Abstract
Antimicrobial resistance is one of the major threats to Public Health worldwide. Understanding the transfer and maintenance of antimicrobial resistance genes mediated by mobile genetic elements is thus urgent. In this work, we focus on the ColE1-like plasmid family, whose distinctive replication and multicopy nature has given rise to key discoveries and tools in molecular biology. Despite being massively used, the hosts, functions, and evolutionary history of these plasmids remain poorly known. Here, we built specific Hidden Markov Model (HMM) profiles to search ColE1 replicons within genomes. We identified 1,035 ColE1 plasmids in five Orders of γ-Proteobacteria, several of which are described here for the first time. The phylogenetic analysis of these replicons and their characteristic MOBP5/HEN relaxases suggest that ColE1 plasmids have diverged apart, with little transfer across orders, but frequent transfer across families. Additionally, ColE1 plasmids show a functional shift over the last decades, losing their characteristic bacteriocin production while gaining several antimicrobial resistance genes, mainly enzymatic determinants and including several extended-spectrum betalactamases and carbapenemases. Furthermore, ColE1 plasmids facilitate the intragenomic mobilization of these determinants, as various replicons were identified co-integrated with large non-ColE1 plasmids, mostly via transposases. These results illustrate how families of plasmids evolve and adapt their gene repertoires to bacterial adaptive requirements.
Author summary
The extraordinary adaptability of bacteria and the massive prevalence of mobile genetic elements within populations has turned antimicrobial resistance into a growing threat to Public Health. Among all the mobile genetic elements, plasmids have been the focus of attention as these extrachromosomal molecules of DNA are able to mobilize several antimicrobial resistance genes at once through conjugation. However, although small mobilizable and non-conjugative replicons have been traditionally overlooked when analyzing plasmid-mediated antimicrobial resistance, they have recently been described as important carriers of AMR genes. In this work, we have analyzed the ColE1-like plasmid family, whose study has been neglected even if they are one of the main groups of small plasmids in natural populations of Proteobacteria. We observed that these plasmids have evolved for a long time within γ-Proteobacteria acquiring different genetic features in specific hosts, being major players in the spread of antimicrobial resistance determinants.
Introduction
Plasmids are extrachromosomal self-replicating molecules of DNA able to transfer between bacteria mainly by conjugation [1]. They play a crucial role in bacterial evolution as they are key drivers of horizontal gene transfer, the major process of gene repertoire variation in prokaryotes [2]. Moreover, plasmids usually encode antimicrobial resistance determinants among their cargo genes and are considered to be the main spreaders of resistance in clinical environments [3].
Among their extraordinary diversity, there is a family of plasmids that has become very popular due to its widespread use in biotechnology since the 1970s: the ColE1-like plasmids (ColE1 plasmids hereinafter) [4]. Their history is closely related to the history of colicin-like bacteriocins, as pColE1 got its name by being the first plasmid characterized encoding the colicin E1 [5]. Since then, the ColE1-like group of replicons refers to every plasmid whose mechanism of replication resembles the original plasmid pColE1, most of which have been related to colicin production. All these plasmids share the same characteristics, traditionally described as small, multicopy and mobilizable replicons [6], generally associated to the MOBP5/HEN family of relaxases [7]. Recently, we showed that these small multicopy plasmids are encapsidated in phages with up to 10,000 times more efficiency than large plasmids, suggesting that phages could be major vectors of antimicrobial resistance genes borne in ColE1 plasmids [8].
The extended popularity of ColE1 plasmids in biotechnology lies in their ability to be stably maintained at high copy number within the cell due to their characteristic mechanism of replication mediated by two antisense and overlapping RNAs encoded in the origin of replication or ori [9]. Briefly, the ~550 bp RNA II pre-primer binds to its homologous DNA forming an RNA-DNA hybrid that triggers plasmid replication [10]. This mechanism is regulated by the ~100 bp RNA I, transcript that forms three stem loops complementary to the nascent structure of RNA II, to which it binds forming the kissing complex (RNA I-RNA II). This union modifies the secondary structure of the RNA II, inhibiting its binding to the plasmid DNA, thus, impeding the plasmid replication [11]. Some ColE1 plasmids encode an auxiliary protein called Rop (Repressor of primer) or Rom (RNA One Modulator), which stabilizes the kissing complex [12].
Known ColE1 replicons show a narrow host-range, mostly restricted to the Order Enterobacterales, where they were first described and extensively analyzed in terms of molecular biology [5,9]. In contrast, their role in wild-type populations has remained poorly studied [13], with a few works suggesting their presence in other γ-Proteobacteria [14–20]. Notwithstanding, recent studies have shown that small multicopy plasmids can have an enormous impact on bacterial evolution [21], often related to the dissemination and evolution of antimicrobial resistance determinants [13,15,22–25].
Given the increasing urgence in understanding the vectors of antimicrobial resistance, we have identified and studied the diversity of this overlooked family of plasmids. We combined a ColE1 Hidden Markov Model (HMM) profile of our own with PlasmidFinder [26] to identify ColE1 plasmids within the RefSeq database. We successfully collected 1,035 replicons and explored, for the first time, the evolutionary history of the ColE1 family among different Orders of γ-Proteobacteria focusing on both the ColE1 origin of replication and its MOBP5/HEN relaxase. This revealed the co-evolution of different mechanisms of replication within some ColE1 plasmids and its association with plasmid size. Finally, we examined the functional contribution that ColE1 replicons provide to their host, highlighting their role in the dissemination of antimicrobial resistance.
Results and discussion
ColE1 replicons are spread across five Orders of Proteobacteria
To identify ColE1 plasmids, we constructed two HMM profiles based on the sequence of 81 ColE1 replicons described in the literature (S1 Table). One profile includes the whole ~550 bp ColE1 origin of replication (ori), from the RNA II promoter to the origin of replication site (oriV), whereas the second one includes only the ~100 bp RNA I (Fig 1A). As the origin of replication of ColE1 plasmids from Pasteurellales was still uncharacterized, we studied eight ColE1 replicons from this Order to build specific HMM profiles (S1 Text).
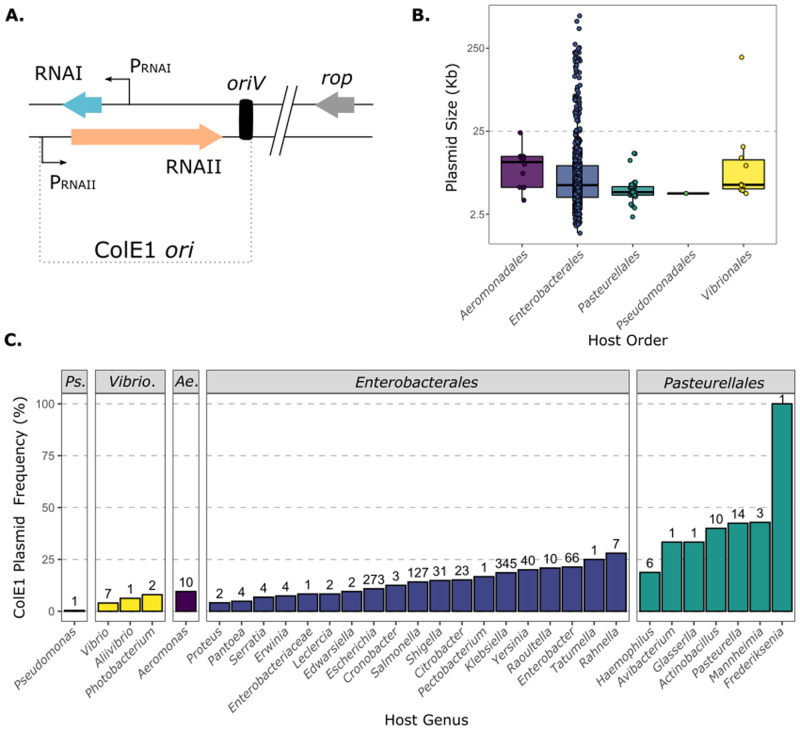
Fig 1. Identification of ColE1 plasmids.
A. The ColE1 origin of replication. Schematic representation of the ColE1 ori, including the RNAI, RNAII, their promoters and the oriV site. The gene encoding the auxiliary protein Rop is also represented. B. Size of ColE1 plasmids. Size of the ColE1 replicons attending to their Order. The y-axis represents the size(Kb) in logarithmic scale. C. Frequency of ColE1 plasmids within the Genus plasmidome. Proportion of ColE1 among plasmids of each Genus of bacteria. The x-axis represents the Genus, whereas the y-axis the frequency of ColE1 plasmids among all the plasmids from the Genus (%). The numbers at the top of the bars indicate the absolute number of ColE1 plasmids identified. The figure includes only the replicons identified in RefSeq. Ps.: Pseudomonadales. Vibrio.: Vibrionales. Ae.: Aeromonadales.
Using the aforementioned HMM profiles and PlasmidFinder (see Materials and methods), we searched the 20,532 plasmids available in RefSeq and identified 1,003 ColE1 plasmids. PlasmidFinder proved to be highly efficient in the identification of these replicons, as 884 out of the 1,003 plasmids were correctly identified as ColE1. Still, our HMM profiles successfully identified 126 additional ColE1 plasmids, substantially increasing the sensitivity of the search. Indeed, they were crucial for broadening the host spectrum of ColE1 replicons, as 96.5% of plasmids outside Enterobacterales were exclusively identified with the HMM profiles. Additionally, 32 ColE1 plasmids used for the construction of the profiles were not present in the RefSeq database. The final dataset has 1,035 ColE1 plasmids, with a mean average size of 14.7 kb and a median of 5.6 kb (Fig 1B and S2 Table).
The replicons were found in 33 different genera, 11 families and 5 Orders of γ-Proteobacteria. Most plasmids were identified in Enterobacterales, with Klebsiella and Escherichia accounting for 60.5% of all presently identified ColE1 plasmids. This is largely due to the over-representation of these bacteria in the database (Fig 1C). ColE1 plasmids represent 18.6% and 10.8% of all known plasmids from Klebsiella spp. and Escherichia spp., whereas they account for almost half of the plasmids in major representatives of Pasteurellales. In Aeromonadales and Vibrionales ColE1 are 9.5% and 6.1% of all plasmids, respectively. Interestingly, two ColE1 plasmids were identified in Pseudomonadales, one in Pseudomonas and another in Acinetobacter. At this stage it is thus unclear if these plasmids are rare in Pseudomonadales or if our method lacks sensitivity to identify them. We conclude that ColE1 plasmids are very abundant across at least four Orders of γ-Proteobacteria, showing particularly high prevalence within Pasteurellales.
