Summary
Background
Sensitive diagnostics are needed for effective management and surveillance of schistosomiasis so that current transmission interruption goals set by WHO can be achieved. We aimed to screen the Schistosoma haematobium secretome to find antibody biomarkers of schistosome infection, validate their diagnostic performance in samples from endemic populations, and evaluate their utility as point of care immunochromatographic tests (POC-ICTs) to diagnose urogenital schistosomiasis in the field.
Methods
We did a biomarker identification study, in which we constructed a proteome array containing 992 validated and predicted proteins from S haematobium and screened it with serum and urine antibodies from endemic populations in Gabon, Tanzania, and Zimbabwe. Arrayed antigens that were IgG-reactive and a select group of antigens from the worm extracellular vesicle proteome, predicted to be diagnostically informative, were then evaluated by ELISA using the same samples used to probe arrays, and samples from individuals residing in a low-endemicity setting (ie, Pemba and Unguja islands, Zanzibar, Tanzania). The two most sensitive and specific antigens were incorporated into POC-ICTs to assess their ability to diagnose S haematobium infection from serum in a field-deployable format.
Findings
From array probing, in individuals who were infected, 208 antigens were the targets of significantly elevated IgG responses in serum and 45 antigens were the targets of significantly elevated IgG responses in urine. Of the five proteins that were validated by ELISA, Sh-TSP-2 (area under the curve [AUC]serum=0·98 [95% CI 0·95–1·00]; AUCurine=0·96 [0·93–0·99]), and MS3_01370 (AUCserum=0·93 [0·89–0·97]; AUCurine=0·81 [0·72–0·89]) displayed the highest overall diagnostic performance in each biofluid and exceeded that of S haematobium-soluble egg antigen in urine (AUC=0·79 [0·69–0·90]). When incorporated into separate POC-ICTs, Sh-TSP-2 showed absolute specificity and a sensitivity of 75% and MS3_01370 showed absolute specificity and a sensitivity of 89%.
Interpretation
We identified numerous biomarkers of urogenital schistosomiasis that could form the basis of novel antibody diagnostics for this disease. Two of these antigens, Sh-TSP-2 and MS3_01370, could be used as sensitive, specific, and field-deployable diagnostics to support schistosomiasis control and elimination initiatives, with particular focus on post-elimination surveillance.
Funding
Australian Trade and Investment Commission and Merck Global Health Institute.
Introduction
Schistosomiasis, caused by infection with parasitic blood flukes of the genus Schistosoma, has a global burden of 1·86 million disability-adjusted life-years.1 Schistosoma haematobium is the most common cause of urogenital schistosomiasis in humans, responsible for infection in approximately half of the estimated 200 million people with the disease throughout the world's tropical and subtropical regions.2 Moreover, S haematobium infection in women substantially increases the risk of acquiring HIV/AIDS,3 and the International Agency for Research on Cancer recognises urogenital schistosomiasis as a group 1 carcinogen because of its association with squamous cell carcinoma of the bladder.4 The focus of the schistosomiasis intervention agenda is shifting from morbidity control to elimination, and a WHO-mandated objective exists to eliminate the disease as a public health concern and interrupt transmission in selected areas.5 It is therefore imperative that methods to detect infection are appropriately sensitive and rapid to diagnose new cases, assess effectiveness of elimination measures, and be applicable to large-scale surveillance, particularly at the post-elimination stage and in children who have not yet been exposed.
Research in context.
Evidence before this study
We searched PubMed from database inception to Feb 1, 2021, for studies on diagnostic biomarkers for urogenital schistosomiasis, using the search terms “schistosomiasis” AND “haematobium” AND (“antibody” OR “serodiagnosis” OR “immunomics” OR “protein microarray” OR “proteome” OR “extracellular vesicle”), without language restrictions. We identified 309 studies, one of which was directly relevant in terms of diagnostic antibody biomarkers against defined protein antigens of Schistosoma haematobium, one for Schistosoma mansoni and one for Schistosoma japonicum. None of these studies took an unbiased approach to screening the secretome of S haematobium for antibody biomarkers of urogenital schistosomiasis. Infection with schistosomes contributes substantially to the global burden of disease and, in the early 2010s, the 56th World Health Assembly called on all schistosomiasis-endemic countries to strengthen disease surveillance and control measures with the aim of eliminating the disease as a public health threat and interrupting transmission in selected areas by 2025. Although undeniable progress towards these goals has been made in some regions, nearly a decade since these objectives were defined, schistosomiasis still remains highly prevalent. Moreover, it is apparent that available diagnostic tools are not sufficiently sensitive, specific, or field-deployable to ensure reliable disease surveillance, thereby limiting the effectiveness of control, and particularly elimination campaigns.
Added value of this study
To respond to the need for novel diagnostics for schistosomiasis, we searched for biomarkers of infection present in the serum and urine of individuals infected with S haematobium, the most common schistosome infecting humans and the cause of urogenital schistosomiasis. We undertook an integrated high-throughput immunomics-guided screen of the S haematobium secretome. The diagnostic performance of selected biomarkers was validated by ELISA using serum and urine from areas of differing schistosomiasis transmission dynamics and, in some cases, outperformed that of S haematobium-soluble egg antigen, a crude parasite extract widely used for immunodiagnosis of the disease. Finally, and with the aim of developing a novel field-deployable diagnostic for schistosomiasis, two of the most sensitive and specific antigens were incorporated into pilot point of care immunochromatographic tests capable of diagnosing S haematobium infection in serum with a sensitivity rate of at least 79%.
Implications of all the available evidence
We have identified new immunodiagnostics for S haematobium that could be sufficiently sensitive and specific in their diagnostic capacity to augment existing schistosomiasis surveillance and elimination strategies, especially in regions where interruption of transmission has been achieved.
Although there is no gold-standard recommended technique for detection of schistosomiasis,6 a widely used method for diagnosing infection involves microscopy-based detection of parasite eggs in urine (S haematobium) or faeces (Schistosoma mansoni and Schistosoma japonicum), which can have poor sensitivity in areas of low transmission,7 limiting its diagnostic value in regions of low endemicity. Compared with microscopy, microhaematuria detection strips offer an alternative for the diagnosis of S haematobium infection; however, results need to be interpreted with caution as microhaematuria can also result from other medical conditions.6 Tests to detect circulating schistosome antigen in the blood or urine are typically more sensitive than traditional microscopy but are not without limitations. For example, an assay to detect circulating cathodic antigen in urine is available as a point of care test that has excellent capability for diagnosing moderate to high level S mansoni infection,6 but reduced performance in detecting S haematobium infection.8 Assays to detect antibodies to crude parasite preparations in urine, such as soluble egg antigen, correlate with urine egg and serum anodic antigen concentrations;9 however, they can have low specificity and reproducibility due to the antigenic complexity of the extract and crossreactivy with other helminthiases. A handful of immunodiagnostics based on recombinant antigens have been developed for schistosomes, as reviewed by Hinz and colleagues.10 However, to our knowledge, diagnostic performance has only been assessed using serum from individuals who are infected.
