Abstract
Although prostate adenocarcinoma lacks distinguishable histopathological subtypes, prostate cancer displays significant inter- and intratumor heterogeneity at the molecular level and with respect to disease prognosis and treatment response. In principle, understanding the basis for prostate cancer heterogeneity can help distinguish aggressive from indolent disease, and help overcome castration-resistance in advanced prostate cancer. In this review, we will discuss recent advances in understanding the cell types of origin, putative cancer stem cells, and tumor plasticity in prostate cancer, focusing on insights from studies of genetically engineered mouse models (GEMMs). We will also outline future directions for investigating tumor heterogeneity using mouse models of prostate cancer.
Keywords: cell of origin, cancer stem cell, lineage plasticity, castration-resistance, genetically engineered mouse model, tumor heterogeneity
1. Introduction
Unlike most other epithelial tumors, prostate cancer lacks distinguishable histopathological subtypes. Almost all prostate cancers are acinar adenocarcinomas, whereas histological variants such as ductal adenocarcinoma, basal cell carcinoma, and neuroendocrine prostate cancer are rare [1, 2]. Nonetheless, prostate adenocarcinoma displays significant inter- and intratumor heterogeneity at the genomic level [3–10], and can have significant differences in disease severity. Notably, patients with low- to intermediate-grade localized primary prostate cancer can have widely different outcomes, ranging from indolent to highly aggressive disease [11, 12].
Since prostate cancer initially relies on ligand-mediated signaling through androgen receptor (AR) for tumor growth, androgen deprivation has been a mainstay of prostate cancer treatment [13, 14]. Although androgen deprivation therapy results in cancer regression, tumors frequently recur through restoration of AR signaling by a variety of molecular mechanisms, including de-regulation of the AR signaling pathway or activation of alternative signaling pathways, resulting in castration-resistant prostate cancer (CRPC) [14]. In recent years, CRPC has been effectively treated by second-generation anti-androgens such as abiraterone and enzalutamide [14, 15]. However, the extent and duration of response are variable, with many tumors developing lineage plasticity and AR-negative phenotypes, including aggressive neuroendocrine prostate cancers (NEPC) that progress to lethal disease [15]. Notably, genomic analyses have revealed that CRPC displays a high degree of inter- and intratumor heterogeneity [9, 10, 16–18]. Thus, a better understanding of the mechanisms that generate inter- and intratumor heterogeneity could help distinguish aggressive from indolent disease and inhibit emergence of castration-resistance.
The origins of inter- and intratumor cancer heterogeneity can potentially be explained by the “cell of origin” and “cancer stem cell” models, respectively. The cell of origin model proposes that differences in the normal cell type that undergoes oncogenic transformation give rise to distinct tumor subtypes that differ in histopathological or molecular properties as well as treatment response or disease outcome (Fig. 1A) [19–22]. However, the relevance of this model to prostate cancer has been unclear since it has been difficult to class prostate adenocarcinoma into distinct tumor subtypes at either the histopathological or molecular level [23]. Furthermore, until recently it has been unclear whether there is considerable cell type heterogeneity in the normal prostate epithelium.
Fig. 1.
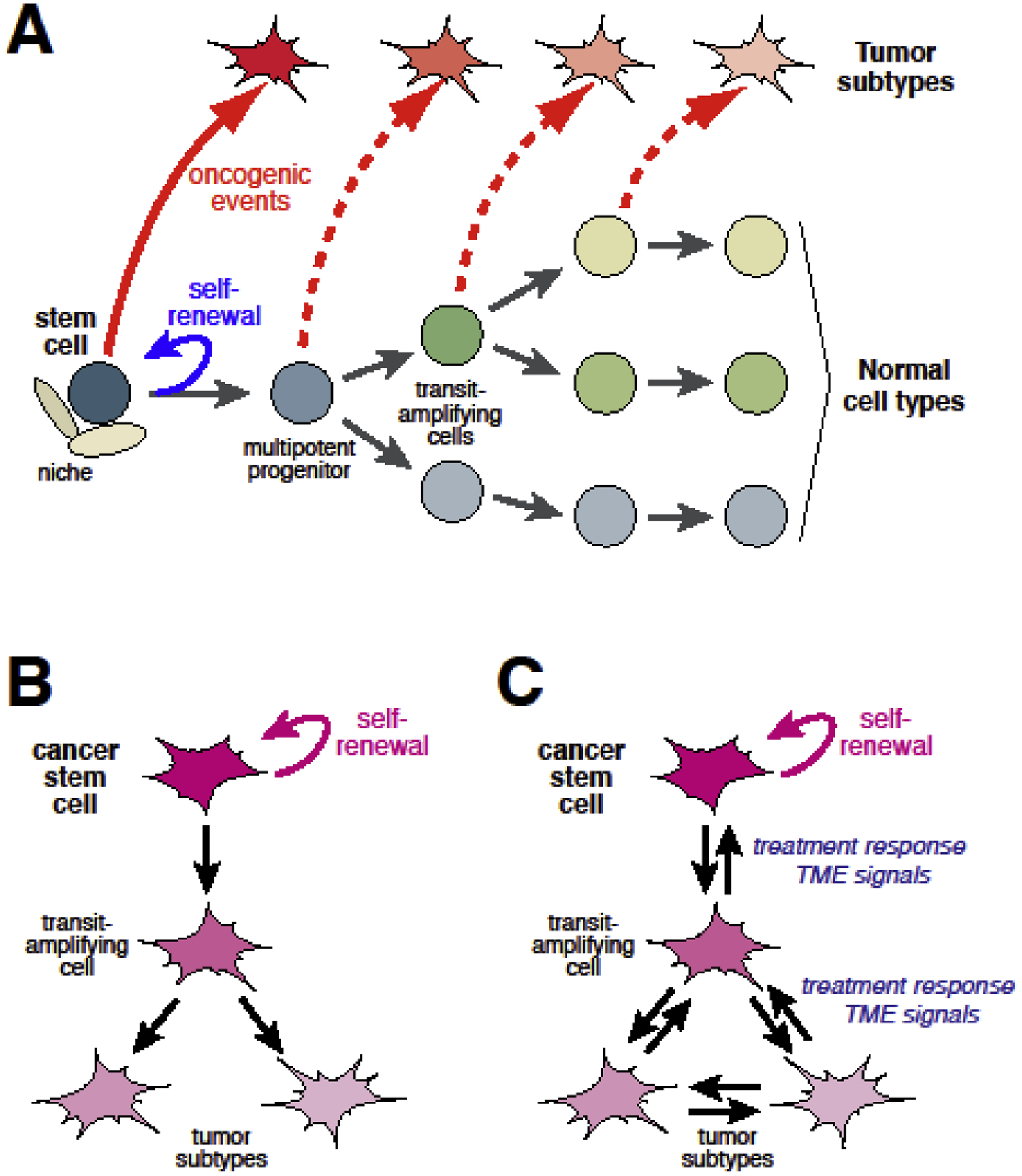
Models of cancer heterogeneity. (A) Cell of origin model. A schematic representation of the lineage hierarchy within an epithelial tissue, at the top of which resides a normal stem cell with properties of multipotency and self-renewal. Different tumor subtypes may arise from oncogenic transformation of the stem cell or non–stem cells within the lineage hierarchy (adapted from [117]. (B) Cancer stem cell model. Upon asymmetric division, a cancer stem cell can give rise to itself and a transit-amplifying cell, which in turn divides and undergoes differentiation to generate non-tumorigenic cancer cells. (C) Cellular plasticity in the cancer stem model. Differentiated non-tumorigenic cancer cells can convert to other tumor cell types via transdifferentiation or can be reprogrammed to cancer stem cells, potentially by modulation from intrinsic and/or extrinsic factors such as oncogenic insults and signals from the tumor microenvironment.
