Abstract
Intracellular trafficking of retroviral RNAs is a potential mechanism to target viral gene expression to specific regions of infected cells. Here we show that the human immunodeficiency virus type 1 (HIV-1) genome contains two sequences similar to the hnRNP A2 response element (A2RE), a cis-acting RNA trafficking sequence that binds to the trans-acting trafficking factor, hnRNP A2, and mediates a specific RNA trafficking pathway characterized extensively in oligodendrocytes. The two HIV-1 sequences, designated A2RE-1, within the major homology region of the gag gene, and A2RE-2, in a region of overlap between the vpr and tat genes, both bind to hnRNP A2 in vitro and are necessary and sufficient for RNA transport in oligodendrocytes in vivo. A single base change (A8G) in either sequence reduces hnRNP A2 binding and, in the case of A2RE-2, inhibits RNA transport. A2RE-mediated RNA transport is microtubule and hnRNP A2 dependent. Differentially labelled gag and vpr RNAs, containing A2RE-1 and A2RE-2, respectively, coassemble into the same RNA trafficking granules and are cotransported to the periphery of the cell. tat RNA, although it contains A2RE-2, is not transported as efficiently as vpr RNA. An A2RE/hnRNP A2-mediated trafficking pathway for HIV RNA is proposed, and the role of RNA trafficking in targeting HIV gene expression is discussed.
Many eucaryotic RNAs have characteristic intracellular trafficking pathways determined by cis-acting elements in the RNA and trans-acting factors in the cell. cis and trans trafficking determinants have been identified for actin mRNA in fibroblasts (38), Vg1 mRNA in frog oocytes (17) and myelin basic protein (MBP) mRNA in oligodendrocytes (11). In the case of MBP mRNA, a 21-nucleotide (nt) cis-acting sequence (GCCAAGGAGCCAGAGAGCATG) mediates RNA trafficking. This sequence, originally termed the RNA trafficking sequence (2), is now referred to as the hnRNP A2 response element (A2RE) because in binds to the trans-acting factor hnRNP A2 in vitro (21), because its RNA trafficking functions are hnRNP A2 dependent (23), and because A2RE mutations that interfere with hnRNP A2 binding abolish RNA trafficking (31).
A2RE and/or hnRNP A2 determinants have been implicated at several different steps in RNA trafficking. In the nucleus, hnRNP A2 associates nonspecifically with newly synthesized RNA transcripts as part of the core hnRNP complex (18) and specifically regulates splice site selection in certain transcripts (29). In Drosophila melanogaster, an hnRNP A2 orthologue, Squid protein, associates with pair-rule transcripts in the nucleus and mediates their subsequent apical localization in the cytoplasm (24). In the silkmoth Chironomus, hnRNP A2 class proteins accompany Balbiani ring mRNA during export through the nuclear pore and into polyribosomes (14, 15, 43). In oligodendrocytes, hnRNP A2 mediates anterograde transport of A2RE-containing RNA granules (6) along microtubules (2, 11), and translation of A2RE-containing RNA is enhanced by hnRNP A2 in vivo and in vitro (23). Since A2RE-like sequences are found in a variety of transported mRNAs (2) and since hnRNP A2 is constitutively expressed in most cell types (22), the A2RE/hnRNP A2 pathway may represent a general RNA trafficking pathway.
Intracellular trafficking of retroviral RNAs is important for viral replication (20). All retroviruses produce a single primary transcript that is processed into various species of mRNA. In the simpler retroviruses, usually only two species of mRNA are made, a singly spliced RNA encoding the envelope protein and an unspliced RNA that functions both as mRNA encoding the Gag and Gag-Pol proteins and as genomic RNA packaged into virions. In more complex retroviruses, such as the human immunodeficiency virus (HIV), processing of the primary transcript is more complicated, generating multiple singly spliced, doubly spliced, or unspliced RNAs. Retroviruses utilize specific mechanisms to allow nuclear export of unspliced and incompletely spliced mRNAs which would otherwise remain in the nucleus since they contain introns (13, 26). In HIV-infected cells, nuclear export of unspliced and singly spliced HIV type 1 (HIV-1) transcripts is mediated by the Rev response element (RRE), a cis-acting sequence in the env gene (28), recognized by the trans-acting viral Rev protein (36). Simpler retroviruses rely on cis-acting RNA signals termed constitutive transport elements (8, 33, 44) that interact directly with cellular proteins to mediate export of intron-containing mRNA (7, 27).
While intranuclear processing and nuclear export of retroviral RNAs have been studied extensively, cytoplasmic trafficking of retroviral RNA is less well understood. In the context of a retrovirus-infected cell, cytoplasmic RNA trafficking could target specific RNAs to discrete subcellular locations, thereby maximizing expression of the encoded proteins at the target location and minimizing ectopic expression elsewhere in the cell. Such a process could, in principle, facilitate viral assembly by steering viral genomic RNA and mRNAs encoding viral structural proteins toward sites of virion assembly.
In this report we test the hypothesis that HIV RNAs contain specific RNA trafficking sequences. We identify two separate A2RE-like sequences in the HIV-1 genome: A2RE-1 in the gag gene and A2RE-2 in a region of overlap between the vpr and tat genes. Both sequences bind to hnRNP A2 in vitro and mediate hnRNP A2-dependent transport of HIV RNAs in oligodendrocytes. The results suggest that intracellular trafficking of HIV RNAs containing these sequences follows the A2RE/hnRNP A2 pathway.
MATERIALS AND METHODS
RNA constructs.
HIV-1 gag RNA (containing the complete 5′ untranslated region [UTR] and gag open reading frame [ORF] but truncated after the gag stop codon) was transcribed from BamHI-linearized pKS-gag. To construct pKS-gag, a fragment (1,855 bp) of HIV-1 proviral DNA from the tat activation (TAR) region to just dowstream of the gag stop codon was PCR amplified using 5′ XhoI and 3′ BamHI primers and inserted into pKSII. HIV-1 gag ΔKLM and HIV-1 gag ΔA14 were transcribed from vectors constructed in a similar way, using fragments of HIV-1 proviral DNA with deletions of the kissing loop (25) and packaging signal (30), respectively. HIV-1 gag ΔA2RE-1 RNA was transcribed from pKS-gag linearized with PstI, which cuts just upstream of A2RE-1. HIV-1 gag ΔUTR RNA was transcribed from pET21cgag ΔUTR. To construct pET21cgag ΔUTR, HIV-1 gag was PCR amplified using 5′ and 3′ BamHI primers and inserted into BamHI-digested pET21c (Novagen). HIV-1 gag hemagglutinin (HA) RNA was transcribed from pKS-gag HA. To construct pKS-gag HA, PCR primers (5′-GGTACCTACCCCTACGACGTGCCCGACTACGACTATGTAGACCGGTTCTATAAA-3′ and 5′-GTAGTCGGGCACGTCGTAGGGGTAGGTACCTATGTCCAGAATGCTGGTAGGGCT-3′) were designed to replace the A2RE-1 sequence with the HA tag. A second round of PCR using flanking XhoI and BamHI primers was performed, and the resultant DNA was cloned into pKSII to generate a gag chimera in which A2RE-1 was replaced with an HA tag.