The upsurge in availability of so-called omics information for the schistosomes over the past decade has set the stage for the development of new antigen discovery technology platforms, notably proteome microarrays, which use a cell-free expression system to produce hundreds or thousands of recombinant proteins that are printed on nitrocellulose arrays coated on glass slides. We and others have previously used array and alternate technologies to study the humoral response to S mansoni11, 12, 13 and S japonicum,14 probing the arrays with serum samples from infected hosts to aid in genome-wide vaccine antigen and serological marker discovery. Our previous efforts focused on proteins secreted by and anchored on the surface tegument of the parasites; since 2015, helminths (including schistosomes) have been shown to secrete extracellular vesicles as a method of host–parasite communication, and their proteomes are a rich source of vaccine and diagnostic candidates, as reviewed by Sotillo and colleagues.15 Of particular relevance to this study is the integral membrane tetraspanins, which are abundant on the extracellular vesicle surface and known to be immunogenic in other helminth infections.
In this study, we aimed to use the S haematobium surface and soluble and vesicular secreted proteomes16, 17 to create the first S haematobium protein microarray, which, containing 992 proteins is the largest created for a multicellular pathogen to our knowledge. In an integrated approach to identify diagnostic markers for urogenital schistosomiasis, we probed the proteome array with the serum and urine of individuals from two geographically distant S haematobium-endemic regions to establish targets of IgG responses, and selected a subset of surface antigens from the extracellular vesicle proteome. Candidates from both selection methods have been validated by ELISA using both diagnostic fluids and, in the early phase transition of our research from bench to field, we documented results from pilot testing of the first point of care immunochromatographic test (POC-ICT) based on two of these antigens.
Methods
Study design and cohorts
We did a biomarker identification study. All samples used in this study were acquired from previous studies and had been stratified on the basis of egg burden as established by microscopy analysis of urine samples (high, ≥50 eggs per 10 mL urine; or light, 1–49 eggs per 10 mL urine). Egg-negative samples were further tested for the presence of circulating anodic antigen (CAA) using the up-converting phosphor lateral flow CAA assay9 and were classified as being egg-negative and CAA-positive or egg-negative and CAA-negative. Protein arrays were probed with serum and urine samples from S haematobium-endemic regions of Gabon and Zimbabwe. ELISAs were done with the same cohorts of serum samples from Gabon and Zimbabwe and the same cohort of urine samples from Zimbabwe only (the Gabon urine cohort had been exhausted from array probing). Additional ELISA validation was done with urine samples from an elimination setting: Pemba and Unguja islands, Zanzibar, Tanzania. Species specificity analysis was done with S japonicum-infected samples from the Philippines, and S mansoni-infected samples from Ethiopia. ICT evaluation was done with serum samples from the Gabon cohort (appendix p 3). A schematic of the study design is shown in figure 1 and a detailed method is described in the appendix (p 8). All samples came from studies with ethical approval. Ethical approval details for both the published and unpublished studies are shown in the appendix (p 8). Written informed consent was required and obtained in the original studies.
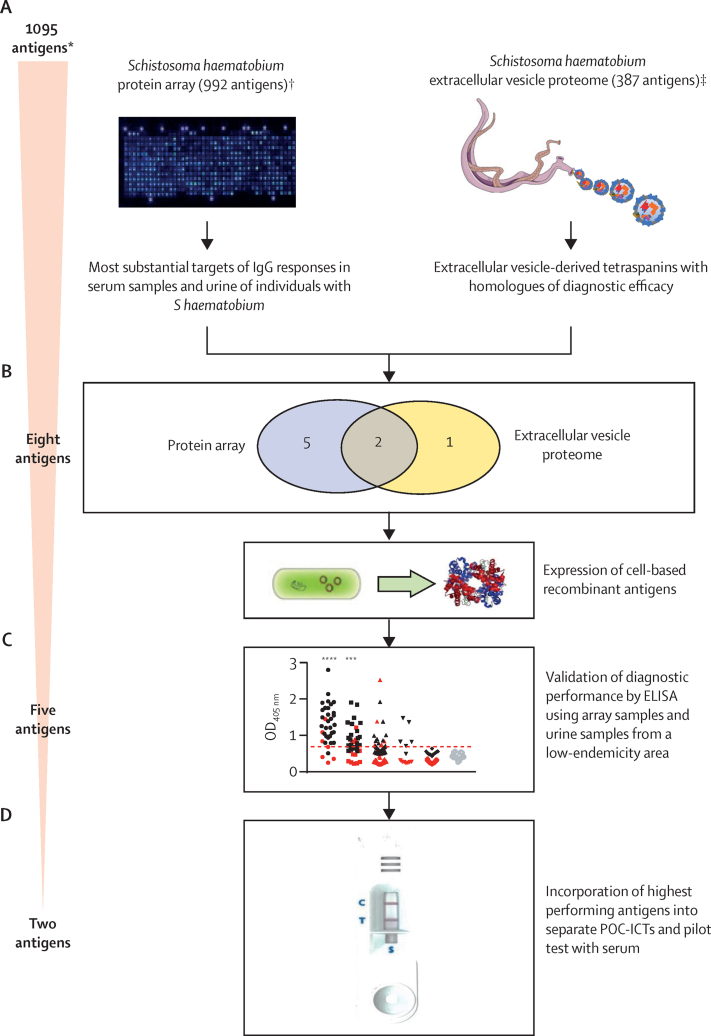
Figure 1.