The cancer stem cell model (Fig. 1B) posits that tumor cells are organized in a hierarchy of cancer stem cells and nontumorigenic cells to generate tumor heterogeneity, thereby accounting for progression and treatment response [22, 24–26]. While considerable evidence supporting the cancer stem cell model exists in hematological malignancies, this model is much more controversial in solid tumors such as prostate. Moreover, recent studies using lineage-tracing in GEMMs of various epithelial tumors have also shown that nontumorigenic cells can display lineage plasticity and acquire properties of stemness or transdifferentiate to other types of nontumorigenic cells in response to treatment or signals from the tumor microenvironment (Fig. 1C), further complicating analyses of putative cancer stem cells [24, 26, 27].
In the review, we will discuss recent advances in understanding prostate cancer heterogeneity and the emergence of castration resistance through the lens of the cell of origin and cancer stem cell models. In particular, we will focus on progress and insights from studies of GEMMs for prostate cancer, and outline directions for future research.
2. Epithelial heterogeneity and plasticity in the normal prostate
The prostate is a male sex accessory gland that produces and secretes fluids that contribute to the ejaculate, and thereby enhances male fertility. In men, the prostate is a walnut-sized tissue surrounding the urethra at the base of the bladder, containing a network of branching ducts with a zonal architecture, corresponding to central, periurethral transition and peripheral zones [28, 29]. In contrast, the mouse prostate consists of multiple lobes that have distinct patterns of ductal branching and histological appearance [30, 31]. Despite these anatomical differences, recent single-cell RNA-sequencing analyses have indicated the conservation of cell populations between the mouse and human prostate [32, 33]. In particular, the mouse lateral lobe is most similar to the human peripheral zone, which harbors the majority of human prostate cancers [33, 34].
In both mouse and human, the prostate contains a pseudostratified epithelium that is composed of three primary cell types, corresponding to luminal cells, basal cells, and rare neuroendocrine cells [23, 35]. Lineage-tracing studies in vivo have shown that differentiated luminal and basal cells are largely maintained by unipotent progenitors during prostate homeostasis [36–38]. Similar conclusions have been provided by lineage reconstruction analyses using patterns of mitochondrial mutations and genome-wide spontaneous somatic mutations in human prostate [39–42]. Interestingly, a recent study has reported an expansion of luminal progenitor cells in the aging mouse and human prostate, raising the possibility that microenvironmental cues may modulate luminal differentiation during aging [43]. During organogenesis, however, both basal and luminal cells can display progenitor features. Bipotent basal progenitors can be detected from birth until approximately four weeks of age [44–46], whereas bipotent luminal progenitors are more transient and are only detectable in the first week postnatally [47]. Similarly, there is evidence for both bipotent basal and luminal progenitors contributing to androgen-mediated regeneration of the regressed prostate following castration [48–54].
Recent single-cell RNA-sequencing studies have demonstrated that luminal cells are heterogenous in both the mouse and human prostate [32, 33, 55–58]; there is also some reported evidence for heterogeneity in the basal population [53]. In the mouse, there is heterogeneity along the proximal-distal axis, with more distal luminal cells displaying the tall columnar secretory morphology typically associated with the luminal phenotype, and the proximal luminal cells displaying a more cuboidal non-secretory phenotype [32, 33, 57, 58] (Fig. 2). These distinct phenotypes are also associated with different progenitor potential in ex vivo assays, as the proximal luminal cells have increased organoid formation and grafting efficiency [32, 33, 57, 58]. Furthermore, these different luminal populations also appear to be conserved in the benign human prostate [32, 33].
Fig. 2.