HIV-1 vpr RNA (containing the vpr ORF) was transcribed from PvuII-linearized pET21c-vpr poly(A)+. This vector is based on pET21c-vpr (5), except that a 30-nt poly(A) tail was added by inserting a SacI/HindIII fragment from sp64polyA+ (Gibco-BRL). HIV-1 vpr ΔA2RE-2 RNA was transcribed from PvuII-linearized pET21c-1-88vpr, which was prepared using 5′ XbaI and 3′ SacI primers to amplify truncated vpr lacking A2RE-2 for insertion into pET21cPolyA+ (described above). HIV-2 vpr RNA was transcribed from an HIV-2 transcription vector (pKS-vpr2) generated by PCR amplification of HIV-2 proviral DNA (HIV-2 ROD; accession number M15390) using 5′ EcoRI and 3′ SalI primers. The PCR fragment was inserted into pKSII to generate pKS-vpr2. HIV-1 vpr A8G RNA was transcribed from pHIV vpr A8G. To construct pHIV vpr A8G, PCR primers (5′-GGG GCT GCA GCG CCG GCC ACC ATG GAA CAA-3′ and 5′-GGG GTC TAG AGC GGA TCT ACT GGC CCC ATT TCT TGC TCT CCT CTG TT-3′) were used to create the A8G mutant vpr sequence, which was digested with XbaI and PstI and cloned into XbaI-PstI-digested pHJ1 (23). A PstI-BsaWI fragment containing the A8G mutant vpr sequence fused in frame to green fluorescent protein (GFP) was cloned into PstI-XmaI-digested pBluescript plasmid, generating pHIV vpr A8G, which was linearized with SpeI for in vitro transcription with T7 polymerase.
HIV-1 tat RNAs were transcribed from HindIII-linearized pKS-based transcription vectors. A SalI-BamHI fragment from pGEM-3/Tatc (34) and a SacI/BamHI fragment from pGEM-3/Ltatc (4) were cloned into pKSII to generate pKS-tat and pKS-Ltat, respectively. HIV-1 tat ΔA2RE-2 RNA, a truncated version of tat RNA lacking the 5′ UTR, was transcribed from pKSTat. To construct pKSTat, SalI- and BamHI-containing 5′ and 3′ primers were used to PCR amplify tat cDNA from just upstream of the tat start codon to the BamHI site downstream of the tat stop codon. The PCR product was recloned into pKSII to generate pKS-TatΔA2RE-2.
GFP RNA was transcribed from pNKT7 (23). GFP A2RE-1 and GFP A2RE-2 RNAs were transcribed from vectors constructed by inserting either A2RE-1 or A2RE-2 into the SalI site of pNKT7.
All constructs were confirmed by restriction analysis and sequencing.
Cell culture and microinjection.
Oligodendrocytes were prepared from newborn mouse brain as described previously (1). Cells were plated on glass coverslips glued to the bottom of plastic culture dishes (MatTek Corp., Ashland, Mass.). Fluorescent RNA, labeled with either Texas red- or AlexaFluor 488-conjugated rUTP, was microinjected into cells as described previously (1). After injection, the cells were returned to the tissue culture incubator for 15 to 30 min to allow time for transport of the injected RNA from the perikaryon to the distal processes and myelin compartment. The injected cells were examined by confocal laser scanning microscopy and scored as either transport positive or transport negative, using criteria described previously (2). Cells were treated with hnRNP A2 sense (TCCGCGATGGAGAGAGAAAAG) or antisense (CTTTTCTCTCTCCATCGCGGA) oligonucleotide to suppress hnRNP A2 expression, as described previously (23). Cells were treated with nocodazole to disrupt microtubules or with cytochalasin to disrupt actin microfilaments as described previously (11).
Single granule ratiometric analysis.
Single granule ratiometric analysis, performed as described previously (6), was used to determine the ratios of different RNAs in individual granules. Oligodendrocytes were coinjected with differentially labeled gag (fluorescein) and vpr (Texas red) RNAs and imaged using dual-channel confocal microscopy. Individual well-resolved granules were selected, and fluorescent intensities in the red and green channels were integrated over an area of nine pixels/granule. The ratio of the integrated intensity in the red channel divided by the sum of the integrated intensities in both channels provides a measure of the ratio of the two RNAs in each granule. For example, a granule containing only vpr RNA with no gag RNA is detected only in the red channel and therefore has a ratio of one, while a granule containing only gag RNA with no vpr RNA is detected only in the green channel and has a ratio of zero. Granules containing both gag and vpr RNAs are detected in both channels and have intermediate ratios. The distribution of ratios for a population of granules provides a measure of the variance in RNA distribution per granule in the population.
hnRNP A2 binding assay.
In vitro binding to RNA sequences immobilized on paramagnetic beads was performed as described previously (21, 31).
RESULTS
HIV RNAs contain A2RE-like sequences.
