Abstract
MYB proteins are highly conserved DNA-binding domains (DBD) and mutations in MYB oncoproteins have been reported to cause aberrant and augmented cancer progression. Identification of MYB molecular biomarkers predictive of cancer progression can be used for improving cancer management. To address this, a biomarker discovery pipeline was employed in investigating deleterious non-synonymous single nucleotide polymorphisms (nsSNPs) in predicting damaging and potential alterations on the properties of proteins. The nsSNP of the MYB family; MYB, MYBL1, and MYBL2 was extracted from the NCBI database. Five in silico tools (PROVEAN, SIFT, PolyPhen-2, SNPs&GO and PhD-SNP) were utilized to investigate the outcomes of nsSNPs. A total of 45 nsSNPs were predicted as high-risk and damaging, and were subjected to PMut and I-Mutant 2.0 for protein stability analysis. This resulted in 32 nsSNPs with decreased stability with a DDG score lower than − 0.5, indicating damaging effect. G111S, N183S, G122S, and S178C located within the helix-turn-helix (HTH) domain were predicted to be conserved, further posttranslational modifications and 3-D protein analysis indicated these nsSNPs to shift DNA-binding specificity of the protein thus altering the protein function. Findings from this study would help in the field of pharmacogenomic and cancer therapy towards better intervention and management of cancer.
Subject terms: Bioinformatics, Cancer genomics, Cancer, Computational biology and bioinformatics, Genetics, Pathogenesis
Introduction
MYB oncoproteins; MYB, MYBL1, and MYBL2 plays important roles in the modulation of cell cycle, and dysregulation in these genes have been implicated with abberant behaviours of the tumour cells. The key functions of MYB proteins are mainly in cell growth and differentiation, thereof mutations within these genes are predicted to be a potential source for oncogenesis1. Numerous studies have reported mutation of MYB proteins toward pathogenesis of human cancers, especially acute lymphoblastic leukaemia (ALL)2, paediatric low-grade gliomas3, cancers of the gastrointestinal tract4–6 and breast cancer7.Growing evidences of MYB oncoproteins and cancers necessitates an in-depth understanding at molecular level in unravelling its pathogenesis towards cancer.
The use of computational predictors to identify damaging non-synonymous single nucleotide polymorphisms (nsSNPs) towards understanding disease-causing role offers a time- and cost-effective alternative8. nsSNPs are changes in an amino acid that could disrupt the structure and stability of protein thus potentially increasing susceptibility towards certain disease9,10. Evaluating the influence of nsSNPs on the protein is important in determining its effect towards characterisation of the disease11. Recent studies employing computational approaches have also revealed the effectiveness of nsSNPs in understanding the molecular mechanisms of numerous diseases12–14.
Considering the pathological role of MYB oncoproteins towards cancer, functional and structural analysis of MYB oncoproteins still remains vague. Therefore, this study sets to examine the role of nsSNPs of MYB, MYBL1, and MYBL2 genes through bioinformatics tools in understanding its pathogenesis toward cancer. This approach enables the differentiation of pathogenic mutations from an abundance of variations, narrowing down to highly significant variant for further investigation using laboratory validation15. In this study, MYB oncoproteins were subjected to multi-level functional and structural analysis in determining their pathogenicity. In brief, this involves (1) nsSNP analysis, where sequence evolutionary conservation information and structure-based information to determine damaging nsSNPs, (2) Prediction of protein stability, which evaluates the energetics of the folded and unfolded state of proteins in determining if the protein is stable, and (3) prediction of post-translational modifications. Each of these tools employs a different machine-learning algorithm to predict the outcomes.
Results
Retrieval of nsSNPs in MYB family genes
NCBI dbSNP was used to retrieve the SNPs of MYB oncoproteins. A total of 51,862 SNPs were extracted from the NCBI database of which; 13,632 of MYB, 17,770 of MYBL1, and 20,460 of MYBL2. Out of these SNPs, 1503 were nsSNPs.
Identification of deleterious nsSNPs in MYB family genes
The nsSNPs were then subjected to five different tools (PROVEAN, SIFT, Polyphen2, SNPs&GO and PhD-SNP) that has different prediction algorithms to identify nsSNP with significant deleterious effects which could affect the biological structure and the function of MYB proteins. Forty-eight nsSNPs were identified as “pathogenic” or “damaging” by all tools, hence classified at “high-risk” (Table 1).
Table 1.
High risk nsSNPs identified in MYB family genes by in silico tools.
| SNP ID | AA change | PROVEAN | SIFT | PolyPhen-2 | SNPs&GO | PhD-SNP | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Preda | Sc | Predb | Sc | Effectc | Sc | Predd | RI | Prede | RI | ||
| MYB | |||||||||||
| rs1302072057 | R73L | Del | − 3.80 | Dmg | 0 | Pro.dmg | 1 | Disease | 4 | Disease | 7 |
| rs1302072057 | R73Q | Del | − 6.65 | Dmg | 0 | Pro.dmg | 1 | Disease | 5 | Disease | 8 |