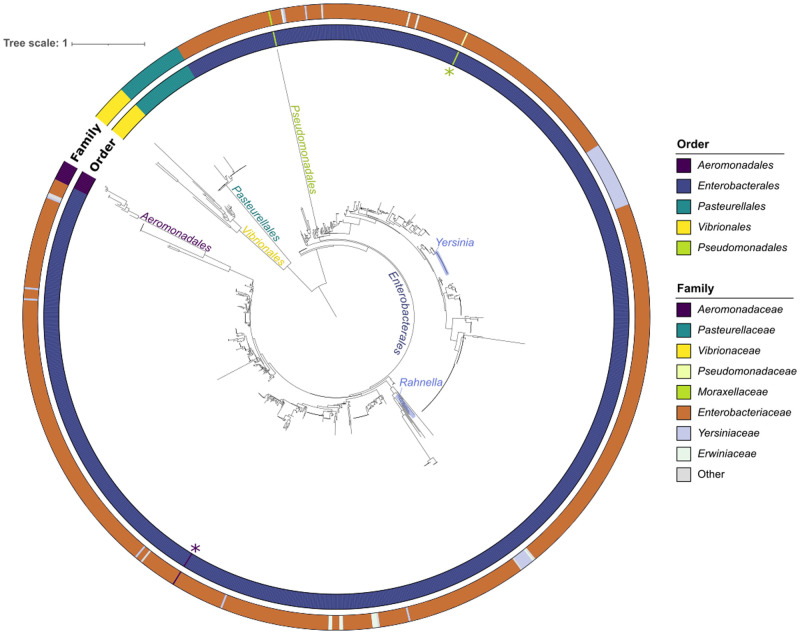
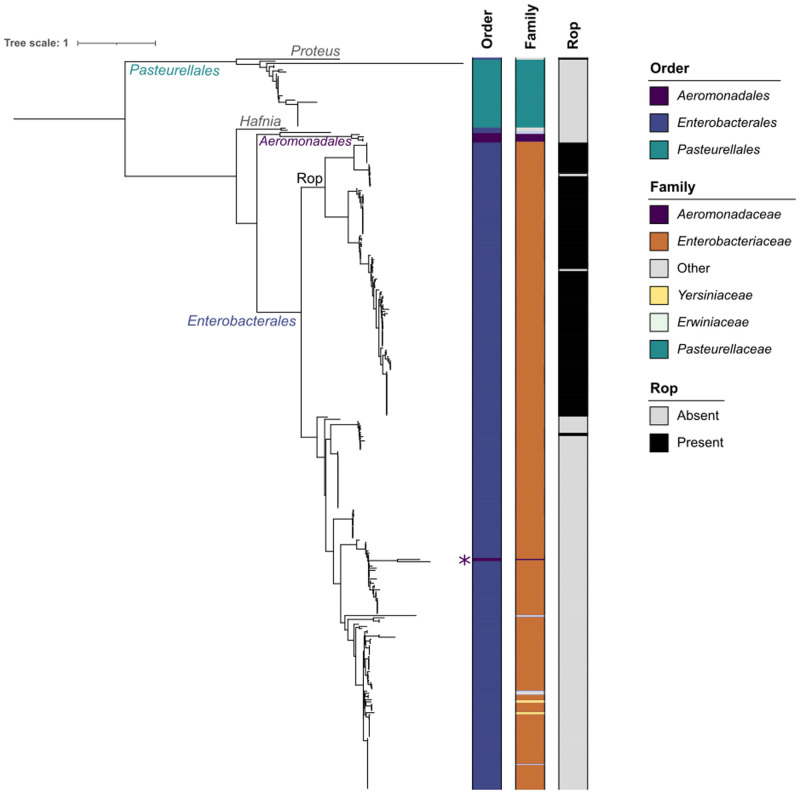
Some distinctions in the ColE1 ori among host clades have already been described. While the RNAs involved in replication generate three stem loops in Enterobacterales [27] and Aeromonadales [20], they generate only two in Vibrionales [16] and Pasteurellales (S1 Text). To assess the evolutionary relations between the ColE1 replicons, we constructed a phylogenetic tree of the 1,035 ColE1 origins of replication, defined as the region encoded from the RNA II promotor to the oriV site. Despite nucleotide sequences being worse phylogenetic markers than proteins [28], the tree was robust enough to observe a clear separation between replicons of different Orders. Even if it clusters the Aeromonadales and Pseudomonadales within the Enterobacterales clade, these correspond to very long branches whose basal position is not very well supported (Fig 2). Only two plasmids out of the 1,035 (0.19%) were classed within other Orders. In contrast, plasmids from different genera were often close together in the phylogenetic tree (S1–S4 Figs), suggesting frequent transfer between bacteria of different genera. Hence, plasmids seem unable to transfer across Orders, but often transfer across genera.
Fig 2. Phylogenetic tree of the 1,035 ColE1 origins of replication.
The colors of the inner circle represent the Order in which the replicon was identified, whereas the outer circle indicates the Family. The two asterisks next to the inner circle shows the two plasmids clustered within the clade of another Order. The phylogenetic tree was inferred following the best-fit model, SYM+R7.
Co-evolution of the ColE1 ori and putative Rep proteins
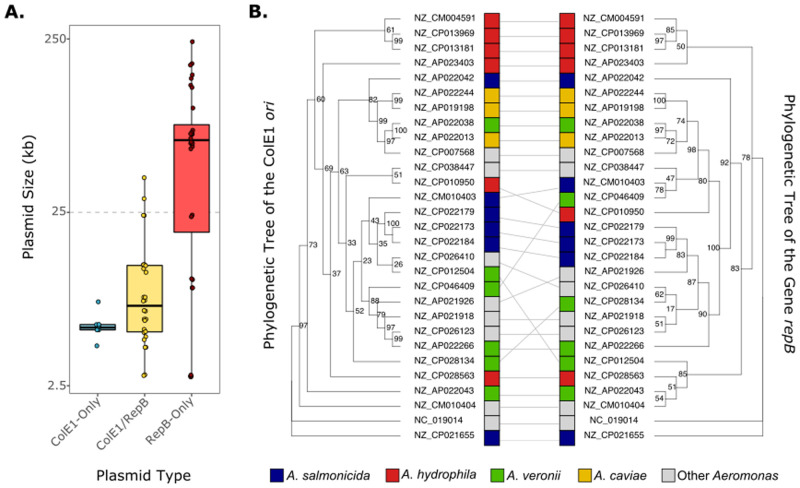
As the ColE1 mechanism of replication does not require any Rep protein, the presence of small ColE1/Rep plasmids in Pasteurellales and Aeromonadales was intriguing (S2 Table). In Pasteurellales, the ColE1 ori association with rep genes seems to have occurred through different independent events (S2 Text). In Aeromonadales, we identified two different plasmids from distinct sources and hosts (A. hydrophila and A. salmonicida) encoding a similar ColE1 ori and a putative RepB protein (81.5% and 82.0%, pairwise identity, respectively). Although this RepB protein is annotated as RepB replication protein in databases (RefSeq reference: WP_103859311.1) and it has been associated to plasmid replication in the literature [29,30], we have not found experimental evidence of its function. We will refer to it as putative RepB. To verify if this combination is a common phenomenon in this Order, and due to their small representation in our 1,035 ColE1 collection, we collected additional plasmids of Aeromonadales from RefSeq Assemblies, filtering those sequences encoding a ColE1 ori and/or the RepB protein. We obtained 4 chromosomes and 68 plasmids (S3 Table). Among plasmids, 8 encode just the ColE1 origin of replication (ColE1-only hereinafter), 32 the repB gene (RepB-only), and 28 both (ColE1/RepB) (Fig 3A). Hence, ColE1 plasmids in Aeromonadales are more frequently found with repB than alone.
Fig 3. ColE1/RepB plasmids in Aeromonadales.
A. Size of the ColE1/RepB plasmids (log scale) per category: ColE1-only (n = 8), ColE1/RepB (n = 30) and RepB-only (n = 28). The horizontal gray dashed line separates the small (<25kb) from the large plasmids (>25kb). B. Tanglegram of the ColE1 origin of replication (left) (best-fit model JTT+G4) and the gene repB (right) (TN+F+I+G4). The bootstrap values of the trees are indicated with numbers next to each node. The accession numbers and host Species of the plasmids are indicated at the tips of the branches.
The 8 ColE1-only plasmids are small (μ = 5,555 bp) (Fig 3A), whereas the RepB-only plasmids tend to be much larger (μ = 71,947 bp). Interestingly, plasmids with both elements are small (μ = 10,448 bp), and only slightly larger than the ColE1-only (t = 2.897, df = 29, p-value = 0.007). This finding denotes that repB is common among small ColE1 and large non-ColE1 plasmids within Aeromonadales. However, the phylogenetic tree of all the ColE1 ori from Aeromonadales separate the ColE1-only from the ColE1/RepB (S5 Fig), suggesting a unique repB acquisition/loss event. Indeed, their combination seems to be important for the plasmid as both elements show a strong genetic linkage, repB being usually in the immediate kilobase upstream the ColE1 ori (S6 Fig). To confirm this hypothesis, we built a tanglegram of the phylogenetic trees of the two genes ColE1/RepB. This analysis revealed their remarkable similarity, highlighting conserved clades of plasmids from A. hydrophila, A. caviae/A. veronii, A. salmonicida and other low-represented species (Figs 3B and S7). Hence, our results suggest that both elements have been co-evolving in plasmids moving between diverse species of Aeromonadales. Therefore, ColE1 plasmids show an alternative evolutionary trajectory within this Order, frequently encoding a putative replication gene but conserving the ColE1 origin of replication itself.