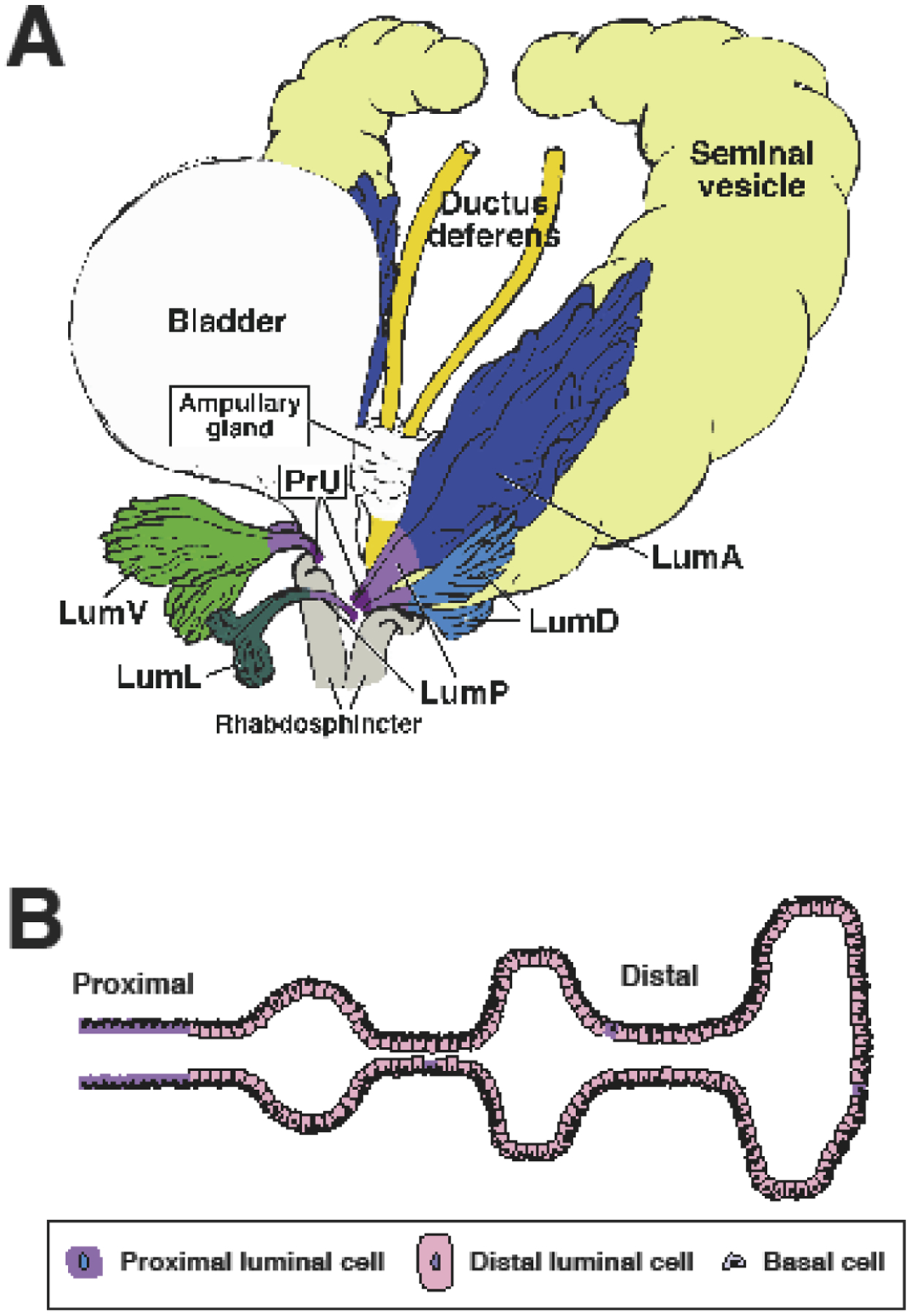
Epithelial heterogeneity in the adult mouse prostate. (A) Schematic depiction of prostate lobes indicating the distribution of luminal epithelial populations (adapted from [33]). (B) Proximal-distal heterogeneity of luminal epithelial cells. Note that cells with properties of proximal luminal cells occur sporadically in distal regions. Lum A: distal luminal cells in anterior lobe; Lum D: distal luminal cells in dorsal lobe; Lum L: distal luminal cells in lateral lobe; Lum V: distal luminal cells in ventral lobe; PrU: periurethral; LumP: proximal luminal cells.
In the adult mouse prostate, both basal and luminal cells can exhibit considerable plasticity in specific physiological contexts. Notably, basal-to-luminal differentiation can occur in contexts of tissue repair following epithelial damage or inflammation, as well as in ex vivo experimental assays involving their isolation from dissociated tissue (Fig. 3). Following cell death of luminal cells induced by deletion of E-cadherin, basal cells can differentiate into luminal cells to repair the prostate epithelium [59]. Similarly, in a mouse model of bacterial prostatitis, tissue damage also triggered basal-to-luminal differentiation [60]. Plasticity can also be observed in luminal cells, as distal luminal cells can acquire a more proximal progenitor-like state after castration, and can regenerate the distal luminal compartment after androgen re-administration [32]. Therefore, the potential plasticity of basal and luminal cells as well as their progenitor capabilities should be considered when investigating the heterogeneity of prostate cancer, particularly with respect to the cell of origin and cancer stem cell models, as described below.
Fig. 3.
