Abstract
Antibody immunodominance refers to the preferential and asymmetric elicitation of antibodies against specific epitopes on a complex protein antigen. Traditional vaccination approaches for rapidly evolving pathogens have had limited success in part because of this phenomenon, as elicited antibodies preferentially target highly variable regions of antigens, and thus do not confer long lasting protection. While antibodies targeting functionally conserved epitopes have the potential to be broadly protective, they often make up a minority of the overall repertoire. Here, we discuss recent protein engineering strategies used to favorably alter patterns of immunodominance, and selectively focus antibody responses toward broadly protective epitopes in the pursuit of next-generation vaccines for rapidly evolving pathogens.
Subject terms: Antibodies, Virology, Protein vaccines
B cell immunodominance
Antibody (Ab) responses raised against complex protein antigens can preferentially target particular epitopes in a reproducible hierarchy, a phenomenon known as immunodominance. These primary targets of Ab responses are often immunologically dominant, while those engaged by minor portions of the overall response are considered immunologically subdominant. This asymmetry contributes to the host-pathogen ‘arms race’, where particular regions of surface-exposed antigens experience the most immune pressure, and are subsequently key sites of antigenic variation1,2. Viral antigens like influenza hemagglutinin (HA) or HIV envelope protein (Env), have conserved structural or functional regions. Abs targeting these epitopes are often broadly neutralizing (bnAbs) or protective (bpAbs), the latter through Fc-mediated effector functions3. However, such antibodies are generally immunologically subdominant, and make up a minority of the overall repertoire. Next-generation vaccines for rapidly evolving pathogens aim to alter patterns of dominance to elicit higher levels of broadly neutralizing or protective responses.
While B cell immunodominance is an incompletely understood phenomenon, there are several key aspects influencing inter-clonal competition in the germinal center (GC) reaction that can be leveraged for rational immunogen design4. (1) Precursor frequency, the number of naïve B cells that engage a specific epitope, is a key limiting factor; if fewer B cells engage an epitope, the greater the likelihood that the subdominant-directed population will be outcompeted by more abundant B cell clones. This is a limiting factor for many bnAb precursors such as VRC01-class Abs targeting the HIV Env CD4 binding site (CD4bs), which are present at very low frequencies in human repertoires5. Precursor frequency may also be influenced by central tolerance as is the case for certain autoreactive HA-stem antibodies; negative selection attempts to remove these autoreactive B cells from the naïve repertoire6,7. The accessibility of a given epitope can also contribute, as an epitope must be accessible to BCRs in order to trigger an antibody response. (2) Precursor affinity for the antigen drives GC establishment or entry, as high affinity for antigen is linked with increased acquisition of antigen and increased density of surface pMHC8. The relationship between precursor frequency and affinity in GC B cells is nonlinear, but even when precursor frequencies are low, B cells can be recruited to GCs if they have sufficiently high affinity9–11. (3) The degree of T cell help during the GC reaction is a limiting factor on GC B cell proliferation and maturation12,13. Increasing the number of T follicular helper (Tfh) cells specific to epitopes on an immunogen may allow B cells into the GC that would otherwise not gain entry14. The modification of even a few helper T cell epitopes to relieve competition between B cell clones can have a marked impact on overall patterns of dominance15.
The structure of a B cell epitope as seen by the BCR likely also plays a role in immunodominance, possibly with subdominant epitopes requiring stringent or stereotyped contacts, but currently there is little experimental evidence directly addressing this topic. Computational analyses of antigen structures has focused primarily on identifying likely B cell epitopes, rather than establishing their relative immunodominance16–18. While immunodominance hierarchies for antigens such as HA and hepatitis C virus E2 have been experimentally mapped, the importance of epitope structure and how it might impact the trajectory of the affinity maturing B cell repertoire remains relatively undefined1,19,20.
In this review, we discuss various protein engineering strategies used to develop immunogens against rapidly evolving pathogens, and how they influence these three rather well-characterized elements of B cell immunodominance to preferentially elicit antibodies to subdominant epitopes.
Consolidating protein engineering strategies into general approaches
The following sections focus on three general approaches of immunogen design. We discuss recent developments in strategies to (1) magnify the overall humoral response, (2) prevent or reduce the elicitation of ‘off-target’ antibody responses, and (3) specifically amplify responses targeting preferred epitopes. Discussion of these strategies focuses on influenza and HIV viral glycoproteins but extend to other viruses including respiratory syncytial virus (RSV), dengue, and Zika. Importantly, the strategies discussed here are not mutually exclusive, and many immunogens will likely influence immunodominance through multiple mechanisms.
Magnification of the overall humoral response
Multimeric display
The repetitive presentation of viral antigens, sensed by the degree of surface Ig-crosslinking, is a key factor to increasing the robustness of B cell responses21. Historically, multimeric display was accomplished using virus-like particles (VLPs) derived from human, insect, or plant viruses such as hepatitis B, flock house virus, and tobacco mosaic virus22–28. This multivalent display mimics the natural presentation of viral epitopes, and can elicit protective responses29–32. However, many of these display platforms are limited to presenting relatively small, linear peptides, and cannot readily present complex conformationally specific epitopes26.
In a key study, ferritin-based nanoparticles were engineered at the three-fold axis to display trimeric influenza HA antigens as genetic fusions. These immunogens elicited higher antigen-specific titers with increased breadth and protection relative to recombinant trimeric HAs33. Importantly, nanoparticle-immunized cohorts had more broadly reactive hemagglutinin inhibition (HAI), neutralization, and higher stem-directed titers, indicating that multimeric display can impact patterns of dominance in favor of cross reactive and subdominant responses33,34. More recently, the use of genetic fusion-based protein nanoparticle immunogens has extended beyond influenza HA; protein scaffolds displaying prefusion stabilized RSV F or HIV Env glycoproteins elicit robust neutralizing titers underscoring the utility of nanoparticle-based display as a general design strategy35,36. Next-generation protein-based nanoparticles involve multi-component constructs that allow for stoichiometrically precise display of multiple antigens37–42. Further computational and rational protein engineering led to the development of nanoparticles with tetrahedral, octahedral, and icosahedral symmetry; allowing for a greater degree of customizable antigen display based on the desired geometry43–45.
Additional multivalent assemblies allow for display of antigens when genetic fusions are not possible or if mixed species are required. Lipid nanoparticles or synthetic liposomes, for example, can be engineered to present multiple antigens, resulting in similar amplification of serum titers relative to recombinant protein46,47. Additionally, recombinantly expressed antigens can be enzymatically ‘clicked’ onto nanoparticle scaffolds (e.g., carbohydrate-based oligomers and VLPs) using the sortase transpeptidase from Staphylococcus aureus48–53. A more recent approach for conjugating antigen to nanoparticle scaffolds involves SpyTag-SpyCatcher54. This technology uses a split fibronectin-binding protein subdomain from Streptococcus pyogenes with a linear peptide tag appended on to the antigen and the remaining protein to the nanoparticle; when combined in vitro a spontaneous covalent linkage occurs via an isopeptide bond54. As a short peptide sequence (13 amino acids), SpyTag is readily appended to nearly any antigen of interest. This platform allows for an easily modifiable ‘plug and display’ approach, where heterologous display can be optimized to present a variety of antigens. Proof of concept studies have used diverse betacoronavirus receptor binding domains (RBDs) as well as influenza HAs55–58.
Despite the significant benefits of these multivalent display strategies, they are not without drawbacks. A caveat of protein-based nanoparticles and assembly domains like SpyCatcher-SpyTag is the lack of precise stoichiometric control of multiple antigens and their own innate immunogenicity. Their use introduces additional epitopes and expands the degree of interclonal competition, potentially skewing the immunodominance hierarchy away from subdominant epitopes of interest. Masking (i.e., hyperglycosylation) of such ‘scaffold’ epitopes may be necessary to limit off-target reactivity. Furthermore, multivalent antigen display on a nanoparticle may result in steric occlusion of desired epitopes due to antigen spacing and overall density; this may ultimately restrict access of potential BCRs from engaging these epitopes leading to a reduction in response despite being presented in greater number than soluble wild-type antigen59.
