Abstract
Lung cancer, one of the most common causes of cancer-related death worldwide, has been associated with high treatment cost and imposed great burdens. The 5-year postoperative survival rate of lung cancer (13%) is lower than many other leading cancers indicating the urgent needs to dissect its pathogenic mechanisms and discover specific biomarkers. Although several proteins have been proposed to be potential candidates for the diagnosis of lung cancer, they present low accuracy in clinical settings. Metabolomics has thus emerged as a very promising tool for biomarker discovery. To date, many lung cancer-related metabolites have been highlighted in the literature but no database is available for scientists to retrieve this information. Herein, we construct and introduce the first Lung Cancer Metabolome Database (LCMD), a freely available online database depositing 2013 lung cancer-related metabolites identified from 65 mass spectrometry-based lung cancer metabolomics studies. Researchers are able to explore LCMD via two ways. Firstly, by applying various filters in the “Browse Metabolites” mode, users can access a list of lung cancer-related metabolites that satisfy the filter specifications. For each metabolite, users can acquire the value of the fold change (cancer/normal), statistical significance (p-value) of the fold change, and the comparative research designs of all the mass spectrometry-based lung cancer metabolomics studies that identify this metabolite. Secondly, by applying various filters in the “Browse Studies” mode, users can obtain a list of mass spectrometry-based lung cancer metabolomics studies that satisfy the filter specifications. For each study, users can view the type of studied specimen, mass spectrometry (MS) method, MS data processing software, and differential analysis method, as well as all the identified lung cancer-related metabolites. Furthermore, the overview of each study is clearly illustrated by a graphical summary. The LCMD (http://cosbi7.ee.ncku.edu.tw/LCMD/) is the first database that brings together the meaningful information of lung cancer-related metabolites. The development of the LCMD is envisioned to promote the biomarker discovery of lung cancer.
Abbreviations: HMDB, Human Metabolome Database; LCMD, Lung Cancer Metabolome Database; NSCLC, Non-Small-Cell Lung Carcinoma; VIP, Variable Importance in Projection
Keywords: Lung cancer, Biomarker, Database, Mass spectrometry, Metabolite, Metabolome, Metabolomics
1. Introduction
Lung cancer is the leading cause of cancer-related death. It imposes the highest treatment cost and burden among all cancers in the United States, Europe, and many other nations [1]. Although the mortality rate of lung cancer was decreasing in recent years, the survival period of fewer than five years was found in 15% of the patients. The possible reason might be the relatively late diagnosis compared with other cancers [2], [3]. The five-year average survival rate could be increased to 55% if patients were successfully diagnosed at an early and localized stage [4]. Current diagnostic approaches for lung cancer are based on medical history, X-rays, and sputum cytology, which is costly and the patients without apparent clinical symptoms can be missed by detection [5]. The utilization of low-dose chest computed tomography (CT) imaging showed a 20% reduction in lung cancer mortality [6]. Yet enormous challenges remain, such as cost, radiation exposure, and the incidence of high false-positive rates (96%). Other radiologic diagnosis techniques, 18F-fluorodeoxyglucose positron emission tomography/computed tomography (FDG PET/CT) for instance, have been proven with a high capability to detect cancerous lesions in the lung; however, these tests are unsuitable for annual screening of general population due to their high cost [7]. Those limitations initiate the discovery of sensitive/specific complementary biomarkers to be used in conjunction with the existing screening process for more accurate diagnosis [8].
Previous studies focused on the early diagnosis of different types and stages of lung cancer using genomics and proteomics strategies. Some serum proteins such as carcinoembryonic antigens (CEA) [5], cytokeratin-19 fragments (CYFRA 21–1) [9], cancer antigen-125 (CA-125), and neuron-specific enolase (NSE) [10] have been discovered as tumor markers in lung cancer. However, none of them has proceeded to clinical usage due to its low specificity and accuracy. The increasingly high demand of cancer biomarkers necessitates more research endeavor contributing to pathogenic mechanisms and clinically relevant information.
It has been reported that a series of early perturbations in cellular metabolism was involved in the process of tumorigenesis [11]. Aside from genomics and proteomics, metabolomics has opened up new perspectives to provide complementary information regarding cellular metabolic processes that drive tumor formation and progression. Metabolic profiling reveals the current physiological state of an individual in response to disease states and environmental factors [12], [13] which cannot be fully predicted from the knowledge of the human genome, transcriptome and proteome [14]. Moreover, because cancer metabolism is highly associated with oncogenic kinase signalling, it has become a vital concept for delineating malignancies and an important hallmark of carcinoma [11], [15]. The first application of metabolomics approach to lung cancer research can be traced back to a decade ago. The growing number of dedicated studies is expected to open a new frontier and give in-depth insights to lung cancer research [16]. Using the keywords ((lung cancer[Title/Abstract]) OR (lung adenocarcinoma[Title/Abstract])) AND ((metabolome[Title/Abstract]) OR (metabolomics[Title/Abstract])) to search PubMed on Nov. 15, 2021, a total of 292 papers on the lung cancer metabolomics was found in the period of 2006–2021 in which about 72% (211/292) of papers were published in recent 5 years.
Currently, there are several freely accessible metabolome databases (HMDB [17], MetaboLights [18], Metabolomics WorkBench [19], GMD@CSB.DB [20], and PRIMe [21]) that contain the information of metabolites, metabolomics experiments, and the associated metadata. They aim to be comprehensive in scale and is suitable for general inquiry when looking at a designated metabolite in the molecular biology research field. However, notwithstanding the dramatical accumulation of lung cancer metabolomics data within the past few years, there is still no way to access to an organized collection of lung cancer metabolomics data. To better explore relevant information from the growing massive metabolomics data referring to a specific type/condition of lung cancer, we have constructed the first Lung Cancer Metabolome Database (LCMD) which covers 2013 metabolites collected from 65 mass spectrometry-based lung cancer metabolomics studies. We expect that LCMD will not only facilitate our understanding of lung cancer metabolism but also promote the development of novel therapeutic strategies or diagnostic markers.