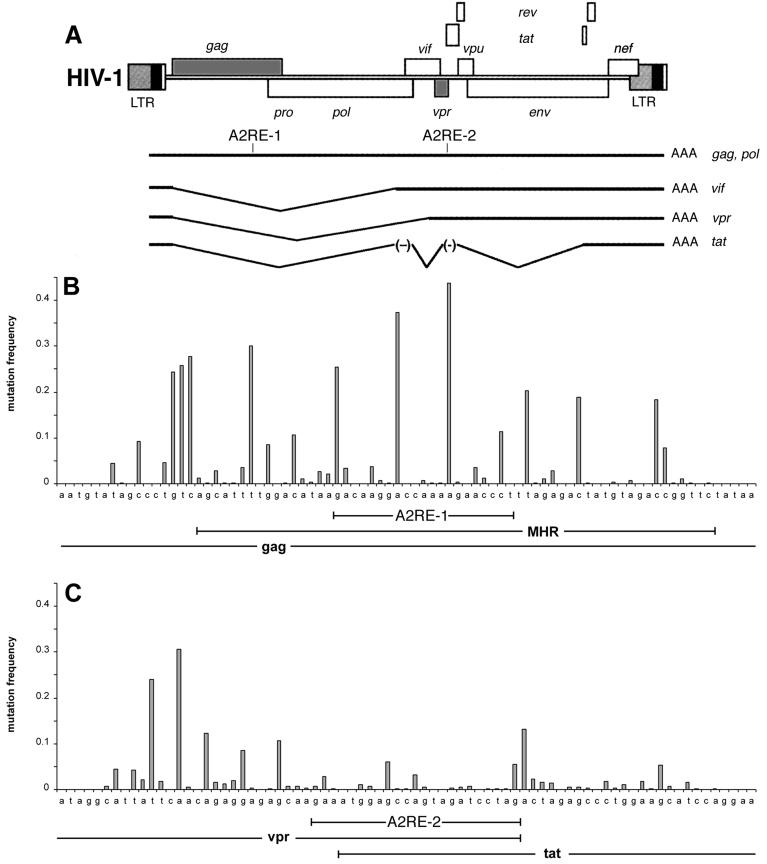
The NCBI nucleotide sequence database was searched for sequences similar to the A2RE. Two A2RE-like sequences were identified in HIV-1 RNA: A2RE-1 in the gag gene and A2RE-2 in a region of overlap between the vpr and tat genes (Fig. 1A). Full-length, unspliced HIV genomic RNA, which is equivalent to gag mRNA, contains both A2RE-1 and A2RE-2. Spliced HIV RNAs encoding vpr, vif, and tat contain A2RE-2 only. Sequence polymorphism in the regions of the genome containing A2RE-1 (Fig. 1B) and A2RE-2 (Fig. 1C) was analyzed in HIV-1 sequences from the NCBI database. A2RE-1 is located within the major homology region (MHR), a highly conserved region of the gag ORF implicated in virion assembly (35, 37, 42). Sequence polymorphism in A2RE-1 is generally less than in flanking regions of the gag gene except for positions 8 and 14, which have almost equivalent frequencies of A and G. A2RE-2 is located in a region of overlap between the 3′ end of the vpr ORF and the 5′ end of the tat ORF, adjacent to a region identified as an exon splicing silencer (ESS2) (3, 40). Sequence polymorphism in A2RE-2 is generally less than in flanking regions of the vpr and tat genes. These results indicate that A2RE-1 and A2RE-2 are relatively conserved sequences in the HIV-1 genome.
FIG. 1.
Identification of A2RE-like sequences in the HIV genome. (A) Diagram of the HIV-1 genome with the positions of the genes indicated. A2RE-1 is located within the MHR in the gag gene and A2RE-2 is located in a region of overlap between the vpr and tat genes. (B) Sequence polymorphism at each position in A2RE-1 and flanking regions in the gag gene. (C) Sequence polymorphism at each position in A2RE-2 and flanking regions of the vpr and tat genes.
To determine if A2RE-like sequences are conserved in other retroviruses, the sequences of corresponding regions of the gag and vpr genes from different retroviruses were compared. Alignments of A2RE-like sequences in the gag genes of 14 different retroviruses and in the vpr genes of 3 different retroviruses are shown in Table 1. Conservation of A2RE-like sequences in the gag gene ranged from 95% between HIV-1 and HIV-2 to 50% between HIV-1 and feline leukemia virus (FeLV). Conservation of A2RE-like sequences in the vpr gene was approximately 50% between HIV-1 and HIV-2 or simian immunodeficiency virus (SIV). These results indicate that A2RE-like sequences are conserved among different retroviruses, suggesting that they are functionally important.
TABLE 1.
A2RE-like sequences in retroviral RNAs
| Virusa | Gene | Positionb | A2RE-like sequence |
|---|---|---|---|
| None | MBP | 1380 | G C C A A G G A G C C A G A G A G C A U G |
| HIV-1 | gag | 1192 | G A / C A A / G G A / C C A / A A A / G A A / C C C / U |
| HIV-2 | gag | 738 | A - / - - - / - - - / - - - / - - - / - - - / - - - / - |
| SIV | gag | 586 | - - / - - G / - - - / - - - / - - - / - - G / - - U / - |
| FIV | gag | 1474 | - - / - - - / - - - / G - U / - - G / - - - / G A U / - |
| EIAV | gag | 833 | - G / - - - / - - - / G - U / - - G / - - - / - - U / - |
| BIV | gag | 1187 | A U / - - G / - - - / - - C / - - G / - - G / - - G / - |
| MPMV | gag | 1613 | A - / - - - / - - - / - - C / G - U / - - G / - - A / - |
| HTLV-1 | gag | 1609 | U C / - - - / - - C / - U G / G - G / - - G / - - U / - |
| GLV | gag | 1950 | U G / - - G / - - - / - - G / G C - / - - - / - - - / C |
| Mo-MuLV | gag | 1429 | C - / - - - / - - G / - - C / - - U / - - G / U - U / C |
| BLV | gag | 1364 | U C / - - - / - - C / - - C / G C C / - - - / A G - / - |
| RSV | gag | 1566 | U G / - - G / - - - / - - - / U C U / - - G / U - - / - |
| ALV | gag | 1790 | C G / - - G / - - - / - - - / U C U / - - G / U - - / - |
| FeLV | gag | 1685 | U G / - - - / - - G / A A - / G - A / - - - / A - G / C |
| Consensus amino acid | R Q G P K E P | ||
| HIV-1 | vpr | 6157 | G A / A A U / G G A / G C C / A G U / A G A / U C C / U |
| HIV-2 | vpr | 5855 | U U / G - A / - - - / - - - / - - A / G A G / C U - / A |
| SIV | vpr | 6174 | - - / - G - / - - - / U - A / G - A / - A U / G U - / A |
| Consensus amino acid | R N G A R E S |
Abbreviations: avian leukosis virus (ALV), bovine immunodeficiency virus (BIV), bovine leukemia virus (BLV), equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV), Gibbon leukemia virus (GLV), human T-cell leukemia virus type 1 (HTLV-1), Mason-Pfizer monkey virus (MPMV), Moloney murine leukemia virus (Mo-MuLV), Rous sarcoma virus (RSV). The accession numbers for the gene sequences are as follows: MBP (NM 010777.1), HIV-1 gag (K02013), HIV-2 gag (L33080), SIV gag (U10899), FIV gag (X57002), EIAV gag (AF170900), BIV gag (M32690), MPMV gag (AF033815), HTLV-1 gag (L03561), GLV gag (U60065), Mo-MuLV gag (AF033811), BLV gag (K02120), RSV gag (AF033808), ALV gag (Z46390), FeLV gag (K01803) HIV-1 vpr (AF133821), HIV-2 vpr (X52223), SIV vpr (AF131870).
Position indicates the number of the first nucleotide in the A2RE-like sequence according to the numbering in the viral sequence from the corresponding accession number.
In all of the retroviral sequences examined, A2RE-like sequences in the gag ORF and the vpr ORF are translated in the same reading frame. Furthermore, many of the nucleotide substitutions in different retroviruses occur in the third position of a codon and therefore do not affect the amino acid sequence of the protein. The consensus amino acid sequences encoded by A2RE-like sequences in the gag gene and vpr genes are 42% identical and 71% similar (Table 1), raising the possibility that amino acid sequences encoded by A2RE-like sequences are important for Gag and Vpr protein function.