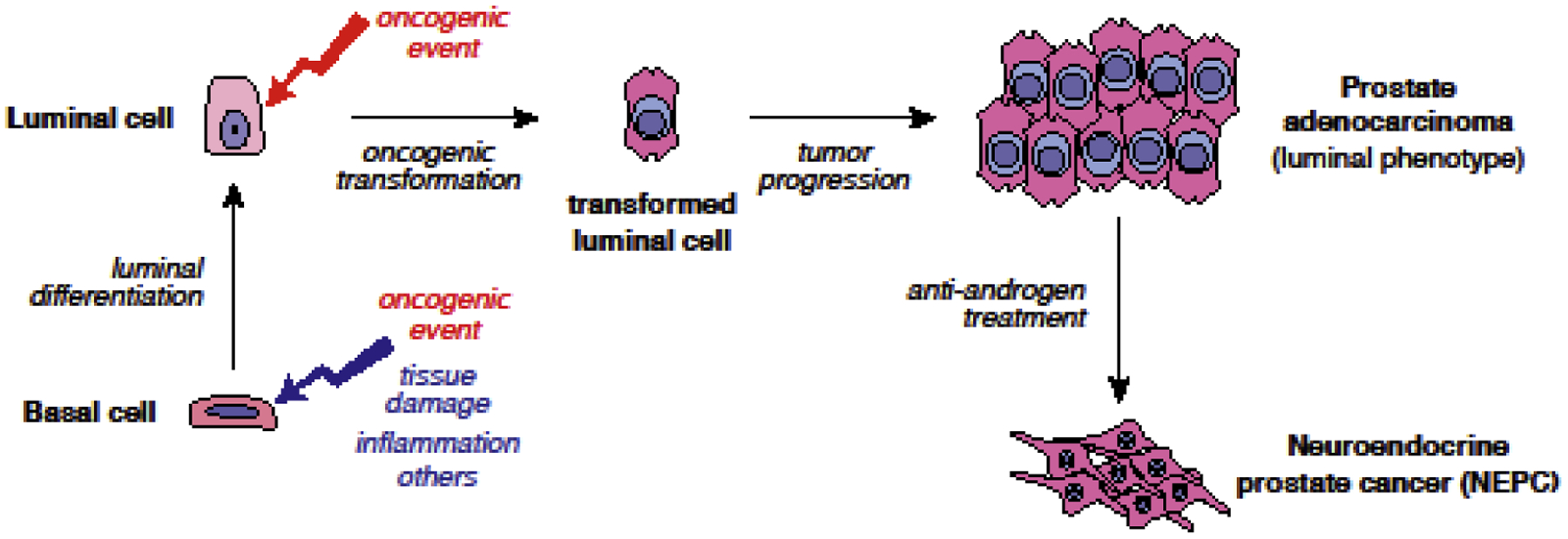
Cellular plasticity in prostate tumorigenesis. Following tissue damage, oncogenic insults, or inflammation and other factors, basal cells can undergo luminal differentiation. Transformed luminal cells can directly form prostate adenocarcinoma whereas basal cells need to undergo basal-to-luminal differentiation to generate luminal adenocarcinomas. During tumor progression and anti-androgen treatment, luminal adenocarcinoma cells can transdifferentiate to neuroendocrine-like cells. NEPC: neuroendocrine prostate cancer.
3. Cell types of origin for prostate cancer
The cell types of origin for prostate cancer have been investigated extensively using genetically-engineered mouse models [19, 23, 61–63]. Multiple studies have performed oncogenic transformation ex vivo followed by organoid culture and/or renal grafting, which have suggested that both basal and luminal cells can be cell types of origin for prostate cancer [64–70]. However, given the plasticity of basal cells in ex vivo assays, these results are potentially difficult to interpret with respect to in vivo contexts.
Thus, to identify cell types of origin in vivo, several groups have used Cre-mediated recombination together with Cre reporter alleles such as R26R-YFP or mTmG to mark and follow the fates of induced tumor cells in GEMMs (Table 1). For example, inducible GEMMs such as CK5-CreERT2;Ptenflox/flox, CK14-CreERT2;Ptenflox/flox, CK8-CreERT2;Ptenflox/flox, PSA-CreERT2;Ptenflox/flox, Tmprss2-CreERT2;Ptenflox/flox, and Nkx3.1-CreERT2; Ptenflox/flox have been used to study the consequences of deletion of the Pten tumor suppressor in adult prostate basal cells or luminal cells [36–38, 71, 72]. Although inducible deletion of Pten in either basal or luminal cells resulted in high-grade PIN and adenocarcinoma, Pten-deleted basal cells undergo a basal-to-luminal differentiation to result in a luminal tumor phenotype [36, 38, 60, 65]. Bioinformatic analyses of expression profiles from these tumors showed that luminal-derived tumors were more aggressive than basal-derived tumors, and identified a molecular signature that could stratify human prostate tumors according to clinical outcome [36]. Related studies have shown that luminal progenitor cells in the regressed prostate, including castration-resistant Nkx3.1-expressing cells (CARNs) and castration-resistant Bmi1-expressing cells (CARBs), can also serve as cells of origin [49–51]. Finally, lineage-tracing analyses in a diverse range of GEMMs, including Nkx3.1+/−;Pten+/−, Hi-Myc, and TRAMP mice, as well as a hormonal carcinogenesis model, have indicated that luminal cells are favored as the cell of origin in each of these tumor models [73]. Consistent with these findings, recent bioinformatic analyses of human prostate tumors have been able to define distinct basal and luminal molecular subtypes that are associated with different clinical outcomes [74].
Table 1.