2. Materials and methods
2.1. Collection of 2013 lung cancer-related metabolites from 65 mass spectrometry-based metabolomics studies
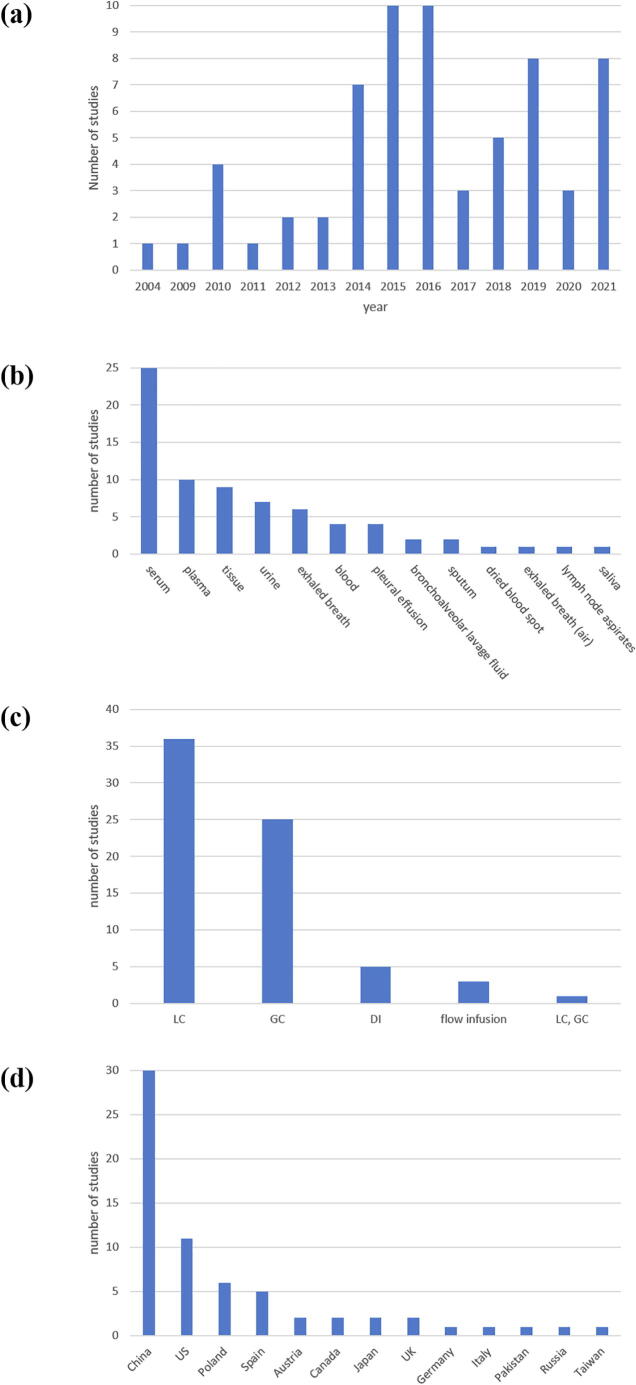
To collect lung cancer-related metabolites from mass spectrometry-based metabolomics studies in the literature, we searched PubMed using the keywords ((lung cancer [Title/Abstract]) OR (lung adenocarcinoma) [Title/Abstract])) AND (mass spec* [Title/Abstract]) AND (metabol* [Title/Abstract]) appeared in the Title/Abstract on Nov. 15, 2021 and found 447 papers. From these 447 papers, we manually checked each paper and kept 65 mass spectrometry-based lung cancer metabolomics studies which aimed to identify metabolite biomarkers for lung cancer in human specimens. For each study (cancer vs. normal), the following information were collected: the type of studied specimen, mass spectrometry (MS) method, MS data processing software, and differential analysis method, as well as all the identified lung cancer-related metabolites. The categorization of these 65 studies based on different characteristics was given in Fig. 1.
Fig. 1.
The categorization of the 65 collected mass spectrometry-based lung cancer metabolomics studies. This figure shows the categorization of the 65 collected studies based on different characteristics: (a) publication year, (b) specimen used, (c) chromatography used, and (d) country.
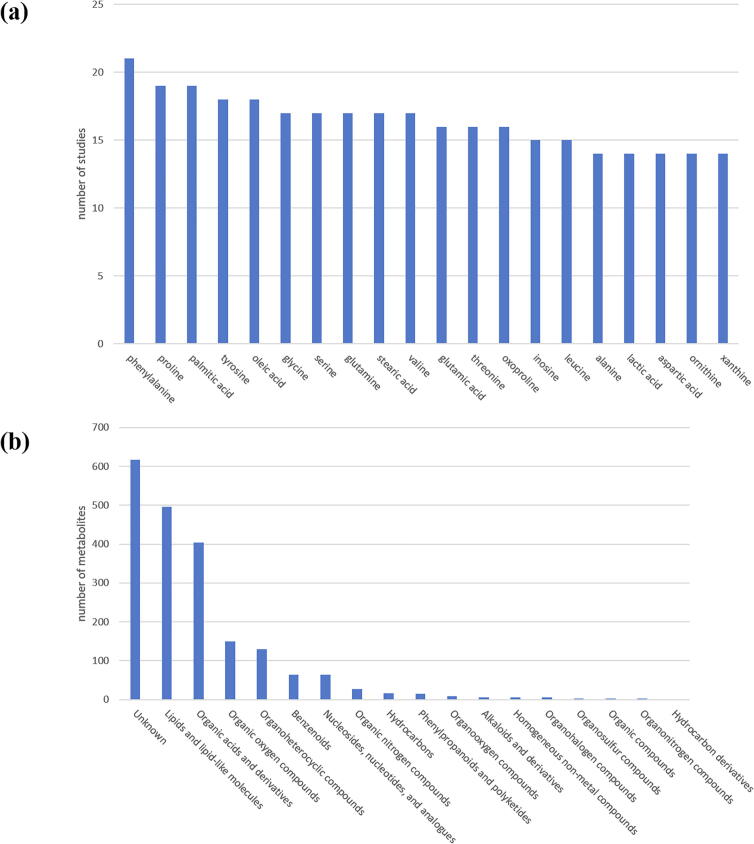
From these 65 mass spectrometry-based lung cancer metabolomics studies, we extracted 2013 lung cancer-related metabolites which were identified in at least one of these 65 studies. For each metabolite, the following information was collected: the value of the fold change (cancer/normal), the statistical significance (p-value) of the fold change, and the comparative research designs of all the studies that identify this metabolite. The categorization of these 2013 metabolites based on different characteristics was given in Fig. 2. For each of the 2013 metabolites in LCMD, we provide the number of studies which identify this metabolite (see the download page of LCMD for details). The higher the number, the higher the confidence of a metabolite as a potential lung cancer biomarker. For example, HMDB0000159 (shown as several different names in different studies: L-phenylalanine, phenylalanine or Phe) has been identified as a lung cancer-related metabolite in 21 mass spectrometry-based metabolomics studies (Fig. 2a). Therefore, HMDB0000159 is the most consistent lung cancer-related metabolite across studies and the most plausible lung cancer biomarker.
Fig. 2.
The categorization of the 2013 collected lung cancer-related metabolites (a) The top 20 most identified lung cancer-related metabolites are shown. (b) The categorization of these 2013 metabolites based on the chemical taxonomy (super class) retrieved from HMDB database is shown.