Schematic overview of the study
We used an integrated approach to identify biomarkers of S haematobium infection. (A) Diagnostic antigens were selected using an immunomics approach to select antigens that were targets of significantly elevated infection-associated IgG responses. (B) Eight antigens were selected for further assessment. (C) Diagnostic performance of five recombinant antigens expressed in Escherichia coli was validated by ELISA. (D) The two highest performing antigens were each incorporated into a separate POC-ICT and their diagnostic performance further assessed. POC-ICT=point of care immunochromatographic test. OD=optical density. *Redundancy between the two antigen groups means that the starting number of antigens does not equal the sum total of the two individual datasets. †Feature selection for the S haematobium protein array was informed by S haematobium proteomic16, 17 and Schistosoma mansoni bioinformatic12 datasets. ‡S haematobium extracellular vesicle proteomic data described by Mekonnen and colleagues.16
Procedures
For protein array construction, probing, and analysis, proteins present in the adult S haematobium tegument, soluble excretory and secretory products and extracellular vesicles, and excretory and secretory products from the egg stage,16, 17 S haematobium orthologues of the S mansoni schistosomula tegument proteome,18 and select proteins from the S mansoni proteome array12 were chosen for printing on the S haematobium array (appendix p 15). Array construction was the same as described previously13 except that open reading frames of arrayed proteins were synthesised instead of amplifying every gene from parasite complementary DNA using PCR. Parasite extracts were included on the array as positive controls.
Serum IgG responses to arrayed antigens were established by probing with human serum samples (one part to 50 parts) in array blocking buffer and 10% Escherichia coli lysate) as previously described13 with the exception that an anti-human IgG-Qdot conjugate (Grace Biolabs, Bend, OR, USA; one part to 100 parts in array blocking buffer) was used as the secondary or detection antibody. Urine IgG responses were established by probing arrays in the same way except that human urine samples were first concentrated 15-fold and buffer-exchanged into phosphate-buffered saline before being diluted one part to five parts in array blocking buffer and 10% E coli lysate.
Statistical analysis
For protein array data analysis and bioinformatics, datasets generated from probing arrays with serum and urine were analysed separately. Significant differences between mean antibody responses in the infected versus non-infected groups were determined by Student's t test. Receiver-operating characteristic (ROC) curves and area under the curve (AUC) values were generated for each antibody response using the ROCR R package, version 4.0.2, and the protein targets of those responses were ranked in order of response significance.
A set of antigens producing an antibody signature that could most effectively discriminate between individuals who were infected and non-infected, using either serum or urine as the diagnostic fluid, was identified. Firstly, for each dataset, all antigens that were the target of an antibody response that was significantly higher in the infected than non-infected populations were selected. From these, antigen targets of responses with a frequency of positivity (reactivity) in less than 30% of the infected population and more than 30% of the non-infected population were also excluded. Antigens in these trimmed datasets were sorted by greatest to least fold change in mean signal intensity between the infected and non-infected populations and frequency of reactivity in the infected population. The most effective antigens in each dataset were used to build a support vector machine classifier, the performance of which was evaluated by Monte Carlo cross-validation.19
For selection of extracellular vesicle-derived tetraspanins, tetraspanins present in the S haematobium extracellular vesicle proteome were sorted by abundance16 (peptide spectrum counting), and the most abundant tetraspanins with homologues of diagnostic efficacy reported in the literature (n=3) were selected for further assessment.
Eight antigens, selected from the immune signature and extracellular vesicle proteomic set (MS3_10385, MS3_10186, MS3_06193, MS3_01466, MS3_05950, MS3_09198, MS3_01370, and Sh-TSP-2), were expressed in E coli as previously described.20 Expression yields of MS3_06193, MS3_01466, and MS3_05950 were at concentrations too low to warrant further development.
IgG responses to E coli-expressed and purified recombinant proteins in each biofluid were measured by ELISA. Plates (Greiner, Kremsmunster, Austria) were coated with antigen, blocked and probed with serum (one part to 50 parts) followed by goat anti-human IgG-horseradish peroxidase (Sigma, St Louis, MO, USA, one part to 5000 parts), and developed with 3,3',5,5'-tetramethylbenzidine. Urine IgG responses to each antigen were measured in a similar way except that urine samples were diluted one part to ten parts with phosphate-buffered saline. Urine IgG responses to multiple antigens were done in the same way and plates were coated with antigen diluted to 2 μg/mL. Species specificity analysis was done as for serum ELISAs. Assays were done in triplicate and blank-corrected values were plotted using Graphpad Prism, version 7. Reactivity cutoffs were determined as the mean plus three SDs of the non-endemic negative group. ROC curves were generated using Graphpad Prism, version 7, and significant differences between mean antibody responses of each infected group and the non-infected group were determined by Student's t test.
For POC test development, a lateral flow ICT was designed (Serve Science, Bangkok, Thailand; appendix p 94). The conjugate pad was coated with 10 optical density (OD) of gold-conjugated mouse anti-human IgG, 1·0 mg/mL of either recombinant MS3_01370, or Sh-TSP-2 (the antigens with the greatest diagnostic capacity as established by ELISA) was sprayed at the test line, and 1·0 mg/mL anti-mouse IgG was sprayed at the control line. Serum (5 μL, diluted one part to ten parts in buffer BS-007) was applied to the sample reservoir, three drops of buffer BS-007 was applied to the sample reservoir, and the test was read after 15 min. For each strip, bands at the test and control lines indicated a positive result, a band at the control line only indicated a negative result, and the test was invalid if there was no band at the control line. Band intensity on positive tests was scored on a four-point scale from most (+4) to least (+1) intense. A score of 0 was given for a negative result. Test results were confirmed by two independent and masked examiners (AL and BAT).
Role of the funding source
The funders of the study had no role in data collection, data analysis, data interpretation, or writing of the report.
