Abstract
The diagnosis of SARS-CoV-2 is based on the use of nucleic acid amplification tests (NAAT), especially rRT-PCR. The latter also allows us to quickly identify variants of concern. However, its use in follow-up of patients and the correlation between Ct value and the viability of the virus is controversial.
Keywords: COVID-19, SARS-CoV-2, NAAT, cell culture
As with other respiratory viruses, the main test for diagnosing COVID-19 is detection of SARS-CoV-2 in respiratory samples using nucleic acid amplification tests (NAATs) [1], mostly real-time reverse transcription polymerase chain reaction (rRT-PCR) [1,2]. Molecular detection is a highly sensitive diagnostic method; however, the results can remain positive for long periods, even when the patient has clinically recovered and the virus has lost its infectivity. The sustained rRTPCR positivity of COVID-19 has complicated the discharge of patients, the transfer of patients between various hospital areas, and the reincorporation of health care workers (HCWs) to their jobs. The use of rRT-PCR as a follow-up tool for SARSCoV-2 infection has led to hypotheses regarding infectivity duration, and even the possibility of reactivation [3].
The performance of rRT-PCR depends on several factors, such as the specimen type, the timing of collection, nucleic acid extraction method, the design of primers and probes and the selection of their viral RNA target, the reagents, and instrument and software used for the rRT-PCR and for the result interpretation.
Regarding specimen type, nasopharyngeal flocked swabs are considered the gold standard for respiratory virus sampling of the upper respiratory tract, while BAL fluid, endotracheal secretions or sputum should be considered when lower tract infection is suspected [4].
In the absence of diagnostic methods with reliable quantification, the cycle threshold (Ct) value obtained in the amplification has been employed as a semiquantitative measure and has been proposed as a parameter for elaborating approaches for removing patients from isolation [5]. Establishing a reliable cut-off Ct value is difficult, given the large number of available rRT-PCR-based diagnostic tests (which amplify different viral regions generally in a multiplex format), the need to use more than one molecular test in most clinical laboratories to meet growing demand, the inclusion of an automated system based on real-time transcription-mediated amplification (which does not provide Ct values), and the use of different types of samples during patient follow-up.
INTER-ASSAY AND INTRA-ASSAY VARIABILITY
We have analyzed the qualitative results obtained in four NAAT assays, three of them based on rRT-PCR, testing 200 respiratory samples obtained during follow-up of patients. We consider a result as true positive when this result was obtained in at least two assays (n = 198 samples).
Table 1 shows the NAATs compared, the regions that each one amplifies, the number of samples with a positive result for each assay, and the agreement obtained with the reference value.
Table 1.
Comparison of qualitative results obtained in four NAAT assays for detection of SARS-CoV-2 RNA
| Assay | |||||||
|---|---|---|---|---|---|---|---|
| Panther-Fusion LDT | Panther TMA | COBAS 6800 | AllplexTM | ||||
| Target gene(s) | E | Orf1ab | E | orf1ab | E | rdRP/S | N |
| Number of positive samples | 200 | 178 | 180 | 173 | 179 | 165 | 172 |
| Agreement (%) | 99 | 89 | 90 | 87 | 90 | 83 | 86 |
When we compare Ct values obtained on assays that amplify E gene (Panther Fusion LDT, COBAS 6800 and AllplexTM) in a subset of 100 samples with Ct values between 30 and 35, according to the results obtained on Panther-Fusion LDT assay, we observe statistically significant differences between median Ct values obtained (Table 2).
Table 2.
Median (IQR) Ct values and statistical significance obtained amplifying SARS-CoV-2
| Assay | ||||
|---|---|---|---|---|
| Panther Fusion LDT | COBAS 6800 | AllplexTM | P value | |
| Median (IQR) | 32.0 (31.0-33.0) |
30.3 (29.1-32.3) |
29.2 (26.8-30.8) |
P < 0.001 |
Since most commercial rRT-PCRs are multiplex assays, we have analyzed the intra-assay variability of COBAS 6800, without finding statistically significant differences in the median of Ct values obtained for the different amplified regions in this case (E gene median Ct value : 29.3 [IQR: 26.6-33.4], orf1ab gene median Ct value: 27.0 [IQR: 25.5-30.5], p: 0.15).