Genesis and evolution of ColE1 co-integrates
Although plasmids containing diverse types of replicons are common [31–33] and co-integration between small and large plasmids is known to occur [34–37], there is limited information available on the genesis and evolution of ColE1 co-integrates with large plasmids. Among the 1,035 ColE1 collection, 64 plasmids were larger than 25 kb (μ = 118.7 kb) (Fig 1B), which suggests a co-integration of the ColE1 plasmid with larger ones. We used PlasmidFinder to identify additional non-ColE1 plasmid types in the 62 “circular” ones. We found them in 33 of the 62 plasmids, mostly from the IncC, IncFIA, IncFIB, IncFII, IncN, IncN2 and IncN3 groups (S2 Table). We evaluated if these plasmids were co-integrates by looking for ColE1 related genes and the ColE1 ori in these larger plasmids. In many cases we identified the auxiliary gene rop, bacteriocin production operons or antimicrobial resistance determinants and transposons typically identified in ColE1-like plasmids (S9 Fig). Therefore, our analysis revealed that the co-integration of ColE1 with other plasmids is frequent. Of note, although many of these plasmids were previously described, their ColE1 origin of replication remained unnoticed [38–43].
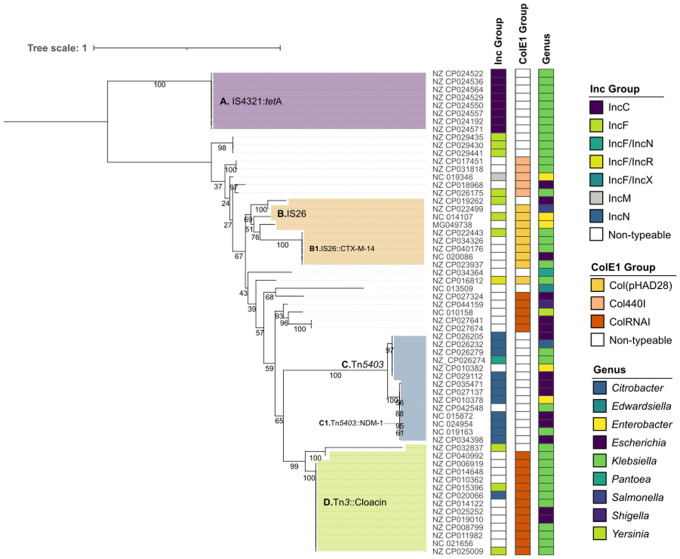
Among the 62 putative ColE1 co-integrates, 61 belonged to the Order Enterobacterales. The exception was a 194,647 bp plasmid from Vibrio campbellii (NZ_CP026317.1), non-typeable by PlasmidFinder. The co-integration in this plasmid occurred immediately upstream an rpn-like endonuclease (S8 Fig), which is the candidate responsible for the recombination event [44]. In Enterobacterales, the tree of the 61 ColE1 origins of replication tends to cluster co-integrates according to the existence of additional replicon types (e.g. the IncC and IncN clades), albeit there are exceptions (e.g. the IncF replicons) (Fig 4). To evaluate if this distribution was the result of unrelated recombination events or a co-evolution process of the ColE1 ori and the additional replicon, we analyzed the most represented clusters of co-integrates (S9 Fig): clade A (ColE1/IncC), clade B (ColE1/IncF and ColE1/NT), clade C (ColE1/IncN) and clade D (mostly ColE1/NT). Each of these clades represent co-integrates generated by different recombinases (Fig 4 and S3 Text).
Fig 4. Phylogenetic tree of the ColE1 origins of replication in large plasmids (>25kb) from Enterobacterales.
The bootstrap values are indicated with a number next to each node. The accession number of the 62 plasmids are indicated at the right of the tree. The color of the columns next to the accession numbers shows, from the left to the right, the Inc group associated to the large plasmid (identified with PlasmidFinder), the ColE1 origin of replication (identified with PlasmidFinder) and the host Genus. The colored shades within the tree (A-D) represent different patterns of co-integration exhibited by the plasmids, further detailed in the S9 Fig. Next to the letter A-D, it is indicated the putative transposon responsible for the recombination event. The phylogenetic tree was inferred following the best-fit model, K2P+I+G4.
The results show varied patterns of genesis and evolution of the co-integrates. In some cases, the conserved genetic environment surrounding the ColE1 ori suggests a co-integration event and subsequent co-evolution of the plasmids over time. These are the cases of the ColE1/IncC (clade A) and ColE1/IncN (clade C). The origin of the former clade seems to be recent, having been produced by a single recombination event involving IS4321s. The latter represents a successful association, as the co-integration through a Tn5403 has been conserved and spread among different hosts (e.g. Escherichia, Klebsiella, Enterobacter, Citrobacter) (Fig 4). In contrast, the genetic environment of the ColE1 ori in other clades suggests that the integration resulted from independent recombination events. That is the case of cluster D, in which the co-integration has occurred in diverse single events with various plasmid types (IncF, IncN, NT) through a Tn3 transposase and mobilizing a whole colicin operon. Lastly, cluster C shows an intermediate situation, in which different recombination events though an IS26 have occurred in different plasmids and hosts, but generating a successful co-integrate that has been evolving in Escherichia and Klebsiella (Cluster B1, Fig 4). Interestingly, most successful associations involve the mobilization of antimicrobial resistance genes (clade A: tetA; clade B1: blaCTX-M-14; clade C: blaNDM-1) and will be further discussed below.
The MOBP5/HEN relaxase has co-evolved with the ColE1 ori among different Orders while influenced by rop
To better understand the evolution of ColE1 plasmids in relation to conjugation, we analyzed its MOBP5/HEN relaxase [7,45]. As it has been noticed that many plasmids lack the relaxase [15], we first investigated its prevalence after discarding the 62 putative co-integrates and the 82 incomplete sequences. Ca. 39% of the ColE1 plasmids encode a relaxase with large differences across Orders: ~90% in Pasteurellales, ~40% in Enterobacterales and Aeromonadales, none in Vibrionales (S2 Table). Among the 348 ColE1 plasmids carrying a relaxase, we identified 352 relaxases of which most were MOBP (n = 339) bearing the characteristic motif III of the MOBP5/HEN group. Two plasmids carried different relaxases (MOBQ and MOBV) and 11 had truncated relaxase genes.
We built a phylogenetic tree of the MOBP5/HEN relaxases encoded in the plasmids (Fig 5). The tree clusters the proteins by the host Order even clearer than the ColE1 ori (Fig 2), implying a different evolutionary trajectory within each Order. Within Orders, the relaxase does not cluster at the genus-level (S10–S12 Figs). The large Enterobacteriaceae clade is divided into two groups, one of them constituted mostly by Escherichia plasmids, whereas the other clade included a diverse group of bacteria. The distinctive characteristic between the two clades is the presence of the auxiliary replication gene rop in the plasmid (Fig 1A), which is encoded in 59% of the plasmids from Enterobacterales (Figs 5 and S12). Although rop is negatively associated with the presence of a relaxase (X2 = 82.057, df = 1, p < 2.2e-16), their combination is not rare in the genus Escherichia, mainly associated to ColRNAI and Col(pHAD28) replicons (S12 Fig). This finding is consistent with previous works that have postulated plasmid recombination events through the oriT and cer sites [46,47], which are located at the opposite ends of the rop-relaxase genetic region [15]. This result implies that the relaxase and rop might be co-evolving within specific genera of Enterobacterales moving between different plasmids and mediating the evolution of this family of replicons.
Fig 5. Phylogenetic tree of the 339 MOBP5/HEN relaxases.
The colors of the columns at the right of the tree indicate, from the left to the right, the host Order, the host Family and the presence or absence of the rop gene in the plasmid. The asterisk next to the first column at the right of the tree represents the only relaxase clustered with plasmids of other Order. The phylogenetic tree was inferred following the best-fit model, JTT+F+I+G4.
The functional contribution of ColE1 plasmids
The ColE1 collection, after discarding plasmids with a “linear” status and putative co-integrates (n = 889), has 3,618 protein genes (μ = 4.07 genes/plasmid) and about 0.62 genes/kb (S4 Table). This is a lower gene density than usually found in bacterial chromosomes, >0.85 genes/kb [48], which may result from the existence of RNA genes or larger regulatory regions in the plasmids. Indeed, we could not identify a known protein coding gene in 50 ColE1 plasmids.
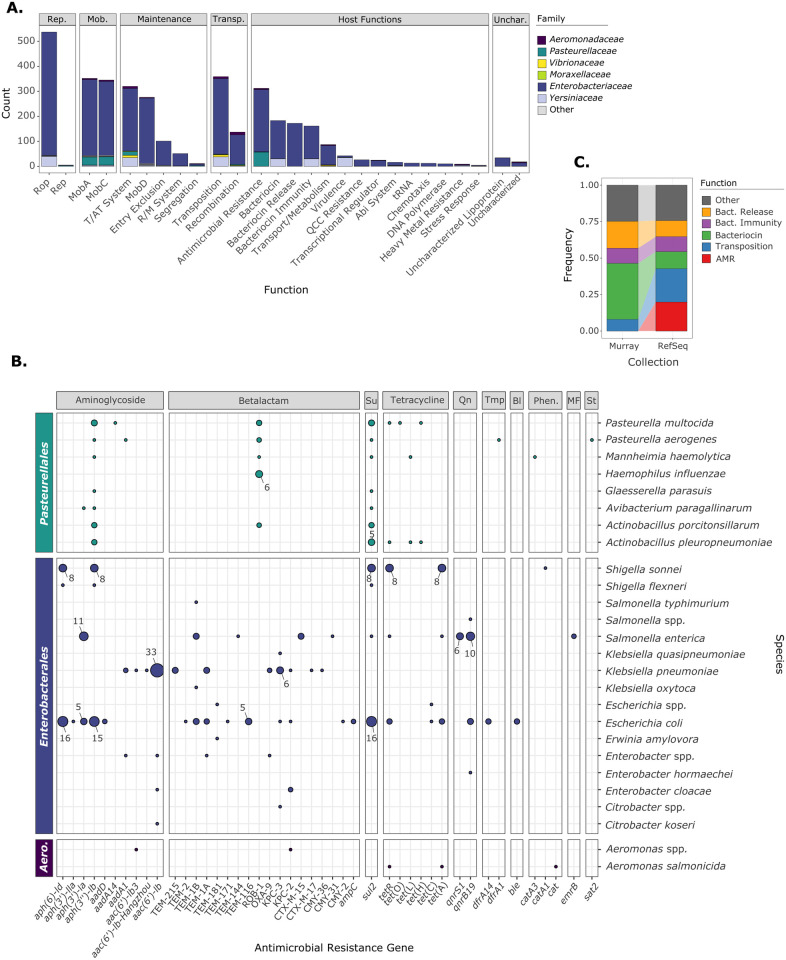
The ColE1 genetic repertoire has 261 different gene families (S4 Table), which we classified into 6 categories and 27 subcategories (Fig 6A). Functions associated with plasmid biology, replication and mobilization, are the most represented(Rop, MobA, MobC, MobD). Toxin/antitoxin systems and transposases are also very frequent, notably the Tn3 family (S4 Table). Nevertheless, ColE1 plasmids also present a variety of genes providing potential advantages to their host, some related to cell metabolism, virulence, defense from phages or heavy metal resistance. It was not unexpected to identify the production of colicin-like bacteriocins as one of the major functions provided by ColE1 plasmids [49], with 173 colicin-encoding plasmids from 9 different genera, despite these genes being restricted to plasmids of Enterobacterales. In contrast, it was surprising to find that antimicrobial resistance is the most frequent accessory function present in ColE1 plasmids (Fig 6A).