Results
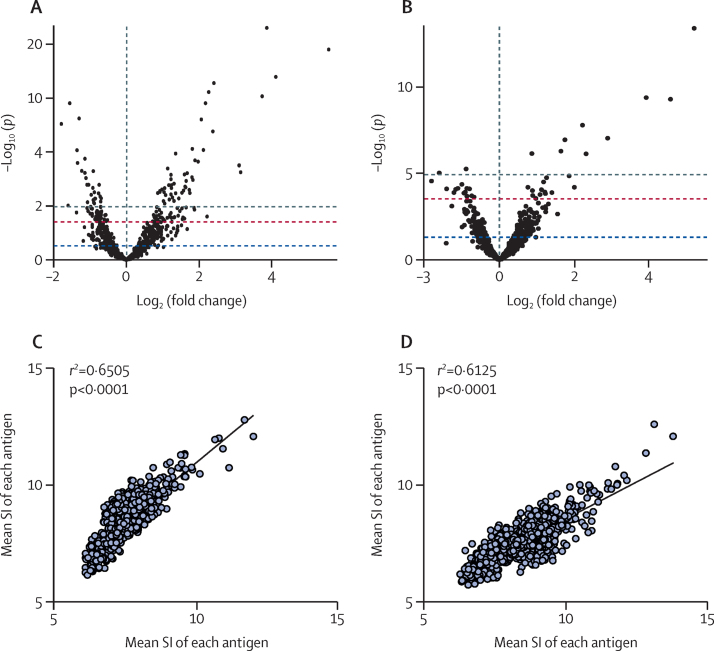
Serum and urine IgG responses to numerous arrayed antigens were significantly elevated in the infected population (ie, samples from all infected cohorts in Gabon and Zimbabwe) when compared with the non-infected population (ie, the non-infected urine samples came from Australia [non-endemic area] and the non-infected serum samples come from the Netherlands [non-endemic area]). 208 antigens were the targets of significantly elevated IgG responses in the serum of individuals who were infected of 243 total serum samples and 45 antigens were the targets of significantly elevated IgG responses in the urine of individuals who were infected of 117 urine samples (figure 2A, B, appendix pp 40, 67). The antigens producing the 20 most substantial responses in each dataset are listed (appendix p 4) with most of these detected in proteomic studies and, of these, at least half in each dataset were identified from the extracellular vesicle portion of the parasite proteome. Seven (35%) of the 20 antigens were shared between the two datasets. A significant correlation was seen between serum and urine IgG responses (r2=0·651; p<0·0001) from all samples used to probe the arrays (serum n=243; urine n=117), supported by a significant correlation between the responses from a subset of matched (n=17) serum and urine samples (r2=0·613; p<0·0001; figure 2C, D). For urine, IgG signal intensity was significantly correlated with infection intensity (r2=0·234; p=0·014) but there was no correlation between infection intensity and serum IgG signal intensity (r2=0·087; p=0·18; appendix p 95).
Figure 2.
Serum and urine IgG responses resulting from probing of Schistosoma haematobium protein arrays
Volcano plot showing fold change and significance of IgG responses between infected (S haematobium-endemic) and non-infected populations with serum (A) or urine (B). Each individual spot denotes a single arrayed antigen. Coloured lines represent different probability thresholds (blue p<0·05; red p<0·01; grey p<0·001). Scatterplot showing correlation of serum and urine IgG responses for all samples (serum n=242; urine n=117; C) and matched serum and urine samples (n=17; D). SI=signal intensity.
An antibody signature was identified that could most effectively discriminate between infected and non-infected populations by using either serum or urine as the diagnostic fluid (appendix p 6). From this set, it was established that a minimum of four antigens were capable of producing an antibody signature with a diagnostic accuracy (AUC) of 0·98 in either diagnostic sample (appendix p 96). Antigens that were the targets of this response included IPSE/alpha-1 (MS3_10186), serpin (MS3_10385), two tetraspanins (MS3_09198 and MS3_01370), and a calcium-binding protein (MS3_05950); three (60%) of the five were identified from the S haematobium extracellular vesicle proteome.
To validate the diagnostic performance of these antigens, five from either the immune signature or extracellular vesicle proteomic set (MS3_10385, MS3_10186, MS3_09198, MS3_01370, and Sh-TSP-2) were assessed by ELISA (figure 3). Urine IgG responses were further assessed using samples from a region of low transmission.21
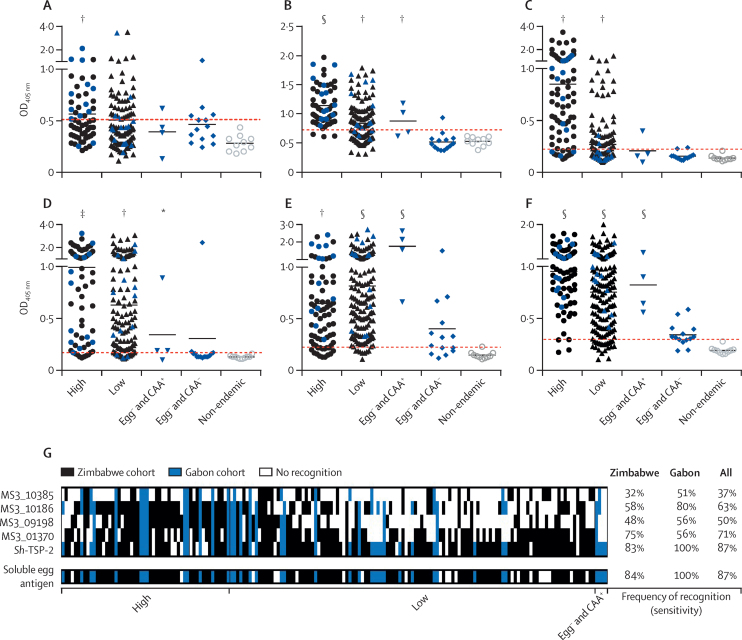
Figure 3.
IgG antibody responses to Escherichia coli-expressed recombinant versions of the most sensitive and specific proteins and Schistosoma haematobium-soluble egg antigen in serum samples from S haematobium-endemic populations
Anti-MS3_10385 (A), anti-MS3_10186 (B), anti-MS3_09198 (C), anti-MS3_01370 (D), anti-Sh-TSP-2 (E), and anti-S haematobium-soluble egg antigen (F). Individuals who were egg-positive were characterised (WHO stratification) as either having a high (≥50 eggs per 10 mL urine) or low (1–49 eggs per 10 mL urine) intensity infection. Egg− and CAA+ indicates egg-negative individuals who were classified as positive (infected) by the more sensitive CAA detection test. Egg− and CAA− indicates egg-negative individuals who were confirmed as antigen-negative by the CAA detection test. Plotted data represent the responses of both the Zimbabwe (black symbols) and Gabon (blue symbols) cohorts. Reactivity cutoffs were determined as the mean plus three SDs of the values of the non-endemic negative group (red dotted line). Significance was analysed by Student's t test. Frequency of recognition patterns were established with serum samples from individuals who were infected resident in S haematobium-endemic populations (G). Samples are sorted from left to right by decreasing egg burden. To facilitate proper comparison, the dataset has been trimmed to exclude any samples not assayed for all five recombinant antigens and S haematobium-soluble egg antigen (n=175). CAA=circulating anodic antigen. OD=optical density. *p≤0·05. †p≤0·01. ‡p≤0·001. §p≤0·0001.