| rs866246271 | C78Y | Del | − 10.45 | Dmg | 0 | Pro.dmg | 0.999 | Disease | 7 | Disease | 9 |
| rs1246761830 | K84E | Del | − 3.80 | Dmg | 0.001 | Pro.dmg | 0.998 | Disease | 5 | Disease | 4 |
| rs1231582413 | P94R | Del | − 8.55 | Dmg | 0.001 | Pos.dmg | 0.608 | Disease | 3 | Disease | 1 |
| rs1776412940 | W95L | Del | − 12.35 | Dmg | 0 | Pro.dmg | 1 | Disease | 4 | Disease | 7 |
| rs1361650612 | G111S | Del | − 5.69 | Dmg | 0 | Pro.dmg | 1 | Disease | 5 | Disease | 7 |
| rs1316378738 | A158E | Del | − 4.74 | Dmg | 0 | Pos.dmg | 0.577 | Disease | 7 | Disease | 6 |
| rs1583273308 | R176Q | Del | − 3.80 | Dmg | 0 | Pro.dmg | 1 | Disease | 6 | Disease | 8 |
| rs1179275735 | N183S | Del | − 4.58 | Dmg | 0 | Pro.dmg | 1 | Disease | 7 | Disease | 5 |
| rs1335964521 | R191Q | Del | − 3.75 | Dmg | 0 | Pro.dmg | 1 | Disease | 2 | Disease | 5 |
| rs1444007668 | K192E | Del | − 3.75 | Dmg | 0 | Pro.dmg | 0.999 | Disease | 7 | Disease | 6 |
| rs1777011832 | D286Y | Del | − 6.18 | Dmg | 0 | Pro.dmg | 1 | Disease | 4 | Disease | 7 |
| rs1247338239 | W406C | Del | − 5.30 | Dmg | 0.001 | Pro.dmg | 0.985 | Disease | 0 | Disease | 5 |
| rs1247579811 | P574S | Del | − 6.02 | Dmg | 0 | Pro.dmg | 1 | Disease | 0 | Disease | 5 |
| rs775717051 | A594P | Del | − 3.37 | Dmg | 0.001 | Pro.dmg | 1 | Disease | 1 | Disease | 6 |
| rs756830286 | G603S | Del | − 4.53 | Dmg | 0.007 | Pro.dmg | 1 | Disease | 0 | Disease | 5 |
| MYBL1 | |||||||||||
| rs1367098628 | R68C | Del | − 7.60 | Dmg | 0 | Pro.dmg | 1 | Disease | 3 | Disease | 7 |
| rs767355502 | R68H | Del | − 4.75 | Dmg | 0 | Pro.dmg | 0.996 | Disease | 1 | Disease | 6 |
| rs1472109411 | P83T | Del | − 7.60 | Dmg | 0 | Pos.dmg | 0.811 | Disease | 3 | Disease | 3 |
| rs766676175 | G122S | Del | − 5.69 | Dmg | 0.002 | Pro.dmg | 0.975 | Disease | 3 | Disease | 5 |
| rs768073245 | R156W | Del | − 6.55 | Dmg | 0.002 | Pro.dmg | 1 | Disease | 2 | Disease | 5 |
| rs1299187617 | R160H | Del | − 4.74 | Dmg | 0 | Pro.dmg | 0.990 | Disease | 2 | Disease | 5 |
| rs866260709 | S175Y | Del | − 3.62 | Dmg | 0 | Pro.dmg | 1 | Disease | 0 | Disease | 2 |
| rs1281804000 | R185Q | Del | − 3.30 | Dmg | 0.003 | Pro.dmg | 0.978 | Disease | 0 | Disease | 3 |
| rs1809689475 | E265K | Del | − 3.34 | Dmg | 0.005 | Pro.dmg | 0.957 | Disease | 2 | Disease | 6 |
| rs1225867137 | M276T | Del | − 4.79 | Dmg | 0.006 | Pro.dmg | 0.992 | Disease | 2 | Disease | 6 |
| rs1808814297 | P512R | Del | − 6.44 | Dmg | 0 | Pro.dmg | 1 | Disease | 1 | Disease | 6 |
| rs1361362325 | C514W | Del | − 5.98 | Dmg | 0.002 | Pro.dmg | 1 | Disease | 1 | Disease | 6 |
| rs770108773 | A562E | Del | − 3.54 | Dmg | 0.001 | Pro.dmg | 1 | Disease | 0 | Disease | 6 |
| rs777095803 | A565P | Del | − 2.90 | Dmg | 0.003 | Pro.dmg | 1 | Disease | 1 | Disease | 7 |
| rs777150665 | G596R | Del | − 3.07 | Dmg | 0.001 | Pro.dmg | 1 | Disease | 0 | Disease | 5 |
| rs1281394929 | G721E | Del | − 5.73 | Dmg | 0.007 | Pro.dmg | 1 | Disease | 5 | Disease | 6 |
| MYBL2 | |||||||||||
| rs748449655 | L57R | Del | − 4.37 | Dmg | 0.001 | Pro.dmg | 0.999 | Disease | 8 | Disease | 2 |
| rs1228232756 | R64C | Del | − 6.64 | Dmg | 0 | Pro.dmg | 1 | Disease | 8 | Disease | 5 |
| rs867195152 | G102D | Del | − 6.10 | Dmg | 0 | Pro.dmg | 0.995 | Disease | 8 | Disease | 6 |
| rs1164247754 | R116Q | Del | − 3.35 | Dmg | 0.022 | Pro.dmg | 1 | Disease | 8 | Disease | 6 |
| rs1323182096 | R124C | Del | − 7.06 | Dmg | 0 | Pro.dmg | 1 | Disease | 8 | Disease | 7 |
| rs1300383239 | N127D | Del | − 4.52 | Dmg | 0.006 | Pro.dmg | 0.971 | Disease | 2 | Disease | 4 |
| rs1295676923 | G166V | Del | − 8.00 | Dmg | 0 | Pos.dmg | 0.946 | Disease | 8 | Disease | 7 |
| rs968286439 | R167K | Del | − 2.63 | Dmg | 0 | Pro.dmg | 0.999 | Disease | 7 | Disease | 5 |
| rs1271670254 | D169G | Del | − 5.87 | Dmg | 0 | Pro.dmg | 1 | Disease | 7 | Disease | 7 |
| rs1438994955 | S178C | Del | − 4.01 | Dmg | 0 | Pro.dmg | 1 | Disease | 3 | Disease | 3 |
| rs781229138 | G188C | Del | − 6.21 | Dmg | 0.003 | Pro.dmg | 0.966 | Disease | 6 | Disease | 5 |
| rs1171631148 | E552V | Del | − 5.52 | Dmg | 0.001 | Pro.dmg | 1 | Disease | 7 | Disease | 2 |
| rs776972688 | G530R | Del | − 5.97 | Dmg | 0.001 | Pro.dmg | 1 | Disease | 7 | Disease | 2 |
| rs779332836 | G669R | Del | − 6.13 | Dmg | 0 | Pro.dmg | 1 | Disease | 6 | Disease | 3 |
| rs776117094 | R682W | Del | − 5.78 | Dmg | 0 | Pro.dmg | 1 | Disease | 5 | Disease | 1 |
AA amino acid, Pred prediction, TI tolerance index, Sc score, Del deleterious, Dmg damaging, Pro.dmg probably damaging, Pos.dmg possibly damaging, RI reliability index.
aPROVEAN: Del (Sc < − 2.5).
bSIFT: Dmg (Sc ≤ 0.05).
cPolyPhen-2: Pos.dmg (0.453 ≤ Sc ≤ 0.956), Pro.dmg (0.957 ≤ Sc ≤ 1.0).
dSNPs&GO: Disease (Probability > 0.5).
ePhD-SNP: Disease (Probability > 0.5).
Verification of high risk nsSNPs by PMut
The selected damaging nsSNPS were then submitted to PMut server to determine the probability score and the status of prediction of the resultant protein due to mutations. Table 2 shows the prediction scores and statuses. All MYBL2 nsSNPs were predicted as high-risk, whereas 16 nsSNPs and 14 nsSNPs from MYB and MYBL1 genes were also identified as “disease”. The “disease” status indicates that the mutated proteins are predicted to be pathogenic.
Table 2.
Predictions of high risk nsSNPs in MYB oncoproteins by PMut, I-Mutant 2.0, and ConSurf.