A2RE-like sequences mediate transport of HIV RNAs.
To determine if A2RE-like sequences in HIV mediate RNA transport, fluorescently tagged RNAs containing these sequences were microinjected into oligodendrocytes, and the intracellular distribution of the injected RNA was analyzed by confocal laser scanning microscopy. Oligodendrocytes, instead of a more common HIV host cell type, were used for the transport assay because their extended and ramified morphology facilitates the visualization of intracellular RNA trafficking intermediates. Truncated HIV RNAs, instead of full-length HIV RNAs, were used in order to measure the RNA trafficking functions of ARE-1 and -2 independently of each other and also to minimize confounding effects of sequence context and potential non-A2RE trafficking signals in the HIV genome. Cells were scored as transport positive if the injected RNA was transported past the first branch point in at least one process. Previous work has shown that cells injected with RNA containing a functional A2RE (such as a full-length MBP mRNA) are 70 to 80% transport positive, while cells injected with RNA lacking a functional A2RE (such as GFP RNA, globin RNA, or MBP RNA with the A2RE deleted) are less than 20% transport positive. Thus, the dynamic range of the RNA transport assay is from 20 to 70%. The structures of the RNAs tested are diagrammed in Fig. 2A. The percentage of transport-positive cells for each RNA is given in Fig. 2B. Images of representative cells injected with each RNA are shown in Fig. 2C.
FIG. 2.
Transport of HIV RNAs in oligodendrocytes. Fluorescent RNAs were microinjected into oligodendrocytes, and the subcellular distribution of the injected RNA was visualized by confocal microscopy. (A) Structures of the various RNA constructs tested. ORFs are indicated as open bars; UTRs are indicated as solid bars. The positions of A2RE-like sequences are indicated. (B) Percentage of transport-positive cells in cells injected with each of the constructs in panel A. For each RNA tested, at least 50 cells were analyzed in at least three separate experiments. (C) Distribution of injected RNA in representative cells for each construct. If the RNA was transported past the first branch point in the processes, the cell was scored as transport positive. Scale bar, 10 μm.
HIV-1 gag RNA, containing the gag 5′ UTR and ORF including A2RE-1 but lacking the 3′ UTR, was transported in >80% of injected cells, indicating that it contains a functional RNA transport signal. Deletion of various regions from the 5′ UTR of HIV-1 gag RNA (HIV-1 gag ΔKLM, HIV-1 gag ΔA14, and HIV-1 gag Δ5′ UTR) did not affect transport, indicating that the 5′ UTR does not contain RNA transport signals. Deletion of A2RE-1 and downstream sequences from HIV-1 gag RNA (HIV-1 gag ΔA2RE-1) reduced transport to <25% of injected cells. Replacement of A2RE-1 with an HA tag (HIV-1 gag HA) reduced transport to 5% of injected cells. These results indicate that A2RE-1 is necessary for transport of gag RNA.
HIV-1 vpr RNA, containing the vpr ORF, including A2RE-2 but lacking 5′ and 3′ UTRs, was transported in >75% of injected cells, indicating that it contains a functional RNA transport signal. Deletion of A2RE-2 from HIV-1 vpr RNA (HIV-1 vpr ΔA2RE-2) reduced transport to <25% of injected cells, indicating that A2RE-2 is necessary for RNA transport. A single point mutation (A8G) in A2RE-2 (HIV-1 vpr A8G) abolished transport of vpr RNA (<25% transport positive). Since this mutation is in the third position of a codon, it alters the nucleotide sequence of vpr RNA without affecting the amino acid sequence of Vpr protein, indicating that the RNA trafficking function of A2RE-2 is nucleotide sequence dependent rather than amino acid sequence dependent. HIV-2 vpr RNA, which is 50% homologous to HIV-1 in the region of A2RE-2, was transported in >75% of injected cells, indicating that the RNA trafficking function of A2RE-2 is conserved between HIV-1 and HIV-2.
HIV-1 tat RNA, which also contains A2RE-2, was transported less efficiently (50% transport positive) than HIV-1 vpr RNA (>75% transport positive), indicating that the transport activity of A2RE-2 is sequence context dependent. Deletion of A2RE-2 from HIV-1 tat RNA (HIV-1 tat ΔA2RE-2) abolished transport of tat RNA (25% transport positive), indicating that transport of tat RNA, albeit less efficient than that of vpr RNA, is nevertheless A2RE-2 dependent. Inclusion of a portion (58 bp) of the tat 5′ UTR (HIV-1 Ltat) abolished transport (<20% transport positive), indicating that this 5′ UTR sequence interferes with the RNA transport function of A2RE-2.
A2RE-1 or A2RE-2 were inserted into GFP mRNA, which by itself is retained in the perikaryon in oligodendrocytes. Both GFP A2RE-1 RNA and GFP A2RE-2 RNA were transported to the periphery (>75% transport positive in both cases), indicating that either A2RE-1 or A2RE-2 is sufficient to mediate RNA transport. Furthermore, since these sequences were inserted into the 3′ UTR of GFP RNA, while they are located within the ORF in gag RNA and vpr RNA, the RNA transport functions of A2RE-1 and A2RE-2 are position independent.
A2RE-mediated transport of vpr and gag RNA is hnRNP A2 dependent.
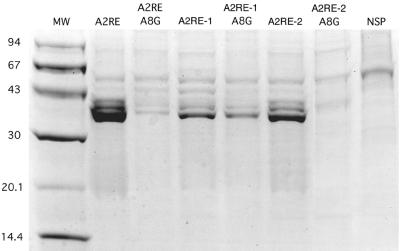
To determine if A2RE-like sequences from HIV-1 bind to hnRNP A2, the A2RE-1 and A2RE-2 sequences were immobilized on paramagnetic beads and incubated with a protein extract from rat brain containing high levels of hnRNP A2. The beads were isolated magnetically, and the bound protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 3). The spectrum of bound proteins was identical for A2RE-1, A2RE-2, and A2RE from MBP mRNA. In each case there were several minor bands and a single predominant band with an apparent molecular mass of 36 kDa, identified previously as hnRNP A2 (21). These results indicate that both A2RE-1 and A2RE-2 bind hnRNP A2 in vitro. The in vitro protein binding assays were performed with rat brain extract, while the in vivo RNA transport assays were performed with mouse oligodendrocytes. However, since rat and mouse hnRNP A2 proteins are >99% identical at the amino acid level, the results of the protein binding experiments suggest that hnRNP A2 is the cognate trans-acting factor for A2RE-mediated RNA transport.
FIG. 3.