Of the responses to recombinant antigens, the most substantially reactive serum antibody in the egg-negative and CAA-positive group (the cohort with the lowest amount of infection) was to Sh-TSP-2. Of the antibody responses to purified recombinant antigens, those serum antibodies with the greatest ability to discriminate between infected and non-infected populations were against MS3_01370 with an AUC of 0·93 (95% CI 0·89–0·97); and Sh-TSP-2 with an AUC of 0·98 (0·95–1·00; appendix p 7). A frequency of recognition pattern analysis among the infected populations revealed that, consistent with their high AUC values, MS3_01370 and Sh-TSP-2 were the two antigens most frequently recognised and, especially for Sh-TSP-2, this finding was due to greater recognition by individuals with a lower infection intensity (figure 3G). Specificity for all recombinant antigens was 100% in all cohorts tested due to the stringent reactivity cutoff set for all assays. Of the three most sensitive and specific tetraspanins (Sh-TSP-2, MS3_01370, and MS3_09198), there was minimal sequence identity across the exposed large extracellular loop (appendix p 99), suggesting that immunological cross-reactivity is unlikely.
Urine IgG responses to all recombinant antigens in the high infection intensity group were significantly reactive. Additionally, anti-Sh-TSP-2 responses in both low and egg-negative and CAA-positive infection intensity groups were also significantly elevated (figure 4). From the five tested, the antigens with the greatest diagnostic value in urine were Sh-TSP-2 (AUC 0·96 [95% CI 0·93–0·99]) and MS3_01370 (AUC 0·81 [0·72–0·89]), which exceeded that of S haematobium-soluble egg antigen in urine (AUC 0·79 [0·69–0·90]).
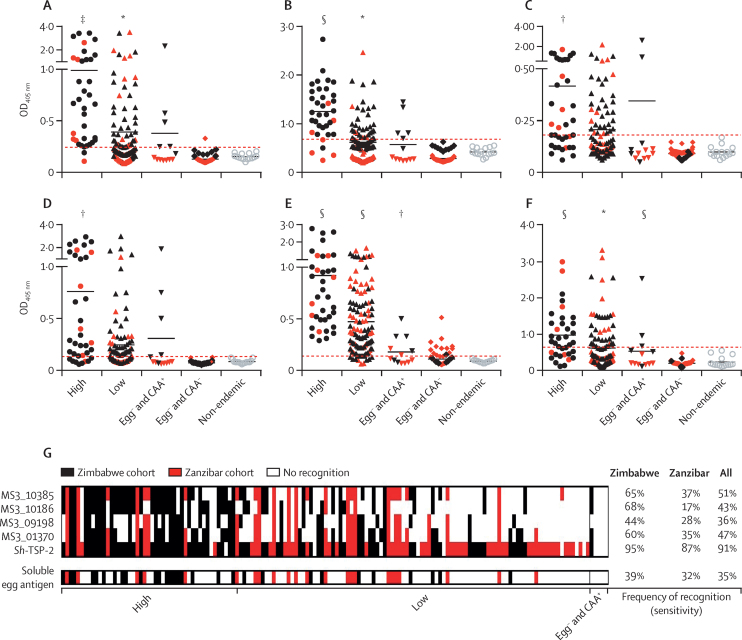
Figure 4.
IgG antibody responses to Escherichia coli-expressed recombinant versions of the most sensitive and specific proteins and Schistosoma haematobium-soluble egg antigen in urine from S haematobium-endemic populations
Anti-MS3_10385 (A), anti-MS3_10186 (B), anti-MS3_09198 (C), anti-MS3_01370 (D), anti-Sh-TSP-2 (E), and anti-S haematobium-soluble egg antigen (F). See figure 3 legend for details. Plotted data represent the responses of both the Zimbabwe (black symbols) and Zanzibar (red symbols) cohorts. Reactivity cutoffs were determined as the mean plus three SDs of the values of the non-endemic negative group (red dotted line). Significance was analysed by Student's t test. Frequency of recognition patterns were established using urine from individuals who were infected resident in S haematobium-endemic populations (G). Samples have been sorted from left to right by decreasing egg burden. To facilitate proper comparison, the dataset has been trimmed to exclude any samples not assayed for all five recombinant antigens and S haematobium-soluble egg antigen (n=148). OD=optical density. *p≤0·05. †p≤0·01. ‡p≤0·001. §p≤0·0001.
AUC values for the responses to all recombinant antigens were high (>0·89) in the Zimbabwe cohort and modest in the Zanzibar cohort (range 0·57–0·69) for all recombinant antigens except Sh-TSP-2 (AUC 0·93 [95% CI 0·87–0·99]; appendix p 7). For urine, the high diagnostic performance of Sh-TSP-2 was reflected in the frequency of recognition analysis, which showed the superior recognition of this antigen in the low infection intensity group (figure 4G). Given the differences in frequency of recognition patterns of the molecules, we tested combinations of the antigens to see if these combinations would elicit higher positivity among the infected population. All antigen combinations resulted in significant responses from all infected cohorts compared with controls (appendix p 97). The use of these combinations did not result in an increase of AUC or frequency of recognition value in the infected population compared with Sh-TSP-2 alone (appendix pp 99–100). As with the serum ELISAs, specificity for all recombinant antigens in all cohorts tested was absolute.
We also sought to establish the recognition of Sh-TSP-2 and MS3_01370 in serum samples from individuals mono-infected with either S mansoni or S japonicum. Both antigens were recognised to a substantially lesser degree by serum antibodies from S japonicum infections and, in the case of Sh-TSP-2, S mansoni infections (appendix p 101). Multiple sequence alignments of the five most sensitive and specific antigens with their closest homologues from human, S japonicum, and S mansoni revealed varying degress of identity ranging from 31–99%. As expected, identity was generally higher between the two African species (S haematobium and S mansoni) compared with the Asian species, S japonicum (appendix p 102).