| Protein | nsSNP ID | Mutation | PMut | I-Mutant 2.0 | ConSurf | ||||
|---|---|---|---|---|---|---|---|---|---|
| Score and percentage | Predictiona | Stability | RI | DDG (kcal/mol) | Conservation scoreb | Prediction | |||
| MYB | rs1302072057 | R73L | 0.86 (91%) | Disease | Decrease | 9 | − 0.90 | 9 | Highly conserved and exposed (f) |
| rs1302072057 | R73Q | 0.73 (87%) | Disease | Decrease | 9 | − 1.38 | 9 | Highly conserved and exposed (f) | |
| rs866246271 | C78Y | 0.79 (89%) | Disease | Decrease | 2 | − 0.07 | 9 | Highly conserved and buried (s) | |
| rs1246761830 | K84E | 0.52 (79%) | Disease | Decrease | 1 | − 0.30 | 9 | Highly conserved and exposed (f) | |
| rs1231582413 | P94R | 0.72 (86%) | Disease | Decrease | 7 | − 0.42 | 9 | Highly conserved and exposed (f) | |
| rs1776412940 | W95L | 0.86 (91%) | Disease | Decrease | 7 | − 0.99 | 9 | Highly conserved and exposed (f) | |
| rs1361650612 | G111S | 0.80 (89%) | Disease | Decrease | 9 | − 1.03 | 9 | Highly conserved and exposed (f) | |
| rs1316378738 | A158E | 0.82 (90%) | Disease | Decrease | 7 | − 1.28 | 9 | Highly conserved and buried (s) | |
| rs1583273308 | R176Q | 0.86 (91%) | Disease | Decrease | 8 | − 0.51 | 9 | Highly conserved and exposed (f) | |
| rs1179275735 | N183S | 0.82 (90%) | Disease | Decrease | 4 | − 0.04 | 9 | Highly conserved and exposed (f) | |
| rs1335964521 | R191Q | 0.77 (88%) | Disease | Decrease | 9 | − 1.13 | 9 | Highly conserved and exposed (f) | |
| rs1444007668 | K192E | 0.55 (80%) | Disease | Decrease | 5 | − 0.95 | 9 | Highly conserved and exposed (f) | |
| rs1777011832 | D286Y | 0.64 (84%) | Disease | Increase | 2 | 0.11 | 2 | Exposed | |
| rs1247338239 | W406C | 0.65 (84%) | Disease | Decrease | 6 | − 1.49 | 7 | Buried | |
| rs1247579811 | P574S | 0.63 (84%) | Disease | Decrease | 9 | − 1.01 | 9 | Highly conserved and exposed (f) | |
| rs775717051 | A594P | 0.65 (84%) | Disease | Increase | 2 | − 1.01 | 9 | Highly conserved and buried (s) | |
| rs756830286 | G603S | 0.50 (82%) | Neutral | Decrease | 6 | − 1.52 | 9 | Highly conserved and exposed (f) | |
| MYBL1 | rs767355502 | R68C | 0.39 (86%) | Neutral | Decrease | 3 | − 0.73 | 9 | Highly conserved and exposed (f) |
| rs767355502 | R68H | 0.86 (91%) | Disease | Decrease | 7 | − 1.31 | 9 | Highly conserved and exposed (f) | |
| rs1472109411 | P83T | 0.81 (89%) | Disease | Decrease | 4 | 0.08 | 9 | Highly conserved and exposed (f) | |
| rs766676175 | G122S | 0.76 (88%) | Disease | Decrease | 7 | − 0.52 | 9 | Highly conserved and exposed (f) | |
| rs768073245 | R156W | 0.82 (90%) | Disease | Decrease | 7 | − 1.04 | 9 | Highly conserved and exposed (f) | |
| rs1299187617 | R160H | 0.77 (88%) | Disease | Decrease | 9 | − 1.17 | 9 | Highly conserved and exposed (f) | |
| rs866260709 | S175Y | 0.76 (88%) | Disease | Increase | 2 | − 0.64 | 9 | Highly conserved and buried (s) | |
| rs1281804000 | R185Q | 0.67 (85%) | Disease | Decrease | 9 | − 1.44 | 4 | Highly conserved and exposed (f) | |
| rs1809689475 | E265K | 0.63 (83%) | Disease | Decrease | 5 | − 0.38 | 6 | Exposed | |
| rs1225867137 | M276T | 0.32 (89%) | Neutral | Decrease | 5 | − 0.62 | 9 | Exposed | |
| rs1808814297 | P512R | 0.86 (91%) | Disease | Decrease | 9 | − 1.25 | 6 | Highly conserved and exposed (f) | |
| rs1361362325 | C514W | 0.63 (84%) | Disease | Decrease | 5 | − 1.06 | 9 | Buried | |
| rs770108773 | A562E | 0.78 (88%) | Disease | Decrease | 5 | − 0.76 | 9 | Highly conserved and buried (s) | |
| rs777095803 | A565P | 0.53 (80%) | Disease | Increase | 0 | − 1.85 | 8 | Highly conserved and exposed (f) | |
| rs777150665 | G596R | 0.53 (80%) | Disease | Decrease | 7 | − 1.87 | 9 | Highly conserved and exposed (f) | |
| rs1281394929 | G721E | 0.82 (90%) | Disease | Increase | 2 | − 0.27 | 9 | Highly conserved and exposed (f) | |
| MYBL2 | rs748449655 | L57R | 0.66 (85%) | Disease | Decrease | 6 | − 0.76 | 9 | Highly conserved and buried (s) |
| rs1228232756 | R64C | 0.78 (88%) | Disease | Decrease | 4 | − 0.66 | 9 | Highly conserved and exposed (f) | |
| rs867195152 | G102D | 0.86 (91%) | Disease | Decrease | 5 | − 1.45 | 9 | Highly conserved and exposed (f | |
| rs1164247754 | R116Q | 0.88 (92%) | Disease | Decrease | 7 | 0.13 | 9 | Highly conserved and exposed (f) | |
| rs1323182096 | R124C | 0.89 (92%) | Disease | Decrease | 6 | 0.41 | 9 | Highly conserved and exposed (f) | |
| rs1300383239 | N127D | 0.85 (91%) | Disease | Decrease | 6 | − 0.73 | 9 | Highly conserved and exposed (f) | |
| rs1295676923 | G166V | 0.88 (92%) | Disease | Increase | 0 | − 0.89 | 9 | Highly conserved and exposed (f) | |
| rs968286439 | R167K | 0.87 (91%) | Disease | Decrease | 9 | − 0.98 | 9 | Highly conserved and exposed (f) | |
| rs1271670254 | D169G | 0.79 (89%) | Disease | Decrease | 6 | − 1.04 | 9 | Highly conserved and exposed (f) | |
| rs1438994955 | S178C | 0.53 (80%) | Disease | Decrease | 2 | − 1.33 | 9 | Highly conserved and exposed (f) | |
| rs781229138 | G188C | 0.80 (89%) | Disease | Decrease | 8 | − 2.28 | 5 | Exposed | |
| rs1171631148 | G530R | 0.76 (88%) | Disease | Decrease | 4 | − 1.69 | 9 | Highly conserved and exposed (f) | |
| rs776972688 | E552V | 0.71 (86%) | Disease | Increase | 2 | 0.08 | 9 | Highly conserved and exposed (f) | |
| rs779332836 | G669R | 0.82 (90%) | Disease | Decrease | 7 | − 1.24 | 9 | Highly conserved and exposed (f) | |
| rs776117094 | R682W | 0.82 (90%) | Disease | Decrease | 5 | 0.65 | 9 | Highly conserved and exposed (f) | |
RI reliability index, DDG free energy change value, (f) predicted functional residue (highly conserved and exposed), (s) predicted structural residue (highly conserved and buried).
aPMut: Disease (Score > 0.5), Neutral (Score ≤ 0.5).
bConSurf: highly variable (1), highly conserved (9).
Determination of protein structural stability by I-Mutant 2.0
The structural stability of resultant proteins was predicted by I-Mutant 2.0. The output of I-Mutant 2.0 was expressed in free energy change value (DDG) and reliability index (RI). In total, 41 nsSNPs were confirmed to cause decrease in stability to the resultant proteins, however only 32 nsSNPs were predicted to have a DDG value < − 0.5, indicating its greater impact towards the proteins.
Evolutionary conservation analysis by ConSurf
The evolutionary conservation was determined through subjecting the mutated protein sequences to the ConSurf web server. A total of thirty-six nsSNPs of MYB family genes were identified as functional, highly conserved with exposed amino acid residues, whereas six nsSNPs were predicted as structural, highly conserved and buried. On the contrary, a total of four nsSNPs were predicted to be exposed but not functional, while two nsSNPs were predicted as buried and not functional (Table 2).