Representative GEMMs used in studies of prostate cancer heterogeneity and plasticity
| Type | Name | Description | References |
|---|---|---|---|
| Basal cell origin and prostate cancer | CK5-CreERT2;Ptenflox/flox | Conditional deletion of Pten in keratin 5 expressing basal cells. Basal cell-derived prostate lesions exhibited PIN and luminal adenocarcinoma phenotype. | [36, 38] |
| CK5-CreERT2; Nkx3.1+/−;Pten+/− | Compound germline deletion of Pten and Nkx3.1. Keratin 5 expressing basal cells are lineage-marked and are not favored as cells of origin for prostate cancer. | [73] | |
| CK5-CreERT2; Hi-Myc | c-Myc driven by ARR2PB promoter. Keratin 5 expressing basal cells are lineage-marked and are not favored as cells of origin for prostate cancer. | [73] | |
| CK5-CreERT2; TRAMP | SV40 large tumor antigen (Tag) driven by a minimal rat probasin promoter (rPB). Keratin 5 expressing basal cells are lineage-marked and are not favored as cells of origin for prostate cancer. | [73] | |
| CK14-CreERT2;Ptenflox/flox | Conditional deletion of Pten in keratin 14 expressing basal cells. Basal cell-derived prostate lesions exhibited PIN and luminal adenocarcinoma phenotype. | [37] | |
| Luminal cell origin and prostate cancer | CK8-CreERT2;Ptenflox/flox | Conditional deletion of Pten in keratin 8 expressing luminal cells. Induction of PIN and prostatic luminal adenocarcinoma. | [36–38] |
| CK8-CreERT2;Nkx3.1+/−;Pten+/− | Compound germline deletion of Pten and Nkx3.1. Keratin 8 expressing luminal cells are lineage-marked and are favored as cells of origin for prostate cancer. | [73] | |
| CK8-CreERT2;Hi-Myc | c-Myc driven by ARR2PB promoter. Keratin 8 expressing luminal cells are lineage-marked and are favored as cells of origin for prostate cancer. | [73] | |
| CK8 -CreERT2;TRAMP | SV40 large tumor antigen (Tag) driven by a minimal rat probasin promoter (rPB). Keratin 8 expressing luminal cells are lineage-marked and are favored as cells of origin for prostate cancer. | [73] | |
| PSA-CreERT2;Nkx3.1+/−;Pten+/− | Compound germline deletion of Pten and Nkx3.1. PSA expressing luminal cells are lineage-marked and are favored as cells of origin for prostate cancer. | [73] | |
| PSA-CreERT2;Hi-Myc | c-Myc driven by ARR2PB promoter. PSA expressing luminal cells are lineage-marked and are favored as cells of origin for prostate cancer. | [73] | |
| PSA -CreERT2;TRAMP | SV40 large tumor antigen (Tag) driven by a minimal rat probasin promoter (rPB). PSA expressing luminal cells are lineage-marked and are favored as cells of origin for prostate cancer. | [73] | |
| PSA-CreERT2;Ptenflox/flox | Conditional deletion of Pten in PSA expressing luminal cells. Induction of PIN and prostatic luminal adenocarcinoma. | [71] | |
| Tmprss2-CreERT2;Ptenflox/flox | Conditional deletion of Pten in Tmprss2 expressing luminal cells. Induction of PIN and prostatic luminal adenocarcinoma. | [72] | |
| Nkx3.1-CreERT2;Ptenflox/flox | Conditional deletion of Pten in distal luminal cells under homeostasis or Nkx3.1-marked luminal progenitor cells after castration. Induction of prostatic luminal adenocarcinoma in both conditions. | [36, 49, 51] | |
| Bmi1-CreERT2;Ptenflox/flox | Conditional deletion of Pten in Bmi1-marked luminal progenitor cells after castration. Induction of prostatic luminal adenocarcinoma. | [50] | |
| Cancer stem cell and prostate cancer | Pb-Cre4; Pten flox/flox | Conditional deletion of Pten in the prostate driven by a minimal probasin promoter driving Cre recombinase. Basal- and luminal-like CSCs were isolated as Sca-1+CD49fhigh and Sca-1+CD49fmed cells, respectively. | [77–79] |
| Probasin-PRL (Pb-PRL) | Prolactin transgene driven by the short probasin promoter. Luminal-like CSCs were isolated as Sca-1+CD49fhigh cells. | [79] | |
| Hi-Myc | c-Myc driven by ARR2PB promoter. Luminal-like CSCs were isolated as Sca-1+CD49fmed cells. | [79] | |
| Pb-Cre4; Trp53flox/flox; Ptenflox/flox | Conditional deletion of Pten and p53 in the prostate driven by a minimal probasin promoter driving Cre recombinase. Luminal-like CSCs were isolated as Epcam+CD49med/loProm1+ cells. | [80] | |
| Cellular plasticity and prostate cancer | Pb-Cre4;Ptenflox/flox;Rosa26LSL-MYCN | Conditional deletion of Pten and ectopic induction of N-Myc expression in the prostate driven by a minimal probasin promoter driving Cre recombinase. Development of poorly differentiated, invasive prostate cancer that is molecularly similar to human NEPC. | [101] |
| Pb-Cre4;Ptenflox/flox;Rb1flox/flox;Trp53flox/flox | Conditional deletion of Pten, Rb, and p53 in the prostate driven by a minimal probasin promoter driving Cre recombinase. Development of poorly differentiated, invasive prostate cancer that is molecularly similar to human NEPC. | [100, 102] | |
| Nkx3.1-CreERT2;Ptenflox/flox; Trp53flox/flox | Conditional deletion of Pten and p53 in distal luminal cells by Nkx3.1 promoter driving Cre recombinase. NEPC emerges from luminal adenocarcinoma in castrated mice with anti-androgen treatment. | [103] |
Notably, prostate inflammation promotes basal cell plasticity following deletion of Pten or Nkx3.1, resulting in increased basal to luminal differentiation (Fig. 3) [60, 75]. The plasticity of basal cells ex vivo is likely to account for the finding that basal cells can give rise to human prostate cancer after oncogenic transformation in culture followed by renal grafting methods [64–67]. Thus, although luminal cells are favored as the cell type of origin for prostate cancer, the plasticity of basal cells in response to tissue damage can potentially render them competent as cells of origin via basal-to-luminal differentiation. Events such as inflammation and tissue damage could therefore contribute to prostate cancer heterogeneity and correlate with distinct disease outcomes.
4. Identification of putative prostate cancer stem cells
The functional identification of cancer stem cells (CSCs) depends on their ability to self-renew and produce both tumorigenic and nontumorigenic tumor cell progeny through asymmetric cell divisions, thereby generating tumor heterogeneity [24–26]. Consequently, therapeutic targeting of CSCs could block prostate cancer progression and potentially overcome castration-resistance. Many studies have sought to identify prostate CSCs using established human cancer cell lines as well as xenografts, which have been summarized in several reviews [19, 22, 63, 76]. However, there have been more limited studies of CSCs in prostate GEMMs in vivo to date.
Interestingly, putative CSCs have been identified in prostate cancer GEMMs with either basal-like and luminal-like phenotypes [19] (Table 1). Notably, basal-like CSCs have been isolated as Sca-1+CD49fhigh cells from Pb-Cre4; Ptenflox/flox mouse tumors, and were shown to have tumor-initiating properties in tissue reconstitution assays [77]. Subsequent work showed that a subset of Sca-1+CD49fhigh CSCs that have CD166 expression harbor increased tumor-initiating and other CSC properties [78], consistent with enrichment of basal-like CSCs.