2.2. Graphical summaries extracted from the 65 mass spectrometry-based lung cancer metabolomics studies
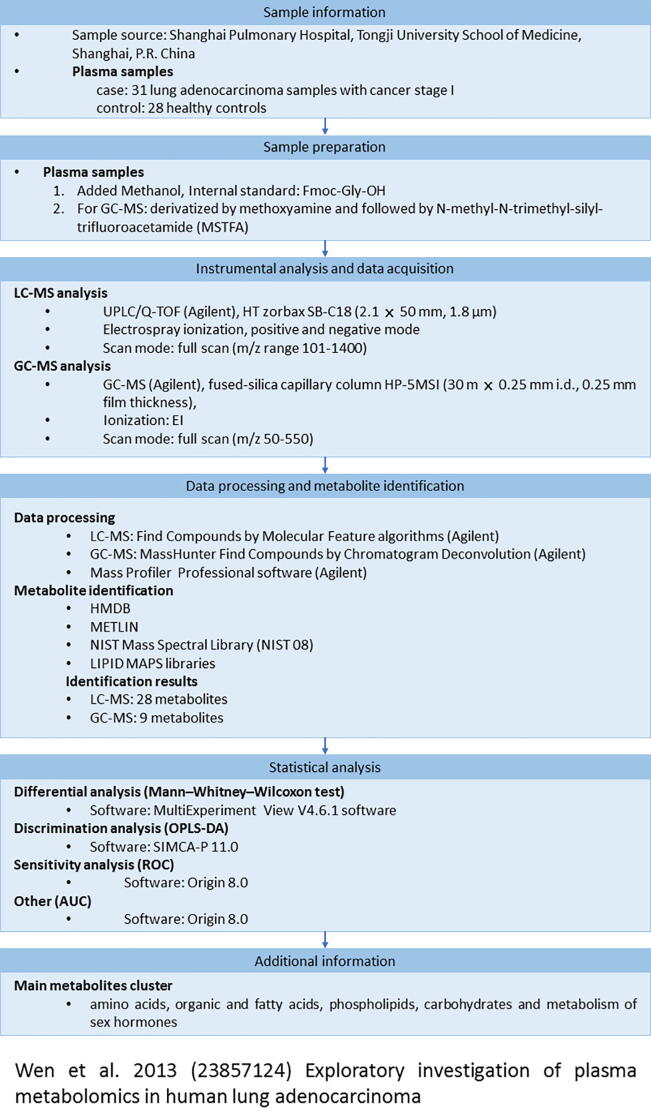
To allow users to quickly gain an understanding of the comparative research designs (cancer vs. normal) of the collected studies, we provide a concise graphical summary for each study (Fig. 3). The graphical summary contains the following information: (i) sample information, (ii) sample preparation, (iii) instrumental analysis and data acquisition, (iv) data processing and metabolite identification, (v) statistical analysis, and (vi) additional information.
Fig. 3.
The graphical summary of a mass spectrometry-based lung cancer metabolomics study. The graphical summary contains the following information: (i) sample information, (ii) sample preparation, (iii) instrumental analysis and data acquisition, (iv) data processing and metabolite identification, (v) statistical analysis, and (vi) additional information.
2.3. Implementation of LCMD website
The web interface of the LCMD was developed in Python using the Django MTV framework. The detailed information of the collected 2013 metabolites and 65 mass spectrometry-based lung cancer metabolomics studies were deposited in MySQL. All tables in the website were produced by the JavaScript and feature-rich JavaScript libraries (jQuery and DataTables). Apart from the main website (http://cosbi7.ee.ncku.edu.tw/LCMD/), one backup site (http://cosbi4.ee.ncku.edu.tw/LCMD/) is also available.
3. Results and discussion
3.1. Database interface
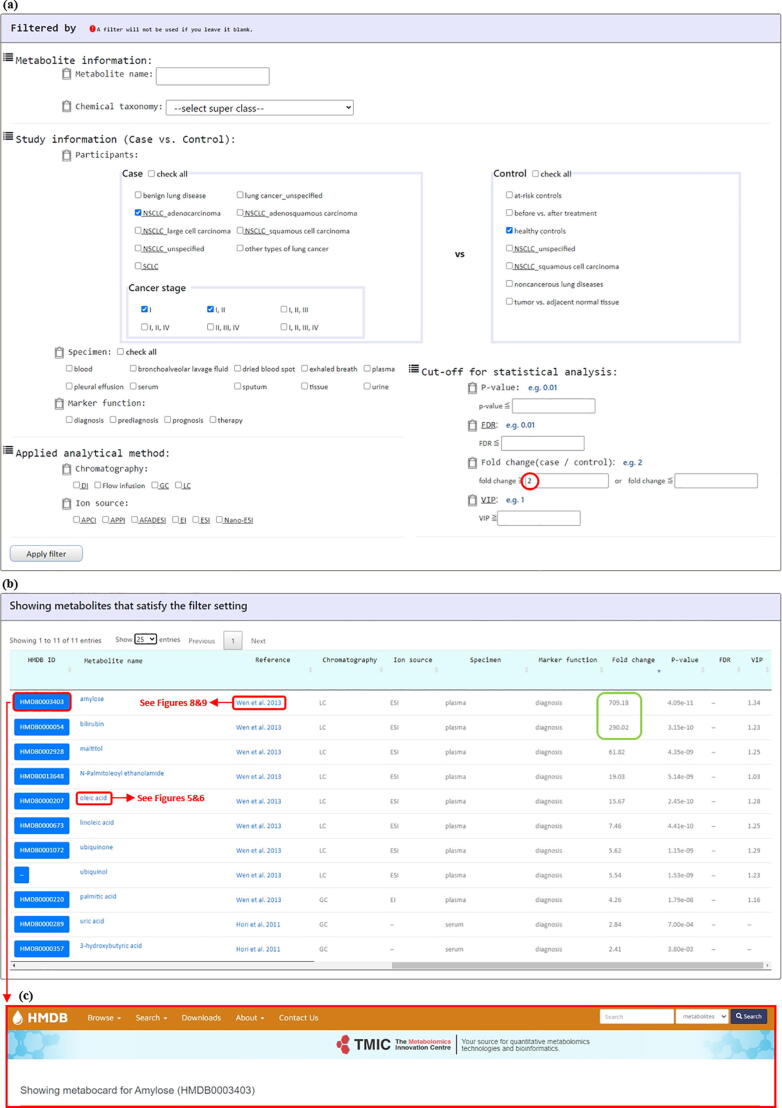
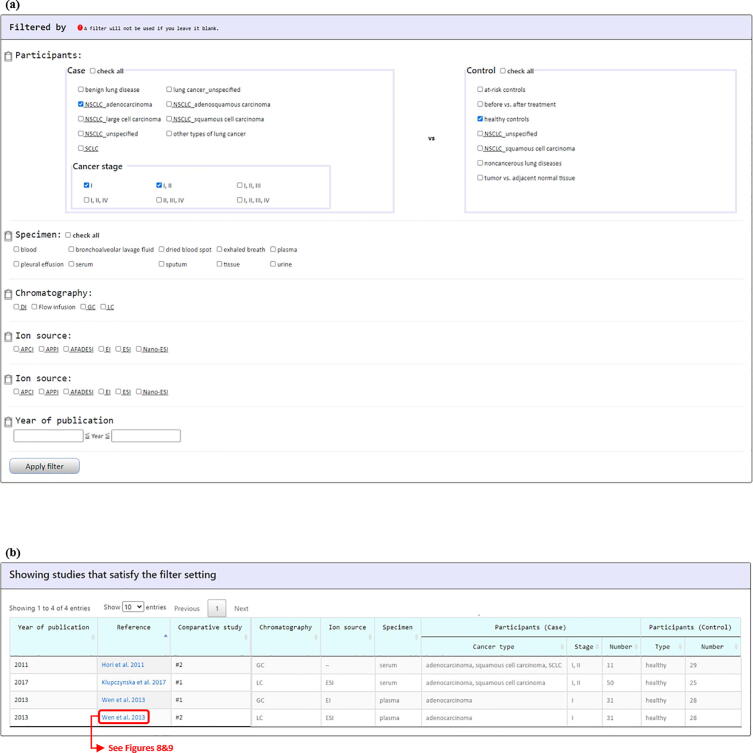
The LCMD provides two browse modes (“Browse Metabolites” and “Browse Studies”). Using the “Browse Metabolites” mode, users can browse metabolites in LCMD by applying 11 kinds of filters (metabolite name, chemical taxonomy, participants, specimen, marker function, chromatography, ion source, p-value, FDR, fold change, and VIP; Fig. 4a). Users then can access to a list of lung cancer-related metabolites that satisfy the filter specifications and receive the summary information of each metabolite (Fig. 4b). By clicking on the “HMDB ID” (e.g. HMDB0003403), users will be directed to the HMDB site (Fig. 4c). By clicking on the “Reference” (e.g. Wen et al. 2013), users will be directed to a page containing the detailed information of the selected mass spectrometry-based lung cancer metabolomics studies. The details of this page will be introduced later.