The R73L, R73Q, K84E, P94R, W95L, G111S, R176Q, N183S, R191Q, K192E, P574S, R68H, P83T, G122S, R156W, R160H, R185Q, P512R, G596R, R64C, G102D, R116Q, R124C, N127D, R167K, D169G, S178C, G530R, G669R, and R682W mutations were predicted to be pathogenic, highly conserved and exposed with decreased protein stability, indicating the most significant damaging effect. Hence, these 30 high risk nsSNPs were proceeded with post-translational modification sites prediction.
Prediction of post-translational modification (PTM) sites
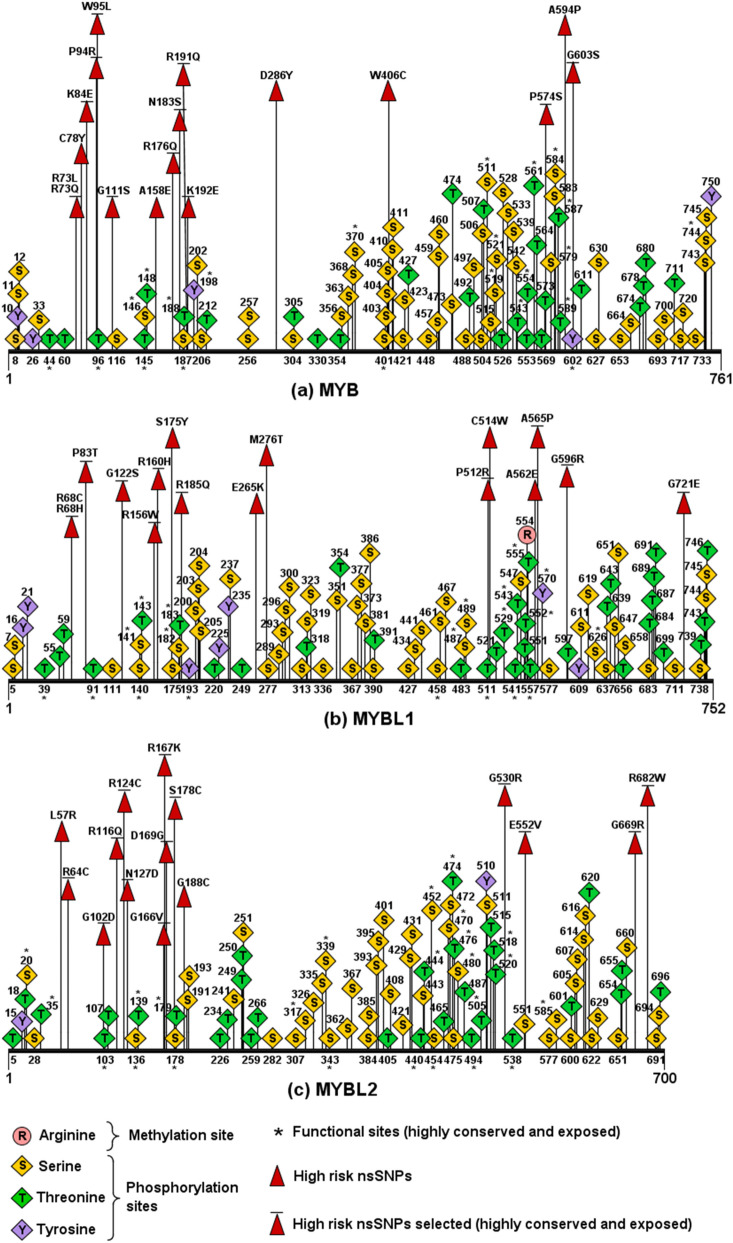
Post-translational modification (PTM) refers to the process where proteins undergo chemical modification to become functional and participate in respective cellular activities16. Putative methylation sites in the MYB family proteins and the 30 high risk nsSNPs were predicted using MusiteDeep and GPS-MSP 1.0. Only the wild-type R554 in MYBL1 was predicted as the common site (Fig. 1). Phosphorylation sites in the native and 30 mutated proteins were predicted using NetPhos 3.1 and GPS 5.0. NetPhos 3.1 predicted 268 residues in the three proteins to have phosphorylation potentials. A total of 386 residues in the proteins were found to be phosphorylated using the GPS 5.0, and 261 phosphorylation sites in all proteins were identified using both the tools, which consisted of 154 serines, 93 threonines, and 14 tyrosines (Fig. 1). However, only two wild-type reisudes (S175 and S178) and four mutant residues (S111, S183, S574, and S122) were the common sites present in the high risk nsSNPs. The ubiquitylation predictors employed in this study were BDM-PUB and UbiNet 2.0. BDM-PUB found 110 ubiquitylation sites at lysine residues in the family proteins. Whereas, only the wild-type K84 and K109 were predicted by UbiNet 2.0 in MYB. Both BDM-PUB and UbiNet 2.0 did not have common findings in all three proteins. After assembling the results from ConSurf and various PTM tools, a total of five high risk nsSNPs; G111S, N183S, P574S, G122S, and S178C containing putative phosphorylation sites were selected to proceed with comparative 3D modelling (Table 3).
Figure 1.
Putative PTM sites and high risk nsSNPs in MYB family proteins. (a) MYB had 90 common phosphorylation sites (Ser:56, Thr:29, Tyr:5), of which 22 were functional sites. (b) MYBL1 had 1 common methylation site and 90 common phosphorylation sites (Ser:50, Thr:33, Tyr:7), of which 21 were functional sites. (c) MYBL2 had 81 common phosphorylation sites (Ser:48, Thr:31, Tyr:2), of which 24 were functional si.
Table 3.
High risk nsSNPs selected by considering ConSurf and PTM predictions.
| Protein | SNP ID | Mutation | ConSurf | PTM | |
|---|---|---|---|---|---|
| Conservation scorea | Prediction | ||||
| MYB | rs1361650612 | G111S | 9 | e, f | Phosphorylation |
| rs1179275735 | N183S | 9 | e, f | Phosphorylation | |
| rs1247579811 | P574S | 9 | e, f | Phosphorylation | |
| MYBL1 | rs766676175 | G122S | 9 | e, f | Phosphorylation |
| MYBL2 | rs1438994955 | S178C | 9 | e, f | Phosphorylation |
b buried residue, s predicted structural residue (highly conserved and buried), e exposed residue, f predicted functional residue (highly conserved and exposed).
aHighly variable (1), highly conserved (9).