Restricting ‘off-target’ responses
Complex protein antigens elicit diverse GCs containing B cells that recognize a range of epitopes60,61. In addition to magnifying the overall humoral response, immunogen design approaches are leveraged to modulate responses to target different epitopes on a given antigen. The many epitopes present on a single immunogen result in inter-clonal competition between B cells targeting both conserved epitopes of interest, and those engaging ‘off-target’ epitopes. Decreasing the size of the competing B cell pool can influence patterns of immunodominance and is accomplished in two major ways: the outright removal or steric occlusion of undesired epitopes.
Removal of undesired epitopes
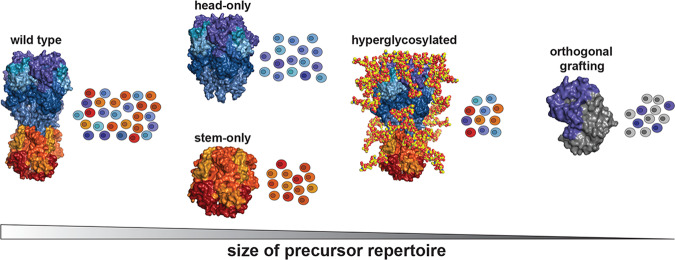
The physical removal of undesired epitopes precludes the elicitation of responses against them, in effect modulating precursor frequency by reducing the overall number of competing B cells (Fig. 1). Many conserved epitopes recognized by B cells are conformation specific, composed of multiple segments proximal in structural space but separated in sequence space that are difficult to selectively present62. However, many viral proteins such as influenza HA, RSV F protein, or HIV Env can be truncated into domain-based constructs in order to minimize the inclusion of off-target epitopes while still maintaining the integrity of the target epitope63–72. Subsequent iterations of these constructs, such as the headless, stem-only domain ‘mini-HAs’, or the ‘engineered outer domain’ (eOD) Env constructs continue to expand the available repertoire of truncated immunogens73,74.
Fig. 1. Modulating precursor repertoire with various immunogen design strategies.
Schematic representation of how precursor frequency may be altered using different protein engineering strategies to remove ‘off-target’ competing clones using influenza hemagglutinin (HA) as a representative example. Colors refer to theoretical epitopes on full length HA; shades of blue and purple correspond to head-directed epitopes; shades of yellow, orange, and red correspond to stem-directed epitopes. For orthogonal grafting, a hypothetical scaffold is shown in gray with purple region illustrating a grafted HA head epitope. All images created in PyMol using PDB 5UGY.
Occlusion of off-target epitopes
Complementary to removing undesired epitopes is the steric occlusion of off-target ones, which can be done in a reversible or irreversible manner. The formation of immune complexes between antibodies and an antigen can mask epitopes; subsequent immunization with these immune complexes can sterically shield epitopes and thus bias antibody responses. This was shown for flavivirus, influenza, and HIV glycoproteins75–78. Recent refinement of this approach has both decreased the size of the shielding to a single chain variable fragment (scFv), or generated covalently stabilized complexes through chemical cross-linking or expression of an antigen-antibody genetic fusion79–81. Analogous to epitope removal, the steric shielding of off-target epitopes reduces the size of the precursor B cell pool able to bind the antigen, thus reducing the pool of potential competitors.
A commonly used method to restrict off-target responses is to introduce novel predicted N-linked glycosylation (PNG) sites on viral surface proteins to “shield” or occlude undesired epitopes. Glycans are naturally present on viral envelope proteins and play key roles in stability, pathogenicity, and immunogenicity, as well as escape from immune surveillance82–90. Overall glycosylation patterns can affect antigen processing, delivery into GCs, and breadth of elicited responses91–101. The variation in these natural glycosylation patterns in response to immune pressure provides a guide to introduce glycans on rationally designed immunogens100,102,103.
Recent work has underscored the effectiveness of glycan masking to restrict off-target responses across a variety of viral glycoproteins including Zika and dengue E, hepatitis C E2, influenza HA, HIV Env, and RSV F protein36,103–107. For flavivirus-based immunogens (e.g., Zika, dengue E), glycan shielding is critical to reduce the likelihood of antibody-dependent enhancement (ADE) mediated by cross-reactive yet non-neutralizing antibodies104. For influenza HA and HIV Env, the glycosylated immunogens focus responses to broadly cross reactive, yet relatively subdominant epitopes such as the HA head interface, the HA stem region, and the Env CD4bs103,106,107. This serum focusing is likely due to an increased proportion of the serum repertoire targeting the epitope of interest; hyperglycosylation generally does not impact overall serum titer, even for heavily glycosylated immunogens103,107,108. More recently, the combination of hyperglycosylation and covalent stabilization of HA trimers, where only the RBS epitope remains exposed, decreased overall serum titers without focusing to the subdominant RBS108. Thus, while hyperglycosylation can enrich antibody responses against desired epitopes, it is not always successful in altering immunodominance.
Amplification of ‘on-target’ responses
In contrast to the occlusion or removal of off-target epitopes, alternative immunogen design strategies aim to expand responses towards desired epitopes. These approaches manipulate both precursor frequency and affinity, as well as epitope accessibility to preferentially target broadly conserved or protective epitopes of interest. General approaches involve the stabilization of preferred antigen conformations, glycan ‘unmasking’ of shielded epitopes, computationally inferred consensus antigens, germline targeting, and epitope resurfacing.
Stabilization of prefusion conformation
Viral envelope glycoproteins undergo significant conformational rearrangements between their prefusion and post-fusion states. To elicit immune responses that specifically recognize the prefusion state present on the invading virus, the glycoproteins are stabilized to prevent spontaneous structural rearrangement. This general concept was first demonstrated for influenza HA to characterize its conformational rearrangements necessary for membrane fusion either through introducing prolines or non-native cysteines to stabilize or covalently “staple” the prefusion state (Fig. 2a)109,110. Subsequent application of these principles for HIV Env led to the development of stabilized ‘SOSIP’ trimers through inter-subunit disulfides and conformation-locking isoleucine to proline mutations111,112. The initial successes with this stabilization approach has resulted in broadly applicable guidelines and even automated the design of prefusion stabilized Env antigens across viral clades113–115. These stabilized constructs have been instrumental in the study of HIV, and form the basis of many current Env immunogens in clinical trials116–118. The general strategy of prefusion stabilization has been widely implemented for other viral antigens and has informed diverse vaccine development efforts119,120. Specifically, the introduction of stabilizing proline residues was effective for RSV F, hMPV F, Lassa virus glycoprotein complex, ebola and Marburg glycoproteins, and several coronavirus spike proteins121–127. For the latter, several new iterations including ‘S-2P’ and ‘HexaPro’ in SARS-CoV-2 spike have improved efficacy and form the basis of the currently approved vaccines127. Such stabilization approaches help limit the overall size of the precursor pool by ensuring the prefusion conformation is dominant, and limit responses against epitopes displayed less frequently on the circulating virus.
Fig. 2. Design strategies to enhance ‘on-target’ epitope responses.
a Stem- and head-only HAs indicating locations of engineered prolines (left, red spheres) and inter-HA cysteines (right, yellow spheres) to stabilize the prefusion conformation. b Selective removal of native glycans to expose neoepitopes (red circle). c Computational design of HA antigens based on overall subtype diversity increases cross-reactive responses. To illustrate the COBRA approach, HA is arbitrarily colored in gray shading to show variation in amino acid identity that ultimately contributes to the consensus sequence; for a complete description see Huang et al.131. d Optimization of a single epitope for a specific class of B cell precursors increases initial antigen affinity. HA receptor binding site (RBS) is shown in green as an example. e Resurfacing of a complex conformational-specific epitope allows for heterologous boosting of subdominant responses within memory. S1–S4 segments of the grafted RBS shown in red, orange, green, and blue, respectively. f Chimeric HAs where head domain (purple) of an antigenically distinct non-circulating HA is transplanted onto a conserved circulating stem domain (gray) to preferentially target stem-directed responses. All images created in PyMol using PDB 5UGY.
‘Un-masking’ of normally occluded epitopes
The removal of naturally occurring glycans introduces new targets for antibody responses (Fig. 2b). For HIV Env and influenza HA, viral proteins with extensive surface glycosylation, selective removal of glycosylation sites around the CD4bs or HA stem epitope modulated responses to these epitopes and increased the overall breadth of elicited antibodies128,129. However, in some cases, glycan removal to expose neoepitopes was not sufficient to elicit broader responses, indicating that other factors influencing immunodominance are present130. Even if a glycan is not sterically occluding the epitope itself, it can impact the elicitation of antibodies against adjacent epitopes, as demonstrated by the increased breadth of serum responses raised against selectively de-glycosylated Env and HA antigens106,128,129,131.