Fig. 4.
The “Browse Metabolites” mode. (a) Users can browse metabolites in LCMD by applying 11 kinds of filters. (b) Users then can access to a list of lung cancer-related metabolites that satisfy the filter specifications and receive the summary information of each metabolite. (c) By clicking on the “HMDB ID” (e.g. HMDB0003403), users will be directed to the HMDB website.
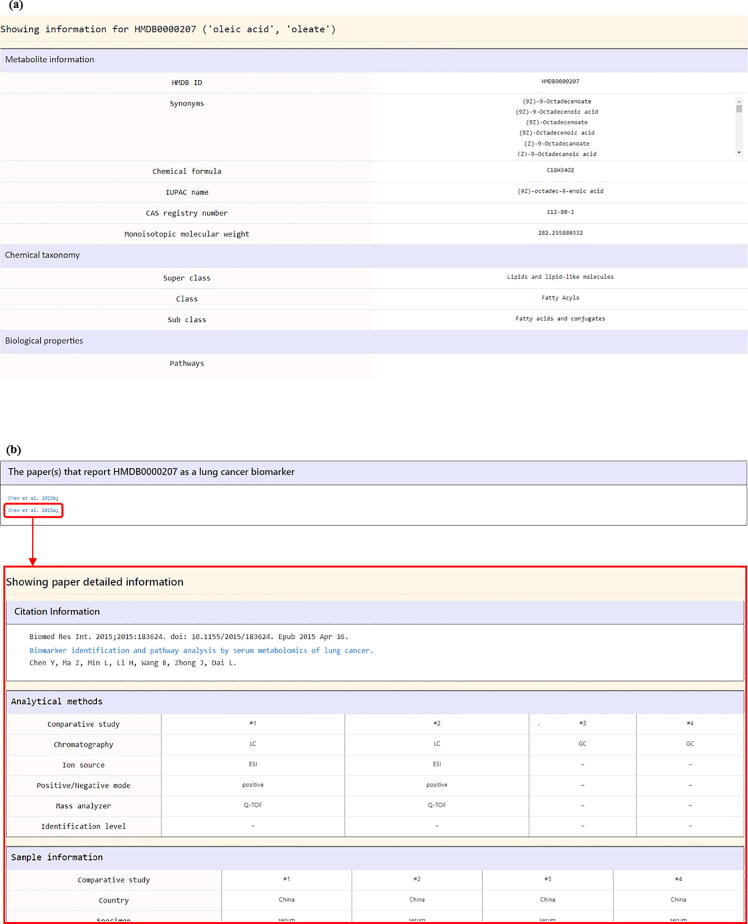
By clicking on the metabolite name (e.g. oleic acid), users will be directed to a page containing the detailed information of this metabolite. This page can be divided into three parts. The first part is the metabolite’s basic information including HMDB ID, synonyms, chemical formula, monoisotopic molecular weight, chemical taxonomy, and pathways (Fig. 5a). The second part refers to the studies that particularly mentioned that this metabolite could serve as a lung cancer biomarker (Fig. 5b). The third part refers to all the mass spectrometry-based lung cancer metabolomics studies that have identified this metabolite. Users can know the detailed (cancer vs. normal) comparative design of each study including (i) sample information (Fig. 6a), (ii) analytical methods (Fig. 6b), (iii) data processing (Fig. 6c), and (iv) statistical analysis (Fig. 6d). It should be noted that the identification of a metabolite does not necessarily mean that this metabolite is a useful biomarker. However, users can still judge whether this metabolite may be a potential biomarker based on the detailed comparative research designs and differential analysis results provided by the LCMD.
Fig. 5.
The metabolite detail page (the first two parts). (a) The first part is the metabolite’s basic information including HMDB ID, synonyms, chemical formula, monoisotopic molecular weight, chemical taxonomy, and pathways. (b) The second part refers to the studies that particularly mentioned that this metabolite could serve as a lung cancer biomarker.