Comparative modelling of wild-type MYB family proteins and their mutant structures
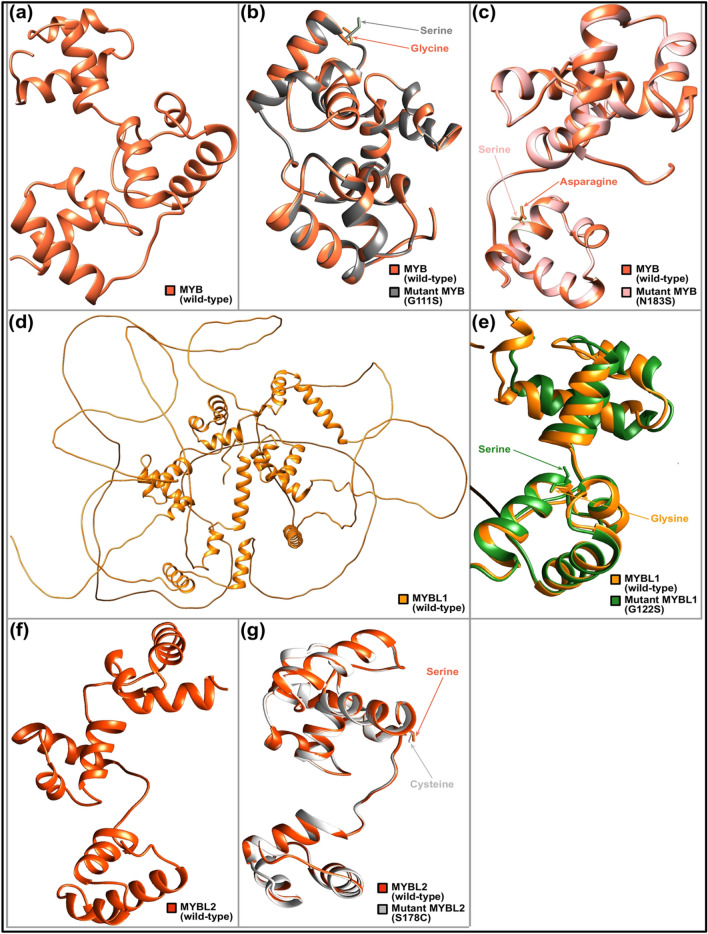
To investigate if the five high risk nsSNPs substantially alter the resultant proteins, predictive 3D modelling was performed along with the structural comparisons between wild-type and mutant models. The c1h88C and c1mseC templates were used to predict the wild-type MYB family proteins and their mutant models, excluding the P574S structure as this mutant residue was not covered in either template. TM-align revealed all mutant models had values of TM-score = 0 and RMSD = 1, showing no structural variations from their wild-type forms (Table 4). SWISS-MODEL was used to construct the 3D models for the wild-type proteins and their mutants. The best template used for the MYB family protein structures was 1h88.1.C as most 3D models can be generated based on this template. The generated mutant models were validated by ERRAT, and models with the highest possible GMQE scores, QMEAN Z-scores, and ERRAT values were selected for structural comparisons (Table 4). These models were visualised in Chimera 1.15 and the corresponding mutation positions induced by the nsSNPs were affirmed (Fig. 2). The structural integrity of generated wild-type and mutant protein structures were further validated with Ramachandran plot through the dihedral angles calculated.
Table 4.
TM-score, RMSD value, GMQE score, QMEAN Z-score, ERRAT value, and PROCHECK Ramachandran plot analysis of the selected protein models.
| Protein | Model | TM-align | SWISS-MODEL | ERRAT | PROCHECK Ramachandran plot analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| TM-scorea | RMSD | GMQE score | QMEAN Z-scoreb | ERRAT value (overall quality factor)c | Residues in most favoured regionsd | Residues in additional allowed regionsd | Residues in generously allowed regionsd | Residues in disallowed regionsd | ||
| MYB | + | Nil | Nil | Nil | Nil | 97.222 | 120 (87.6%) | 17 (12.4%) | 0 (0.0%) | 0 (0.0%) |
| G111S | 1 | 0 | 0.15 | 0.55 | 99.3056 | 122 (88.4%) | 15 (10.9%) | 0 (0.0%) | 1 (0.7%) | |
| N183S | 1 | 0 | 0.16 | 0.72 | 98.6111 | 122 (89.1%) | 15 (10.9%) | 0 (0.0%) | 0 (0.0%) | |
| MYBL1 | + | Nil | Nil | Nil | Nil | 97.9167 | 123 (89.1%) | 15 (10.9%) | 0 (0.0%) | 0 (0.0%) |
| G122S | 1 | 0 | 0.15 | − 0.02 | 98.6014 | 123 (89.1%) | 15 (10.9%) | 0 (0.0%) | 0 (0.0%) | |
| MYBL2 | + | Nil | Nil | Nil | Nil | 96.5278 | 119 (86.2%) | 19 (13.8%) | 0 (0.0%) | 0 (0.0%) |
| S178C | 1 | 0 | 0.16 | 0.19 | 96.5278 | 118 (85.5%) | 20 (14.5%) | 0 (0.0%) | 0 (0.0%) | |
TM-score template modelling-score, RMSD root-mean-square deviation, GMQE global model quality estimation, QMEAN qualitative model energy analysis, “+” Wildtype.
aRandom structural similarity (0.0 < TM-score < 0.30), both structures are within the same fold (0.50 < TM-score < 1.00).
bLow quality model (QMEAN Z-score ≤ − 4.0).
cReliable model (ERRAT value > 85%).
dNumber of residues (percentage of residues).
Figure 2.
Structural comparison of wild-type MYB family proteins with their mutant forms. (a) 3D model of wild-type MYB protein. (b) Superimposed structures of wild-type MYB protein and its mutant having mutation from Glycine to Serine at position 111. (c) Superimposed structures of wild-type MYB protein and its mutant having mutation from Asparagine to Serine at position 183. (d) 3D model of wild-type MYBL1 protein. (e) Superimposed structures of wild-type MYBL1 protein and its mutant having mutation from Glycine to Serine at position 122. (f) 3D model of wild-type MYBL2 protein. (g) Superimposed structures of wild-type MYBL2 protein and its mutant having mutation from Serine to Cysteine at position 178.This figure was generated using UCSF Chimera 1.15 (https://www.cgl.ucsf.edu/chimera/download.html).
Wild-type and mutant PDB inputs were subjected to PROCHECK for analysis. The wildtype MYB has 120 residues (87.6%) in the most favored region, 17 residues (12.4%) in the additional allowed region. The more damaging mutants, G111S has 122 residues (88.4%) in the most favored region, 15 residues (10.9%) in the additional allowed region and 1 residue (0.7%) in the disallowed region, followed by N183S possesses 122 residues (89.1%) in the most favored region, 15 residues (10.9%) in the additional allowed region. In MYBL1, both G122S mutant and wildtype possess the same amino acid residue patterns, 123 residues (89.1%) in the most favoured region and 15 residues (10.9%) in the additional allowed region, indicating no significant changes in the alteration in the structure. Wildtype MYBL2 has 119 residues (86.2%) in the most favoured region and 19 residues (13.8%) in the additional allowed region. Mutant S178C in MYBL2 has 118 residues (85.5%) in the most favoured region and 20 residues (14.5%) in the additional allowed region as shown as in Table 4.
Discussion
This study has successfully identified high-risk pathogenic nsSNPs in the MYB oncoproteins towards understanding its association with human cancer using an in-silico approach. MYB oncoproteins play crucial roles in multiple signalling pathways for cellular activities. A study by Andersson and colleagues17 showed that overexpressed wild-type MYB genes are normally benign, however, overexpression accompanied by gene alteration, dysregulated gene rearrangement or the incorrect oncoprotein binding onto enhancer region could promote tumorigenesis1. Despite the mutations in MYB oncoproteins being reported frequently in numerous cancers, the precise mechanisms of tumour initiations and/or maintenance remains vague. Therefore, examining the outcomes of deleterious nsSNPs of the MYB oncoproteins could potentially pave ways into a better understanding thus revealing its deleterious effects. Therefore, the aim of this study is to develop a bioinformatics pipeline in determining the most damaging nsSNPs and their effects on the structure and function of MYB, MYBL1, and MYBL2 proteins.