In other studies, luminal-like CSCs have been isolated as Sca-1+CD49fmed cells from Pb-Cre4; Ptenflox/flox, Probasin-PRL (Pb-PRL), and Hi-Myc mouse models, and were shown to have highly proliferative and tumor-initiating properties [79]. Interesting, another population of luminal-like CSCs has been described as Epcam+CD49med/loProm1+ cells in the Pb-Cre4; Trp53flox/flox; Ptenflox/flox model [80]. These cells can generate tumor organoids in culture that display multi-lineage (luminal and basal) as well as luminal-only lineage differentiation, and form tumors with adenosquamous histology and adenocarcinoma, respectively, after transplantation into immunodeficient mice. These findings suggest the existence of two types of luminal CSCs that can be arranged in a hierarchical relationship, with multipotent CSCs giving rise to unipotent luminal-committed progenitors [80, 81].
5. Cellular plasticity in advanced prostate cancer
Many differentiated cell types can display some degree of plasticity by altering their fates in response to physiological stresses such as inflammation and tissue damage [82–85]. Plasticity is more prevalent in cancer, where the genetic and epigenetic constraints upon the differentiated state are weakened and stresses such as inflammation are accentuated. Notably, plasticity of cancer cells also contributes to tumor heterogeneity and provides mechanisms for tumor cells to evade immune surveillance and acquire metastatic potential [82, 85].
Following treatment with second-generation anti-androgens, recurrent CRPC can display a range of phenotypes with differing levels of expression for AR as well as neuroendocrine markers such as synaptophysin and chromogranin A [14, 15, 86, 87]. This heterogeneity of AR expression in CRPC is likely related to different responses to castration and enzalutamide treatment, as AR+ CRPC xenografts are enzalutamide-sensitive whereas AR−/lo CRPC xenografts are resistant [88]. Notably, CRPC can be classified into distinct entities known as ARPC (AR-expressing prostate cancer) that lacks neuroendocrine marker expression, amphicrine (AMPC) that expresses both AR and neuroendocrine markers, DNPC (double-negative prostate cancer) that does not express either AR or neuroendocrine markers, and NEPC (neuroendocrine prostate cancer) that is AR-negative and expresses neuroendocrine markers [89–93]. While the relationships of AMPC, DNPC, and NEPC to each other are poorly understood at present, their frequent co-occurrence in proximity to ARPC suggests a close lineage relationship [15]. Furthermore, although de novo NEPC is rare in primary prostate cancer [1], neuroendocrine differentiation is much more common in CRPC [10, 15, 94–96], suggesting that AR down-regulation promotes neuroendocrine differentiation.
Several studies have explored the origin of neuroendocrine cells in prostate development and cancer. In studies of normal prostate organogenesis, lineage-tracing has been used to conclude that neuroendocrine cells arise from basal progenitors [44] as well as from caudal neural crest [97], but the basis for these apparently conflicting results has not yet been resolved. In addition, cell culture studies have shown that human prostate basal cells can be reprogrammed to neuroendocrine-like cancer cells by a common set of defined oncogenic drivers [98, 99]. In the case of NEPC, several GEMMs with neuroendocrine phenotypes have been described, including those with combined loss of the Trp53, Rb1, and/or Pten tumor suppressors, or together with activation of N-myc. For example, Pten deletion together with N-Myc overexpression or with deletion of Rb1 and Trp53 in the Pb-Cre4;Ptenflox/flox;Rosa26LSL-MYCN and Pb-Cre4;Ptenflox/flox;Rb1flox/flox;Trp53flox/flox GEMMs induced prostate adenocarcinoma followed by emergence of NEPC [100–102] (Table 1). Notably, the NEPC phenotype in these and other GEMMs arises from prostate adenocarcinoma with a luminal phenotype, suggesting that tumor cell plasticity gives rise to neuroendocrine-like cells [51, 100–106]. Importantly, lineage-tracing studies in a GEMM of prostate cancer have directly demonstrated that neuroendocrine tumor cells arise by transdifferentiation from luminal adenocarcinoma cells [103] (Fig. 3).
6. Concluding remarks and future directions
The cell of origin and cancer stem cell models provide plausible explanations for the generation and maintenance of prostate cancer heterogeneity, but require much more investigation to establish their general validity. GEMMs for prostate cancer will remain central in future studies centered on these models.
With respect to the cell of origin model, the recent identification of different luminal populations by single-cell RNA-sequencing has raised the question of whether they represent distinct cell types of origin for prostate cancer. Notably, however, both distal as well as proximal luminal cells can give rise to prostate tumors following Pten deletion [36, 57], suggesting that further phenotypic and molecular analyses will be necessary to distinguish whether these tumors differ in aggressiveness as a consequence of their cell type of origin. Furthermore, new GEMMs for lineage-tracing of these luminal populations as well as cross-species analyses to validate their relevance for human prostate cancer will be essential to dissect the relevance of the cell of origin. Genetic barcoding approaches in vivo [107, 108] may be particularly useful to identify stem/progenitor cells in prostate tumors, for example to investigate the cellular plasticity that gives rise to neuroendocrine differentiation.
Although putative cancer stem cells have been identified in GEMMs of prostate cancer, widely used candidate CSC markers have generated inconsistent results in differing experimental systems [19]. Importantly, definitive studies using direct lineage-tracing have not yet been performed to demonstrate that CSCs can generate both tumorigenic and non-tumorigenic cells, and that ablation of CSCs inhibit prostate cancer progression and improve treatment response. Such lineage-tracing approaches have been successfully used in other tumor types to define CSC populations [109–111]. At the technical level, such studies might benefit from improved GEMMs that utilize alternative recombination systems together with the Cre-loxP system, such as the Dre-rox system [112, 113]. In addition, the use of organoid approaches may permit lineage-tracing of CSCs in human prostate cancer [114]. Such future studies will undoubtedly provide rigorous tests of the CSC model for generating prostate cancer heterogeneity and guiding prostate cancer treatment.