Computationally optimized broadly reactive antigens
While many immunogen design strategies are designed to target a single epitope, computationally optimized broadly reactive antigens (COBRAs) use a consensus sequence-based approach to optimize the entire antigen to simultaneously present multiple cross-reactive epitopes (Fig. 2c). First developed for H5 influenza HAs, COBRAs improved upon previous classes of consensus-based immunogens by removing biases from overrepresentation of certain sequence clades due to uneven sequence availability132. Iterative rounds of consensus sequence generation within, and then between, clades yielded HA immunogens that conferred higher HAI titers and protection against diverse H5 isolates even relative to cocktail immunizations of members from multiple H5 clades133,134. The COBRA approach translates to other human (e.g., H1, H2, H3), avian (e.g., H7), and swine (e.g., H1) HAs, as well as other viral antigens such as dengue virus E protein. Importantly these immunogens work in both naïve animal models and in the setting of preexisting immunity135–147. The exact mechanism of enhanced breadth elicited by COBRA immunogens is unknown; one possibility is increased recruitment of B cells against antigenic sites with greater cross-reactive potential leads to the enhanced diversity and breadth of responses148,149.
Germline-targeting antigens
Germline biases for specific epitopes may be observed if the amino acids encoded by the naïve sequence has a threshold affinity for the antigen such that it results in a competitive advantage. For influenza HA, stem-binding antibodies are predominantly enriched for VH1–69, but VH6-1, VH1–18, and VH3–30 have also been observed; sialic acid-mimicking HA receptor binding site (RBS)-directed Abs are enriched for JH6 genes150–158. Similar gene enrichment is seen in responses to other viral glycoproteins, including Zika E (e.g., VH3–23/VK1–5), hepatitis C virus E2 (e.g., VH1–69), and SARS-CoV-2 spike (e.g., VH3–53, VH1–2, VH3–9, and VH3–30)159–163. While germline-encoded affinity can dramatically alter patterns of dominance if the precursor B cells are present with high enough frequency, this natural advantage is often insufficient in the setting of rare precursors164. For example, high-affinity VRC01-class precursors that target the HIV Env CD4bs are predicted to have a frequency of 1 in 0.9 million naïve B cells, and despite nanomolar affinity for rationally designed immunogens targeting these precursors, they remained minor components of the overall repertoire when present in animal models at these levels5.
Germline-targeting immunogens are used to preferentially bias responses towards these rare high-affinity clones (Fig. 2d). In initial studies, nanoparticles were multimerized with HIV Env eOD antigens designed to engage inferred germline precursors for VRC01-class antibodies; these immunogens activated both germline and mature VRC01 expressing cell lines74. Studies using VRC01 germline knock-in mice showed that elicited antibodies have VRC01-class characteristics, with varying affinity and breadth for native HIV Envs165,166. Subsequent generations of VRC01-class germline-targeting nanoparticles allowed for isolation and characterization of human VRC01 precursors present in naïve individuals167. Applying similar principles led to the development of immunogens able to engage germline-precursors for HIV Env targeting PGT121- and CH235-like broadly neutralizing antibodies, as well as those from cross-group stem-reactive influenza HA antibodies168–170.
Recently, germline targeting designs have been generalized to target larger pools of B cell precursors making CDRH3-mediated contacts with a given epitope171. This represents a conceptual shift in the approach: rather than attempting to engage the exact precursors within the repertoire, immunogens now engage a wider set of precursors that have the potential to mature breadth over the course of multiple immunizations. However, key questions remain about how to ‘guide’ BCR maturation along the desired evolutionary path, and what are the key structural characteristics of immunogens necessary for eliciting antibodies with the desired breadth172,173. Performing these studies in the context of adoptively transferred germline-presenting B cells, rather than a complete knock-in model, will also be necessary to understand the effects of inter-clonal competition on the ability of a rationally designed immunogen to selectively enrich for desired responses174.
Epitope grafting or resurfacing
For antigenically diverse, yet structurally similar antigens, epitope ‘grafting’ or ‘resurfacing’ can be used to target subdominant responses from immunologic memory. Desired epitopes can be transplanted onto antigenically distinct scaffolds (Fig. 2e)175. These resurfaced constructs are thought to increase responses towards the grafted epitope due to preferential recall of memory responses against the graft versus de novo responses against the scaffold. Recently the H1 HA RBS, a complex conformationally specific epitope, was grafted onto the non-circulating avian H4 and H14 HAs. Binding to a panel of H1 RBS-directed antibodies confirmed the successful recapitulation of the grafted epitope175. Immunization with the resurfaced HA in mice primed with H1 HA increased the breadth of RBS-directed B cells against historical H1 isolates175.
A similar grafting approach was used for the HA stem region to create chimeric HAs (cHAs) whereby antigenically distinct and often non-circulating HA heads have been swapped onto stems of circulating HA subtypes (Fig. 2f)176. Prime-boost immunizations with these cHAs presenting head domains to which there is no preexisting immunity, but with conserved stem domains, elicited cross-reactive stem-directed antibodies capable of protecting via Fc-effector functions177–184. In clinical trials, cHAs show evidence of eliciting cross-reactive serum antibodies that target the HA stem in human subjects185. More recently, an analogous approach was used to design chimeric coronavirus spike immunogens186.
A complementary type of epitope resurfacing involves mutating ‘off-target’ regions to disrupt recognition of undesired epitopes. This approach has been used as a screening tool to isolate HIV Env CD4bs-directed antibodies, and flavivirus E immunogens with lower cross-reactive but non-neutralizing titers187–189. For example, resurfaced dengue E domain III (EDIII) immunogens preserved the broadly neutralizing 4E11 epitope while peripheral epitopes were mutated to disrupt recall responses against these regions188,189. Similar to masking off-target EDIII epitopes with glycans, resurfacing off-target epitopes may help prevent antibody-dependent enhancement as cross-reactive yet non-neutralizing responses are limited.
A further extension of this approach is the grafting of desired epitopes onto orthogonal protein scaffolds that lack structural homology with the wild-type antigen. In initial studies, multiple neutralizing epitopes of RSV F protein were transplanted onto orthogonal protein scaffolds. The antibody responses elicited by orthogonally-scaffolded immunogens were specific to the grafted epitopes, and had increased titers against the grafted sites despite lower titers to RSV F protein overall190,191. While the difficulty in designing orthogonal scaffolds significantly increases with the complexity of the target epitope, this approach completely avoids eliciting antigen-specific off-target responses.
A primary drawback of epitope resurfacing is the introduction of a significant number of neoepitopes. For example, grafting the H1 HA RBS onto an H4 full length soluble ectodomain (FLsE) HA scaffold, or grafting an antigenically distinct HA head onto a conserved HA stem domain, expands inter-clonal competition via the introduction of novel immunodominant HA head epitopes175,176. Likewise, while orthogonal grafting does not present any epitopes shared by that class of antigens (e.g., RSV F protein), it still presents neoepitopes which will have their own intrinsic immunodominance hierarchy. Overall, while resurfaced immunogens can elicit antibodies against desired epitopes with increased frequency or breadth, current iterations do not optimally alter patterns of dominance. The combination of epitope grafting with other design strategies is likely needed to fully realize the epitope immune-focusing potential of such constructs.
Increasing the breadth of antibody responses
The development of next-generation vaccines is further complicated by the fact that engagement of a conserved epitope does not guarantee breadth192. Additional steps are therefore needed to ensure broad reactivity even if elicited antibodies engage the desired epitope. Immunogen design strategies to increase the breadth of responses can be broadly classified as single- or multi-step approaches.
Prime and boost formulations to focus from within memory
Initial exposures to a pathogen can influence subsequent responses, a phenomenon termed “original antigenic sin”193,194. Inferred germline antibodies or unmutated common ancestors (UCAs) of broadly neutralizing influenza HA antibodies isolated from human donors show high affinity for strains circulating during the donor’s early childhood, suggesting that early exposures imprint responses that are subsequently matured and refined upon re-exposure195–197. Repeated exposure in the form of seasonal vaccination against influenza does not appear to alter the patterns of dominance established towards particular HA subtypes175,198–200. However, recall and maturation of HA stem-directed responses has been observed following immunization with divergent wild-type or chimeric strains, where the stem region is the conserved epitope between exposures201–205. Even if overall patterns of dominance are not altered, subdominant antibodies can show increased breadth upon re-exposure to antigenically similar epitopes175.