A2RE-1 and A2RE-2 bind to hnRNP A2 in vitro. Biotin-conjugated oligoribonucleotides were immobilized on streptavidin-coated paramagnetic beads and incubated with a protein extract from rat brain. The beads were separated magnetically and washed extensively, and the proteins bound to the beads were eluted and analyzed by SDS-PAGE. The predominant protein, with a molecular mass of approximately 36 kDa, has been previously identified as hnRNP A2. The sequences of the oligoribonucleotides tested were as follows: A2RE, GCCAAGGAGCCAGAGAGCAUG; A2RE A8G, GCCAAGGGGCC; A2RE-1, GACAAGGACCAAAAGAACCCU; A2RE-1 A8G, GACAAGGGCCAAAAGAACCCU; A2RE-2, GAAAUGGAGCCAGTAGAUCCUAG; A2RE-2 A8G, GAAAUGGGGCCAGTAGAUCCUAG; and NSP, CAAGCACCGAACCGGCAACUG.
In the MBP A2RE a single point mutation (A8G) completely abolished hnRNP A2 binding in vitro and RNA transport in vivo (31). An analogous A8G mutation was introduced into A2RE-1 and A2RE-2 and tested for hnRNP A2 binding by the paramagnetic bead binding assay (Fig. 3). The A8G mutation in A2RE-1 decreased hnRNP A2 binding approximately twofold and in A2RE-2 abolished hnRNP A2 binding completely, indicating that hnRNP A2 binding is nucleotide sequence specific. Since the A8G mutation in A2RE-2 also abolished transport of vpr RNA in vivo (Fig. 2), this suggests that the RNA trafficking function of A2RE-2 requires hnRNP A2 binding.
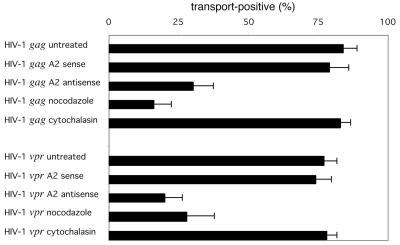
To determine if the transport of gag and vpr RNAs is hnRNP A2 dependent, oligodendrocytes were treated with antisense oligonucleotide to suppress hnRNP A2 expression prior to analysis of RNA transport. Previous work demonstrated that the treatment of oligodendrocytes with antisense oligonucleotide reduced hnRNP A2 levels at least 10-fold and inhibited the transport of A2RE-containing RNA, while the sense oligonucleotide had little effect (23). The percent transport of HIV-1 gag and vpr RNAs in sense- and antisense-treated cells is shown in Fig. 4. In sense-treated cells, the transport of gag and vpr RNA was unaffected (79 and 75% transport positive, respectively). In antisense-treated cells, the transport of both gag RNA and of vpr RNA was inhibited (30 and 20% transport positive, respectively), indicating that transport of gag and vpr RNA is hnRNP A2 dependent.
FIG. 4.
Transport of HIV-1 gag and vpr RNA is hnRNP A2 dependent and requires intact microtubules. Prior to microinjection oligodendrocytes were treated with hnRNP A2 sense oligonucleotide as a control, hnRNP A2 antisense oligonucleotide to reduce hnRNP A2 expression, nocodazole to disrupt microtubules, or cytochalasin to disrupt microfilaments. The percentages of transport-positive cells for HIV-1 gag and vpr RNA are shown.
Transport of MBP RNA by the A2RE/hnRNP A2 RNA trafficking pathway is microtubule dependent. To determine if the transport of HIV-1 gag and vpr RNAs was microtubule dependent, oligodendrocytes were treated with nocodazole, to disrupt microtubules, or with cytochalasin, to disrupt microfilaments, prior to microinjection. The percent transport of HIV-1 gag and vpr RNAs in nocodazole- and cytochalasin-treated cells is shown in Fig. 4. In nocodazole-treated cells, the transport of both gag and vpr RNAs was inhibited (16 and 28% transport positive, respectively), while in cytochalasin-treated cells, the transport was unaffected (83 and 78% transport positive, respectively). These results indicate that transport of gag and vpr RNA is microtubule dependent.
Coassembly of gag and vpr RNAs into granules.
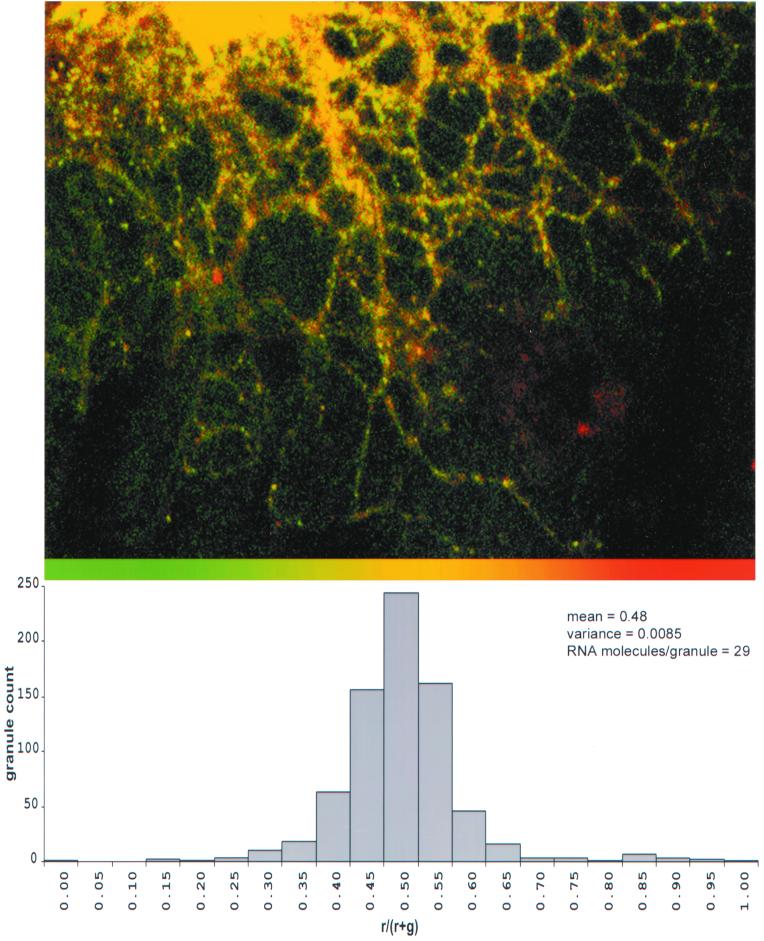
Assembly into RNA granules is an integral step in RNA trafficking (6). To determine if HIV gag and vpr RNAs are coassembled into the same granules, differentially labeled gag (fluorescein) and vpr (Texas red) RNAs were coinjected into oligodendrocytes and analyzed by dual-channel confocal microscopy (Fig. 5, top panel). In the merged image, virtually all of the granules appeared yellow, indicating colocalization of gag (green) and vpr (red) RNAs. The relative distribution of gag and vpr RNAs in individual granules was analyzed by single granule ratiometric analysis (Fig. 5, bottom panel). The histogram shows that most granules contained both gag and vpr RNA molecules. Since both gag and vpr RNAs follow the A2RE/hnRNP A2 trafficking pathway, this suggests that RNAs with similar trafficking pathways are coassembled into the same granules.