A total of 51,862 SNPs were extracted from the NCBI dbSNP for the MYB family genes, of which 1503 were nsSNPs. Structural analysis using (PROVEAN, PolyPhen-2, SIFT, SNPs&GO, and PhD-SNP) and functional analysis using PMut resulted in 45 “high-risk” pathogenic nsSNPs. Next, the stability of these nsSNPs were determined using I-Mutant 2.0, where 41 nsSNPs were identified with “decreased stability”. Protein stability is one of the key features to determine if a protein is biologically active and functional18. Proteins with decreased stability due to mutation might give rise to tumorigenesis as the fitness level for normal proteins dropped and conferred the fitness for tumorigenic proteins19.
Evolutionary conservation of MYB protein residues were calculated using ConSurf. The evolutionary conservation of an amino acid indicates its natural tendency for mutation to take place and highly conserved and exposed amino acids that undergo mutations can be expected to be most deleterious20. Thirty-six nsSNPs were identified to be highly conserved and exposed by ConSurf with the conservation score of 9. Next, these nsSNPs were subjected to PTMs analysis to determine its effect on regulating functions and structures of proteins. The G111S, N183S, P574S, G122S, and S178C showed to harbour putative phosphorylation sites. These nsSNPs coinciding with the putative phosphorylation sites may cause functional impairment and destabilisation of the corresponding proteins, thereby enhancing PTM impairment. PTM plays a pivotal role in modulating various protein functions and expressions, therefore mutations in PTM sites could lead towards malfunctions of the protein’ regulatory mechanisms, contributing to cellular dysfunctions such as transformation into cancer cells21. Several studies showed that mutated residues at phosphorylation sites had led to detrimental alterations in the expressions and functions of MYB22,23 and MYBL224–26. Among the four nsSNPs, G111S mutation in MYB was reportedly associated with uterine leiomyosarcomas27. Analysis using methylation and ubiquitylation predictors were also performed on the selected high risk nsSNPs. Only the R554 consensus methylation site was identified in MYBL1. As the results of methylation and ubiquitylation sites prediction were not in agreement for MYB and MYBL2, it was considered that both PTM sites were not predicted in these two proteins.
Structural alterations on the resulting proteins were constructed using the Phyre2 homology modelling tool. Protein 3-dimensional analysis offers a detailed insight into the associated molecular changes28. Two templates (c1h88C and c1mseC) were utilised to construct the protein models. These templates were selected based on high sequence similarities and high GMQE value, providing a high coverage. The TM-scores and RMSD values obtained for the four mutants suggest that the nsSNPs might not have a significant structural consequence on the proteins for TM-align to detect. Protein structure homology-modelling tool SWISS-MODEL was conducted to remodel the four nsSNPs for structure and function prediction. The template (1h88.1.C) was used as it has high sequence identity and desired coverage range. GMQE scores of the models were estimated to be around 0.15–0.16, indicating that the models cover only 15–16% of the targeted sequence. All models have QMEAN Z-scores greater than − 4.0, indicating the high quality of the models. Finally, ERRAT evaluation gives overall quality factors that approximately hundred to all models, indicating high quality models were built.
The G111S and G122S were identified to be located in the helix-turn-helix (HTH) myb-type 2 domain and S178C and N183S in the HTH myb-type 3 domain. These domains are important for the binding of DNA sequences and gene expression. This could cause activation and overexpression of the gene through loss of the C-terminal negative regulatory domain (C-myb)29. Previous studies have also reported that these could lead towards loss of the 3′ UTR binding sites thus negatively regulating MYB mRNA stability and translation30,31.
Thus, the nsSNPs identified within these regions may have a complete shift in the DNA-binding specificity, resulting in a pathogenic protein synthesis32.
Methods
Retrieving nsSNPs
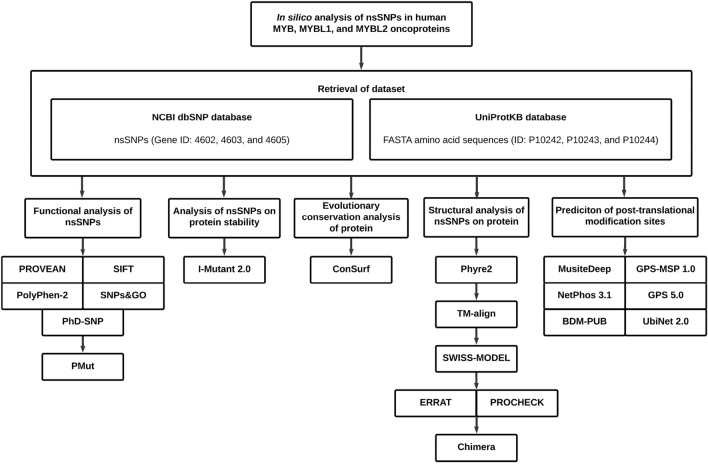
nsSNPs of MYB (gene ID: 4602), MYBL1 (gene ID: 4603), and MYBL2 (gene ID: 4605) were extracted from NCBI (National Center for Biological Information) dbSNP database [https://www.ncbi.nlm.nih.gov/snp/]33. DNA sequences as well as other information related to the nsSNPs of each gene, including the SNP IDs, allele changes, positions, protein accession numbers, residue changes, and global minor allele frequencies (MAFs) were also retrieved from this database. A total of 490 MYB, 483 MYBL1, and 530 MYBL2 nsSNPs were extracted respectively. The amino acid sequences of these genes (UniProtKB ID: P10242, P10243, and P10244) were obtained from the UniProtKB (Universal Protein Knowledgebase) database [https://www.uniprot.org/uniprot/] in FASTA format. Overview of the whole methodological approach is summarised in a schematic diagram (Fig. 3).
Figure 3.
Diagrammatic representation of methodology.
Identifying deleterious nsSNPs
The functional effect of the nsSNPs were predicted through five bioinformatics tools; PROVEAN (Protein Variation Effect Analyser) embedded with SIFT (Sorting Intolerant From Tolerant) [http://provean.jcvi.org/genome_submit_2.php?species=human]34–36, PolyPhen-2 (Polymorphism Phenotyping v2) [http://genetics.bwh.harvard.edu/pph2/bgi.shtml]37, and SNPs&GO (Single Nucleotide Polymorphisms and Gene Ontology) embedded with PhD-SNP (Predictor of human Deleterious Single Nucleotide Polymorphisms) [https://snps.biofold.org/snps-and-go/snps-and-go.html]38. Those nsSNPs which were predicted to be deleterious by all five in silico tools were considered as “high-risk” nsSNPs and selected for further downstream analysis39. This ensured the stringency and accuracy of the results by incorporating the scores of all five computational tools to increase the precision of prediction.
Validating the high risk nsSNPs
PMut [http://mmb.irbbarcelona.org/PMut/] was resorted to validate the pathological nature of the selected high risk nsSNPs40. This neural network-based tool includes 27,203 harmful and 38,078 benign mutations for 12,141 proteins. Prediction score ranging from 0 to 1 was computed along with the prediction percentage. The nsSNPs with a score of ≤ 0.5 are classified as neutral, whereas those with > 0.5 are predicted as disease-associated40.
Determining protein stability
Protein stability of the nsSNPs were determined through I-Mutant 2.0 [https://folding.biofold.org/i-mutant/i-mutant2.0.html]41. This tool determines the increase decrease of stability change in mutated protein, and simultaneously estimates the corresponding values of free energy change (DDG). I-Mutant 2.0 uses a support vector machine method and a ProTherm-derived dataset, which is the most collective databank containing experimental thermodynamic data of free energy changes in mutated protein stability41. Along with these predictions, a reliability index (RI) ranging from 0 (lowest reliability) to 10 (highest reliability) was also computed by this web server.
