Abstract
A duplex LightCycler PCR assay targeting the mecA gene and a Staphylococcus aureus-specific marker was used to test 165 S. aureus strains and 80 strains of other bacterial species. Within an assay time of 60 min plus 10 min for sample preparation, S. aureus as well as the presence or absence of the mecA gene was correctly identified.
Staphylococcus aureus represents one of the most significant pathogens causing nosocomial and community-acquired infections. Beta-lactam antibiotics are the preferred drugs for serious S. aureus infections (11). Since the introduction of methicillin into clinical use in 1961, the occurrence of methicillin-resistant S. aureus (MRSA) strains has increased steadily, and nosocomial infections have become a serious problem worldwide (1, 11, 14, 19, 21). Therefore, the detection of methicillin resistance has important implications for therapy and management of patients. In the clinical laboratory, S. aureus is identified by growth characteristics and the subsequent detection of catalase and coagulase activities or specific surface constituents. The DNase and thermostable endonuclease tests have been used as confirmatory tests for inconclusive or negative coagulase tests. Conventional susceptibility testing of S. aureus reliably detects resistance to methicillin or oxacillin if agar dilution or agar screening methods are used according to NCCLS standards (9). Methicillin resistance is associated with the production of a penicillin-binding protein, encoded by the mecA gene (3, 5), or, in rare cases, with the hyperproduction of β-lactamase.
Conventional identification methods are time-consuming and may yield false-positive or false-negative results, and misclassifications with automated susceptibility testing systems or commercially available latex agglutination kits have been reported recently (16, 17, 18, 23). The rapid and sensitive PCR-based detection of the mecA gene has evolved as the method of choice for the definitive identification of MRSA and other methicillin-resistant staphylococci (2, 4, 6, 7, 10, 13, 20, 22). Since S. aureus can resemble coagulase-negative staphylococci (CoNS) on visual examination of agar plates and the coagulase status is not always easily established in a timely fashion, the inclusion of a species-specific marker is favorable (8).
Here we describe the development and evaluation of a duplex PCR assay for the simultaneous detection of the mecA gene and a recently described S. aureus-specific genomic fragment (12). In order to meet the requirements of true rapid diagnostics, the novel LightCycler device (Roche Molecular Biochemicals, Mannheim, Germany) was used for ultrarapid thermal cycling and online monitoring of the amount of specific PCR product present in the amplification mixture.
All the strains used in this study were maintained on Columbia blood agar and identified by colony morphology, Gram stain characteristics, catalase reaction, coagulase production, and the results of the API Staph System (bioMerieux Vitek, Inc., Hazelwood, Mo.). Oxacillin susceptibility was determined by the agar screening method with Mueller-Hinton agar containing 2% NaCl and 6 mg of oxacillin per liter for S. aureus or 0.5 mg of oxacillin per liter for CoNS (9).
Template DNA was prepared by a simple and rapid boiling procedure (15). Briefly, portions of individual bacterial colonies were suspended in 200 μl of lysis buffer containing 1% Triton X-100, 0.5% Tween 20, 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA and incubated in a screw-cap reaction tube for 10 min in a boiling water bath. After centrifugation for 2 min at 10,000 × g to sediment the debris, a 2-μl aliquot of the clear supernatant was directly transferred to the PCR apparatus. Alternatively, a High Pure PCR Template Preparation Kit (Roche) was used to obtain pure genomic DNA, which can be stored at −20°C for several months. The efficiencies of the commercial kit and the boiling procedure were found to be comparable for the extraction of amplifiable S. aureus DNA.