FIG. 5.
Colocalization of HIV-1 gag and vpr RNAs. Oligodendrocytes were coinjected with fluorescein-labeled gag RNA and Texas red-labeled vpr RNA. The distributions of the two RNAs were visualized by dual-channel confocal microscopy. (A) Distribution of gag RNA in the green channel and vpr RNA in the red channel. The overlap of red and green appears yellow. The ratio of gag and vpr RNA in each granule was determined by single granule ratiometric analysis. (B) The distribution of ratios for a population of 748 granules is shown as a histogram. The mean is 0.48, and the variance is 0.0085, from which a sample size of 29 RNA molecules/granule was calculated.
DISCUSSION
Two cis-acting RNA trafficking sequences, A2RE-1 and A2RE-2, have been identified in the HIV genome. Both sequences bind to the trans-acting factor hnRNP A2 in vitro and mediate hnRNP A2-dependent transport of HIV RNAs in oligodendrocytes in vivo. Since A2RE-like sequences are found in a variety of different RNAs (2) and since hnRNP A2 is expressed in many different cell types (22), it is likely that the A2RE/hnRNP A2 pathway is a constitutive RNA trafficking pathway in most cells. Therefore, A2RE-containing HIV RNAs may follow the A2RE/hnRNP A2 trafficking pathway in HIV host cells.
In HIV-infected cells, unspliced 9-kb HIV-1 RNA, which serves as both genomic RNA and gag mRNA, contains both A2RE-1 and A2RE-2, while singly spliced vpr and vif mRNAs and multiply spliced tat mRNA contain A2RE-2 alone. Most of the HIV RNAs analyzed in this work lacked the 5′ and/or 3′ UTRs found in HIV RNAs expressed in infected cells. If the missing regions contain sequences that modulate A2RE-1 or A2RE-2 function or non-A2RE RNA trafficking signals, the trafficking pathways for HIV RNAs in infected cells might be different from the trafficking pathways reported here for truncated HIV RNAs in oligodendrocytes.
The sequence context does affect the trafficking function of A2RE-2 in tat RNA which was transported less efficiently than vpr RNA. One possibility is that the RNA trafficking function of A2RE-2 is position dependent. A2RE-2 is located near the 3′ end of the ORF in vpr RNA and near the 5′ end of the ORF in tat RNA, which may affect its function. This is unlikely because the A2RE in MBP RNA is located in the 3′ UTR, whereas A2RE-2 in vpr RNA is located within the ORF and also functions when inserted into the 3′ UTR of GFP RNA. A second possibility is that the tat ORF and/or 5′ UTR contain perikaryon retention signals that are epistatic to the transport function of A2RE-2, causing retention of tat RNA in the perikaryon, despite the presence of A2RE-2. This is also unlikely because the 5′ UTR sequences that are present in tat RNA, which is not transported efficiently, are also present in gag RNA, which is transported efficiently. A third possibility is that the tat ORF and/or 5′ UTR contain sequences that interfere with the RNA transport function of A2RE-2. RNA transport requires binding of the trans-acting ligand hnRNP A2 to A2RE. RNA secondary structure in the region of the A2RE could interfere with hnRNP A2 binding, thereby reducing RNA transport efficiency. The potential secondary structure in the A2RE-2 region of vpr and tat RNA was examined using the MFOLD program in the GCG Sequence Analysis Package (University of Wisconsin) (data not shown). In vpr RNA no stable secondary structure that might interfere with hnRNP A2 binding to A2RE-2 was predicted. In tat RNA, on the other hand, the A2RE-2 region was predicted to base pair with a downstream region of the ORF to form a stable stem-loop structure that could potentially interfere with hnRNP A2 binding and reduce the transport efficiency of tat RNA compared to vpr RNA. Furthermore, a complicated secondary structure consisting of several stem-loops has been predicted in the 5′ UTR of tat RNA (30). Juxtaposition of this 5′ UTR secondary structure with A2RE-2 in the tat ORF could reduce hnRNP A2 binding, thereby inhibiting transport of tat RNA. Differences in RNA secondary structure resulting in differences in hnRNP A2 binding to A2RE-2 could explain the differential transport efficiencies for vpr and tat RNAs.
A single base change at the A8 position in A2RE-1 or -2 was sufficient to inhibit hnRNP A2 binding and, in the case of A2RE-2, to abrogate vpr RNA transport. Sequence polymorphism at the A8 position in A2RE-1 and A2RE-2 was analyzed in HIV-1 sequences from the NCBI database. The A8G mutation in A2RE-1, which reduced but did not abolish hnRNP A2 binding, was present in ca. 40% of HIV-1 isolates (Fig. 1), indicating that it represents a frequently occurring sequence polymorphism in the HIV-1 genome. This suggests that this mutation does not have significant deleterious phenotypic consequences for the virus, implying that the residual hnRNP A2 binding observed with this mutation is sufficient for A2RE-1 function. On the other hand, the A8G mutation in A2RE-2, which abolished hnRNP A2 binding and abrogated vpr RNA transport, was found in only three HIV-1 sequences (all from Australia) out of 1,074 different HIV-1 sequences in the database. One sequence (accession number U61889) was isolated from a long-term nonprogressor (39), a second sequence with several variants characterized by high replication in macrophages (accession numbers AF133405, AF133410, AF133415, and AF133420) was isolated from an individual with category IV AIDS (32), and a third sequence (accession number U41708) was isolated from an intravenous drug user (19). Since each of these isolates carries additional sequence polymorphisms in other regions of the genome, their phenotypes are not necessarily attributable to the A8G mutation in A2RE-2. Nevertheless, the infrequent occurrence of the A8G mutation in the HIV genome implies that hnRNP A2 binding is important for A2RE-2 function.
Identification of A2RE-like RNA trafficking signals in the HIV genome suggests that in infected cells HIV RNAs follow the constitutive A2RE/hnRNP A2 RNA trafficking pathway, as outlined in Fig. 6. According to this model, hnRNP A2 binds to A2RE-containing HIV RNAs in the nucleus and remains associated throughout subsequent trafficking. Although not analyzed in this study, nuclear processing of HIV RNAs may be affected by hnRNP A2 and A2RE determinants. Exon splicing silencing by cis-acting ESS sequences in viral and cellular RNAs is mediated by hnRNP A2 (10, 16), and A2RE-2 is immediately adjacent to ESS2 in HIV RNA. A recent model describing coupled HIV-1 RNA splicing and RNA transport (27) proposes that several RNA-binding proteins are involved in premature spliceosome dissolution on unspliced and singly spliced HIV-1 transcripts destined for Rev-mediated nuclear export. Splicing and transport of HIV RNAs could be coupled via A2RE and hnRNP A2 determinants.