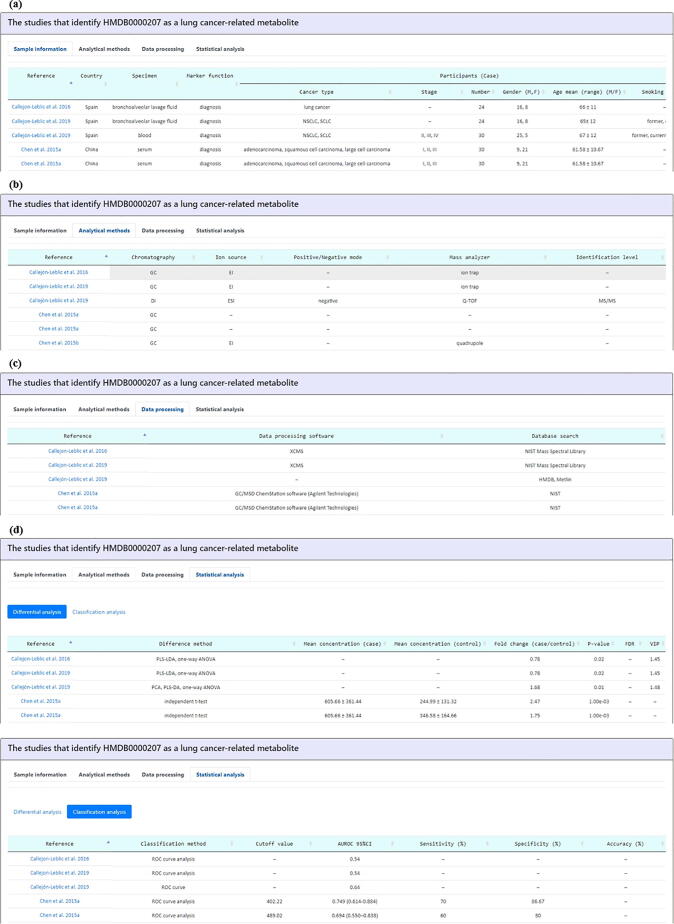
Fig. 6.
The metabolite detail page (the third part). The third part refers to all the mass spectrometry-based lung cancer metabolomics studies that have identified this metabolite. Users can know the detailed (cancer vs. normal) comparative research design of each study including (a) sample information, (b) analytical methods, (c) data processing, and (d) statistical analysis (including differential analysis and classification analysis).
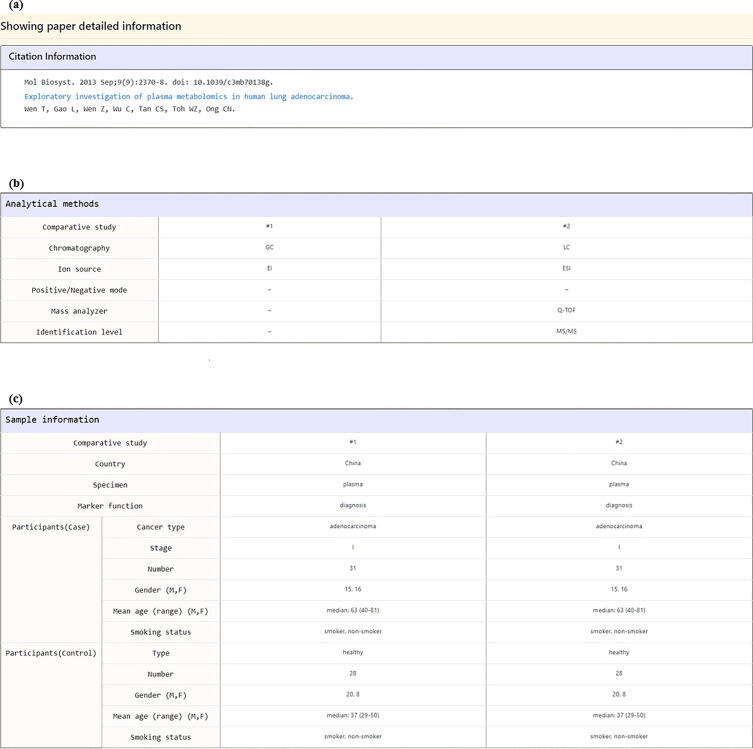
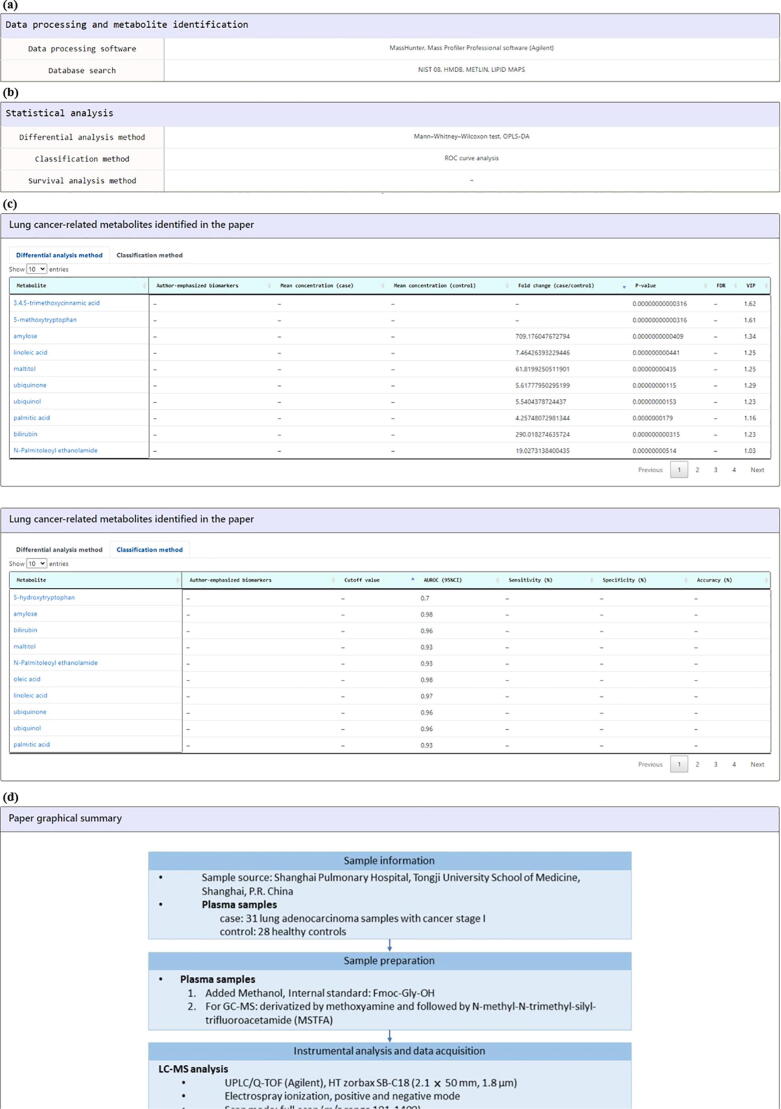
The second way for exploring the LCMD is using “Browse Studies” mode. Users can browse studies in the LCMD by applying 5 kinds of filters (participants, specimen, chromatography, ion source, and year of publication; Fig. 7a). Users then can obtain a list of mass spectrometry-based metabolomics studies that satisfy the filter specifications (Fig. 7b). By clicking on the “Reference” (e.g. Wen et al. 2013), users will be directed to a page containing the detailed information of the selected study. This page can be divided into 7 parts. First, “Citation information” provides the authors’ names, paper titles, journal names, and links to the PubMed (Fig. 8a). Second, “Analytical methods” provides the details of the mass spectrometry being used (Fig. 8b). Third, “Sample information” provides the details of the specimen and participants (Fig. 8c). Fourth, “Data processing and metabolite identification” provides the details of the software and database search engines being used to identify the metabolites from the mass spectrometry data (Fig. 9a). Fifth, “Statistical analysis” provides the details of the differential analysis, classification, and survival analysis methods being used (Fig. 9b). Sixth, “Lung cancer-related metabolites identified in the paper” provides the details of all the metabolites identified in the paper derived from the differential analysis and classification analysis (Fig. 9c). Seventh, “Paper graphical summary” provides a summary of the study design of the paper (Fig. 9d or Fig. 3).