Protein evolutionary conservation analysis
ConSurf [https://consurf.tau.ac.il/] was used to predict the evolutionary conservation of each residue position in the native MYB proteins41. The prediction is based on an empirical Bayesian algorithm and the phylogenetic relations between close homologous sequences. For each amino acid position, a colorimetric conservation score between 1 and 9 is calculated by the tool and then classified as either a variable (1–4), intermediately conserved (5–6), or highly conserved residue (7–9). The exposed (on protein surface) or buried (inside protein core) status of each residue position in the protein structure is also determined. A functional residue is predicted when it is highly conserved and exposed, whereas a structural residue is predicted if it is highly conserved and buried20,42.
Post‑translational modification sites prediction
The putative methylation sites at arginine and lysine residues in each MYB protein, were predicted using MusiteDeep [https://www.musite.net/]43 and GPS-MSP 1.0 (Group-based Prediction System-Methyl-group Specific Predictor Version 1.0) [http://msp.biocuckoo.org/online.php]44. Using a default cut-off of 0.5, the deep learning-based MusiteDeep predicts and labels the desired PTM sites in the sequence according to the confidence threshold43. As for GPS-MSP 1.0, types of mono, symmetrical di-, and asymmetrical di-methylation specific to arginines, as well as mono, di- and tri-methylation types specific for lysines were predicted 45. Phosphorylation sites in each MYB protein at serines, threonines, and tyrosines were predicted using NetPhos 3.1 [https://services.healthtech.dtu.dk/service.php?NetPhos-3.1]46. and GPS 5.0 (Group-based Prediction System Version 5.0) [http://gps.biocuckoo.cn/index.php]47. A higher score in GPS 5.0 indicates higher probability of residues getting phosphorylated. Then, BDM-PUB (Prediction of Ubiquitination Sites with Bayesian Discriminant Method) [http://bdmpub.biocuckoo.org/prediction.php]48 and UbiNet 2.0 [https://awi.cuhk.edu.cn/~ubinet/index.php]49 were employed to predict putative protein ubiquitination sites at the lysines in MYB family proteins. A balanced cut-off option and a threshold of 0.3 were selected for the BDM-PUB server to perform the prediction based upon Bayesian Discriminant Method (BDM)50.
Examining the effects of nsSNPs with 3D protein modelling
The nsSNPs that were predicted as pathogenic, highly conserved with decreased protein stability, and possessing PTM sites were chosen to proceed with 3D protein modelling using 1h88.1.C template. To construct the 3D structures for wild-type and mutants MYB proteins, two distinct homology-modelling tools were employed: Phyre2 (Protein Homology/analogy Recognition Engine V 2.0) [http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index]51 and SWISS-MODEL [https://swissmodel.expasy.org/]52. Each nsSNP was individually substituted into the respective sequence of each MYB protein and then submitted to Phyre2 for the creation of 3D mutant models based on selected templates. TM-align (Template Modelling-align) [https://zhanggroup.org/TM-align/]53 was utilised to investigate the similarities between the modelled wild-type and mutant protein structures by computing template modelling-score (TM-score) and root-mean-square deviation (RMSD) values. TM-score yields a result from 0 to 1, where 1 denotes a perfect match between both structures54. Precisely, 0.0 < TM-score < 0.30 indicates random structural similarity, whereas 0.50 < TM-score < 1.00 implies that both structures are within the same fold55,56. A lower TM-score and a higher RMSD value indicate a greater structural deviation of mutant models from those of wild-type57. To build the 3D models in SWISS-MODEL, templates were analysed and selected based on coverage, sequence identity, qualitative model energy analysis (QMEAN) Z-score, and global model quality estimation (GMQE) score. A QMEAN Z-score of ≤ − 4.0 denotes a low quality model58. The GMQE score, which ranges from 0 to 1, indicates the likely accuracy of the model constructed with that alignment and the target coverage59. Therefore, templates with higher sequence similarities and a higher GMQE value were prioritized, concurrent with the coverage of the mutation site in that template, thus, template 1h88.1.C was selected. The built models were then validated by ERRAT [https://saves.mbi.ucla.edu/] and PROCHECK Ramachandran plot analysis [https://saves.mbi.ucla.edu/] to estimate their structural quality. Then, the validated structures were viewed and superimposed using Chimera 1.15 [https://www.cgl.ucsf.edu/chimera/download.html].
Conclusion
MYB family members are often aberrantly expressed in human cancers, suggesting that they could be important for tumour initiation and/or maintenance. In this study, a total of 30 nsSNPs were predicted as high-risk pathogenic, conserved with decreased stability, suggesting potential deleterious effect on the protein structure. Further PTM and 3D protein modeling indicated rs1361650612 (G111S), rs1179275735 (N183S), rs766676175 (G122S), and rs1438994955 (S178C) located within the helix-turn-helix (HTH) myb-type 2 and myb-type 3 domains were identified pathogenic with the ability to potentially cause great functional and stability impairment on the proteins. This study concise confidence that these findings could serve as a benchmark towards potential diagnostic and therapeutic interventions.
Author contributions
L.S.W., T.J.K.K., L.E.C. conducted the analysis and L.S.W., T.J.K.K. and M.S. wrote the manuscript text. M.A.O. and L.K.S. reviewed and edited the manuscript. Y.W.S. and N.M.A.N.A.R. designed, conceptualised and provided funding acquisition. All authors reviewed the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wai-Sum Yap, Email: wsyap@ucsiuniversity.edu.my.
Nik Abd Rahman Nik Mohd Afizan, Email: m.afizan@upm.edu.