Fig 6. Functional classification of ColE1 plasmids.
A. Functions encoded on the ColE1 plasmids. Only plasmids with a status “Complete” and “Circular” were retrieved. Putative co-integrates were discarded. Rep.: Replication. Mob.: Mobilization. Transp.: Transposition. Unchar.: Uncharacterized. T/AT System: Toxin/Antitoxin System. R/M System: Restriction/Modification System. QCC Resistance: Quaternary Cation Compound Resistance. Abi System: Abortive-Infection System. B. Antimicrobial resistance determinants in ColE1 plasmids. The circles within the figure indicate the presence of the gene, their size being proportional to the number of genes. When a gene was identified 5 or more times, the exact number is indicated next to the circle. Aero.: Aeromonadales. Su.: Sulphonamide. Qn.: Quinolone. Tmp.: Trimethoprim. Bl.: Bleomycin. MF.: MFS transporter. St.: Streptothricin. C. Shift in the cargo genes of ColE1 plasmids from the Pre-antibiotic Era (Murray Collection) to the current one (RefSeq collection). Only the functions classified as “Host Functions” and “Transposition” are represented.
Antimicrobial resistance encoded in ColE1 plasmids
The analysis of the ColE1-associated resistome showed that 20% (n = 182) of ColE1 plasmids harbor at least one antimicrobial resistance determinant for a total of 312 genes related to antimicrobial resistance (μ = 1.71 genes/plasmid). Eleven plasmids encode five or more antimicrobial resistance genes, frequently providing multidrug-resistance genotypes despite their small size. Interestingly, resistance determinants are preferentially encoded in ColE1 plasmids without relaxases (χ2 = 6.305, df = 1, p = 0.012), where they show a higher density (μ = 1.88 genes/plasmid) than in the relaxase-encoding plasmids (μ = 1.34 genes/plasmid).
The ColE1-associated resistome is represented by 45 different genes conferring resistance against 9 classes of antimicrobials (Fig 6B), mostly aminoglycosides (n = 127) and betalactams (n = 73). The majority of these genes encode for enzymatic determinants, such as betalactamases, aminoglycoside phosphotransferases or aminoglycoside acetyltransferases. Genes coding for enzymatic determinants show a dose-dependent phenotype and could benefit from the high copy numbers of ColE1 plasmids as they will amplify their expression [50]. Even more, betalactamases and aminoglycoside enzymatic determinants exhibit a wide range of variants [51,52] and multicopy plasmids have been demonstrated to potentiate the evolution of their plasmid-encoded genes as they provide with higher supply of mutations [23,53]. Therefore, it raises the question of whether they could be involved in the wide range of variants within these families of resistance genes.
Our analysis also revealed that ColE1 replicons are associated with the emergence of antimicrobial resistant species categorized by the WHO as “high priority pathogens for the research and development of new antibiotics” [54]: ampicillin-resistant H. influenzae (blaROB-1), fluoroquinolone-resistant Salmonella (qnrS and qnrB1), and carbapenem-resistant and ESBL-producing Enterobacteriaceae (Fig 6B). Among the ESBLs and carbapenemases, we identified KPC-2, KPC-3, CTX-M-5, CTX-M-17, OXA-9, CMY-2, CMY-31 and CMY-36 encoded on ColE1 plasmids (Fig 6B). This study corroborates the growing evidence connecting the small multicopy replicons with ESBL and carbapenemase production observed in diverse isolates over the last years [13,55–59].
Although ColE1 co-integrates have not been included in the functional analysis above, their role in the evolution of antimicrobial resistance is worth mentioning. The co-integration of a CTX-M-17-encoding ColE1 plasmid with a large replicon has been already identified in a clinical E. coli from Vietnam and, furthermore, a recent study has observed that the 80% of KPC-3 producing K. pneumoniae outside hospital environments in Portugal carried the betalactamase in ColE1/IncF co-integrates [36,60]. Our results reveal that the co-integration of ColE1 plasmids is a widespread phenomenon in Enterobacterales, in many cases mediated by ESBL/carbapenemase-encoding transposons (NDM-1, CTX-M-14, KPC-2, KPC-3) (S9 Fig and S3 Text). Among these, the ColE1/IncN2 encoding NDM-1 and the ColE1/NT encoding CTX-M-14 seem to be the most relevant from a clinical perspective, as they have been identified in isolates from diverse Genera of Enterobacterales and spread across different continents [38,41,42]. These results demonstrate that ColE1 plasmids are key players in the mobility of antimicrobial resistance determinants within and between bacteria.
As ColE1 plasmids have been traditionally identified encoding bacteriocins, we wondered if AMR genes were recently acquired by this plasmid family. To do so, we compared our ColE1 collection against 115 ColE1 replicons identified within the Murray Collection (S5 Table, Materials and methods), Enterobacteriaceae isolates from the Pre-Antibiotic Era [61]. Their phylogenetic analysis cluster the ColE1 replicons from the Murray Collection together within our ColE1 collection (S13 Fig), showing little differences in terms of the ColE1 ori. However, none of the ColE1 plasmids from the Murray Collection encoded antimicrobial resistance (S6 Table), hence, the acquisition of AMR genes in ColE1 plasmids supposes a major recent shift in their cargo genes (Fig 6C) most certainly due to the increased selection pressure for acquisition of antibiotic resistance during the last decades.
Conclusions and perspectives
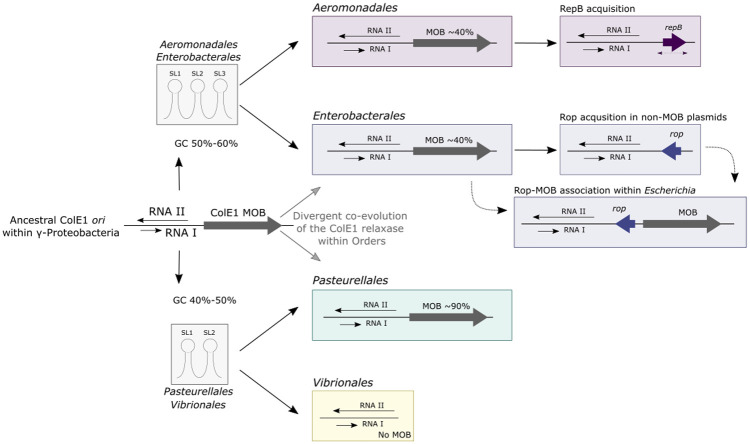
The present work provides new insights into the origin, evolution and current role of the ColE1-like plasmid family. The phylogeny of the ColE1 ori (Fig 2) and its MOBP5/HEN relaxase (Fig 5) denotes key differences according to the Order in which they have been described. Their GC contents differ between clades because they resemble those of their hosts. For instance, the average GC in Pasteurellales is 41.5% for the ColE1 ori, 43.1% for the relaxase and 40.3% for their genome, whereas in Enterobacterales it is 52.8%, 57.7% and 53.0%, respectively (S2 Table). This is consistent with the phylogenetic evidence and indicates that the ColE1 origin of replication originated some time ago in the Class γ-Proteobacteria, where it has been divergently co-evolving with the MOBP5/HEN relaxase within Orders but with little transfer across them (Fig 7). During this process, the secondary structure of the kissing complex has been modified and additional genes have been acquired in some taxa, such as repB in Aeromonadales or rop in Enterobacterales, the latter further associated with the relaxase in Escherichia (Fig 7). Nevertheless, our phylogenetic analysis suggests that although the ColE1 ori are specific to each Order, plasmids transfer much more freely across Genera (S1–S4 and S10–S12 Figs).
Fig 7. Evolutionary history of the ColE1 plasmids.
The schematic representation of the ColE1 ori is illustrated with two antisense black arrows for both the RNAI and RNAII, and the ColE1 relaxase with a thick grey arrow annotated as MOB. The divergent evolution of the ColE1 ori within Aeromonadales/Enterobacterales and Pasteurellales/Vibrionales, respectively, is represented with black arrows ending in two grey boxes, each of them showing three (SL1-3) or two stem loops (SL1-2). At the right of the figure, the genetic elements related to the ColE1 plasmids within each Order are represented. The rop gene in Enterobacterales is represented with a blue arrow; the repB gene in Aeromonadales with a purple arrow; and the dashed purple arrow below the repB gene represents its variable location within the plasmid.