ASSESSMENT OF VIRAL VIABILITY
Previous studies [6,7] have shown prolonged viral shedding in patients with severe COVID-19 and its relation to high viral loads. Although we observed a significant positive correlation between low Ct values and the presence of viable virus, this viral load estimate appears insufficient for discriminating samples harboring infective virus. We showed that, in immunocompetent patients with severe forms of COVID-19, viral replication can be detected even with moderate or low viral loads during prolonged periods [8].
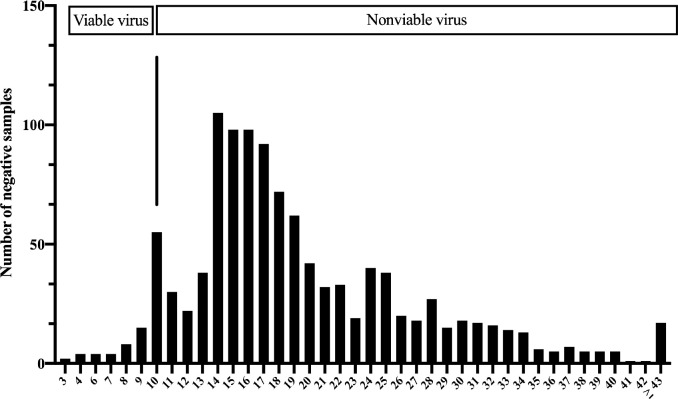
Figure 1 shows the discordance between timing for rRTPCR to become negative and timing for viable virus clearance in nasopharyngeal samples. This is a cohort of health workers (HCW) with mild COVID-19, infected in the first pandemic wave, who were prospectively followed until the rRT-PCR was negative. The virus remains viable for up to 10 days after the onset of symptoms.
Figure 1.
Time (days) for rRT-PCR to becomes negative after the onset of symptoms in a HCW cohort during the first pandemic wave.
SARS-COV-2 VARIANTS SURVEILLANCE
A fast and extensive strategy for detecting SARS-CoV-2 variants of concern (VOC) and variants of interest (VOI) is achieved by testing of all RT-PCR SARS-CoV-2 positive samples with subsequent variant RT-PCR [9]. This approach can have a positive impact on adequate and timely contact tracing, and could facilitate targeted public health measures. In comparison to whole genome sequencing (WGS) this PCR-based screening method is easy to implement in molecular diagnostic laboratories.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
References
- 1.Safiabadi Tali SH, LeBlanc JJ, Sadiq Z, Oyewunmi OD, Camargo C, Nikpour B,et al. Tools and techniques for severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin Microbiol Rev. 2021; 34: e00228-20. doi: 10.1128/CMR.00228-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK,et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25: 2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sethuraman N, Jeremiah SS, Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020;323: 2249-2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 4.Charlton CL, Babady E, Ginocchio CC, Hatchette TF, Jerris RC, Li Y, et al. Practical Guidance for Clinical Microbiology Laboratories: Viruses Causing Acute Respiratory Tract Infections. Clin Microbiol Rev. 2018; 32: e00042-18. doi: 10.1128/CMR.00042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tom MR, Mina MJ. To Interpret the SARS-CoV-2 Test, Consider the Cycle Threshold Value. Clin Infect Dis. 2020;71: 2252-2254. doi: 10.1093/cid/ciaa619. PMid: ; PMCID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020: 581: 465-469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious severe acute respiratory syndrome Corona-virus 2 from diagnostic samples. Clin Infect Dis. 2020;71: 2663-2666. doi: 10.1093/cid/ciaa638. PMid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folgueira MD, Luczkowiak J, Lasala F, Pérez-Rivilla A, Delgado R. Prolonged SARS-CoV-2 cell culture replication in respiratory samples from patients with severe COVID-19. Clin Microbiol Infect. 2021; 27: 886-891. doi: 10.1016/j.cmi.2021.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong DSY, Koeleman JGM, Vaessen N, Breijer S, Paltansing S, de Man P. Rapid screening method for the detection of SARS-CoV-2 variants of concern. J Clin Virol. 2021;141:104903. doi: 10.1016/j.jcv.2021.104903. [DOI] [PMC free article] [PubMed] [Google Scholar]