Oligonucleotide primers and fluorescence-labeled hybridization probes, designed for amplification and sequence-specific detection of a 408-bp fragment within the mecA gene and a 179-bp fragment within an S. aureus-specific genomic marker (12), were obtained from Tib Molbiol (Berlin, Germany). Nucleotide sequences and positions are listed in Table 1. Amplification mixtures contained 2 μl of 10× LightCycler FastStart DNA Master Hybridization Probes mixture (Roche), 5 mM MgCl2 (total concentration), 1 μM each Mec-F and Mec-R primer oligonucleotide, 0.25 μM each Sa442-F and Sa442-R primer oligonucleotide, 0.2 μM each hybridization probe oligonucleotide, and 2 μl of template DNA in a final volume of 20 μl. Following an initial denaturation at 95°C for 10 min to activate the FastStart Taq DNA polymerase, the 40-cycle amplification profile consisted of heating at 20°C/s to 95°C with a 10-s hold, cooling at 20°C/s to 50°C with a 10-s hold, and heating at 20°C/s to 72°C with a 20-s hold. Fluorescence values of each capillary were measured at 640 and 705 nm (dual-color option).
TABLE 1.
Oligonucleotide primers and LightCycler hybridization probes used in the PCR assay
| Oligonucleotides | Sequencea | Target gene | Nucleotide positions | GenBank accession no. | Source |
|---|---|---|---|---|---|
| Mec-F | CAAGATATGAAGTGGTAAATGGT | mecA | 1471–1493 | X52593 | This study |
| Mec-R | TTTACGACTTGTTGCATACCATC | mecA | 1879–1857 | X52593 | This study |
| Mec-HP-1 | CAGGTTACGGACAAGGTGAAATACTGATT-[FAM] | mecA | 1690–1718 | X52593 | This study |
| Mec-HP-2 | [Red 640]-ACCCAGTACAGATCCTTTCAATCTATAGCG-Ph | mecA | 1720–1739 | X52593 | This study |
| Sa442-F | GTCGGGTACACGATATTCTTCACG | Sa442 (12) | 12–34 | AF033191 | This study |
| Sa442-R | CTCTCGTATGACCAGCTTCGGTAC | Sa442 (12) | 191–168 | AF033191 | This study |
| Sa442-HP-1 | TACTGAAATCTCATTACGTTGCATCGGAA-[FAM] | Sa442 (12) | 95–123 | AF033191 | This study |
| Sa442-HP-2 | [Red 705]-ATTGTGTTCTGTATGTAAAAGCCGTCTTG-Ph | Sa442 (12) | 126–154 | AF033191 | This study |
[FAM], fluorescein; [Red 640], LightCycler-Red 640-N-hydroxysuccinimide ester; [Red 705], LightCycler-Red 705-phosporamidite; Ph, 3′-phosphate.
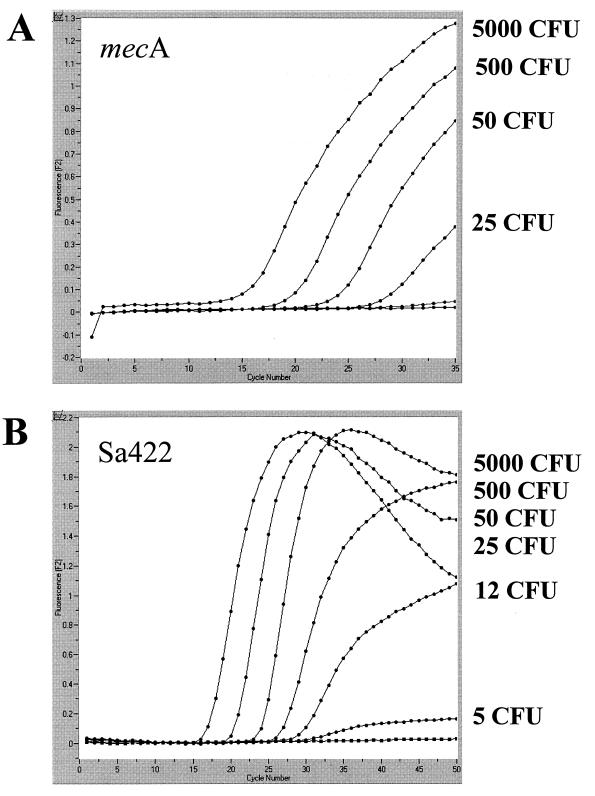
PCR experiments performed with MRSA strain ATCC 33592 (American Type Culture Collection, Manassas, Va.), grown and serially diluted in phosphate-buffered saline as described previously (12), revealed a detection limit of approximately 25 CFU (25 genome equivalents) per assay for the mecA-specific primer pair Mec-F–Mec-R in combination with hybridization probes Mec-HP-1 and Mec-HP-2 (Fig. 1A). For amplification and detection of an S. aureus-specific genomic fragment (12), a detection limit of approximately 5 CFU of S. aureus per assay was determined with primer pair Sa442-F–Sa442-R and hybridization probes Sa442-HP-1 and Sa442-HP-2 (Fig. 1B). The duplex approach, containing four different primer oligonucleotides and four different hybridization probes within a single capillary, revealed identical detection limits (data not shown). Significant formation of primer dimers or secondary structures or other cross-reactions between oligonucleotide components, which frequently interfere with the analytical sensitivity of multiplex PCR approaches, are therefore unlikely in this particular assay.