References
- 1.Cicirò Y, Sala A. MYB oncoproteins: Emerging players and potential therapeutic targets in human cancer. Oncogenesis. 2021;10:1–15. doi: 10.1038/s41389-021-00309-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansour MR, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandopadhayay P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 2016;48:273–282. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams BB, et al. Induction of T cell-mediated immunity using a c-Myb DNA vaccine in a mouse model of colon cancer. Cancer Immunol. Immunother. 2008;57:1635–1645. doi: 10.1007/s00262-008-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsay R, et al. Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ. 1992;3:723–723. [PubMed] [Google Scholar]
- 6.Hugo H, et al. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer. 2006;45:1143–1154. doi: 10.1002/gcc.20378. [DOI] [PubMed] [Google Scholar]
- 7.Yang R-M, et al. MYB regulates the DNA damage response and components of the homology-directed repair pathway in human estrogen receptor-positive breast cancer cells. Oncogene. 2019;38:5239–5249. doi: 10.1038/s41388-019-0789-3. [DOI] [PubMed] [Google Scholar]
- 8.Wolf Pérez A-M, Lorenzen N, Vendruscolo M, Sormanni P. Therapeutic Antibodies. Springer; 2022. pp. 57–113. [DOI] [PubMed] [Google Scholar]
- 9.Stefl S, Nishi H, Petukh M, Panchenko AR, Alexov E. Molecular mechanisms of disease-causing missense mutations. J. Mol. Biol. 2013;425:3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamaraj B, Rajendran V, Sethumadhavan R, Kumar CV, Purohit R. Mutational analysis of FUS gene and its structural and functional role in amyotrophic lateral sclerosis 6. J. Biomol. Struct. Dyn. 2015;33:834–844. doi: 10.1080/07391102.2014.915762. [DOI] [PubMed] [Google Scholar]
- 11.Nzabonimpa GS, Rasmussen HB, Brunak S, Taboureau O, Consortium I. Investigating the impact of missense mutations in hCES1 by in silico structure-based approaches. Drug Metab. Personal. Ther. 2016;31:97–106. doi: 10.1515/dmpt-2015-0034. [DOI] [PubMed] [Google Scholar]
- 12.Rajendran V, Gopalakrishnan C, Sethumadhavan R. Pathological role of a point mutation (T315I) in BCR-ABL1 protein—A computational insight. J. Cell. Biochem. 2018;119:918–925. doi: 10.1002/jcb.26257. [DOI] [PubMed] [Google Scholar]
- 13.Kumar A, Purohit R. Computational screening and molecular dynamics simulation of disease associated nsSNPs in CENP-E. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2012;738:28–37. doi: 10.1016/j.mrfmmm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Rajendran V, Sethumadhavan R. Drug resistance mechanism of PncA in Mycobacteriumtuberculosis. J. Biomol. Struct. Dyn. 2014;32:209–221. doi: 10.1080/07391102.2012.759885. [DOI] [PubMed] [Google Scholar]
- 15.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramazi S, Zahiri J. Posttranslational modifications in proteins: Resources, tools and prediction methods. Database. 2021 doi: 10.1093/database/baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson MK, et al. ATR is a MYB regulated gene and potential therapeutic target in adenoid cystic carcinoma. Oncogenesis. 2020;9:1–10. doi: 10.1038/s41389-020-0194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromiha MM. Protein Bioinformatics: From Sequence to Function. Academic Press; 2010. [Google Scholar]
- 19.Wilcken R, Wang G, Boeckler FM, Fersht AR. Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc. Natl. Acad. Sci. 2012;109:13584–13589. doi: 10.1073/pnas.1211550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karve TM, Cheema AK. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids. 2011 doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bies J, Sramko M, Wolff L. Stress-induced phosphorylation of Thr486 in c-Myb by p38 mitogen-activated protein kinases attenuates conjugation of SUMO-2/3. J. Biol. Chem. 2013;288:36983–36993. doi: 10.1074/jbc.M113.500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa K, et al. Substitution of Thr572 to Ala in mouse c-Myb attenuates progression of early erythroid differentiation. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-71267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werwein E, Cibis H, Hess D, Klempnauer K-H. Activation of the oncogenic transcription factor B-Myb via multisite phosphorylation and prolyl cis/trans isomerization. Nucleic Acids Res. 2019;47:103–121. doi: 10.1093/nar/gky935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartsch O, Horstmann S, Toprak K, Klempnauer KH, Ferrari S. Identification of cyclin A/Cdk2 phosphorylation sites in B-Myb. Eur. J. Biochem. 1999;260:384–391. doi: 10.1046/j.1432-1327.1999.00191.x. [DOI] [PubMed] [Google Scholar]
- 26.Werwein E, Biyanee A, Klempnauer KH. Intramolecular interaction of B-MYB is regulated through Ser-577 phosphorylation. FEBS Lett. 2020;594:4266–4279. doi: 10.1002/1873-3468.13940. [DOI] [PubMed] [Google Scholar]
- 27.da Costa LT, et al. The mutational repertoire of uterine sarcomas and carcinosarcomas in a Brazilian cohort: A preliminary study. Clinics. 2021 doi: 10.6061/clinics/2021/e2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar A, et al. Computational SNP analysis: Current approaches and future prospects. Cell Biochem. Biophys. 2014;68:233–239. doi: 10.1007/s12013-013-9705-6. [DOI] [PubMed] [Google Scholar]
- 29.Wefers AK, et al. Isomorphic diffuse glioma is a morphologically and molecularly distinct tumour entity with recurrent gene fusions of MYBL1 or MYB and a benign disease course. Acta Neuropathol. 2020;139:193–209. doi: 10.1007/s00401-019-02078-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung EY, et al. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol. Ther. 2008;7:1758–1764. doi: 10.4161/cbt.7.11.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YC, et al. c-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol. Biol. Evol. 2008;25:2189–2198. doi: 10.1093/molbev/msn165. [DOI] [PubMed] [Google Scholar]
- 32.Caramori G, Ruggeri P, Mumby S, Atzeni F, Adcock IM. Transcription factors. eLS. 2019 doi: 10.1002/9780470015902.a0005278.pub3. [DOI] [Google Scholar]
- 33.Sherry ST, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi Y, Chan AP. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genom. Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 36.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 37.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;76:7.20.21–27.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capriotti E, et al. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013;14:1–7. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamaraj B, Purohit R. Computational screening of disease-associated mutations in OCA2 gene. Cell Biochem. Biophys. 2014;68:97–109. doi: 10.1007/s12013-013-9697-2. [DOI] [PubMed] [Google Scholar]
- 40.López-Ferrando V, Gazzo A, De La Cruz X, Orozco M, Gelpí JL. PMut: A web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res. 2017;45:W222–W228. doi: 10.1093/nar/gkx313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D, et al. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020;48:W140–W146. doi: 10.1093/nar/gkaa275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue Y, et al. GPS: A comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–W187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A, Thakur M, Singh SK, Sharma LK, Chandra K. Exploring the effect of nsSNPs in human YPEL3 gene in cellular senescence. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-72333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mans B, Neitz A. Der-p2 (Dermatophagoidespteronyssinus) allergen-like protein from the hard tick Ixodesricinus-a novel member of ML (MD-2-related lipid-recognition) domain protein family. Nat. Lond. 1998;391:753–754. doi: 10.1017/S0031182009992083. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, et al. GPS 5.0: An update on the prediction of kinase-specific phosphorylation sites in proteins. Genom. Proteom. Bioinform. 2020;18:72–80. doi: 10.1016/j.gpb.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, A., Gao, X., Ren, J., Jin, C. & Xue, Y. BDM-PUB: Computational prediction of protein ubiquitination sites with a Bayesian discriminant method. In BDM-PUB: Computational Prediction of Protein Ubiquitination Sites with a Bayesian Discriminant Method (2009).
- 49.Li Z, et al. UbiNet 2.0: A verified, classified, annotated and updated database of E3 ubiquitin ligase–substrate interactions. Database. 2021 doi: 10.1093/database/baab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lira SS, Ahammad I. A comprehensive in silico investigation into the nsSNPs of Drd2 gene predicts significant functional consequences in dopamine signaling and pharmacotherapy. bioRxiv. 2021;583:195. doi: 10.1038/s41598-021-02715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Modelos C. Trabajo práctico No 13. Varianzas en función de variable independiente categórica. Nat. Protoc. 2016;10:845–858. [Google Scholar]
- 52.Andrew W, et al. Heer Florian T, de Beer Tjaart A P, Rempfer Christine, Bordoli Lorenza, Lepore Rosalba, Schwede Torsten. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hossain MS, Roy AS, Islam MS. In silico analysis predicting effects of deleterious SNPS of human rassf5 gene on its structure and functions. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-71457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karthik L, et al. Protease inhibitors from marine actinobacteria as a potential source for antimalarial compound. PLoS ONE. 2014;9:e90972. doi: 10.1371/journal.pone.0090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arshad M, Bhatti A, John P. Identification and in silico analysis of functional SNPs of human TAGAP protein: A comprehensive study. PLoS ONE. 2018;13:e0188143. doi: 10.1371/journal.pone.0188143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res, 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.