Finally, cellular plasticity plays a central role in regulating prostate cancer progression and treatment response. For example, more differentiated tumor cells might display plasticity in de-differentiating into CSCs to compensate for the loss of pre-existing CSCs or to escape drug treatment [24, 115, 116]. In the case of CRPC, neuroendocrine differentiation represents a major mechanism of resistance to anti-androgen treatment. Understanding this form of lineage plasticity will likely require the generation of more sophisticated GEMMs that recapitulate specific molecular subtypes of CRPC, as well as the investigation of regulators of prostate cancer cell plasticity. Such advances will be essential for the development of new therapeutic approaches to overcome lineage plasticity and castration-resistance in prostate cancer.
Acknowledgements
We apologize to many colleagues whose work has not been cited due to space constraints. Work on prostate development and cancer in the Shen laboratory is supported by grants from the National Institutes of Health (R01CA238005, R01CA251527) and the Prostate Cancer Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
Nothing to declare
References
- [1].Grignon DJ, Unusual subtypes of prostate cancer, Mod Pathol 17(3) (2004) 316–27. [DOI] [PubMed] [Google Scholar]
- [2].Humphrey PA, Histopathology of prostate cancer, Cold Spring Harb Perspect Med 7(10) (2017) a030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL, Integrative genomic profiling of human prostate cancer, Cancer Cell 18(1) (2010) 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, Macdonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA, Punctuated evolution of prostate cancer genomes, Cell 153(3) (2013) 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, Macdonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA, Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer, Nat Genet 44(6) (2012) 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP, Fox NS, Xie M, Prokopec SD, Berlin A, Lalonde E, Ahmed M, Trudel D, Luo X, Beck TA, Meng A, Zhang J, D’Costa A, Denroche RE, Kong H, Espiritu SM, Chua ML, Wong A, Chong T, Sam M, Johns J, Timms L, Buchner NB, Orain M, Picard V, Hovington H, Murison A, Kron K, Harding NJ, P’ng C, Houlahan KE, Chu KC, Lo B, Nguyen F, Li CH, Sun RX, de Borja R, Cooper CI, Hopkins JF, Govind SK, Fung C, Waggott D, Green J, Haider S, Chan-Seng-Yue MA, Jung E, Wang Z, Bergeron A, Dal Pra A, Lacombe L, Collins CC, Sahinalp C, Lupien M, Fleshner NE, He HH, Fradet Y, Tetu B, van der Kwast T, McPherson JD, Bristow RG, Boutros PC, Genomic hallmarks of localized, non-indolent prostate cancer, Nature 541(7637) (2017) 359–364. [DOI] [PubMed] [Google Scholar]
- [7].Espiritu SMG, Liu LY, Rubanova Y, Bhandari V, Holgersen EM, Szyca LM, Fox NS, Chua MLK, Yamaguchi TN, Heisler LE, Livingstone J, Wintersinger J, Yousif F, Lalonde E, Rouette A, Salcedo A, Houlahan KE, Li CH, Huang V, Fraser M, van der Kwast T, Morris QD, Bristow RG, Boutros PC, The evolutionary landscape of localized prostate cancers drives clinical aggression, Cell 173(4) (2018) 1003–1013 e15. [DOI] [PubMed] [Google Scholar]
- [8].Network CGAR, The molecular taxonomy of primary prostate cancer, Cell 163(4) (2015) 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM, Integrative clinical genomics of advanced prostate cancer, Cell 161(5) (2015) 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, Cieslik M, Benelli M, Robinson D, Van Allen EM, Sboner A, Fedrizzi T, Mosquera JM, Robinson BD, De Sarkar N, Kunju LP, Tomlins S, Wu YM, Nava Rodrigues D, Loda M, Gopalan A, Reuter VE, Pritchard CC, Mateo J, Bianchini D, Miranda S, Carreira S, Rescigno P, Filipenko J, Vinson J, Montgomery RB, Beltran H, Heath EI, Scher HI, Kantoff PW, Taplin ME, Schultz N, deBono JS, Demichelis F, Nelson PS, Rubin MA, Chinnaiyan AM, Sawyers CL, Genomic correlates of clinical outcome in advanced prostate cancer, Proc Natl Acad Sci USA 116(23) (2019) 11428–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thompson IM Jr., Goodman PJ, Tangen CM, Parnes HL, Minasian LM, Godley PA, Lucia MS, Ford LG, Long-term survival of participants in the prostate cancer prevention trial, N Engl J Med 369(7) (2013) 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020, CA Cancer J Clin 70(1) (2020) 7–30. [DOI] [PubMed] [Google Scholar]
- [13].Scher HI, Sawyers CL, Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis, J Clin Oncol 23(32) (2005) 8253–8261. [DOI] [PubMed] [Google Scholar]
- [14].Watson PA, Arora VK, Sawyers CL, Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer, Nat Rev Cancer 15(12) (2015) 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, Kelly K, Lin D, Dicker A, Arnold J, Hecht T, Wicha M, Sears R, Rowley D, White R, Gulley JL, Lee J, Diaz Meco M, Small EJ, Shen M, Knudsen K, Goodrich DW, Lotan T, Zoubeidi A, Sawyers CL, Rudin CM, Loda M, Thompson T, Rubin MA, Tawab-Amiri A, Dahut W, Nelson PS, The role of lineage plasticity in prostate cancer therapy resistance, Clin Cancer Res 25(23) (2019) 6916–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, Tomlins SA, Nanus DM, Tagawa ST, Van Allen EM, Elemento O, Sboner A, Garraway LA, Rubin MA, Demichelis F, Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer, Nat Med 22(3) (2016) 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, Kivinummi K, Goody V, Latimer C, O’Meara S, Dawson KJ, Isaacs W, Emmert-Buck MR, Nykter M, Foster C, Kote-Jarai Z, Easton D, Whitaker HC, Group IPU, Neal DE, Cooper CS, Eeles RA, Visakorpi T, Campbell PJ, McDermott U, Wedge DC, Bova GS, The evolutionary history of lethal metastatic prostate cancer, Nature 520(7547) (2015) 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hong MK, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, Alexandrov LB, Sloggett C, Cmero M, Marass F, Tsui D, Mangiola S, Lonie A, Naeem H, Sapre N, Phal PM, Kurganovs N, Chin X, Kerger M, Warren AY, Neal D, Gnanapragasam V, Rosenfeld N, Pedersen JS, Ryan A, Haviv I, Costello AJ, Corcoran NM, Hovens CM, Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer, Nature Communications 6 (2015) 6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li JJ, Shen MM, Prostate stem cells and cancer stem cells, Cold Spring Harb Perspect Med 9(6) (2019) a030395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Visvader JE, Cells of origin in cancer, Nature 469(7330) (2011) 314–322. [DOI] [PubMed] [Google Scholar]