FIG. 1.
Analytical sensitivity of the PCR assay for the mecA gene (A) and the S. aureus-specific genomic fragment Sa442 (B), determined with serial dilutions of a cultured MRSA strain. Detection limits in terms of numbers of CFU, determined by a standard plating procedure, are given next to the corresponding amplicon curves.
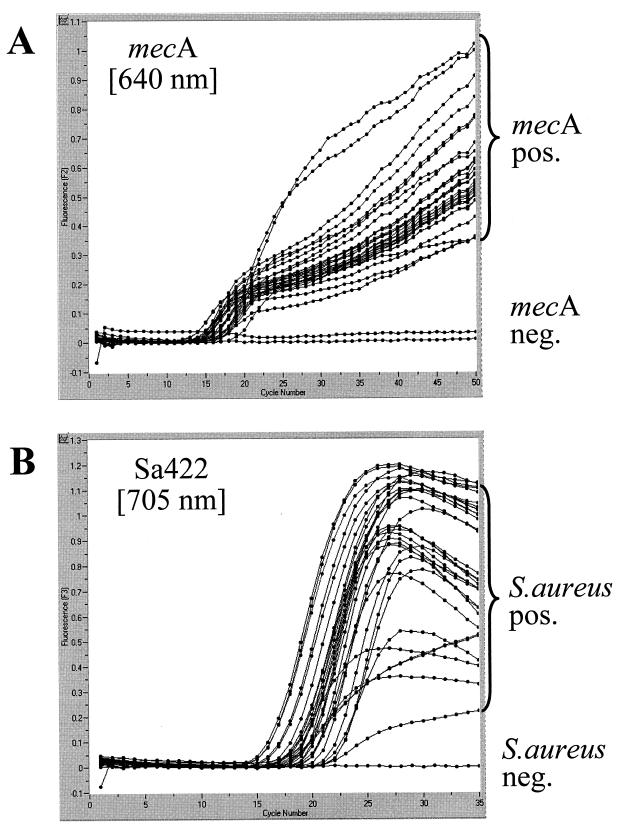
For specificity testing, type strains of methicillin-susceptible S. aureus (MSSA) (ATCC 35696, ATCC 27704, ATCC 23361, ATCC 15752, and ATCC 27695) and MRSA (ATCC 43300, ATCC 33591, ATCC 33592, ATCC 33593, and ATCC 33594) and 155 clinical isolates of S. aureus, including 55 MRSA isolates with different patterns in pulsed-field gel electrophoresis, were examined. Figure 2 shows the results for a representative panel of strains. Visual examination of the plots generated by the LightCycler software (cycle number versus fluorescence) allowed for a clear discrimination between positive and negative samples. All 165 strains were correctly amplified and detected with the S. aureus-specific set of primers and hybridization probes, and all 60 methicillin-resistant strains were identified with the mecA-specific set of primers and hybridization probes. Compared with the results of conventional identification and susceptibility testing, the PCR assay showed 100% specificity and sensitivity for species identification and detection of methicillin resistance for the 165 S. aureus strains investigated.
FIG. 2.