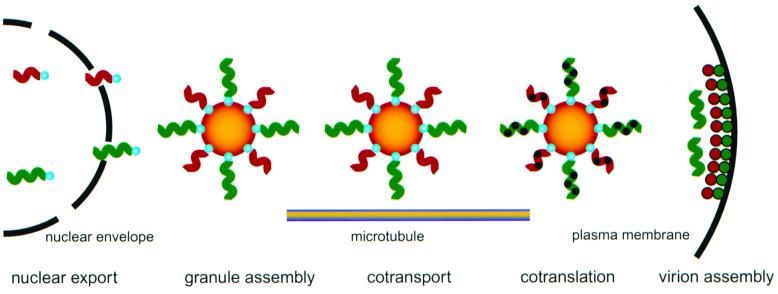
FIG. 6.
Model for hnRNP A2/A2RE-mediated intracellular trafficking of HIV RNAs. gag RNA (green squiggles) and vpr RNA (red squiggles) both contain A2RE-like sequences that bind to hnRNP A2 (blue spheres) in the nucleus. In the cytoplasm, multiple gag and vpr RNA molecules, with hnRNP A2 associated, coassemble into granules (orange spheres). Granules containing both gag and vpr RNA are transported to the plus ends of microtubules in the periphery of the cell where the Gag protein (green spheres) and the Vpr protein (red spheres) are cotranslated. Gag and Vpr proteins coassemble with HIV genomic RNA (green squiggles) into virions at the plasma membrane.
Nuclear export of unspliced and singly spliced HIV transcripts is mediated by the cis-acting RRE (28), which is recognized by the trans-acting viral Rev protein (36). In the cytosol, RRE-containing RNAs are either translated on polyribosomes or, in the case of full-length, unspliced HIV RNA, encapsidated into progeny virions. There are certain parallels between the previously characterized Rev/RRE pathway and the hnRNP A2/A2RE pathway described here. Both Rev and hnRNP A2 bind to their respective target RNAs in the nucleus, remain associated during export through the nuclear pore, and enhance translation in the cytoplasm (9, 23, 43). Since unspliced and singly spliced HIV RNAs contain both RRE and A2RE-1 and/or A2RE-2, trafficking of these RNAs may be mediated by the combined actions of Rev and hnRNP A2. The Rev/RRE pathway determines which HIV RNAs are exported from the nucleus. The hnRNP A2/A2RE pathway may determine where these RNAs are transported in the cytoplasm.
In the perikaryon, HIV RNAs, containing either A2RE-1 or A2RE-2, are coassembled into granules for subsequent trafficking. Coassembly into the same granules ensures that A2RE-containing RNAs are cotransported and cotranslated in the same region of the cell. The HIV RNAs analyzed here each contained a single A2RE-like sequence. In HIV-infected cells, unspliced gag RNA contains two A2RE-like sequences, A2RE-1 and A2RE-2, while singly spliced vpr and vif RNAs contain only A2RE-2. If the stoichiometry of assembly into granules is determined by the number of A2RE-like sequences in each RNA, then unspliced gag mRNA may be assembled into granules more efficiently than singly spliced vpr and vif RNAs. This could result in proportionately higher levels of gag RNA compared to vpr and vif RNAs in each granule, leading to higher levels of Gag protein expression compared to the Vpr and Vif proteins. The stoichiometries of different viral RNAs in each granule could provide a mechanism for coordinating the expression of the corresponding viral proteins.
The total number of RNA molecules per granule can be calculated from the distribution of ratios shown in Fig. 5. If the number of RNA molecules in each granule corresponds to a sample of size N, from a binomial population of vpr (red) and gag (green) RNA molecules, then the ratio for each granule corresponds to the mean for that sample, and the distribution of ratios represents the sampling distribution of the means for samples of size N. According to the central limit theorem, as the sample size increases, the distribution of the sample means approaches normality. The variance, V, in the distribution of ratios is inversely proportional to the sample size, N, according to the following equation: V = RG/N, where R is the mean proportion of red molecules (vpr RNA) and G is the mean proportion of green molecules (gag RNA) in the population. The calculated variance for the distribution of ratios of vpr RNA and gag RNA was 0.0085. This corresponds to an estimated mean sample size of 29, which means that each granule contains approximately 29 RNA molecules.
This estimate is based on several assumptions. One assumption is that the observed variance is entirely statistical with no experimental component. Experimental factors that could potentially contribute to the variance include bias in granule selection, misregistration between the red and green channels due to a chromatic aberration in the optical path, and differences in signal-to-noise, sensitivity, or nongranule background fluorescence values between the two channels. These experimental factors could either increase or decrease the observed variance, leading to underestimation or overestimation of the actual number of RNA molecules per granule. A second assumption is that the number of RNA molecules is the same for all granules. Granules with lower intensity will contribute disproportionately more to the observed variance in the population, leading to underestimation of the mean number of RNA molecules per granule, while granules with higher intensity will contribute less to the variance, leading to overestimation. A third assumption is that each granule contains exclusively gag and vpr RNAs. Oligodendrocytes are known to express several endogenous A2RE-containing RNAs, such as MBP mRNA and MOBP mRNA (2), that may coassemble into the same granules as gag and vpr RNAs. Since endogenous A2RE-containing RNAs are unlabeled, the actual number of RNA molecules per granule may be greater than the estimate made from the ratios of labeled gag and vpr RNAs. Since these assumptions are difficult to test directly, there is some uncertainty in the estimated number of RNA molecules per granule.
Granules containing HIV RNAs with either A2RE-1 and/or -2 are transported toward the plus ends of microtubules in the periphery of the cell, where translation is activated and virion assembly occurs. A2RE-mediated transport of HIV RNAs may affect localization of the encoded proteins. Tat RNA, which is not transported, encodes a protein destined for the nucleus, while gag and vpr RNAs, which are transported, encode proteins that are incorporated into virions. Vif RNA, which was not tested in the transport assay, also contains A2RE-2, and Vif protein colocalizes with Gag protein in infected cells (41). A2RE-mediated RNA transport may steer the expression of Gag and Vpr (and possibly Vif) proteins to sites of virion assembly, while retention of tat RNA in the cell body may restrict expression of Tat protein to the perikaryon.
Translation of A2RE-containing RNA is activated through an hnRNP A2-dependent mechanism. In other systems translation is repressed while the RNA is in transit, preventing ectopic expression of the encoded proteins. If translation of HIV gag and vpr RNA is repressed until the RNA is localized to the periphery of the cell, this would restrict the expression of Gag and Vpr proteins to the periphery of the cell, close to the plasma membrane where virion assembly occurs. Both Gag and Vpr proteins contain strong nuclear import signals, which might cause them to be imported into the nucleus if the proteins were expressed at high levels in the perikaryon. Restricting the translation of gag and vpr RNAs to the cell periphery could provide a mechanism to effectively steer expression of Gag and Vpr proteins away from the nucleus and toward the sites of virion assembly.