Additionally, we have observed a functional shift of cargo genes between ColE1 plasmids identified prior to the extended use of antibiotics and those identified more recently (Fig 6C). This shift from bacteriocin production to antimicrobial resistance is presumably due to the high selective pressure undergone within bacterial population for antibiotic resistance, although it remains unclear why bacteriocins are now less frequent in ColE1 plasmids. The same ColE1 backbones are identified either as bacteriocin-producing ColE1, as antimicrobial resistant ColE1 or as cryptic. Indeed, most ColE1 plasmids do encode neither bacteriocins nor AMR. This means that the functional shift may not be mediated by direct replacement of bacteriocin-producing loci by antimicrobial resistant genes, but by a genetic turnover of functions that currently tends to increase the frequency of AMR genes because of natural selection for this trait. Interestingly, the acquisition of AMR genes was concomitant with an increase in genes encoding transposases (Fig 6C), which may have facilitated their acquisition by ColE1 plasmids. Overall, this analysis supports early works, in which it was suggested that the acquisition of AMR genes after the antibiotic use was mediated by the same plasmids from the pre-antibiotic era [62].
Our results also raise intriguing and challenging questions that could be the aim of future research lines. (i) Are ColE1 plasmids present in other Orders of Proteobacteria? Our analysis revealed that the only plasmid already identified in Pseudomonadales [13] was phylogenetically distant from the remaining replicons. This suggests that ColE1 plasmids might be circulating within Pseudomonadales in underrepresented genera or in minor prevalence. Indeed, due to the divergent evolution of ColE1 replicons and the source of our HMM profiles, the circulation of distant variants of ColE1 cannot be discarded. (ii) What are the consequences of the frequent occurrence of repB in ColE1 plasmids from Aeromonadales? The extended co-occurrence of both the ColE1 ori and repB suggests that rather than switching the plasmid’s mechanism of replication, there might be a synergistic effect between both, making these replicons more successful within this Order. (iii) Is the ColE1 ori functional when the replicon co-integrates with large plasmids? If the ColE1 ori increases the plasmid copy number (PCN) of the large one, it could make the co-integrate unstable due to its higher fitness cost. This could have happened after the integration of the ColE1 plasmid pIP843 with an additional replicon in the co-integrate pE66An [36], where the ColE1 ori was truncated after the recombination event. In contrast, if the PCN is not modified, the genesis of ColE1 co-integrates could increase mobility via conduction, but also affect gene expression, fitness cost, and evolvability of the multicopy plasmid.
The frequent co-integration of ColE1 plasmids with additional replicons within Enterobacterales suggests they may have a more determinant role in the evolution of the bacterial plasmidome than previously envisaged. The shift in their cargo genes from bacteriocin production to antimicrobial resistance suggests these plasmids are becoming important drivers of the spread of antibiotic resistance.
Materials and methods
Collection of ColE1 plasmids characterized in the literature
To create a collection of ColE1 plasmids, we first looked for the ColE1 replicons that had been already described in the literature, examining every published work deposited in Pubmed (last accessed in July, 2020) using as query “ColE1”. We only retrieved those plasmids whose ColE1 origin of replication had been annotated, either the RNAI or the RNA II, obtaining a total number of 74 ColE1 plasmids (S1 Table). While examining the bibliography, it was noticed that ColE1 plasmids had been described in the Order Pasteurellales but, to date, no work has characterized their origin of replication. Therefore, we selected from literature and characterized 8 putative ColE1 plasmids described in Pasteurellales to include ColE1 replicons from this taxon in our analysis. This way, our initial ColE1 collection was constituted by 81 plasmids, representing our reference dataset (S1 Table).
Characterization of ColE1 replicons from Pasteurellales
The eight plasmids selected for the description of their ColE1 origin of replication in Pasteurellales were pB1000 (DQ840517), pIG1 (NC_001774.1), pLS88 (L23118), pAB2 (Z21724), pB1002 (JQ773456), pB1005 (NC_012215.1), pB1006 (NC_012216.1) and pB1000’ (NC_019177.1). To characterize their origin of replication we followed different approaches: i) the current literature available on their origin of replication; ii) multiple sequence alignments of the origin of replication among the different plasmids; iii) data from an RNA-Seq analysis of H. influenzae RdKW20 carrying pB1000 available in the European Nucleotide Archive under the Accession Number PRJEB44283; and iv) an in silico analysis of the secondary structure of the putative ColE1 RNAs (S14 Fig). Detailed information on this analysis is available in the S1 Text.
Still, to validate our results: i) we corroborated that the elements of the ColE1 origin of replication were conserved among the 8 plasmids; ii) we demonstrated that ColE1 plasmids that have been described coexisting within a cell show key mutations in their RNAs allowing their compatibility [19,24]; and iii) we verified that mutations in specific nucleotides of the RNAs modify the plasmid copy number [63], as it has been demonstrated in ColE1 plasmids from Enterobacterales [9] (S1 Text and S15 Fig).
Construction of HMM profiles for ColE1 plasmids
For the construction of Hidden Markov Model (HMM) profiles, we first performed multiple sequence alignments (MSA) of the 81 ColE1 plasmids collected in our reference dataset using MAFFT [64], version 7.450, options—globalpair and—maxiterate 1000 and examined the results with Geneious Prime (2019.0.4) for the detection of artifacts. We performed two different MSAs, the first one was specific for the RNAI sequence of the ColE1 plasmids, obtained from their respective published works (S1 Table). The second MSA was broader, including the whole ori region, between the RNAII promoter and the oriV site, thus including both RNAs and their promoters.
Once we had the alignments, we used HMMer (http://hmmer.org, last accessed December 2020), version 3.1b2 and built the HMM profiles with hmmbuild. Due to the remarkable sequence disagreements between ColE1 plasmids from different Orders, we constructed specific profiles for each Order of bacteria in addition to a profile including all the ColE1 plasmids. All the HMM profiles were used in our ColE1 search in order to increase our sensitivity.
Identification of ColE1 plasmids
For the identification of ColE1 plasmids, we used the dataset of complete bacterial genomes from NCBI RefSeq (last accessed in September, 2019). We retrieved the 20,523 plasmids following the classification of the replicon as “plasmid” or “chromosome” within the GenBank file. For the detection of ColE1 plasmids, we combined the search for the aforementioned HMM profiles using the HMMer tool hmmsearch with a parallel search using PlasmidFinder [26]. To increase the specificity of our search, only those plasmids identified with both HMM profiles (ori and RNAI) were retrieved for further analysis. When using the RNAI HMM profile, the E-value threshold was augmented to 0.01 due to its short sequence (~100 bp) following the recommendations of the authors, whereas in the complete ori HMM profile, the E-value threshold was maintained at the default 0.001.
The 1,056 plasmids identified during the search were examined using Geneious Prime (2019.0.4) to verify their ColE1 origin of replication (presence of both the RNA I, RNA II and oriV site). After this inspection, 53 ColE1 replicons were discarded from different reasons: 15 sequences were incomplete plasmids with a ColE1 ori partially sequenced, 17 were actual cloning vectors and 21, although identified during the search, did not show the characteristic ColE1 origin of replication when manually inspected. To the 1,003 remaining plasmids, we added 32 additional replicons employed for the construction of the HMM profiles and absent within RefSeq, reaching 1,035 plasmids. These 32 elements obtained from the literature, but absent from RefSeq, were not used to estimate the ColE1 frequency within genera. All the plasmids were characterized according to the Inc/rep typing and the MOB typing, respectively. For the Inc/rep typing we used PlasmidFinder [26], version 2.0.1, with a minimum identity threshold of 95% and a minimum coverage threshold of 60%, with both the Enterobacteriaceae and Gram positive databases (last update on January 1st, 2021). We considered the following results as ColE1-like representatives: ColRNAI, Col(pHAD28), Col(YF27601), Col440I and Col440II. For the MOB-typing we used the online version of MOBscan [65] (last accessed on January, 2021) with the default parameters, which employed the program hmmscan, version 3.3, and the MOBfamDB database. Detailed information on the plasmids is available in the S2 Table.
Functional analysis of ColE1 plasmids
Among the 1,035 ColE1 plasmids– 1,003 identified in RefSeq plus 32 additional plasmids described in the literature but absent from RefSeq–only the 958 circular sequences were considered for the analysis of plasmid size. Among them, just the 889 canonical ColE1 plasmids (<25kb) were further used for the functional analysis. The sequences were annotated using Prokka, version 1.13 [66], and the results were manually curated using Geneious Prime(2019.0.4). The resulting genes were classified in 6 groups and 27 subgroups attending to their functions and frequency (S4 Table). As certain sections of this work focus on specific functions of these plasmids such as antimicrobial resistance and bacteriocin production, we further validated the antimicrobial resistance genes using ResFinder version 4.0 [67], with a minimum identity threshold of 90% and a minimum coverage threshold of 60%. The genes encoding bacteriocins identified with Prokka were further validated with the online tool blastx (https://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed December 2020), annotating the bacteriocin according to the best match from the RefSeq database of Reference Proteins.
Phylogenetic analysis
The phylogenetic analysis of the 1,035 ColE1 plasmids was performed from a MSA of the ColE1 origin of replication (ori) region, defined as the region between the RNA II promoter to the oriV site, using MAFFT [64], version 7.450, options—globalpair and—maxiterate 1000 and examined the result with Geneious Prime (2019.0.4). The phylogenetic tree was inferred by maximum-likelihood using IQ-Tree [68], version 1.6.1, with 1000 ultrafast bootstrap experiments [69] and the ModelFinder function [70], being the selected model indicated in the legend of each figure. The visualization of the inferred tree was performed with iTOL [71], version 5.7. Additional phylogenetic analyses were performed in this work focusing on the relaxase protein (n = 339), the ColE1 origin of replication within large plasmids in Enterobacterales (n = 61) and the ColE1 ori and RepB protein within Aeromonadales (n = 29). All these analyses followed the same procedure described for the 1,035 ColE1 origins of replication. The Software Dendroscope [72], version 3.7.3, was used for the tanglegram analysis of both the ColE1 ori and RepB phylogenetic trees and its visualization. All the phylogenetic trees in Newick format have been included in the supplementary material (S4 Text).