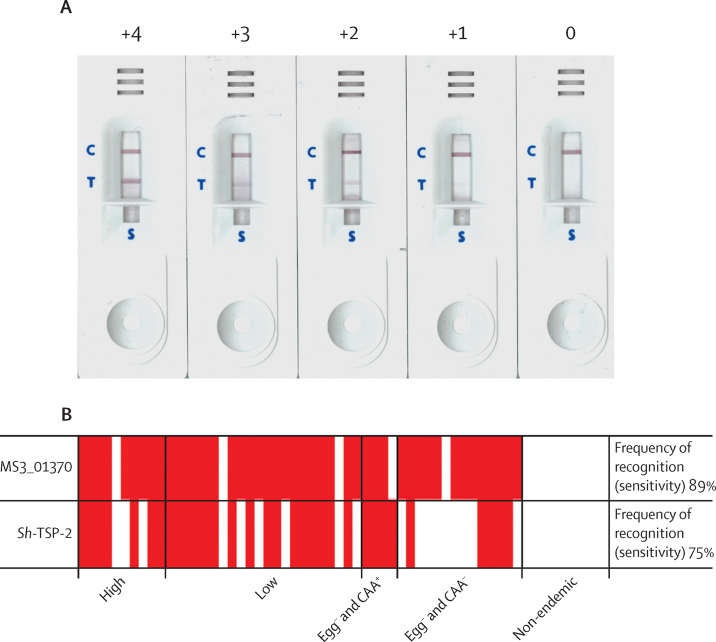
Finally, we assessed whether the diagnostic performance of Sh-TSP-2 and MS3_01370 could be translated into a field-compatible format. ICTs coated with either Sh-TSP-2 or MS3_01370 detected antibodies at every level of infection intensity from the cohort sampled (sensitivity 75% for Sh-TSP-2 and 89% for MS3_01370), even in individuals who were egg-negative by urine filtration and were only positive for CAA. Both sets of ICTs had 100% specificity (figure 5).
Figure 5.
Pilot development of a point of care immunochromatographic test for diagnosis of urogenital schistosomiasis using serum
(A) Representative tests showing positive results of differing band intensities. (B) Schematic showing positive (red boxes) and negative (white boxes) results for MS3_01370 or Sh-TSP-2 immunochromatographic test using ELISA-validated serum samples from the Gabon cohort (high, n=10; low, n=22; egg− and CAA+, n=4; egg− and CAA−, n=14; non-endemic, n=10). Strips were coated with either MS3_01370 or Sh-TSP-2 at the test line to facilitate capture and detection of anti-MS3_01370 or anti-Sh-TSP-2 IgG in serum added to the sample reservoir. Appearance of a band at the test and control lines was considered a positive result and a band at the control line only was considered a negative result. Test bands were given a score from least (+1) to most (+4) intense and a score of 0 was given to a negative result. Two independent readers had to agree on a test result. Every test performed was valid, as confirmed by the appearance of a band at the control line. Samples have been sorted from left to right by decreasing egg burden.
Discussion
Current urogenital schistosomiasis diagnostics are not without disadvantages, and the suite of sensitive and specific determinants of infection available for this purpose is poor. We took an integrated approach to identify novel biomarkers of infection with this carcinogenic parasite by doing a high-throughput screen of the S haematobium predicted immunome with serum and urine antibody responses of individuals who are infected from geographically distinct regions and targeting select tetraspanins from the extracellular vesicle proteome. Proteins from the parasite secretome were selected on the basis of their deployment at the host–parasite interface, making them likely to be targets of an antibody response to infection. The diagnostic performance of a subset of antigens was then validated by ELISA, probing purified recombinant antigens with the same samples used to interrogate arrays as well as additional cohorts of urine from a schistosomiasis elimination setting. The ELISA for the two most sensitive and specific antigens had sensitivities of 97% for Sh-TSP-2 and 93% for MS3_01370, and a signature of the four most sensitive and specific antigens had a sensitivity of 98%. Finally, the cognate antigens of the two highest performing validated biomarkers were incorporated into pilot POC-ICTs to determine their effectiveness as cheap, field-deployable tools for the diagnosis of urogenital schistosomiasis.
Responses to many immunoreactive antigens were shared between the two fluids, probably because of serum leakage into the bladder caused by the transit of parasite eggs.22 Historical accounts of urogenital schistosomiasis describe the urine of patients who are heavily infected as having a deep claret colour23 and the presence of haematuria remains a diagnostic parameter for detection of urogenital schistosomiasis.6
Antigens targeted by IgG responses that were most substantially elevated in the infected population belonged to distinct protein families including a serpin that interacts with the host complement and coagulation systems;24 a saposin domain-containing protein, homologues of which are haemolytic and likely to be involved in blood-feeding;25 the interleukin-4-inducing protein IPSE, which infiltrates host cells to exert its immunomodulatory effects;26 and tetraspanins, which are major membrane constituents of parasite extracellular vesicles and mediate their communication with host cells.15 A common feature of these proteins is their identification in the extracellular vesicle proteome, and there are numerous accounts of schistosome proteins with diagnostic potential that have been found subsequently to derive from extracellular vesicles.27, 28, 29 Extracellular vesicles are constitutively secreted from schistosomes and their relative abundance and stability in biofluids is likely to result in sustained stimulation of the host anti-schistosome extracellular vesicle immune response, making their constituent proteins effective tools for diagnosis.
For each antigen that was validated by ELISA, the AUC values for both biofluids were similar to those generated by array probing, validating our approach to biomarker selection. The highest AUC values and sensitivities (combined Gabon and Zimbabwe cohorts) in serum samples were generated to MS3_01370 and Sh-TSP-2, with the diagnostic performance of Sh-TSP-2 equalling S haematobium-soluble egg antigen. AUC values and sensitivities in urine for all antigens except Sh-TSP-2 were significantly lower in the Zanzibar than the Zimbabwe cohorts, presumably due to the low infection intensity in this elimination setting. Sh-TSP-2 displayed equally high diagnostic performance in both cohorts. The diagnostic performance of all other recombinant proteins was high in the Zanzibar urine cohort, indicating their potential diagnostic utility for high-endemicity populations.
The diagnostic performance of Sh-TSP-2 in the urine samples surveyed exceeded that of S haematobium-soluble egg antigen, likely to be due to the non-specific anti-S haematobium-soluble egg antigen response in non-endemic samples. Non-specific reactivity with samples from non-endemic settings or cross-reactivity with samples from endemic settings in individuals who are poly-parasitised can lead to false-positive results, as documented with soluble egg antigen and other extracts.10 Even defined recombinant antigens of similar diagnostic performance to S haematobium-soluble egg antigen, such as MS3_01370 in urine, have advantages over the crude extract as they can be manufactured to produce a standardised diagnostic reagent. Sh-TSP-2 was the most reactive of all the antigens by ELISA with samples from endemic settings that were both egg-negative and CAA-negative, likely to indicate infections in the past 6–12 months or active but very low intensity infections. Although this finding might indicate slow antibody decay rates post-treatment and make it harder to establish true infection status, we envisage an ultimate POC-ICT test that contains multiple antigens, including those with an accelerated antibody decay rate such as MS3_01370. Empirical testing before and after praziquantel therapy in a controlled environment is warranted as these tests evolve and undergo validation.