Computational studies suggest that there are optimal antigenic distances between primary and secondary immunogens that will help direct responses toward conserved and broadly protective epitopes206–209. Too little antigenic distance, and memory responses against variable epitopes will dominate the repertoire; too great a distance, and de novo responses against variable epitopes dominate. Understanding how antigenic distance impacts preferential recall of imprinted responses will be critical for designing optimized immunogens capable of expanding subdominant populations from within the memory compartment.
Eliciting cross-reactive responses from single immunizations
Immunization with a cocktail of HA homotypic nanoparticles has been shown to elicit broader serum titers than individual nanoparticles34. However, the breadth of these serum responses is due to the summation of individual strain-specific responses rather than broad reactivity from specific antibodies. The simultaneous presentation of multiple antigens within a single immunogen, however, elicits broader antibodies, and in certain cases focuses antibody responses on conserved epitopes40,41,55,56. While the precise mechanism of how heterologous display focuses responses to cross-reactive epitopes has not yet been determined, evidence suggests that such an immunogen confers a competitive avidity advantage to cross-reactive B cells40,41.
However, with heterologous display, especially when presenting an array of wild-type antigens, there is often a gradient of epitope conservation that ranges from unique, to partially conserved, to fully conserved across the set of antigens being displayed. Thus, there appears to be a ‘sliding scale’ of benefit to all antigen-specific B cells that is combined with the preexisting immunodominance hierarchy to determine the overall epitope distribution (Fig. 3). If a goal is to focus antibody responses to a specific epitope, heterologous display could be optimized such that only a single epitope is present multiple times, while all other epitopes are present only once. This ‘epitope-enriched’ immunogen would, in theory, confer an avidity advantage specifically to B cells engaging the desired epitope, and thus maximize their relative competitive advantage. Heterologous display of epitope-resurfaced antigens, where antigenically distinct scaffolds present a common grafted epitope, would likely result in enhanced immune focusing.
Fig. 3. Proposed use of heterologous antigen display to focus responses toward desired epitopes.
A proposed spectrum of epitope focusing immunogens incorporating various multimeric or heterologous display strategies is shown; unique colors represent antigenically distinct components. As the relative overall enrichment of the target epitope increases, so does the degree of epitope focusing. Heterologous display within a subtype presents a wider gradient of conserved epitopes due to the smaller antigenic distance between presented components. Heterologous display of antigenically distinct antigens enriches for conserved epitopes, but there are still other epitopes that are conserved between various combinations of component antigens. The endpoint of this spectrum is an epitope-enriched immunogen, where a single epitope is presented as multiple copies with all other epitopes presented once. All images created in PyMol using PDBs 5UGY and 3BVE.
Future questions
Despite significant advancements in recent years, understanding immunodominance and what is required to create a vaccine with long-term efficacy against a rapidly evolving pathogen is necessary. Some key fundamental questions that remain include, but are not limited to:
If immunodominance hierarchies are altered by next-generation vaccines, will previously conserved subdominant epitope(s) nevertheless remain conserved in response to significant immune pressure? In other words, by focusing immune responses to a conserved epitope, will escape (i.e., mutation) occur? While many conserved regions on viral glycoproteins play important functional roles (e.g., receptor binding, membrane fusion), key residues are often a fraction of the entire eptiope210–212.
Can B cell populations targeting a particular conserved epitope be guided towards breadth through serial immunizations with rationally designed constructs, or is the potential for breadth intrinsic to a specific subset of clones? This has significant implications for pathogens such as HIV, where many bnAbs have structural features (e.g., CDR length) that greatly restrict the size of precursor populations.
Are there structural, epitope-specific factors that influence immunodominance hierarchies? The major components of precursor frequency and affinity, T cell help, and epitope accessibility do not account for the impact of the epitope itself as a substrate during affinity maturation. For example, an HA receptor-mimicking RBS-directed antibody makes contacts within the sialic acid binding pocket, a recessed epitope surrounded by a highly variable periphery, where acquired mutations in CDRs can affect or completely abrogate binding154. In contrast, an HA lateral patch-binding antibody makes contacts with a mostly continuous hydrophilic surface, where the similar CDR mutations are less likely to disrupt binding213. In effect, the affinity maturing antibodies may follow evolutionary paths, determined by the targeted epitope. Over time, the relative frequency of beneficial, neutral, or deleterious mutations impact the size of competing clonal B cell pools, and thus the trajectory of inter-clonal competition.
Conclusion
Current protein engineering strategies, like those described above, allow tailoring of an immunogen to elicit the desired antibody response. Importantly, many of the described approaches take orthogonal and therefore complementary approaches to immune-focusing. For example, a germline-targeting immunogen, with hyperglycosylated periphery, displayed on a nanoparticle leverages three different strategies: (1) the multimeric display, (2) steric shielding of ‘off-target’ and nanoparticle scaffold epitopes, and (3) epitope optimization to engage a set of precursor B cells. Multimeric display increases the overall size of the precursor B cell pool and the avidity of antigen interactions, steric shielding of undesired responses leads to the reduction of inter-clonal competition from undesired clones, and germline targeting ensures a select set of precursors have increased affinity and thus a competitive edge in the GC reaction. While the intrinsic immunodominance hierarchies for specific antigens will likely determine the optimal approach needed to expand broadly protective responses, the above strategies allow for selective manipulation of the major contributing factors to immunodominance.
These protein engineering approaches can be similarly used to generate probes to deconvolute the GC reaction. Selective resurfacing or occlusion of epitopes, the modulation of endogenous/exogenous T cell epitopes, display on oligomeric scaffolds, and other rational design strategies, provide an opportunity to perform mechanistic experiments investigating underlying B-cell biology. Simultaneous basic and translational advancements will allow for iterative improvement of pre-clinical vaccine candidates in the pursuit of next-generation viral vaccines for rapidly evolving pathogens.
Acknowledgements
We acknowledge support from the NIH for R01AI146779 (A.G.S.), P01AI89618-A1 (A.G.S.), and T32 GM007753 (T.M.C.). This research has been funded in whole or part with federal funds under a contract from the National Institute of Allergy and Infectious Diseases, NIH contract 75N93019C00050 (A.G.S.).