Fig. 7.
The “Browse Studies” mode. (a) Users can browse studies in the LCMD by applying 5 kinds of filters (participants, specimen, chromatography, ion source, and year of publication). (b) Users then can obtain a list of mass spectrometry-based metabolomics studies that satisfy the filter specifications.
Fig. 8.
The study detail page (the first three parts). (a) The first part “Citation information” provides the authors’ names, paper titles, journal names, and links to the PubMed. (b) The second part “Analytical methods” provides the details of the mass spectrometry being used. (c) The third part “Sample information” provides the details of the specimen and participants.
Fig. 9.
The study detail page (the last four parts). The last four parts of the study detail page are as follows. (a) “Data processing and metabolite identification” provides the details of the software and database search engines being used to identify the metabolites from the mass spectrometry data. (b) “Statistical analysis” provides the details of the differential analysis, classification, and survival analysis methods being used. (c) “Lung cancer-related metabolites identified in the paper” provides the details of all the metabolites identified in the paper derived from the differential analysis or classification analysis. (d) “Paper graphical summary” provides a summary of the comparative research design of the paper.
3.2. A case study: using LCMD to find metabolites associated with early diagnosis of lung adenocarcinoma
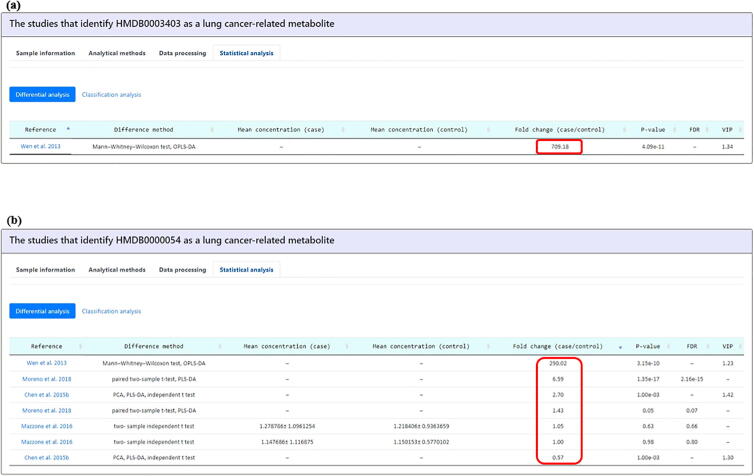
The feasibility of the LCMD was demonstrated by searching potential biomarkers for a case of early diagnosis of lung adenocarcinoma. By studying this case, we are able to systematically sieve out the metabolites from the comprehensive results of the literature according to the customized needs. Using the “Browse Metabolites” mode, we focused on finding out the differentially expressed (fold change ≥ 2) metabolites between early-stage (I and II) lung adenocarcinoma patients and healthy controls (Fig. 4a). Based on these criteria, a total of 11 metabolites were found (Fig. 4b). Among them, we are interested in two metabolites, namely amylose and bilirubin, of which plasma abundance expressed 709 and 290 times higher in lung adenocarcinoma patients compared to that of healthy controls (Fig. 4b). Checking the metabolite detail page of amylose (Fig. 10a), we observed that amylose was identified in only one lung cancer metabolomics study [22]. It is known that the increase of amylose level has been associated with Alzheimer’s disease [23] but such phenomenon has not yet been well reported in other diseases or cancers. The possible reason for the elevation of plasma amylose is the activation of glucose metabolism found in lung cancer. One of amylose synthesis pathway occurs in starch and sucrose metabolism as well as glycolysis/gluconeogenesis [24]. The growth of non-small-cell lung carcinoma (NSCLC) cells rely on the energy offered by the active glycolysis [25], which may give rise to the amylose level. Besides, amylase, an enzyme that can catalyze the hydrolysis of amylose, was considered to be a sensitive tumor marker for amylase-producing lung adenocarcinoma [26]. Above information suggests that amylose may function as a small-molecule indicator for lung cancer. Since the role of amylose in other lung cancer types remains unclear, it will be worthy to conduct the experiments to address these issues.
Fig. 10.
The metabolite detail page of amylose and bilirubin. (a) The metabolomics study that identify amylose as a lung cancer-related metabolite is shown. (b) The metabolomics studies that identify bilirubin as a lung cancer-related metabolite are shown.
Another metabolite caught our attention is bilirubin. Bilirubin showed 290 times the abundance in plasma from stage I lung adenocarcinoma patients compared to that from healthy controls (Fig. 4b). Checking the metabolite detail page of bilirubin (Fig. 10b), we noticed that, for the squamous cell carcinoma (stage I, II, and III), bilirubin showed higher level (6-fold) in tumor tissues than in adjacent normal tissues [27]. The above two studies [22], [27] indicated that a higher level of bilirubin may be related to the pathogenesis of lung cancer especially the early stage (stage I) of lung adenocarcinoma. Although in the previous clinical studies, increased serum bilirubin has been associated with lower incidence of lung cancer, chronic obstructive lung disease, and lung cancer mortality [28], [29], their result did not take the cancer stage into consideration. It has been reported that the increase level of bilirubin measured at the early stage in NSCLC may due to its antioxidant and anti-inflammatory properties [30], which suggested that biomarkers are associated with specific cancer stages. Besides, the bilirubin level in lung cancer patients was found to be higher (2.7 fold) than that in the patients after treatment (operation) [2]. Moreover, the bilirubin level does not change between adenocarcinoma/squamous cell carcinoma patients (stage I, II, and III) and at-risk controls [31]. Therefore, for the application of this marker, further targeted analysis should be conducted to verify the association between bilirubin and different stages of different lung cancer types. To sum up, checking all the mass spectrometry-based metabolomics studies of a lung cancer-related metabolite provided by LCMD can not only help researchers quickly judge whether this metabolite is a potential biomarker but also disclose which knowledge of this metabolite have been known and which still needs further investigation.