HIV genomic RNA contains both A2RE-1 and -2 and therefore presumably binds to hnRNP A2 and is assembled into granules for transport to sites of virion assembly. However, hnRNP A2 is not detected in mature virus particles (unpublished observations). This means that genomic RNA molecules must dissociate from hnRNP A2 and be released from granules prior to encapsidation. The mechanism(s) for the dissociation of HIV genomic RNA from hnRNP A2 and release from granules is not known.
In summary, the HIV genome contains specific sequences that can mediate intracellular trafficking of HIV RNAs through the A2RE/hnRNP A2 RNA pathway. Intracellular RNA trafficking could potentially affect various aspects of HIV gene expression including splicing, nuclear export, cytoplasmic transport, translation, and virion assembly. The A2RE/hnRNP A2 RNA pathway may play an important role in the HIV life cycle.
ACKNOWLEDGMENTS
This work was supported by NIH grant NS15190 to J.H.C.; NIH grant NS19943 and National Multiple Sclerosis Society grant RG2843 to E.B.; Australian National Health and Medical Research Grant to R.S.; NIH grant AI47008 to D.R.; Shoppers DrugMart Research Grant, Canadian Foundation for AIDS Research, to A.J.M.; and a Medical Research Council of Canada grant to E.A.C.
We thank Andrew Lever and Michael Laughrea for DNA constructs. Frank Morgan (University of Connecticut Health Center, Farmington) developed specialized computer programs for analysis of sequence polymorphism and single granule ratiometric analysis and also helped in the preparation of figures. Confocal imaging was performed in the Center for Biomedical Imaging Technology at the University of Connecticut Health Center (Farmington).
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill S J, Barry C, Barbarese E, Carson J H. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ainger K, Avossa D, Diana A S, Barry C, Barbarese E, Carson J H. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt B A, Si Z-H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arya S K, Guo C, Josephs S F, Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III. Science. 1985;229:69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- 5.Bachand F, Yao X J, Hrimech M, Rougeau N, Cohen E A. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J Biol Chem. 1999;274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 6.Barbarese E, Koppel D E, Deutscher M P, Smith C L, Ainger K, Morgan F, Carson J H. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- 7.Braun I, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell L H, Borg K T, Haines R T, Moon D R, Schoenberg D R, Arrigo S J. Human immunodeficiency virus type 1 Rev is required in vivo for binding of poly(A)-binding protein to Rev-dependent RNAs. J Virol. 1994;68:5433–5438. doi: 10.1128/jvi.68.9.5433-5438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi M, Mayeda A, Krainer A D, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson J H, Worboys K, Ainger K, Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil Cytoskeleton. 1997;38:318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Carson J H, Kwon S, Barbarese E. RNA trafficking in myelinating cells. Curr Opin Neurosci. 1998;8:607–612. doi: 10.1016/s0959-4388(98)80088-3. [DOI] [PubMed] [Google Scholar]
- 13.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 14.Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 15.Daneholt B. Pre-mRNP particles: from gene to nuclear pore. Curr Biol. 1999;9:R412–R415. doi: 10.1016/s0960-9822(99)80256-5. [DOI] [PubMed] [Google Scholar]
- 16.Del Gatto-Konczak F, Gesnel M C, Olive M, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshler J O, Highett M I, Schnapp B J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 18.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 19.Ge Y C, Wang B, Dwyer D E, Xiang S-H, Cunningham A L, Saksena N. Length polymorphism of the viral protein R of human immunodeficiency virus type 1 strains. AIDS Res Hum Retrovir. 1996;12:351–354. doi: 10.1089/aid.1996.12.351. [DOI] [PubMed] [Google Scholar]
- 20.Hammarskjöld M-L. Regulation of retroviral RNA transport. Semin Cell Dev Biol. 1997;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- 21.Hoek K S, Kidd G J, Carson J H, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 22.Kamma H, Horiguchi H, Wan L, Matsui M, Fujiwara M, Fujimoto M, Yazawa T, Dreyfuss G. Molecular characterization of the hnRNP A2/B1 proteins: tissue specific expression and novel isoforms. Exp Cell Res. 1999;246:399–411. doi: 10.1006/excr.1998.4323. [DOI] [PubMed] [Google Scholar]
- 23.Kwon S, Barbarese E, Carson J H. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 function to enhance cap-dependent translation. J Cell Biol. 1999;147:247–256. doi: 10.1083/jcb.147.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lall S, Francis-Lang H, Flament A, Norvell A, Schupbach T, Ish-Horowicz D. Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell. 1999;98:171–180. doi: 10.1016/s0092-8674(00)81012-0. [DOI] [PubMed] [Google Scholar]
- 25.Laughrea M, Kleiman L, Jette L, Liang C, Mak J, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–83. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Reddy T R, Tang H, Rose D W, Mullen T M, Wong-Staal F, Westberg C. A role for RNA helicase A in post-transcriptional regulation of HIV 1. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim M H, Maizel J V, Hauber J, Cullen B R, Le S Y. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–2577. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 29.Mayeda A, Munroe S H, Caceres J F, Krainer A R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miele G, Mouland A, Harrison G P, Cohen E, Lever A M. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munro T P, Magee R J, Kidd G J, Carson J H, Barbarese E, Smith L M, Smith R. Mutational analysis of a heterogeneous ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- 32.Naif H M, Li S, Alali M, Chang J, Mayne C, Sullivan J, Cunningham A L. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocyte-derived macrophages by using twins. J Virol. 1999;73:4866–4881. doi: 10.1128/jvi.73.6.4866-4881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkin N T, Cohen E A, Darveau A, Rosen C, Haseltine W, Sonenberg N. Mutational analysis of the 5′ non′ coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patarca R, Haseltine W A. A major retroviral core protein related to EPA and TIMP. Nature. 1985;318:390. doi: 10.1038/318390a0. [DOI] [PubMed] [Google Scholar]
- 36.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1999;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 37.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immonodeficiency virus type 1 gag gene: effects on virion particle assembly, release and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross A, Oleynikov Y, Kislauskis E, Taneja K, Singer R. Characterization of a beta actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saksena N K, Ge Y C, Wang B, Xiang S H, Dwyer D E, Randle C, Palasanthiran P, Ziegler J, Cunningham A L. An HIV-1 infected long-term non-progressor (LTNP): molecular analysis of HIV-1 strains in the vpr and nef genes. Ann Acad Med Singapore. 1996;25:848–854. [PubMed] [Google Scholar]
- 40.Si Z, Amendt B A, Stoltzfus C M. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon J H M, Carpenter E A, Fouchier R A M, Malim M H. Vif and the P55 Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizating membrane free cytoplasmic complexes. J Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srinivasakumar N, Hammarskjold M L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and gag-pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 44.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]