Evaluation of the PCR assay with clinical samples. A representative set of clinical samples was simultaneously tested for the mecA gene (A) and the S. aureus-specific marker (B). Amplicon curves representing mecA-positive (pos.) isolates are indicated by brackets. The mecA-negative (neg.) samples of this panel were two MSSA strains. The S. aureus-negative sample in this panel was a methicillin-resistant strain of S. epidermidis.
Specificity was further confirmed by testing a panel of 40 gram-negative and 40 gram-positive bacterial species, including methicillin-resistant isolates of Staphylococcus epidermidis (n = 3), Staphylococcus hominis (n = 2), and Staphylococcus pasteuri (n = 1) as well as methicillin-susceptible isolates of S. hominis (n = 1), Staphylococcus warneri (n = 1), Staphylococcus capitis (n = 1), and Staphylococcus haemolyticus (n = 1). Among the CoNS, methicillin-resistant isolates were positive for the mecA gene but negative for the S. aureus-specific marker (e.g., the negative curve in Fig. 2B representing a methicillin-resistant S. epidermidis strain). All other bacterial isolates were found negative for both parameters of the duplex PCR assay. These findings were in accordance with the results of Martineau et al. (12), who reported 100% sensitivity and specificity for this particular S. aureus target sequence.
Taq DNA polymerase inhibition was investigated with 64 samples, which were randomly selected from DNA preparations obtained with both extraction methods. Each of the samples was spiked with 300 pg of human DNA. PCR was performed using the components of the LightCycler Control Kit (Roche), allowing for specific detection of the human β-globin gene. Inhibition events were observed with none of the DNA preparations tested.
Showing a detection limit of at least 25 MRSA genome equivalents, this PCR assay should also be sensitive enough for direct detection and identification of MRSA in clinical specimens. However, direct detection of MRSA is restricted to clinical specimens from normally sterile sites where the presence of only a single bacterial species is expected. If a clinical specimen contains, for example, a mixture of MSSA and methicillin-resistant CoNS (even in trace amounts in cases of contamination), PCR performed directly on this particular specimen will be positive for both S. aureus and mecA. Due to this dilemma, our PCR assay was routinely applied to bacterial colonies on agar plates judged to be pure.
Since procedures for sequence-specific detection of amplicons have evolved as the “gold standard” in the field of diagnostic PCR, we concentrated on the LightCycler HybProbe concept to avoid the application of time-consuming and laborious postamplification procedures, such as Southern blotting, solid-phase hybridization, or DNA sequencing. Due to its compact and reliable nature, our duplex PCR assay proved to be a valuable tool for the rapid identification of MRSA isolates in the environment of a routine microbiological laboratory setting. In combination with the simple boiling protocol for template DNA preparation, it can be easily integrated into the work flow of any diagnostic laboratory. Once the growth of staphylococci is observed on agar plates, a portion of the colony can be transferred to PCR. After 10 min of physical manipulation necessary for DNA extraction and completion of the reaction mixture, results are available within 60 min.
Acknowledgments
We thank Markus Heep and Stefan Lukas for active support and Vanessa Bennett and Jeffrey Emch for critically reading the manuscript. We gratefully acknowledge the excellent technical assistance of Michaela Hien and Xiaojun Wu during the study.