Identification of ColE1/RepB plasmids in Aeromonadales
The identification of further ColE1 plasmids within Aeromonadales, as well as plasmids encoding the repB gene described in pAsa10 (NZ_ MF621616.1) and p2_045096 (NZ_CP028563.1), was performed within the Assemblies database of NCBI RefSeq (accessed November, 2020), retrieving the 515 entries belonging to Aeromonadales. First, for the identification of ColE1 plasmids we used the aforementioned HMM profiles specific for Aeromonadales, with the HMMer tool hmmsearch, following the same procedure as previously described. Among the 217 sequences harboring a ColE1 origin of replication, only the 40 circular ones were selected for further analysis: 36 plasmids (μ = 9,36 kb) and 4 chromosomes (μ = 4,88 Mb).
Among the latter 40 sequences, 16 encoded an homolog to pAsa10 and p2_045096 repB. Therefore, we used the 16 repB genes for the construction of a new HMM profile, performing an MSA using MAFFT, options—maxiterate 1000 and—global-pair. The MSA was visualized with Geneious Prime (2019.0.4). Then, the RepB HMM profile was built with the HMMer tool hmmbuild and used for the identification of the gene within the same RefSeq database, using the HMMer tool hmmsearch (default, E-value < 0.001). Among the 467 sequences with the gene, only the 63 circular were selected for the analysis: 60 plasmids (μ = 43,24 kb) and 3 chromosomes (μ = 4,90 Mb). A total of 28 sequences were identified in the searches for both ColE1 ori and repB.
Identification of ColE1 plasmids from the Murray Collection
Raw Illumina Sequencing data from 370 isolates of the Murray Collection was downloaded from the European Nucleotide Archive, available under the accession number PRJEB3255 [61]. We performed a quality control using the software FastQC [73], version 0.11.9, and trimmed the reads using fastp [74], version 0.20.1. Putative plasmids were assembled from the Illumina reads using PlasmidSPAdes [75], version 3.15.2, with the default parameters. We obtained a total number of 40,138 contigs, with an average size of 2,243 bp. Then, we used our Enterobacterales ColE1 HMM profile and PlasmidFinder to identify ColE1 replicons within the contigs, following the same conditions as previously specified. A total number of 173 sequences were retrieved although 58 were afterwards discarded due to various reasons (S5 Table): 18 were ColE1 replicons that presented partially sequenced the ori, 6 were too short sequences (μ = 413,2 bp) and 34 were not actual ColE1 replicons after manual inspection. The functional contribution of the 115 ColE1 plasmids (μ = 8,667.6 bp) identified was analyzed using Prokka and ResFinder, with the aforementioned parameters (S6 Table).
Statistics and data visualization
The different statistical tests used during this work (ANOVA, Student t-test, correlation test, Chi-squared test, Fisher’s exact test) were performed with the default R package stats in RStudio, version 3.6.1. Most data visualization was performed with the R package ggplot2 with few aforementioned exceptions. Plasmid representations were drawn with Easyfig [76], version 2.2.5.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(TXT)
Collection of plasmids already described in the literature and used for building the HMM profiles.
(XLSX)
Collection of 1,035 ColE1 plasmids identified in RefSeq with both the HMM profiles and PlasmidFinder.
(XLSX)
Collection of plasmids identified in Aeromonadales encoding a ColE1 ori and/or repB.
(XLSX)
Annotation of the complete canonical ColE1 plasmids (<25kb) from the 1,035 ColE1 collection.
(XLSX)
ColE1 replicons identified within the ENA project PRJEB44283.
(XLSX)
Annotation of the ColE1 plasmids identified within the ENA project PRJEB44283.
(XLSX)
Phylogenetic tree of the 12 ColE1 origins of replication identified in Aeromonadales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, K2P+G4.
(TIFF)
Phylogenetic tree of the 20 ColE1 origins of replication identified in Vibrionales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, TIM3e+G4.
(TIFF)
Phylogenetic tree of the 38 ColE1 origins of replication identified in Pasteurellales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase, either MOBV or MOBP. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, HKY+F+G4.
(TIFF)
Phylogenetic tree of the 964 ColE1 origins of replication identified in Enterobacterales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase, either MOBP, MOBQ or truncated (ΔMOB). The legend is at the right of the figure. The bootstrap values are indicated with the nodes and branch colors. Bootstraps under 50 are represented in black, whereas bootstraps over 50 follow the legend at the right. The phylogenetic tree was inferred following the best-fit model, SYM+R7.
(TIFF)
The colors of the column at the right of the tree represent if the plasmid is ColE1-only or ColE1/RepB. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, TVMe+R3.
(TIFF)
At the left, it is represented the phylogenetic tree of the ColE1 ori from the ColE1/RepB plasmids, as indicated in Fig 3. At the end of every branch it is indicated the host Species (colored square, legend at the bottom), the Accession Number and the plasmid size. At the right of the figure, it is represented the genetic content of the ColE1/RepB plasmids. The ColE1 ori is represented with a purple rectangle whereas repB with a purple arrow. The remaining genes are represented with colored arrows, being the legend at the bottom of the figure.
(TIFF)
Comparison of the phylogenetic trees of the ColE1 origin of replication (left) (best-fit model JTT+G4) and the gene repB (right) (TN+F+I+G4). The color of the branches represents the comparison metric. The legend is shown at the bottom of the figure. A score of 1 denotes the subtree structure of the node is identical to the subtree structure of its best corresponding node. The figure was performed with the phylo.io tool [77].
(TIFF)
Schematic representation of the ColE1 co-integrate identified in V. campbellii (NZ_CP026317.1). The complete plasmid (194 kb) is represented at the top of the figure. The purple square indicates the genetic environment of the ColE1 origin of replication, which is represented at the bottom of the figure. The ColE1 ori and remaining genes are represented with colored squares and arrows, being the legend at the bottom of the figure.
(TIFF)
Schematic representation of the genetic environment of the ColE1 ori in the most-represented clades of ColE1 co-integrates from Enterobacterales (Fig 4). The ColE1 ori and remaining genes are represented with colored squares and arrows, being the legend at the top of the figure. The Accession Number and size of each plasmid is indicated at the middle of the figure.
(TIFF)
Phylogenetic tree of the ColE1 relaxases from Aeromonadales. The colors of the first column represent the Genus and the second column represents the Species. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, JTT+F+G4.
(TIFF)
Phylogenetic tree of the ColE1 relaxases from Pasteurellales. The colors of the first column represent the Genus and the second column represents the Species. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, VT+G4.
(TIFF)
Phylogenetic tree of the ColE1 relaxases from Enterobacterales. The colors of the first column represent the Genus, the second column represents the presence or absence of rop and the third one indicates the PlasmidFinder result. The legend is at the right of the figure. The phylogenetic tree was inferred following the best-fit model, JTT+F+I+G4.
(TIFF)
The colors of the column represent the origin of the plasmid, Murray Collection or RefSeq. The legend is at the right of the figure. The phylogenetic tree was inferred following the best-fit model, SYM+R8.
(TIFF)
Schematic representation of the secondary structure of the putative RNA I from eight ColE1 plasmids from Pasteurellales. The name and Accession Number of each plasmid is indicated at the top of each sequence. The color of the nucleotides indicates the base-pair probabilities, from 0 to 1, being the legend next to each sequence. The secondary structure and probabilities were inferred with RNAfold WebServer.
(TIFF)
Schematic representation of the secondary structure of the putative RNA I from different ColE1 plasmids from Pasteurellales described coexisting within the cell. The different plasmid combinations are represented separated in the boxes A, B, C and D, being indicated the Species isolate, the plasmid names and the published reference. The Accession Number of each plasmid is indicated at the top of the sequence. The letters and arrows show the dissimilarities identified among the coexisting plasmids, in black those affecting the loop and in grey those affecting the stem. The color of the nucleotides indicates the base-pair probabilities, from 0 to 1, being the legend at the top right of each box. The secondary structure and probabilities were inferred with RNAfold WebServer.