Author contributions
T.M.C. and A.G.S. wrote and edited the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Timothy M. Caradonna, Email: caradonna@crystal.harvard.edu
Aaron G. Schmidt, Email: aschmidt@crystal.harvard.edu
References
- 1.Angeletti D, et al. Defining B cell immunodominance to viruses. Nat. Immunol. 2017;18:456–463. doi: 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman MO, Angeletti D, Yewdell JW. Antibody immunodominance: the key to understanding influenza virus antigenic drift. Viral Immunol. 2018;31:142–149. doi: 10.1089/vim.2017.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanderven HA, Kent SJ. The protective potential of Fc-mediated antibody functions against influenza virus and other viral pathogens. Immunol. Cell Biol. 2020;98:253–263. doi: 10.1111/imcb.12312. [DOI] [PubMed] [Google Scholar]
- 4.Angeletti D, Yewdell JW. Understanding and manipulating viral immunity: antibody immunodominance enters center stage. Trends Immunol. 2018;39:549–561. doi: 10.1016/j.it.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Havenar-Daughton, C. et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med.10.1126/scitranslmed.aat0381 (2018). [DOI] [PMC free article] [PubMed]
- 6.Bajic G, et al. Autoreactivity profiles of influenza hemagglutinin broadly neutralizing antibodies. Sci. Rep. 2019;9:3492. doi: 10.1038/s41598-019-40175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khurana, S., Hahn, M., Klenow, L. & Golding, H. Autoreactivity of broadly neutralizing influenza human antibodies to human tissues and human proteins. Viruses10.3390/v12101140 (2020). [DOI] [PMC free article] [PubMed]
- 8.Yeh C-H, Nojima T, Kuraoka M, Kelsoe G. Germinal center entry not selection of B cells is controlled by peptide-MHCII complex density. Nat. Commun. 2018;9:928–928. doi: 10.1038/s41467-018-03382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dosenovic P, et al. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl Acad. Sci. USA. 2018;115:4743–4748. doi: 10.1073/pnas.1803457115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbott RK, et al. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity. 2018;48:133–146.e136. doi: 10.1016/j.immuni.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang D, et al. B cells expressing authentic naive human VRC01-class BCRs can be recruited to germinal centers and affinity mature in multiple independent mouse models. Proc. Natl Acad. Sci. USA. 2020;117:22920–22931. doi: 10.1073/pnas.2004489117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwickert TA, et al. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J. Exp. Med. 2011;208:1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shulman Z, et al. T follicular helper cell dynamics in germinal centers. Science. 2013;341:673–677. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, J. H. et al. Modulating the quantity of HIV Env-specific CD4 T cell help promotes rare B cell responses in germinal centers. J. Exp. Med.10.1084/jem.20201254 (2021). [DOI] [PMC free article] [PubMed]
- 15.Woodruff MC, Kim EH, Luo W, Pulendran B. B cell competition for restricted T cell help suppresses rare-epitope responses. Cell Rep. 2018;25:321–327.e323. doi: 10.1016/j.celrep.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivalingam GN, Shepherd AJ. An analysis of B-cell epitope discontinuity. Mol. Immunol. 2012;51:304–309. doi: 10.1016/j.molimm.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sela-Culang I, et al. Using a combined computational-experimental approach to predict antibody-specific B cell epitopes. Structure. 2014;22:646–657. doi: 10.1016/j.str.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brasher NA, Adhikari A, Lloyd AR, Tedla N, Bull RA. Hepatitis C virus epitope immunodominance and B cell repertoire diversity. Viruses. 2021;13:983. doi: 10.3390/v13060983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amitai A, et al. Defining and manipulating B cell immunodominance hierarchies to elicit broadly neutralizing antibody responses against influenza virus. Cell Syst. 2020;11:573–588.e579. doi: 10.1016/j.cels.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachmann MF, Zinkernagel RM. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 1997;15:235–270. doi: 10.1146/annurev.immunol.15.1.235. [DOI] [PubMed] [Google Scholar]
- 22.Nuzzaci M, et al. Cucumber mosaic virus as a presentation system for a double hepatitis C virus-derived epitope. Arch. Virol. 2007;152:915–928. doi: 10.1007/s00705-006-0916-7. [DOI] [PubMed] [Google Scholar]
- 23.Yusibov V, et al. Peptide-based candidate vaccine against respiratory syncytial virus. Vaccine. 2005;23:2261–2265. doi: 10.1016/j.vaccine.2005.01.039. [DOI] [PubMed] [Google Scholar]
- 24.Pushko P, Pumpens P, Grens E. Development of virus-like particle technology from small highly symmetric to large complex virus-like particle structures. Intervirology. 2013;56:141–165. doi: 10.1159/000346773. [DOI] [PubMed] [Google Scholar]
- 25.Roldão A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. Virus-like particles in vaccine development. Expert Rev. Vaccines. 2010;9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 26.Schneemann A, et al. A virus-like particle that elicits cross-reactive antibodies to the conserved stem of influenza virus hemagglutinin. J. Virol. 2012;86:11686–11697. doi: 10.1128/JVI.01694-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skrastina D, et al. Chimeric derivatives of hepatitis B virus core particles carrying major epitopes of the rubella virus E1 glycoprotein. Clin. Vaccine Immunol. 2013;20:1719–1728. doi: 10.1128/CVI.00533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koudelka KJ, Pitek AS, Manchester M, Steinmetz NF. Virus-based nanoparticles as versatile nanomachines. Annu. Rev. Virol. 2015;2:379–401. doi: 10.1146/annurev-virology-100114-055141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan FR, et al. A chimaeric plant virus vaccine protects mice against a bacterial infection. Microbiology. 1999;145:2061–2067. doi: 10.1099/13500872-145-8-2061. [DOI] [PubMed] [Google Scholar]
- 30.Koo M, et al. Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proc. Natl Acad. Sci. USA. 1999;96:7774–7779. doi: 10.1073/pnas.96.14.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rennermalm A, et al. Antibodies against a truncated Staphylococcus aureus fibronectin-binding protein protect against dissemination of infection in the rat. Vaccine. 2001;19:3376–3383. doi: 10.1016/s0264-410x(01)00080-9. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, et al. Expression of foot-and-mouth disease virus epitopes in tobacco by a tobacco mosaic virus-based vector. Vaccine. 2003;21:4390–4398. doi: 10.1016/s0264-410x(03)00428-6. [DOI] [PubMed] [Google Scholar]
- 33.Kanekiyo M, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darricarrère, N. et al. Development of a Pan-H1 Influenza Vaccine. J. Virol.10.1128/jvi.01349-18 (2018). [DOI] [PMC free article] [PubMed]
- 35.Marcandalli J, et al. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell. 2019;176:1420–1431.e1417. doi: 10.1016/j.cell.2019.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson, K. A. et al. A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain. Sci. Immunol.10.1126/sciimmunol.aba6466 (2020). [DOI] [PubMed]
- 37.Antanasijevic A, et al. Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens. PLoS Pathog. 2020;16:e1008665. doi: 10.1371/journal.ppat.1008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouwer PJM, et al. Immunofocusing and enhancing autologous Tier-2 HIV-1 neutralization by displaying Env trimers on two-component protein nanoparticles. NPJ Vaccines. 2021;6:24. doi: 10.1038/s41541-021-00285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgiev IS, et al. Two-component ferritin nanoparticles for multimerization of diverse trimeric antigens. ACS Infect. Dis. 2018;4:788–796. doi: 10.1021/acsinfecdis.7b00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanekiyo M, et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyoglu-Barnum, S. et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature10.1038/s41586-021-03365-x (2021). [DOI] [PMC free article] [PubMed]
- 42.Brouwer PJM, et al. Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection. Cell. 2021;184:1188–1200.e1119. doi: 10.1016/j.cell.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsia Y, et al. Design of a hyperstable 60-subunit protein icosahedron. Nature. 2016;535:136–139. doi: 10.1038/nature18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueda G, et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. eLife. 2020;9:e57659. doi: 10.7554/eLife.57659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokatlian T, et al. Enhancing humoral responses against HIV envelope trimers via nanoparticle delivery with stabilized synthetic liposomes. Sci. Rep. 2018;8:16527. doi: 10.1038/s41598-018-34853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Portnoff, A. D. et al. Influenza hemagglutinin nanoparticle vaccine elicits broadly neutralizing antibodies against structurally distinct domains of H3N2 HA. Vaccines10.3390/vaccines8010099 (2020). [DOI] [PMC free article] [PubMed]
- 48.Schwarz B, et al. Symmetry controlled, genetic presentation of bioactive proteins on the P22 virus-like particle using an external decoration protein. ACS Nano. 2015;9:9134–9147. doi: 10.1021/acsnano.5b03360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S, Gupta K, Shukla S, Sampathkumar S-G, Roy RP. Sortase-click strategy for defined protein conjugation on a heptavalent cyclodextrin scaffold. PLoS ONE. 2019;14:e0217369. doi: 10.1371/journal.pone.0217369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patterson D, et al. Sortase-mediated ligation as a modular approach for the covalent attachment of proteins to the exterior of the bacteriophage P22 virus-like Particle. Bioconjug. Chem. 2017;28:2114–2124. doi: 10.1021/acs.bioconjchem.7b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Q, et al. Sortase A-mediated multi-functionalization of protein nanoparticles. Chem. Commun. 2015;51:12107–12110. doi: 10.1039/c5cc03769g. [DOI] [PubMed] [Google Scholar]