3.3. Comparison with existing metabolome databases
Curated databases with reference data and chemical structures are critical for studying biological metabolites. Major resources such as Human Metabolome Database (HMDB) [17], MetaboLights [18], Metabolomics workbench [19], Golm Metabolome Database (GMD@CSB.DB) [20], and PRIMe [21] provide general information about metabolites and metabolomics researches. However, these databases are not useful for lung cancer researchers. The reasons are as follows. First, users have no way to retrieve any lung cancer-related metabolites from Metabolomics workbench, GMD@CSB.DB, and PRIMe. Second, MetaboLights does not clearly annotate any metabolites as lung cancer-related metabolites. Despite using “lung cancer” as a keyword in compound search returns 192 metabolites where descriptions contain “lung cancer”, MetaboLights does not provide any further information to explain why these metabolites may be related to lung cancer. Third, users can only find 42 metabolites related to lung cancer in HMDB by browsing lung cancer diseases in HMDB. Although HMDB points out the lung cancer studies for these 42 metabolites, HMDB does not outline the detailed comparative research designs of these lung cancer studies.
On the contrary, our LCMD is a very useful resource for lung cancer researchers. The LCMD collected 2013 lung cancer-related metabolites from 65 mass spectrometry-based lung cancer metabolomics studies. Using the “Browse Metabolites” mode, users can browse the lung cancer-related metabolites of interest by applying 11 kinds of filters (metabolite name, chemical taxonomy, participants, specimen, marker function, chromatography, ion source, p-value, FDR, fold change, and VIP; Fig. 4a). Using the “Browse Studies” mode, users can browse lung cancer metabolomics studies of interest by applying 5 kinds of filters (participants, specimen, chromatography, ion source, and year of publication; Fig. 7a). In summary, the LCMD is the most comprehensive repository for lung cancer-related metabolites and provides various advanced filters for users to retrieve both the metabolites and studies of interest.
4. Conclusion
In this study, we constructed the first Lung Cancer Metabolome Database (LCMD) which deposits 2013 lung cancer-related metabolites retrieved from 65 mass spectrometry-based lung cancer metabolomics studies in the literature. The LCMD provides various filters for users to efficiently browse both the lung cancer-related metabolites and metabolomics studies of interest. In the case study, we showed that by using several filters, users can easily find 11 metabolites that are differentially expressed (fold change ≥ 2) between early-stage (I and II) lung adenocarcinoma patients and healthy controls. Among these 11 metabolites, two metabolites (amylose and bilirubin) were further discussed and suggested to be potential biomarkers for early diagnosis of lung adenocarcinoma. We believe that the LCMD is a useful resource for lung cancer research. Our research group will keep updating the LCMD to include any newly published mass spectrometry-based lung cancer metabolomics studies in the future.
CRediT authorship contribution statement
Wei-Sheng Wu: Conceptualization, Investigation, Supervision, Project administration, Visualization, Writing – original draft, Writing – review & editing. Hsin-Yi Wu: Investigation, Writing – original draft. Pin-Hsuan Wang: Investigation. Ting-Yu Chen: Investigation, Software, Visualization. Kuan-Ru Chen: Software, Visualization. Chih-Wei Chang: Investigation. Dong-En Lee: Visualization. Bo-Heng Lin: Software, Visualization. William Chih-Wei Chang: Investigation, Writing – review & editing. Pao-Chi Liao: Conceptualization, Investigation, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors gratefully acknowledge the use of ICP00401 and MS004000 equipment belonging to the Core Facility Center of National Cheng Kung University. We thank National Cheng Kung University and Ministry of Science and Technology of Taiwan for their support.
Funding
This work was supported by Ministry of Science and Technology of Taiwan [MOST 105-2923-M-006-005-MY3, MOST 107-2221-E-006-225-MY3, MOST 108-2628-E-006-004-MY3, MOST 109-2113-M-006-015, MOST 110-2113-M-006-014, and MOST 110-2221-E-006-198-MY3].
Authors’ contributions
WSW and PCL conceived the research topic and provided essential guidance. PHW and CWC collected the 2013 metabolite information from the 65 mass spectrometry-based lung cancer metabolomics studies and prepared the graphical summaries. TYC, KRC, BHL, and DEL constructed the website. KRC and DEL prepared all the figures in the manuscript. WSW, CWC and HYW wrote the manuscript. WSW, HYW, PHW, and CWC tested the website. CWC prepared the demo video. All authors read, edited and approved the final manuscript.
Data availability
LCMD is freely available at http://cosbi7.ee.ncku.edu.tw/LCMD/ or http://cosbi4.ee.ncku.edu.tw/LCMD/. The complete metabolite tables and study tables can be downloaded from the Download page of LCMD website. We also deposit all the downloadable files in a public repository at Github (https://github.com/cosbi-nckuee/LCMD). Demo video can be accessed at https://youtu.be/xUb1nHDMxyY.
References
- 1.Wong M.C.S., Lao X.Q., Ho K.F., Goggins W.B., Tse S.L.A. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7(1):14300. doi: 10.1038/s41598-017-14513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Ma Z., Li A., Li H., Wang B., Zhong J., et al. Metabolomic profiling of human serum in lung cancer patients using liquid chromatography/hybrid quadrupole time-of-flight mass spectrometry and gas chromatography/mass spectrometry. J Cancer Res Clin Oncol. 2015;141(4):705–718. doi: 10.1007/s00432-014-1846-5. [DOI] [PubMed] [Google Scholar]
- 3.Lokhov P.G., Kharybin O.N., Archakov A.I. Diagnosis of lung cancer based on direct-infusion electrospray mass spectrometry of blood plasma metabolites. Int J Mass Spectrom. 2012;309:200–205. [Google Scholar]
- 4.American Cancer Society. Cancer Facts and Figures 2017. Atlanta: American Cancer Society; 2017.