(TIFF)
Acknowledgments
We would like to thank Carlos Serna for the technical help with the Murray Collection availability.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
B.G.Z. was supported by grants from the European Joint Program One Health EJP, ARDIG Grant Agreement No 77380; from the European Union’s Horizon 2020 research and innovation program. M.A.A. was supported by the Universidad Complutense de Madrid (Grant ID CT27/16-CT28/16 and EB14/19). E.P.C.R. acknowledges the financial support of Equipe FRM (EQU201903007835) and the Laboratoire d’Excellence IBEID (ANR-10-LABX-62-IBEID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zechner EL, Moncalián G, de la Cruz F. Relaxases and Plasmid Transfer in Gram-Negative Bacteria. In: Backert S, Grohmann E, eds. Type IV Secretion in Gram-Negative and Gram-Positive Bacteria. Vol 413. Current Topics in Microbiology and Immunology. Springer International Publishing; 2017:93–113. doi: 10.1007/978-3-319-75241-9_4 [DOI] [PubMed] [Google Scholar]
- 2.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7(1):e1001284. doi: 10.1371/journal.pgen.1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol IJMM. 2013;303(6–7):298–304. doi: 10.1016/j.ijmm.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 4.Hershfield V, Boyer HW, Yanofsky C, Lovett MA, Helinski DR. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974;71(9):3455–9. doi: 10.1073/pnas.71.9.3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeWitt W, Helinski DR. Characterization of colicinogenic factor E1 from a non-induced and a mitomycin C-induced Proteus strain. J Mol Biol. 1965;13(3):692–703. doi: 10.1016/S0022-2836(65)80136-X [DOI] [Google Scholar]
- 6.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73(5):1121–37. doi: 10.1093/jac/dkx488 [DOI] [PubMed] [Google Scholar]
- 7.Francia MV, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28(1):79–100. doi: 10.1016/j.femsre.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Rubio L, Serna C, Ares-Arroyo M, Matamoros BR, Delgado-Blas JF, Montero N, et al. Extensive antimicrobial resistance mobilization via multicopy plasmid encapsidation mediated by temperate phages. J Antimicrob Chemother. 2020;75(11):3173–80. doi: 10.1093/jac/dkaa311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camps M. Modulation of ColE1-like Plasmid Replication for Recombinant Gene Expression. Recent Pat DNA Gene Seq. 2010;4(1):58–73 doi: 10.2174/187221510790410822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masukata H, Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986;44(1):125–36. doi: 10.1016/0092-8674(86)90491-5 [DOI] [PubMed] [Google Scholar]
- 11.Cesareni G, Helmer-Citterich M, Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet TIG. 1991;7(7):230–5. doi: 10.1016/0168-9525(91)90370-6 [DOI] [PubMed] [Google Scholar]
- 12.Castagnoli L, Scarpa M, Kokkinidis M, Banner DW, Tsernoglou D, Cesareni G. Genetic and structural analysis of the ColE1 Rop (Rom) protein. EMBO J. 1989;8(2):621–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez MS, Iriarte A, Reyes-Lamothe R, Sherratt DJ, Tolmasky ME. Small Klebsiella pneumoniae Plasmids: Neglected Contributors to Antibiotic Resistance. Front Microbiol. 2019;10. doi: 10.3389/fmicb.2019.02182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brovedan M, Repizo GD, Marchiaro P, Viale AM, Limansky A. Characterization of the diverse plasmid pool harbored by the blaNDM-1-containing Acinetobacter bereziniae HPC229 clinical strain. PLoS One. 2019;14(11):e0220584. doi: 10.1371/journal.pone.0220584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ares-Arroyo M, Bernabe-Balas C, Santos-Lopez A, Baquero MR, Prasad KN, Cid D, et al. PCR-Based Analysis of ColE1 Plasmids in Clinical Isolates and Metagenomic Samples Reveals Their Importance as Gene Capture Platforms. Front Microbiol. 2018;9:469. doi: 10.3389/fmicb.2018.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Roux F, Davis BM, Waldor MK. Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res. 2011;39(3):1004–13. doi: 10.1093/nar/gkq852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan L, Leung PC, Gu J-D. A new ColE1-like plasmid group revealed by comparative analysis of the replication proficient fragments of Vibrionaceae plasmids. J Microbiol Biotechnol. 2010;20(8):1163–78. [DOI] [PubMed] [Google Scholar]
- 18.San Millan A, Escudero JA, Catalan A, Nieto S, Farelo F, Gibert M, et al. β-Lactam Resistance in Haemophilus parasuis Is Mediated by Plasmid pB1000 Bearing blaROB-1. Antimicrob Agents Chemother. 2007;51(6):2260–4. doi: 10.1128/AAC.00242-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Millan A, Escudero JA, Gutierrez B, Hidalgo L, Garcia N, Llagostera M, et al. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother. 2009;53(8):3399–404. doi: 10.1128/AAC.01522-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent AT, Emond-Rheault J-G, Barbeau X. Attéré SA, Frenette M, Lagüe P, et al. Antibiotic resistance due to an unusual ColE1-type replicon plasmid in Aeromonas salmonicida. Microbiology. 2016;162(6):942–53. doi: 10.1099/mic.0.000286 [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Beltrán J, DelaFuente J, León-Sampedro R, MacLean RC, San Millán Á. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Microbiol. Published online January 19, 2021. doi: 10.1038/s41579-020-00497-1 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Beltran J, Hernandez-Beltran JCR, DelaFuente J, Escudero JA, Fuentes-Hernandez A, MacLean RC, et al. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat Ecol Evol. 2018;2(5):873–81. doi: 10.1038/s41559-018-0529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol 2016;1(1):10. doi: 10.1038/s41559-016-0010 [DOI] [PubMed] [Google Scholar]
- 24.Santos-Lopez A, Bernabe-Balas C, Ares-Arroyo M, Ortega-Huedo R, Hoefer A, San Millan A, et al. A Naturally Occurring Single Nucleotide Polymorphism in a Multicopy Plasmid Produces a Reversible Increase in Antibiotic Resistance. Antimicrob Agents Chemother. 2017;61(2). doi: 10.1128/AAC.01735-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santer M, Uecker H. Evolutionary Rescue and Drug Resistance on Multicopy Plasmids. Genetics. 2020;215(3):847–68. doi: 10.1534/genetics.119.303012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob Agents Chemother. 2014;58(7):3895–903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilly J, Camps M. Mechanisms of Theta Plasmid Replication. Microbiol Spectr. 2015;3(1):PLAS-0029-2014. doi: 10.1128/microbiolspec.PLAS-0029-2014 [DOI] [PubMed] [Google Scholar]
- 28.Patwardhan A, Ray S, Roy A. Phylogenetics & evolutionary biology molecular markers in phylogenetic studies—a review. Phylogenetics Evol Biol. 2014;57 [Google Scholar]
- 29.Attéré SA, Vincent AT, Paccaud M, Frenette M, Charette SJ. The Role for the Small Cryptic Plasmids As Moldable Vectors for Genetic Innovation in Aeromonas salmonicida subsp. salmonicida. Front Genet. 2017;8:211. doi: 10.3389/fgene.2017.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HJ, Jin HM, Park MS, Park W, Madsen EL, Jeon CO. Recovery of Plasmid pEMB1, Whose Toxin-Antitoxin System Stabilizes an Ampicillin Resistance-Conferring β-Lactamase Gene in Escherichia coli, from Natural Environments. Wommack KE, ed. Appl Environ Microbiol 2015;81(1):40–47. doi: 10.1128/AEM.02691-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osborn AM, da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 2000;146(Pt 9):2267–75. doi: 10.1099/00221287-146-9-2267 [DOI] [PubMed] [Google Scholar]
- 32.Qu D, Shen Y, Hu L, Jiang X, Yin Z, Gao B, et al. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:IncpA1763-KPC:IncN1 or IncFIIpHN7A8:IncpA1763-KPC:IncN1. Infect Drug Resist. 2019;12:285–96. doi: 10.2147/IDR.S189168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Y, Shen P, Liang W, Jin J, Jiang X. A putative multi-replicon plasmid co-harboring beta-lactamase genes blaKPC-2, blaCTX-M-14 and blaTEM-1 and trimethoprim resistance gene dfrA25 from a Klebsiella pneumoniae sequence type(ST) 11 strain in China. PLoS One. 2017;12(2):e0171339. doi: 10.1371/journal.pone.0171339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dionisio F, Zilhão R, Gama JA. Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid. 2019;102:29–36. doi: 10.1016/j.plasmid.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 35.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, et al. Insertion Sequence IS26 Reorganizes Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. MBio. 2015;6(3):e00762. doi: 10.1128/mBio.00762-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin DL, Ramirez MS, Tran T, Tolmasky ME. A cointegrate-like plasmid that facilitates dissemination by conjugation of the extended-spectrum β-lactamase CTX-M-17. Antimicrob Agents Chemother. 2013;57(10):5191–2. doi: 10.1128/AAC.01365-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson BC, Hashimoto H, Rownd RH. Cointegrate formation between homologous plasmids in Escherichia coli. J Bacteriol. 1982;151(3):1086–94. doi: 10.1128/jb.151.3.1086-1094.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y-T, Lin A-C, Siu LK, Koh TH. Sequence of closely related plasmids encoding bla(NDM-1) in two unrelated Klebsiella pneumoniae isolates in Singapore. PLoS One. 2012;7(11):e48737. doi: 10.1371/journal.pone.0048737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorrie CL, Mirceta M, Wick RR, Judd LM, Wyres KL, Thomson NR, et al. Antimicrobial-Resistant Klebsiella pneumoniae Carriage and Infection in Specialized Geriatric Care Wards Linked to Acquisition in the Referring Hospital. Clin Infect Dis Off Publ Infect Dis Soc Am. 2018;67(2):161–70. doi: 10.1093/cid/ciy027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindsey RL, Batra D, Rowe L, Loparev VN, Stripling D, Garcia-Toledo L, et al. High-Quality Genome Sequence of an Escherichia coli O157 Strain Carrying an mcr-1 Resistance Gene Isolated from a Patient in the United States. Genome Announc. 2017;5(11). doi: 10.1128/genomeA.01725-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netikul T, Sidjabat HE, Paterson DL, Kamolvit W, Tantisiriwat W, Steen JA, et al. Characterization of an IncN2-type blaNDM-₁-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J Antimicrob Chemother. 2014;69(11):3161–3. doi: 10.1093/jac/dku275 [DOI] [PubMed] [Google Scholar]
- 42.Poirel L, Bonnin RA, Nordmann P. Analysis of the Resistome of a Multidrug-Resistant NDM-1-Producing Escherichia coli Strain by High-Throughput Genome Sequencing. Antimicrob Agents Chemother. 2011;55(9):4224–9. doi: 10.1128/AAC.00165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, et al. Genomic Analysis of Hospital Plumbing Reveals Diverse Reservoir of Bacterial Plasmids Conferring Carbapenem Resistance. MBio. 2018;9(1). doi: 10.1128/mBio.02011-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kingston AW, Ponkratz C, Raleigh EA. Rpn (YhgA-Like) Proteins of Escherichia coli K-12 and Their Contribution to RecA-Independent Horizontal Transfer. J Bacteriol. 2017;199(7). doi: 10.1128/JB.00787-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Millan A, Garcia-Cobos S, Escudero JA, Hidalgo L, Gutierrez B, Carrilero L, et al. Haemophilus influenzae Clinical Isolates with Plasmid pB1000 Bearing blaROB-1: Fitness Cost and Interspecies Dissemination. Antimicrob Agents Chemother. 2010;54(4):1506–11. doi: 10.1128/AAC.01489-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran T, Andres P, Petroni A, Soler-Bistué A, Albornoz E, Zorreguieta A, et al. Small plasmids harboring qnrB19: a model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob Agents Chemother. 2012;56(4):1821–7. doi: 10.1128/AAC.06036-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zakharova MV, Beletskaya IV, Denjmukhametov MM, Yurkova TV, Semenova LM, Shlyapnikov MG, et al. Characterization of pECL18 and pKPN2: a proposed pathway for the evolution of two plasmids that carry identical genes for a Type II restriction-modification system. Mol Genet Genomics MGG. 2002;267(2):171–8. doi: 10.1007/s00438-002-0644-y [DOI] [PubMed] [Google Scholar]