REFERENCES
- 1.Barber M. Methicillin-resistant staphylococci. J Clin Pathol. 1961;14:385–393. doi: 10.1136/jcp.14.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bekkaoui F, McNevin J P, Leung C H, Peterson G J, Patel A, Bhatt R S, Bryan R N. Rapid detection of the mecA gene in methicillin resistant staphylococci using a colorimetric cycling probe technology. Diagn Microbiol Infect Dis. 1999;34:83–90. doi: 10.1016/s0732-8893(99)00012-7. [DOI] [PubMed] [Google Scholar]
- 3.Chambers H F. Coagulase-negative staphylococci resistant to beta-lactam antibiotics in vivo produce penicillin-binding protein 2a. Antimicrob Agents Chemother. 1987;31:1919–1924. doi: 10.1128/aac.31.12.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain Z, Stoakes L, Lannigan R, Longo S, Nancekivell B. Evaluation of screening and commercial methods for detection of methicillin resistance in coagulase-negative staphylococci. J Clin Microbiol. 1998;36:273–274. doi: 10.1128/jcm.36.1.273-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain, Z., L. Stoakes, V. Massey, D. Diagre, V. Fitzgerald, S. El Sayed, and R. Lannigan. Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J. Clin. Microbiol. 38:752–754. [DOI] [PMC free article] [PubMed]
- 8.Kearns A M, Seiders P R, Wheeler J, Freeman R, Steward M. Rapid detection of methicillin-resistant staphylococci by multiplex PCR. J Hosp Infect. 1999;43:33–37. doi: 10.1053/jhin.1999.0631. [DOI] [PubMed] [Google Scholar]
- 9.Kohner P, Uhl J, Kolbert C, Persing D, Cockerill F. Comparison of susceptibility testing methods with mecA gene analysis for determining oxacillin (methicillin) resistance in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus spp. J Clin Microbiol. 1999;37:2952–2961. doi: 10.1128/jcm.37.9.2952-2961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolbert C P, Arruda J, Varga-Delmore P, Zheng X, Lewis M, Kolberg J, Persing D H. Branched-DNA assay for detection of the mecA gene in oxacillin-resistant and oxacillin-sensitive staphylococci. J Clin Microbiol. 1998;36:2640–2644. doi: 10.1128/jcm.36.9.2640-2644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 12.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;36:618–623. doi: 10.1128/jcm.36.3.618-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panlilio A L, Culver D H, Gaynes R P, Banerjee S, Henderson T S, Tolson J S, Martone W J. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975–1991. Infect Control Hosp Epidemiol. 1992;13:582–586. doi: 10.1086/646432. [DOI] [PubMed] [Google Scholar]
- 15.Reischl U, Pulz M, Ehret W, Wolf H. PCR-based detection of mycobacteria in sputum samples using a simple and reliable DNA extraction protocol. BioTechniques. 1994;17:844–845. [PubMed] [Google Scholar]
- 16.Ribeiro J, Vieira F D, King T, D'Arezzo J B, Boyce J M. Misclassification of susceptible strains of Staphylococcus aureus as methicillin-resistant Staphylococcus aureus by a rapid automated susceptibility testing system. J Clin Microbiol. 1999;37:1619–1620. doi: 10.1128/jcm.37.5.1619-1620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruane P J, Morgan M A, Citron D M, Mulligan M E. Failure of rapid agglutination methods to detect oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1986;24:490–492. doi: 10.1128/jcm.24.3.490-492.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarzkopf A, Karch H, Schmidt H, Lenz W, Heesemann J. Phenotypic and genotypic characterization of epidemic clumping factor-negative, oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:2281–2285. doi: 10.1128/jcm.31.9.2281-2285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towner K J, Talbot D C, Curran R, Webster C A, Humphreys H. Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1998;47:607–613. doi: 10.1099/00222615-47-7-607. [DOI] [PubMed] [Google Scholar]
- 20.Vannuffel P, Laterre P F, Bouyer M, Gigi J, Vandercam B, Reynaert M, Gala J L. Rapid and specific molecular identification of methicillin-resistant Staphylococcus aureus in endotracheal aspirates from mechanically ventilated patients. J Clin Microbiol. 1998;36:2366–2368. doi: 10.1128/jcm.36.8.2366-2368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss A, Milatovic D, Wallrauch-Schwarz C, Rosdahl V T, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]
- 22.Wallet F, Roussel-Delvallez M, Courcol R J. Choice of a routine method for detecting methicillin-resistance in staphylococci. J Antimicrob Chemother. 1996;37:901–909. doi: 10.1093/jac/37.5.901. [DOI] [PubMed] [Google Scholar]
- 23.Wilkerson M, McAllister S, Miller J M, Heiter B J, Bourbeau P P. Comparison of five agglutination tests for identification of Staphylococcus aureus. J Clin Microbiol. 1997;35:148–151. doi: 10.1128/jcm.35.1.148-151.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]