- 52.Mao H, Hart SA, Schink A, Pollok BA. Sortase-mediated protein ligation: a new method for protein engineering. J. Am. Chem. Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 53.Warden-Rothman R, Caturegli I, Popik V, Tsourkas A. Sortase-tag expressed protein ligation: combining protein purification and site-specific bioconjugation into a single step. Anal. Chem. 2013;85:11090–11097. doi: 10.1021/ac402871k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl Acad. Sci. USA. 2012;109:E690. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen AA, et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen AA, et al. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS ONE. 2021;16:e0247963. doi: 10.1371/journal.pone.0247963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walls AC, et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e1317. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan TK, et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021;12:542. doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brouwer PJM, et al. Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle. Nat. Commun. 2019;10:4272. doi: 10.1038/s41467-019-12080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuraoka M, et al. Complex antigens drive permissive clonal selection in germinal centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finney J, Yeh C-H, Kelsoe G, Kuraoka M. Germinal center responses to complex antigens. Immunol. Rev. 2018;284:42–50. doi: 10.1111/imr.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 63.Corbett KS, et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor b cell receptors of broadly neutralizing antibody lineages. mBio. 2019;10:e02810–02818. doi: 10.1128/mBio.02810-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 65.Darricarrère N, et al. Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med. 2021;13:eabe5449. doi: 10.1126/scitranslmed.abe5449. [DOI] [PubMed] [Google Scholar]
- 66.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 67.Boyington JC, et al. Structure-based design of head-only fusion glycoprotein immunogens for respiratory syncytial virus. PLoS ONE. 2016;11:e0159709. doi: 10.1371/journal.pone.0159709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang SC, Liao HY, Zhang JY, Cheng TR, Wong CH. Development of a universal influenza vaccine using hemagglutinin stem protein produced from Pichia pastoris. Virology. 2019;526:125–137. doi: 10.1016/j.virol.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 69.Valkenburg SA, et al. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 2016;6:22666. doi: 10.1038/srep22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutton TC, et al. Protective efficacy of influenza group 2 hemagglutinin stem-fragment immunogen vaccines. NPJ Vaccines. 2017;2:35. doi: 10.1038/s41541-017-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang H, et al. Epitope-focused immunogens against the CD4-binding site of HIV-1 envelope protein induce neutralizing antibodies against auto- and heterologous viruses. J. Biol. Chem. 2018;293:830–846. doi: 10.1074/jbc.M117.816447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Lubbe JEM, et al. Mini-HA is superior to full length hemagglutinin immunization in inducing stem-specific antibodies and protection against group 1 influenza virus challenges in mice. Front. Immunol. 2018;9:2350. doi: 10.3389/fimmu.2018.02350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jardine J, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsouchnikas G, et al. Immunization with immune complexes modulates the fine specificity of antibody responses to a flavivirus antigen. J. Virol. 2015;89:7970. doi: 10.1128/JVI.00938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams JG, Tomer KB, Hioe CE, Zolla-Pazner S, Norris PJ. The antigenic determinants on HIV p24 for CD4+ T cell inhibiting antibodies as determined by limited proteolysis, chemical modification, and mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:1560–1569. doi: 10.1016/j.jasms.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 77.Zarnitsyna VI, et al. Masking of antigenic epitopes by antibodies shapes the humoral immune response to influenza. Philos. Trans. R. Soc. B Biol. Sci. 2015;370:20140248. doi: 10.1098/rstb.2014.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang AF, Enyindah-Asonye G, Hioe CE. Immune complex vaccine strategies to combat hiv-1 and other infectious diseases. Vaccines. 2021;9:112. doi: 10.3390/vaccines9020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Galkin A, et al. HIV-1 gp120–CD4-induced antibody complex elicits CD4 binding site–specific antibody response in mice. J. Immunol. 2020;204:1543. doi: 10.4049/jimmunol.1901051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y, et al. An HIV-1 Env-antibody complex focuses antibody responses to conserved neutralizing epitopes. J. Immunol. 2016;197:3982–3998. doi: 10.4049/jimmunol.1601134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weidenbacher PA, Kim PS. Protect, modify, deprotect (PMD): a strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc. Natl Acad. Sci. USA. 2019;116:9947. doi: 10.1073/pnas.1822062116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leemans A, et al. Characterization of the role of N-glycosylation sites in the respiratory syncytial virus fusion protein in virus replication, syncytium formation and antigenicity. Virus Res. 2019;266:58–68. doi: 10.1016/j.virusres.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 83.Altman, M. O. et al. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. mBio10.1128/mBio.00204-19 (2019). [DOI] [PMC free article] [PubMed]
- 84.Zhao, D. et al. Glycosylation of the hemagglutinin protein of H5N1 influenza virus increases its virulence in mice by exacerbating the host immune response. J. Virol.10.1128/jvi.02215-16 (2017). [DOI] [PMC free article] [PubMed]
- 85.Wu CY, et al. Influenza A surface glycosylation and vaccine design. Proc. Natl Acad. Sci. USA. 2017;114:280–285. doi: 10.1073/pnas.1617174114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin Y, et al. Glycosylation deletion of hemagglutinin head in the H5 subtype avian influenza virus enhances its virulence in mammals by inducing endoplasmic reticulum stress. Transbound. Emerg. Dis. 2020;67:1492–1506. doi: 10.1111/tbed.13481. [DOI] [PubMed] [Google Scholar]
- 87.Kosik I, et al. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog. 2018;14:e1006796. doi: 10.1371/journal.ppat.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watanabe Y, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liang Y, et al. Changes in structure and antigenicity of HIV-1 Env trimers resulting from removal of a conserved CD4 binding site-proximal glycan. J. Virol. 2016;90:9224–9236. doi: 10.1128/JVI.01116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Umotoy J, et al. Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity. 2019;51:141–154.e146. doi: 10.1016/j.immuni.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gristick HB, et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.LaBranche CC, et al. HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies. PLoS Pathog. 2018;14:e1007431. doi: 10.1371/journal.ppat.1007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crooks ET, et al. Glycoengineering HIV-1 Env creates ‘supercharged’ and ‘hybrid’ glycans to increase neutralizing antibody potency, breadth and saturation. PLoS Pathog. 2018;14:e1007024. doi: 10.1371/journal.ppat.1007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang D, Zaia J. Why glycosylation matters in building a better flu vaccine. Mol. Cell Proteom. 2019;18:2348–2358. doi: 10.1074/mcp.R119.001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tokatlian T, et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363:649–654. doi: 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tseng YC, et al. Egg-based influenza split virus vaccine with monoglycosylation induces cross-strain protection against influenza virus infections. Proc. Natl Acad. Sci. USA. 2019;116:4200–4205. doi: 10.1073/pnas.1819197116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim JI, et al. Glycosylation generates an efficacious and immunogenic vaccine against H7N9 influenza virus. PLoS Biol. 2020;18:e3001024. doi: 10.1371/journal.pbio.3001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hariharan V, Kane RS. Glycosylation as a tool for rational vaccine design. Biotechnol. Bioeng. 2020;117:2556–2570. doi: 10.1002/bit.27361. [DOI] [PubMed] [Google Scholar]
- 99.Nuñez IA, Ross TM. Human COBRA 2 vaccine contains two major epitopes that are responsible for eliciting neutralizing antibody responses against heterologous clades of viruses. Vaccine. 2020;38:830–839. doi: 10.1016/j.vaccine.2019.10.097. [DOI] [PubMed] [Google Scholar]
- 100.Lavie M, Hanoulle X, Dubuisson J. Glycan shielding and modulation of hepatitis C virus neutralizing antibodies. Front. Immunol. 2018;9:910–910. doi: 10.3389/fimmu.2018.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watanabe Y, Bowden TA, Wilson IA, Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys. Acta Gen. Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Salpini, R. et al. A Hyper-glycosylation of HBV surface antigen correlates with hbsag-negativity at immunosuppression-driven HBV reactivation in vivo and hinders HBsAg recognition in vitro. Viruses10.3390/v12020251 (2020). [DOI] [PMC free article] [PubMed]
- 103.Bajic G, et al. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe. 2019;25:827–835.e826. doi: 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin HH, et al. Dengue and zika virus domain III-flagellin fusion and glycan-masking E antigen for prime-boost immunization. Theranostics. 2019;9:4811–4826. doi: 10.7150/thno.35919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pierce, B. G. et al. Structure-based design of hepatitis C virus E2 glycoprotein improves serum binding and cross-neutralization. J. Virol.10.1128/jvi.00704-20 (2020). [DOI] [PMC free article] [PubMed]
