. E., Cross-correlated relaxation enhanced 1H−13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Bertini I., Luchinat C., Parigi G., Magnetic susceptibility in paramagnetic NMR. Prog. NMR Spectrosc. 40, 249–273 (2002). [Google Scholar]
- 79.Asami S., Kallies W., Gunther J. C., Stavropoulou M., Glaser S. J., Sattler M., Ultrashort broadband cooperative pulses for multidimensional biomolecular NMR experiments. Angew. Chem. Int. Ed. Eng. 57, 14498–14502 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Sattler M., Schleucher J., Griesinger C., Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 34, 93–158 (1999). [Google Scholar]
- 81.Neri D., Szyperski T., Otting G., Senn H., Wuthrich K., Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28, 7510–7516 (1989). [DOI] [PubMed] [Google Scholar]
- 82.Manalastas K., Konarev P. V., Hajizadeh N. R., Kikhney A. G., Petoukhov M. V., Molodenskiy D., Panjkovich A., Mertens H. D., Gruzinov A. Y., Borges C., Jeffries C. M., Svergun D. I., Franke D., ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tria G., Mertens H. D., Kachala M., Svergun D. I., Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2, 207–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richter K., Muschler P., Hainzl O., Reinstein J., Buchner J., Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J. Biol. Chem. 278, 10328–10333 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Hayes D. B., Stafford W. F., SEDVIEW, real-time sedimentation analysis. Macromol. Biosci. 10, 731–735 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N., ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Hawle P., Siepmann M., Harst A., Siderius M., Reusch H. P., Obermann W. M., The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol. Cell. Biol. 26, 8385–8395 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nathan D. F., Lindquist S., Mutational analysis of Hsp90 function: Interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15, 3917–3925 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 90.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992). [Google Scholar]
- 91.Mukrasch M. D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., Zweckstetter M., Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 7, e34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bledsoe R. K., Montana V. G., Stanley T. B., Delves C. J., Apolito C. J., McKee D. D., Consler T. G., Parks D. J., Stewart E. L., Willson T. M., Lambert M. H., Moore J. T., Pearce K. H., Xu H. E., Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110, 93–105 (2002). [DOI] [PubMed] [Google Scholar]
- 93.Bledsoe R. K., Madauss K. P., Holt J. A., Apolito C. J., Lambert M. H., Pearce K. H., Stanley T. B., Stewart E. L., Trump R. P., Willson T. M., Williams S. P., A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J. Biol. Chem. 280, 31283–31293 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4
Figs. S1 to S13
References