To test the diagnostic performance of MS3_01370 and Sh-TSP-2 in a rapid field-deployable format, POC-ICTs were developed for each molecule. Serum was chosen as the diagnostic sample as we reasoned that the increased amount of target antibody in this biofluid would maximise the success of a pilot test, which could then be optimised for use with urine. Both tests could detect infection at all levels of intensity, with the MS3_01370 ICT being diagnostically superior. Nevertheless, we believe that both antigens could be further developed into a POC test, and incorporating both antigens into a single ICT or similar platform could increase sensitivity by capturing a wider anti-schistosome antibody repertoire while still retaining epitope accessibility of both antigens.
In the POC-ICTs, there was antibody reactivity to some of the samples that were both egg-negative and CAA-negative, a result also seen by ELISA. Given that antibodies can persist in the host after the parasite has been eliminated, we posit that the antibody response seen in these individuals is likely to be due to infection in the past 3–12 months. In the absence of corroborating testing, a potential limitation of using antibodies to diagnose infection can be the inability to distinguish between past and current infection; however, the utility of antibody-based diagnosis, particularly in monitoring of transmission in areas where elimination has been achieved and praziquantel treatment effect monitoring is taking place,30 should not be underestimated. Determination of antibody decay rates post-infection could augment diagnostic value in some applications and the monitoring of antibody titres to the antigens in this study in response to praziquantel treatment will be the subject of future investigation.
Given the extensive geographical overlap between S mansoni and S haematobium, a species-specific diagnostic would be sought after. Although the handful of recombinant antigens to diagnose S haematobium infection described so far have shown a favourable absence of cross-reactivity with non-schistosome infections, none of these have displayed species-specific reactivity.10 Of the antigens tested, Sh-TSP-2 was least cross-reactive with S mansoni infections. Sh-TSP-2 probably contains multiple B-cell epitopes, and identification of species-specific epitopes will advance the development of a truly species-specific diagnostic antigen, should it prove desirable. That said, many areas are co-endemic for S mansoni and S haematobium, and treatment and elimination strategies are likely to be the same for both species, so the development of pan-African schistosomiasis diagnostics is of merit.
In summary, we used an integrated immunomics approach to identify numerous novel antibody biomarkers for the diagnosis of urogenital schistosomiasis in serum and urine samples from individuals who were infected. Arrayed antigens that were the target of some of the most diagnostically precise IgG responses, as well as tetraspanins abundantly represented in the extracellular vesicle proteome, were validated by ELISA using both biofluids. Two of the most promising antigens, Sh-TSP-2 and MS3_01370, were each incorporated into a field-deployable POC-ICT to further validate their diagnostic performance using serum. Future studies will address the performace of pilot POC-ICTs using urine. This study represents a truly bench-to-field approach to advance the development of urogenital schistosomiasis diagnostics and, to our knowledge, is the first report documenting the use of recombinant antigens for ICT-based detection of any schistosome infection.
Data sharing
A detailed description of all experimental methodology is available in the appendix (p 8). The full list of S haematobium antigens used in construction of the protein array is available online. Additonal raw data will be made available upon request by contacting the corresponding author.
Declaration of interests
MSP, AL, and JS are inventors on a provisional patent filed by the funding body that captures the data presented in this study. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This study received financial support from Merck Global Health Institute and the Australian Trade and Investment Commission (Australian Tropical Medicine Commercialisation grants programme ATMC50322). Research for the Zanzibar Elimination of Schistosomiasis Transmission project was funded by the University of Georgia Research Foundation, which is funded by the Bill & Melinda Gates Foundation for the Schistosomiasis Consortium for Operational Research and Evaluation projects (prime award number 50816, sub-award number RR374-053/4893206). SK received financial support by sub-award number RR374-053/4893196 and via direct grants from the Gates Foundation (investment identification numbers OPP1191423 and OPP1198086). AL was funded by a National Health and Medical Research Council Senior Principal Research Fellowship (number APP1117504). BAT was funded by a James Cook University Postgraduate Scholarship. GGM was funded by an Australian Institute of Tropical Health and Medicine postgraduate scholarship. Funding was granted to AAA by a European and Developing Countries Clinical Trials Partnership senior fellowship training award (number TA_11_40200). FM was funded by the Thrasher Research Fund (number 12440) and the Wellcome Trust (number 108061/Z/15/Z).
Contributors
MSP, JS, DLD, CP, and AL designed and planned the study. MSP, BAT, GGM, CP, LB, RN, AJ, ASA, SK, DR, SMA, AAA, GvD, PLAMC, TM, FM, SC, PC, and JS collected the data. MSP, BAT, GGM, CP, DLD, MAF, FM, JS, and AL analysed and interpreted the data. AL and BAT verified the data. MSP, JS, and CP prepared the figures. MSP wrote the first draft of the manuscript. JS, DLD, CP, CHH, PLF, ASA, SK, DR, PLAMC, DPM, MEB, SC, TL, and AL reviewed and edited the manuscript. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication. All authors read and approved the final version of the manuscript.
Contributor Information
Mark S Pearson, Email: mark.pearson@jcu.edu.au.
Alex Loukas, Email: alex.loukas@jcu.edu.au.