- 106.Boyoglu-Barnum S, et al. Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat. Commun. 2020;11:791. doi: 10.1038/s41467-020-14579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duan H, et al. Glycan masking focuses immune responses to the HIV-1 CD4-binding site and enhances elicitation of VRC01-class precursor antibodies. Immunity. 2018;49:301–311.e305. doi: 10.1016/j.immuni.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thornlow, D. N. et al. Altering the Immunogenicity of Hemagglutinin Immunogens by Hyperglycosylation and Disulfide Stabilization. Front. Immunol.10.3389/fimmu.2021.737973 (2021). [DOI] [PMC free article] [PubMed]
- 109.Godley L, et al. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 110.Qiao H, et al. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J. Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rawi R, et al. Automated design by structure-based stabilization and consensus repair to achieve prefusion-closed envelope trimers in a wide variety of HIV strains. Cell Rep. 2020;33:108432. doi: 10.1016/j.celrep.2020.108432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rutten L, et al. A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep. 2018;23:584–595. doi: 10.1016/j.celrep.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Joyce MG, et al. Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep. 2017;21:2992–3002. doi: 10.1016/j.celrep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 116.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanders RW, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanders RW, Moore JP. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rey FA, Lok SM. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell. 2018;172:1319–1334. doi: 10.1016/j.cell.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanders RW, Moore JP. Virus vaccines: proteins prefer prolines. Cell Host Microbe. 2021;29:327–333. doi: 10.1016/j.chom.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krarup A, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Battles MB, et al. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat. Commun. 2017;8:1528. doi: 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hastie KM, et al. Structural basis for antibody-mediated neutralization of Lassa virus. Sci.ence. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rutten L, et al. Structure-based design of prefusion-stabilized filovirus glycoprotein trimers. Cell Rep. 2020;30:4540–4550.e4543. doi: 10.1016/j.celrep.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pallesen J, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Walls AC, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e1015. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hsieh CL, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dubrovskaya V, et al. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity. 2019;51:915–929.e917. doi: 10.1016/j.immuni.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu WC, Jan JT, Huang YJ, Chen TH, Wu SC. Unmasking stem-specific neutralizing epitopes by abolishing N-linked glycosylation sites of influenza virus hemagglutinin proteins for vaccine design. J. Virol. 2016;90:8496–8508. doi: 10.1128/JVI.00880-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ringe, R. P. et al. Closing and opening holes in the glycan shield of HIV-1 envelope glycoprotein sosip trimers can redirect the neutralizing antibody response to the newly unmasked epitopes. J. Virol.10.1128/jvi.01656-18 (2019). [DOI] [PMC free article] [PubMed]
- 131.Huang, Y., Owino, S. O., Crevar, C. J., Carter, D. M. & Ross, T. M. N-linked glycans and K147 residue on hemagglutinin synergize to elicit broadly reactive H1N1 influenza virus antibodies. J. Virol.10.1128/jvi.01432-19 (2020). [DOI] [PMC free article] [PubMed]
- 132.Giles BM, Ross TM. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine. 2011;29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giles BM, Bissel SJ, Dealmeida DR, Wiley CA, Ross TM. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin. Vaccin. Immunol. 2012;19:128–139. doi: 10.1128/CVI.05533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Giles BM, et al. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J. Infect. Dis. 2012;205:1562–1570. doi: 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Uno, N. & Ross, T. M. Universal dengue vaccine elicits neutralizing antibodies against strains from all four dengue virus serotypes. J. Virol.10.1128/jvi.00658-20 (2021). [DOI] [PMC free article] [PubMed]
- 136.Crevar CJ, Carter DM, Lee KYJ, Ross TM. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccin. Immunother. 2015;11:572–583. doi: 10.1080/21645515.2015.1012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carter DM, et al. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J. Virol. 2016;90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wong, T. M. et al. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J. Virol.10.1128/jvi.01581-17 (2017). [DOI] [PMC free article] [PubMed]
- 139.Sautto GA, et al. Elicitation of broadly protective antibodies following infection with influenza viruses expressing H1N1 computationally optimized broadly reactive hemagglutinin antigens. Immunohorizons. 2018;2:226–237. doi: 10.4049/immunohorizons.1800044. [DOI] [PubMed] [Google Scholar]
- 140.Fadlallah, G. M. et al. Vaccination with consensus H7 elicits broadly reactive and protective antibodies against eurasian and north american lineage H7 viruses. Vaccines10.3390/vaccines8010143 (2020). [DOI] [PMC free article] [PubMed]
- 141.Reneer, Z. B., Jamieson, P. J., Skarlupka, A. L., Huang, Y. & Ross, T. M. Computationally optimized broadly reactive h2 ha influenza vaccines elicited broadly cross-reactive antibodies and protected mice from viral challenges. J. Virol.10.1128/jvi.01526-20 (2020). [DOI] [PMC free article] [PubMed]
- 142.Skarlupka, A. L., Reneer, Z. B., Abreu, R. B., Ross, T. M. & Sautto, G. A. An Influenza virus hemagglutinin computationally optimized broadly reactive antigen elicits antibodies endowed with group 1 heterosubtypic breadth against swine influenza viruses. J. Virol.10.1128/jvi.02061-19 (2020). [DOI] [PMC free article] [PubMed]
- 143.Reneer ZB, Skarlupka AL, Jamieson PJ, Ross TM. Broadly reactive H2 hemagglutinin vaccines elicit cross-reactive antibodies in ferrets preimmune to seasonal influenza a viruses. mSphere. 2021;6:e00052–00021. doi: 10.1128/mSphere.00052-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carter DM, et al. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J. Virol. 2017;91:e01283–01217. doi: 10.1128/JVI.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Allen JD, Jang H, DiNapoli J, Kleanthous H, Ross TM. Elicitation of protective antibodies against 20 years of future H3N2 cocirculating influenza virus variants in ferrets preimmune to historical H3N2 influenza viruses. J. Virol. 2019;93:e00946–00918. doi: 10.1128/JVI.00946-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Skarlupka AL, et al. Computationally optimized broadly reactive vaccine based upon swine H1N1 influenza hemagglutinin sequences protects against both swine and human isolated viruses. Hum. Vaccin Immunother. 2019;15:2013–2029. doi: 10.1080/21645515.2019.1653743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sautto GA, et al. A computationally optimized broadly reactive antigen subtype-specific influenza vaccine strategy elicits unique potent broadly neutralizing antibodies against hemagglutinin. J. Immunol. 2020;204:375–385. doi: 10.4049/jimmunol.1900379. [DOI] [PubMed] [Google Scholar]
- 148.Allen JD, Ross TM. Next generation methodology for updating HA vaccines against emerging human seasonal influenza A(H3N2) viruses. Sci. Rep. 2021;11:4554. doi: 10.1038/s41598-020-79590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bar-Peled Y, et al. Structural and antigenic characterization of a computationally-optimized H5 hemagglutinin influenza vaccine. Vaccine. 2019;37:6022–6029. doi: 10.1016/j.vaccine.2019.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nakamura G, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 152.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 153.Wyrzucki A, et al. Alternative recognition of the conserved stem epitope in influenza A virus hemagglutinin by a VH3-30-encoded heterosubtypic antibody. J. Virol. 2014;88:7083–7092. doi: 10.1128/JVI.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schmidt AG, et al. Viral receptor-binding site antibodies with diverse germline origins. Cell. 2015;161:1026–1034. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Harshbarger WD, et al. Unique structural solution from a V(H)3-30 antibody targeting the hemagglutinin stem of influenza A viruses. Nat. Commun. 2021;12:559. doi: 10.1038/s41467-020-20879-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Investig. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Robbiani DF, et al. Recurrent potent human neutralizing antibodies to zika virus in Brazil and Mexico. Cell. 2017;169:597–609.e511. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Esswein SR, et al. Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody. Proc. Natl Acad. Sci. USA. 2020;117:9865–9875. doi: 10.1073/pnas.1919269117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yuan M, et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wu NC, et al. An alternative binding mode of IGHV3-53 antibodies to the SARS-CoV-2 receptor binding domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen F, et al. Functional convergence of a germline-encoded neutralizing antibody response in rhesus macaques immunized with HCV envelope glycoproteins. Immunity. 2021;54:781–796.e784. doi: 10.1016/j.immuni.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sangesland M, et al. Germline-encoded affinity for cognate antigen enables vaccine amplification of a human broadly neutralizing response against influenza virus. Immunity. 2019;51:735–749.e738. doi: 10.1016/j.immuni.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jardine JG, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bonsignori M, et al. Inference of the HIV-1 VRC01 antibody lineage unmutated common ancestor reveals alternative pathways to overcome a key glycan barrier. Immunity. 2018;49:1162–1174.e1168. doi: 10.1016/j.immuni.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jardine JG, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Steichen JM, et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.LaBranche CC, et al. Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancestor of CH235 lineage CD4bs broadly neutralizing antibodies. PLoS Pathog. 2019;15:e1008026. doi: 10.1371/journal.ppat.1008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Corbett, K. S. et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio10.1128/mBio.02810-18 (2019). [DOI] [PMC free article] [PubMed]