- 48.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17(10):589–96. doi: 10.1016/s0168-9525(01)02447-7 [DOI] [PubMed] [Google Scholar]
- 49.Cascales E, Buchanan SK, Duché D, Colin Kleanthous C, Lloubès R, Postle K, et al. Colicin biology. Microbiol Mol Biol Rev MMBR. 2007;71(1):158–229. doi: 10.1128/MMBR.00036-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandegren L, Andersson DI. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7(8):578–88. doi: 10.1038/nrmicro2174 [DOI] [PubMed] [Google Scholar]
- 51.Bush K. Past and Present Perspectives on β-Lactamases. Antimicrob Agents Chemother. 2018;62(10):e01076–18, /aac/62/10/e01076-18.atom. doi: 10.1128/AAC.01076-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramirez MS, Tolmasky ME. Amikacin: Uses, Resistance, and Prospects for Inhibition. Mol J Synth Chem Nat Prod Chem. 2017;22(12). doi: 10.3390/molecules22122267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44(7):1771–7. doi: 10.1128/AAC.44.7.1771-1777.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–27. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 55.Barry KE, Wailan AM, Sheppard AE, Crook D, Vegesana K, Stoesser N, et al. Don’t overlook the little guy: An evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid. 2019;103:1–8. doi: 10.1016/j.plasmid.2019.03.005 [DOI] [PubMed] [Google Scholar]
- 56.Cerdeira LT, Lam MMC, Wyres KL, Wick RR, Judd LM, Lopes R, et al. Small IncQ1 and Col-Like Plasmids Harboring blaKPC-2 and Non-Tn4401 Elements (NTEKPC-IId) in High-Risk Lineages of Klebsiella pneumoniae CG258. Antimicrob Agents Chemother. 2019;63(3). doi: 10.1128/AAC.02140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garbari L, Busetti M, Dolzani L, Petix V, Knezevich A, Bressan R, et al. pKBuS13, a KPC-2-encoding plasmid from Klebsiella pneumoniae sequence type 833, carrying Tn4401b inserted into an Xer site-specific recombination locus. Antimicrob Agents Chemother. 2015;59(9):5226–31. doi: 10.1128/AAC.04543-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendrickx APA, Landman F, de Haan A, Dyogo Borst D, Sandra Witteveen S, van Santen-Verheuvel MG, et al. Plasmid diversity among genetically related Klebsiella pneumoniae blaKPC-2 and blaKPC-3 isolates collected in the Dutch national surveillance. Sci Rep. 2020;10(1):16778. doi: 10.1038/s41598-020-73440-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papagiannitsis CC, Dolejska M, Izdebski R, Dobiasova H, Studentova V, Esteves FJ, et al. Characterization of pKP-M1144, a Novel ColE1-Like Plasmid Encoding IMP-8, GES-5, and BEL-1 β-Lactamases, from a Klebsiella pneumoniae Sequence Type 252 Isolate. Antimicrob Agents Chemother. 2015;59(8):5065–8. doi: 10.1128/AAC.00937-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodrigues C, Bavlovič J, Machado E, Amorim J, Peixe L, Novais Â. KPC-3-Producing Klebsiella pneumoniae in Portugal Linked to Previously Circulating Non-CG258 Lineages and Uncommon Genetic Platforms (Tn4401d-IncFIA and Tn4401d-IncN). Front Microbiol. 2016;7. doi: 10.3389/fmicb.2016.01000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker KS, Burnett E, McGregor H, Deheer-Graham A, Boinett C, Langridge GC, et al. The Murray collection of pre-antibiotic era Enterobacteriacae: a unique research resource. Genome Med. 2015;7(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datta N, Hughes VM. Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature. 1983;306(5943):616–7. doi: 10.1038/306616a0 [DOI] [PubMed] [Google Scholar]
- 63.Bernabe-Balas C. Identificación y Caracterización Molecular de Mecanismos de Adaptación Plasmídica a Nuevas Familias Bacterianas (Doctoral Dissertation). Universidad Complutense de Madrid; 2019
- 64.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcillán-Barcia MP, Redondo-Salvo S, Vielva L, de la Cruz F. MOBscan: Automated Annotation of MOB Relaxases. Methods Mol Biol Clifton NJ. 2020;2075:295–308. doi: 10.1007/978-1-4939-9877-7_21 [DOI] [PubMed] [Google Scholar]
- 66.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinforma Oxf Engl. 2014;30(14):2068–9. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 67.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol. 2015;32(1):268–74. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018;35(2):518–22. doi: 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst Biol. 2012;61(6):1061–7. doi: 10.1093/sysbio/sys062 [DOI] [PubMed] [Google Scholar]
- 73.Andrews S. FastQC: A quality control analysis tool for high throughput sequencing data. Published 2019. https://github.com/s-andrews/FastQC
- 74.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. Published online July 27, 2016:btw493. doi: 10.1093/bioinformatics/btw493 [DOI] [PubMed] [Google Scholar]
- 76.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinforma Oxf Engl 2011;27(7):1009–10. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson O, Dylus D, Dessimoz C. Phylo.io: Interactive Viewing and Comparison of Large Phylogenetic Trees on the Web. Mol Biol Evol. 2016;33(8):2163–6. doi: 10.1093/molbev/msw080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(TXT)
Collection of plasmids already described in the literature and used for building the HMM profiles.
(XLSX)
Collection of 1,035 ColE1 plasmids identified in RefSeq with both the HMM profiles and PlasmidFinder.
(XLSX)
Collection of plasmids identified in Aeromonadales encoding a ColE1 ori and/or repB.
(XLSX)
Annotation of the complete canonical ColE1 plasmids (<25kb) from the 1,035 ColE1 collection.
(XLSX)
ColE1 replicons identified within the ENA project PRJEB44283.
(XLSX)
Annotation of the ColE1 plasmids identified within the ENA project PRJEB44283.
(XLSX)
Phylogenetic tree of the 12 ColE1 origins of replication identified in Aeromonadales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, K2P+G4.
(TIFF)
Phylogenetic tree of the 20 ColE1 origins of replication identified in Vibrionales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, TIM3e+G4.
(TIFF)
Phylogenetic tree of the 38 ColE1 origins of replication identified in Pasteurellales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase, either MOBV or MOBP. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, HKY+F+G4.
(TIFF)
Phylogenetic tree of the 964 ColE1 origins of replication identified in Enterobacterales. The colors of the first column represent the Genus in which the replicon was identified, the second column represents the Species and the third column indicates the presence or absence of a relaxase, either MOBP, MOBQ or truncated (ΔMOB). The legend is at the right of the figure. The bootstrap values are indicated with the nodes and branch colors. Bootstraps under 50 are represented in black, whereas bootstraps over 50 follow the legend at the right. The phylogenetic tree was inferred following the best-fit model, SYM+R7.
(TIFF)
The colors of the column at the right of the tree represent if the plasmid is ColE1-only or ColE1/RepB. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, TVMe+R3.
(TIFF)
At the left, it is represented the phylogenetic tree of the ColE1 ori from the ColE1/RepB plasmids, as indicated in Fig 3. At the end of every branch it is indicated the host Species (colored square, legend at the bottom), the Accession Number and the plasmid size. At the right of the figure, it is represented the genetic content of the ColE1/RepB plasmids. The ColE1 ori is represented with a purple rectangle whereas repB with a purple arrow. The remaining genes are represented with colored arrows, being the legend at the bottom of the figure.
(TIFF)
Comparison of the phylogenetic trees of the ColE1 origin of replication (left) (best-fit model JTT+G4) and the gene repB (right) (TN+F+I+G4). The color of the branches represents the comparison metric. The legend is shown at the bottom of the figure. A score of 1 denotes the subtree structure of the node is identical to the subtree structure of its best corresponding node. The figure was performed with the phylo.io tool [77].
(TIFF)
Schematic representation of the ColE1 co-integrate identified in V. campbellii (NZ_CP026317.1). The complete plasmid (194 kb) is represented at the top of the figure. The purple square indicates the genetic environment of the ColE1 origin of replication, which is represented at the bottom of the figure. The ColE1 ori and remaining genes are represented with colored squares and arrows, being the legend at the bottom of the figure.
(TIFF)
Schematic representation of the genetic environment of the ColE1 ori in the most-represented clades of ColE1 co-integrates from Enterobacterales (Fig 4). The ColE1 ori and remaining genes are represented with colored squares and arrows, being the legend at the top of the figure. The Accession Number and size of each plasmid is indicated at the middle of the figure.
(TIFF)
Phylogenetic tree of the ColE1 relaxases from Aeromonadales. The colors of the first column represent the Genus and the second column represents the Species. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, JTT+F+G4.
(TIFF)
Phylogenetic tree of the ColE1 relaxases from Pasteurellales. The colors of the first column represent the Genus and the second column represents the Species. The legend is at the right of the figure. The bootstrap values are indicated with a number next to each node. The phylogenetic tree was inferred following the best-fit model, VT+G4.
(TIFF)
Phylogenetic tree of the ColE1 relaxases from Enterobacterales. The colors of the first column represent the Genus, the second column represents the presence or absence of rop and the third one indicates the PlasmidFinder result. The legend is at the right of the figure. The phylogenetic tree was inferred following the best-fit model, JTT+F+I+G4.
(TIFF)
The colors of the column represent the origin of the plasmid, Murray Collection or RefSeq. The legend is at the right of the figure. The phylogenetic tree was inferred following the best-fit model, SYM+R8.
(TIFF)
Schematic representation of the secondary structure of the putative RNA I from eight ColE1 plasmids from Pasteurellales. The name and Accession Number of each plasmid is indicated at the top of each sequence. The color of the nucleotides indicates the base-pair probabilities, from 0 to 1, being the legend next to each sequence. The secondary structure and probabilities were inferred with RNAfold WebServer.
(TIFF)
Schematic representation of the secondary structure of the putative RNA I from different ColE1 plasmids from Pasteurellales described coexisting within the cell. The different plasmid combinations are represented separated in the boxes A, B, C and D, being indicated the Species isolate, the plasmid names and the published reference. The Accession Number of each plasmid is indicated at the top of the sequence. The letters and arrows show the dissimilarities identified among the coexisting plasmids, in black those affecting the loop and in grey those affecting the stem. The color of the nucleotides indicates the base-pair probabilities, from 0 to 1, being the legend at the top right of each box. The secondary structure and probabilities were inferred with RNAfold WebServer.
(TIFF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.