Supplementary Material
References
- 1.Hay SI, Abajobir AA, Abate KH, et al. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1260–1344. doi: 10.1016/S0140-6736(17)32130-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- 3.Zirimenya L, Mahmud-Ajeigbe F, McQuillan R, Li Y. A systematic review and meta-analysis to assess the association between urogenital schistosomiasis and HIV/AIDS infection. PLoS Negl Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Møller H, Heseltine E, Vainio H. Working group report on schistosomes, liver flukes and Helicobacter pylori. Int J Cancer. 1995;60:587–589. doi: 10.1002/ijc.2910600502. [DOI] [PubMed] [Google Scholar]
- 5.WHO Ending the neglect to attain the Sustainable Development Goals: a roadmap for neglected tropical diseases 2021–2030. https://www.who.int/neglected_diseases/Ending-the-neglect-to-attain-the-SDGs-NTD-Roadmap.pdf?ua=1
- 6.Ochodo EA, Gopalakrishna G, Spek B, et al. Circulating antigen tests and urine reagent strips for diagnosis of active schistosomiasis in endemic areas. Cochrane Database Syst Rev. 2015;3 doi: 10.1002/14651858.CD009579.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engels D, Sinzinkayo E, Gryseels B. Day-to-day egg count fluctuation in Schistosoma mansoni infection and its operational implications. Am J Trop Med Hyg. 1996;54:319–324. doi: 10.4269/ajtmh.1996.54.319. [DOI] [PubMed] [Google Scholar]
- 8.Midzi N, Butterworth AE, Mduluza T, Munyati S, Deelder AM, van Dam GJ. Use of circulating cathodic antigen strips for the diagnosis of urinary schistosomiasis. Trans R Soc Trop Med Hyg. 2009;103:45–51. doi: 10.1016/j.trstmh.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 9.de Dood CJ, Hoekstra PT, Mngara J, et al. Refining diagnosis of Schistosoma haematobium infections: antigen and antibody detection in urine. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.02635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinz R, Schwarz NG, Hahn A, Frickmann H. Serological approaches for the diagnosis of schistosomiasis—a review. Mol Cell Probes. 2017;31:2–21. doi: 10.1016/j.mcp.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Crosnier C, Hokke CH, Protasio AV, et al. Screening of a library of recombinant Schistosoma mansoni proteins with sera from murine and human controlled infections identifies early serological markers. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa329. published online June 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Assis RR, Ludolf F, Nakajima R, et al. A next-generation proteome array for Schistosoma mansoni. Int J Parasitol. 2016;46:411–415. doi: 10.1016/j.ijpara.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Gaze S, Driguez P, Pearson MS, et al. An immunomics approach to schistosome antigen discovery: antibody signatures of naturally resistant and chronically infected individuals from endemic areas. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driguez P, Li Y, Gaze S, et al. Antibody signatures reflect different disease pathologies in patients with schistosomiasis due to Schistosoma japonicum. J Infect Dis. 2016;213:122–130. doi: 10.1093/infdis/jiv356. [DOI] [PubMed] [Google Scholar]
- 15.Sotillo J, Robinson MW, Kimber MJ, et al. The protein and microRNA cargo of extracellular vesicles from parasitic helminths—current status and research priorities. Int J Parasitol. 2020;50:635–645. doi: 10.1016/j.ijpara.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Mekonnen GG, Tedla BA, Pickering D, et al. Schistosoma haematobium extracellular vesicle proteins confer protection in a heterologous model of schistosomiasis. Vaccines. 2020;8:e416. doi: 10.3390/vaccines8030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotillo J, Pearson MS, Becker L, et al. In-depth proteomic characterization of Schistosoma haematobium: towards the development of new tools for elimination. PLoS Negl Trop Dis. 2019;13 doi: 10.1371/journal.pntd.0007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sotillo J, Pearson M, Becker L, Mulvenna J, Loukas A. A quantitative proteomic analysis of the tegumental proteins from Schistosoma mansoni schistosomula reveals novel potential therapeutic targets. Int J Parasitol. 2015;45:505–516. doi: 10.1016/j.ijpara.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Proietti C, Zakrzewski M, Watkins TS, et al. Mining, visualizing and comparing multidimensional biomolecular data using the genomics data miner (GMine) web-server. Sci Rep. 2016;6 doi: 10.1038/srep38178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson MS, Pickering DA, McSorley HJ, et al. Enhanced protective efficacy of a chimeric form of the schistosomiasis vaccine antigen Sm-TSP-2. PLoS Negl Trop Dis. 2012;6 doi: 10.1371/journal.pntd.0001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knopp S, Person B, Ame SM, et al. Evaluation of integrated interventions layered on mass drug administration for urogenital schistosomiasis elimination: a cluster-randomised trial. Lancet Glob Health. 2019;7:e1118–e1129. doi: 10.1016/S2214-109X(19)30189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JH, Christie JD. The pathobiology of Schistosoma haematobium infection in humans. Hum Pathol. 1986;17:333–345. doi: 10.1016/s0046-8177(86)80456-7. [DOI] [PubMed] [Google Scholar]
- 23.Chute HM. Bilharzia haematobia, Distomum haematobium, endemic haematuria. S Afr Med J. 1888;10:85–87. [Google Scholar]
- 24.Blanton RE, Licate LS, Aman RA. Characterization of a native and recombinant Schistosoma haematobium serine protease inhibitor gene product. Mol Biochem Parasitol. 1994;63:1–11. doi: 10.1016/0166-6851(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 25.Don TA, Oksov Y, Lustigman S, Loukas A. Saposin-like proteins from the intestine of the blood-feeding hookworm, Ancylostoma caninum. Parasitology. 2007;134:427–436. doi: 10.1017/S003118200600148X. [DOI] [PubMed] [Google Scholar]
- 26.Pennington LF, Alouffi A, Mbanefo EC, et al. H-IPSE is a pathogen-secreted host nucleus-infiltrating protein (Infiltrin) expressed exclusively by the Schistosoma haematobium egg stage. Infect Immun. 2017;85:e00301–e00317. doi: 10.1128/IAI.00301-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köster B, Hall MR, Strand M. Schistosoma mansoni: immunoreactivity of human sera with the surface antigen Sm23. Exp Parasitol. 1993;77:282–294. doi: 10.1006/expr.1993.1086. [DOI] [PubMed] [Google Scholar]
- 28.Tanigawa C, Fujii Y, Miura M, et al. Species-specific serological detection for schistosomiasis by serine protease inhibitor (SERPIN) in multiplex assay. PLoS Negl Trop Dis. 2015;9 doi: 10.1371/journal.pntd.0004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Zhang Y, Lin D, et al. Serodiagnosis of Schistosoma japonicum infection: genome-wide identification of a protein marker, and assessment of its diagnostic validity in a field study in China. Lancet Infect Dis. 2014;14:489–497. doi: 10.1016/S1473-3099(14)70067-2. [DOI] [PubMed] [Google Scholar]
- 30.Rollinson D, Knopp S, Levitz S, et al. Time to set the agenda for schistosomiasis elimination. Acta Trop. 2013;128:423–440. doi: 10.1016/j.actatropica.2012.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A detailed description of all experimental methodology is available in the appendix (p 8). The full list of S haematobium antigens used in construction of the protein array is available online. Additonal raw data will be made available upon request by contacting the corresponding author.