- 171.Steichen, J. M. et al. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science10.1126/science.aax4380 (2019). [DOI] [PMC free article] [PubMed]
- 172.Lin YR, et al. HIV-1 VRC01 germline-targeting immunogens select distinct epitope-specific B cell receptors. Immunity. 2020;53:840–851.e846. doi: 10.1016/j.immuni.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Scharf, L. et al. Structural basis for germline antibody recognition of HIV-1 immunogens. Elife10.7554/eLife.13783 (2016). [DOI] [PMC free article] [PubMed]
- 174.Huang D, et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun. 2020;11:5850. doi: 10.1038/s41467-020-19650-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bajic G, et al. Structure-guided molecular grafting of a complex broadly neutralizing viral epitope. ACS Infect. Dis. 2020;6:1182–1191. doi: 10.1021/acsinfecdis.0c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hai R, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J. Virol. 2012;86:5774. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Choi A, et al. Chimeric hemagglutinin-based influenza virus vaccines induce protective stalk-specific humoral immunity and cellular responses in mice. Immunohorizons. 2019;3:133–148. doi: 10.4049/immunohorizons.1900022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Asthagiri Arunkumar G, et al. Vaccination with viral vectors expressing NP, M1 and chimeric hemagglutinin induces broad protection against influenza virus challenge in mice. Vaccine. 2019;37:5567–5577. doi: 10.1016/j.vaccine.2019.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.McMahon M, et al. Vaccination With Viral Vectors Expressing Chimeric Hemagglutinin, NP and M1 Antigens Protects Ferrets Against Influenza Virus Challenge. Front Immunol. 2019;10:2005. doi: 10.3389/fimmu.2019.02005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Isakova-Sivak I, et al. Broadly protective anti-hemagglutinin stalk antibodies induced by live attenuated influenza vaccine expressing chimeric hemagglutinin. Virology. 2018;518:313–323. doi: 10.1016/j.virol.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 181.Nachbagauer, R., Krammer, F. & Albrecht, R. A. A live-attenuated prime, inactivated boost vaccination strategy with chimeric hemagglutinin-based universal influenza virus vaccines provides protection in ferrets: a confirmatory study. Vaccines10.3390/vaccines6030047 (2018). [DOI] [PMC free article] [PubMed]
- 182.Liu, W. C. et al. Chimeric hemagglutinin-based live-attenuated vaccines confer durable protective immunity against influenza A Viruses in a preclinical ferret model. Vaccines10.3390/vaccines9010040 (2021). [DOI] [PMC free article] [PubMed]
- 183.Liu WC, et al. Sequential immunization with live-attenuated chimeric hemagglutinin-based vaccines confers heterosubtypic immunity against influenza A viruses in a preclinical ferret model. Front. Immunol. 2019;10:756. doi: 10.3389/fimmu.2019.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nachbagauer R, et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines. 2017;2:26. doi: 10.1038/s41541-017-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bernstein DI, et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 2020;20:80–91. doi: 10.1016/S1473-3099(19)30393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martinez David R, et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Frei, J. C. et al. Engineered dengue virus domain III proteins elicit cross-neutralizing antibody responses in mice. J. Virol.10.1128/jvi.01023-18 (2018). [DOI] [PMC free article] [PubMed]
- 189.Remmel JL, et al. Combinatorial resurfacing of dengue envelope protein domain iii antigens selectively ablates epitopes associated with serotype-specific or infection-enhancing antibody responses. ACS Comb. Sci. 2020;22:446–456. doi: 10.1021/acscombsci.0c00073. [DOI] [PubMed] [Google Scholar]
- 190.Sesterhenn F, et al. Boosting subdominant neutralizing antibody responses with a computationally designed epitope-focused immunogen. PLoS Biol. 2019;17:e3000164. doi: 10.1371/journal.pbio.3000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sesterhenn, F. et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science10.1126/science.aay5051 (2020). [DOI] [PMC free article] [PubMed]
- 192.Zost SJ, et al. Identification of antibodies targeting the H3N2 hemagglutinin receptor binding site following vaccination of humans. Cell Rep. 2019;29:4460–4470.e4468. doi: 10.1016/j.celrep.2019.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Francis T. On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 1960;104:572–578. [Google Scholar]
- 194.Vatti A, et al. Original antigenic sin: a comprehensive review. J. Autoimmun. 2017;83:12–21. doi: 10.1016/j.jaut.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 195.Schmidt AG, et al. Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 2015;13:2842–2850. doi: 10.1016/j.celrep.2015.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dugan, H. L. et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med.10.1126/scitranslmed.abd3601 (2020). [DOI] [PMC free article] [PubMed]
- 197.Arevalo CP, et al. Original antigenic sin priming of influenza virus hemagglutinin stalk antibodies. Proc. Natl Acad. Sci. USA. 2020;117:17221–17227. doi: 10.1073/pnas.1920321117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nachbagauer R, et al. Defining the antibody cross-reactome directed against the influenza virus surface glycoproteins. Nat. Immunol. 2017;18:464–473. doi: 10.1038/ni.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Abreu, R. B., Kirchenbaum, G. A., Clutter, E. F., Sautto, G. A. & Ross, T. M. Preexisting subtype immunodominance shapes memory B cell recall response to influenza vaccination. JCI Insight10.1172/jci.insight.132155 (2020). [DOI] [PMC free article] [PubMed]
- 200.Jegaskanda, S. et al. Hemagglutinin head-specific responses dominate over stem-specific responses following prime boost with mismatched vaccines. JCI Insight10.1172/jci.insight.129035 (2019). [DOI] [PMC free article] [PubMed]
- 201.Joyce MG, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell. 2016;166:609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nachbagauer R, et al. Pandemic influenza virus vaccines boost hemagglutinin stalk-specific antibody responses in primed adult and pediatric cohorts. NPJ Vaccines. 2019;4:51. doi: 10.1038/s41541-019-0147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kirchenbaum GA, Carter DM, Ross TM. Sequential infection in ferrets with antigenically distinct seasonal H1N1 influenza viruses boosts hemagglutinin stalk-specific antibodies. J. Virol. 2016;90:1116–1128. doi: 10.1128/JVI.02372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Carter DM, et al. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J. Virol. 2013;87:1400–1410. doi: 10.1128/JVI.02257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Andrews SF, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sprenger KG, Louveau JE, Murugan PM, Chakraborty AK. Optimizing immunization protocols to elicit broadly neutralizing antibodies. Proc. Natl Acad. Sci. USA. 2020;117:20077. doi: 10.1073/pnas.1919329117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shaffer JS, Moore PL, Kardar M, Chakraborty AK. Optimal immunization cocktails can promote induction of broadly neutralizing Abs against highly mutable pathogens. Proc. Natl Acad. Sci. USA. 2016;113:E7039. doi: 10.1073/pnas.1614940113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Conti S, et al. Design of immunogens to elicit broadly neutralizing antibodies against HIV targeting the CD4 binding site. Proc. Natl Acad. Sci. USA. 2021;118:e2018338118. doi: 10.1073/pnas.2018338118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Louie RHY, Kaczorowski KJ, Barton JP, Chakraborty AK, McKay MR. Fitness landscape of the human immunodeficiency virus envelope protein that is targeted by antibodies. Proc. Natl Acad. Sci. USA. 2018;115:E564. doi: 10.1073/pnas.1717765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Roubidoux, E. K. et al. Mutations in the hemagglutinin stalk domain do not permit escape from a protective, stalk-based vaccine-induced immune response in the mouse model. mBio10.1128/mBio.03617-20 (2021). [DOI] [PMC free article] [PubMed]
- 211.Wu NC, et al. Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses. Science. 2020;368:1335. doi: 10.1126/science.aaz5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Doud MB, Lee JM, Bloom JD. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 2018;9:1386. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Raymond DD, et al. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl Acad. Sci. USA. 2018;115:168–173. doi: 10.1073/pnas.1715471115. [DOI] [PMC free article] [PubMed] [Google Scholar]