- 5.Okada M., Nishio W., Sakamoto T., Uchino K., Yuki T., Nakagawa A., et al. Prognostic significance of perioperative serum carcinoembryonic antigen in non-small cell lung cancer: analysis of 1,000 consecutive resections for clinical stage I disease. Ann Thorac Surg. 2004;78(1):216–221. doi: 10.1016/j.athoracsur.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 6.National Lung Screening Trial Research Team, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365(5):395-409. [DOI] [PMC free article] [PubMed]
- 7.Callejón-Leblic B., García-Barrera T., Pereira-Vega A., Gómez-Ariza J.L. Metabolomic study of serum, urine and bronchoalveolar lavage fluid based on gas chromatography mass spectrometry to delve into the pathology of lung cancer. J Pharm Biomed Anal. 2019;163:122–129. doi: 10.1016/j.jpba.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 8.Pass H.I., Beer D.G., Joseph S., Massion P. Biomarkers and molecular testing for early detection, diagnosis, and therapeutic prediction of lung cancer. Thorac Surg Clin. 2013;23(2):211–224. doi: 10.1016/j.thorsurg.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Kulpa J., Wójcik E., Reinfuss M., Kołodziejski L. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21–1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem. 2002;48(11):1931–1937. [PubMed] [Google Scholar]
- 10.Ferrigno D., Buccheri G., Giordano C. Neuron-specific enolase is an effective tumour marker in non-small cell lung cancer (NSCLC) Lung Cancer. 2003;41(3):311–320. doi: 10.1016/s0169-5002(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 11.Ward P., Thompson C. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carr D.F., Alfirevic A., Pirmohamed M. Pharmacogenomics: current state-of-the-art. Genes (Basel) 2014;5(2):430–443. doi: 10.3390/genes5020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantae V., Krekels E.H.J., Esdonk M.J.V., Lindenburg P., Harms A.C., Knibbe C.A.J., et al. Integration of pharmacometabolomics with pharmacokinetics and pharmacodynamics: towards personalized drug therapy. Metabolomics. 2017;13(1):9. doi: 10.1007/s11306-016-1143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beger R.D., Dunn W., Schmidt M.A., Gross S.S., Kirwan J.A., Cascante M., et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics. 2016;12(9) doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiagarajan P.S., Wu X., Zhang W., Shi I., Bagai R., Leahy P., et al. Transcriptomic-metabolomic reprogramming in EGFR-mutant NSCLC early adaptive drug escape linking TGFβ2-bioenergetics-mitochondrial priming. Oncotarget. 2016;7(50):82013–82027. doi: 10.18632/oncotarget.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bamji-Stocke S., van Berkel V., Miller D.M., Frieboes H.B. A review of metabolism-associated biomarkers in lung cancer diagnosis and treatment. Metabolomics. 2018;14(6):81. doi: 10.1007/s11306-018-1376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018;46(D1):D608-D617. [DOI] [PMC free article] [PubMed]
- 18.Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48(D1):D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sud M., Fahy E., Cotter D., Azam K., Vadivelu I., Burant C., et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44(D1):D463–D470. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 2005;21(8):1635-1638. [DOI] [PubMed]
- 21.Sakurai T, Yamada Y, Sawada Y, Matsuda F, Akiyama K, Shinozaki K, Hirai MY, Saito K. PRIMe Update: innovative content for plant metabolomics and integration of gene expression and metabolite accumulation. Plant Cell Physiol 2013;54(2):e5. [DOI] [PMC free article] [PubMed]
- 22.Wen T., Gao L., Wen Z., Wu C., Tan C.S., Toh W.Z., et al. Exploratory investigation of plasma metabolomics in human lung adenocarcinoma. Mol Biosyst. 2013;9(9):2370. doi: 10.1039/c3mb70138g. [DOI] [PubMed] [Google Scholar]
- 23.Huang L., Hollingsworth R.I., Castellani R., Zipser B. Accumulation of high-molecular-weight amylose in Alzheimer's disease brains. Glycobiology. 2004;14(5):409–416. doi: 10.1093/glycob/cwh042. [DOI] [PubMed] [Google Scholar]
- 24.Chang T.-S., Liu C.-W., Lin Y.-L., Li C.-Y., Wang A.Z., Chien M.-W., et al. Mapping and comparative proteomic analysis of the starch biosynthetic pathway in rice by 2D PAGE/MS. Plant Mol Biol. 2017;95(4-5):333–343. doi: 10.1007/s11103-017-0652-2. [DOI] [PubMed] [Google Scholar]
- 25.Chang L., Fang S., Gu W. The molecular mechanism of metabolic remodeling in lung cancer. J Cancer. 2020;11(6):1403–1411. doi: 10.7150/jca.31406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Zhang L., Pan S., Gu B., Zhen Y., Yan J., et al. Amylase: sensitive tumor marker for amylase-producing lung adenocarcinoma. J Thorac Dis. 2013;5(4):E167–E169. doi: 10.3978/j.issn.2072-1439.2013.08.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno P., Jiménez‐Jiménez C., Garrido‐Rodríguez M., Calderón‐Santiago M., Molina S., Lara‐Chica M., et al. Metabolomic profiling of human lung tumor tissues - nucleotide metabolism as a candidate for therapeutic interventions and biomarkers. Mol Oncol. 2018;12(10):1778–1796. doi: 10.1002/1878-0261.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsfall L.J., Rait G., Waltes K., Swallow D.M., Pereira S.P., Nazareth I., et al. Serum bilirubin and risk of respiratory disease and death. JAMA. 2011;305(7):691–697. doi: 10.1001/jama.2011.124. [DOI] [PubMed] [Google Scholar]
- 29.Temme E.H., Zhang J., Schouten E.G., Kesteloot H. Serum bilirubin and 10-year mortality risk in a Belgian population. Cancer Causes Control. 2001;12(10):887–894. doi: 10.1023/a:1013794407325. [DOI] [PubMed] [Google Scholar]
- 30.Stocker R., Yamamoto Y., McDonagh A.F., Glazer A.N., Ames B.N. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235(4792):1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 31.Mazzone P.J., Wang X.-F., Beukemann M., Zhang Q.i., Seeley M., Mohney R., et al. Metabolite profiles of the serum of patients with non-small cell carcinoma. J Thorac Oncol. 2016;11(1):72–78. doi: 10.1016/j.jtho.2015.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
LCMD is freely available at http://cosbi7.ee.ncku.edu.tw/LCMD/ or http://cosbi4.ee.ncku.edu.tw/LCMD/. The complete metabolite tables and study tables can be downloaded from the Download page of LCMD website. We also deposit all the downloadable files in a public repository at Github (https://github.com/cosbi-nckuee/LCMD). Demo video can be accessed at https://youtu.be/xUb1nHDMxyY.