- [21].Blanpain C, Tracing the cellular origin of cancer, Nat Cell Biol 15(2) (2013) 126–134. [DOI] [PubMed] [Google Scholar]
- [22].Rycaj K, Tang DG, Cell-of-origin of cancer versus cancer stem cells: assays and interpretations, Cancer Res 75(19) (2015) 4003–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shen MM, Abate-Shen C, Molecular genetics of prostate cancer: new prospects for old challenges, Genes Dev 24(18) (2010) 1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Batlle E, Clevers H, Cancer stem cells revisited, Nat Med 23(10) (2017) 1124–1134. [DOI] [PubMed] [Google Scholar]
- [25].Kreso A, Dick JE, Evolution of the cancer stem cell model, Cell Stem Cell 14(3) (2014) 275–291. [DOI] [PubMed] [Google Scholar]
- [26].Nassar D, Blanpain C, Cancer stem cells: basic concepts and therapeutic implications, Annu Rev Pathol 11 (2016) 47–76. [DOI] [PubMed] [Google Scholar]
- [27].Meacham CE, Morrison SJ, Tumour heterogeneity and cancer cell plasticity, Nature 501(7467) (2013) 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Timms BG, Prostate development: a historical perspective, Differentiation 76(6) (2008) 565–577. [DOI] [PubMed] [Google Scholar]
- [29].Ittmann M, Anatomy and histology of the human and murine prostate, Cold Spring Harb Perspect Med 8(5) (2018) a030346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y, The endocrinology and developmental biology of the prostate, Endocr Rev 8(3) (1987) 338–362. [DOI] [PubMed] [Google Scholar]
- [31].Marker PC, Donjacour AA, Dahiya R, Cunha GR, Hormonal, cellular, and molecular control of prostatic development, Dev Biol 253(2) (2003) 165–174. [DOI] [PubMed] [Google Scholar]
- [32].Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, Carver B, Gopalan A, Abida W, Laudone V, Biton M, Chaudhary O, Xu T, Masilionis I, Manova K, Mazutis L, Pe’er D, Regev A, Sawyers CL, Regenerative potential of prostate luminal cells revealed by single-cell analysis, Science 368(6490) (2020) 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, Li W, Hibshoosh H, Loda M, Rabadan R, Shen MM, A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors, eLife 9 (2020) e59465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Berquin IM, Min Y, Wu R, Wu H, Chen YQ, Expression signature of the mouse prostate, J Biol Chem 280(43) (2005) 36442–36451. [DOI] [PubMed] [Google Scholar]
- [35].Toivanen R, Shen MM, Prostate organogenesis: tissue induction, hormonal regulation and cell type specification, Development 144(8) (2017) 1382–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, Shen MM, Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity, Nat Cell Biol 15(3) (2013) 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Choi N, Zhang B, Zhang L, Ittmann M, Xin L, Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation, Cancer Cell 21(2) (2012) 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lu TL, Huang YF, You LR, Chao NC, Su FY, Chang JL, Chen CM, Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice, Am J Pathol 182(3) (2013) 975–991. [DOI] [PubMed] [Google Scholar]
- [39].Gaisa NT, Graham TA, McDonald SA, Poulsom R, Heidenreich A, Jakse G, Knuechel R, Wright NA, Clonal architecture of human prostatic epithelium in benign and malignant conditions, J Pathol 225(2) (2011) 172–180. [DOI] [PubMed] [Google Scholar]
- [40].Blackwood JK, Williamson SC, Greaves LC, Wilson L, Rigas AC, Sandher R, Pickard RS, Robson CN, Turnbull DM, Taylor RW, Heer R, In situ lineage tracking of human prostatic epithelial stem cell fate reveals a common clonal origin for basal and luminal cells, J Pathol 225(2) (2011) 181–188. [DOI] [PubMed] [Google Scholar]
- [41].Grossmann S, Hooks Y, Wilson L, Moore L, O’Neill L, Martincorena I, Voet T, Stratton MR, Heer R, Campbell PJ, Development, maturation, and maintenance of human prostate inferred from somatic mutations, Cell Stem Cell (2021) S1934–5909(21)00055–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Moad M, Hannezo E, Buczacki SJ, Wilson L, El-Sherif A, Sims D, Pickard R, Wright NA, Williamson SC, Turnbull DM, Taylor RW, Greaves L, Robson CN, Simons BD, Heer R, Multipotent basal stem cells, maintained in localized proximal niches, support directed long-ranging epithelial flows in human prostates, Cell Reports 20(7) (2017) 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Crowell PD, Fox JJ, Hashimoto T, Diaz JA, Navarro HI, Henry GH, Feldmar BA, Lowe MG, Garcia AJ, Wu YE, Sajed DP, Strand DW, Goldstein AS, Expansion of luminal progenitor cells in the aging mouse and human prostate, Cell Reports 28(6) (2019) 1499–1510 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ousset M, Van Keymeulen A, Bouvencourt G, Sharma N, Achouri Y, Simons BD, Blanpain C, Multipotent and unipotent progenitors contribute to prostate postnatal development, Nat Cell Biol 14(11) (2012) 1131–1138. [DOI] [PubMed] [Google Scholar]
- [45].Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, Blanpain C, Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells, Genes Dev 30(11) (2016) 1261–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tika E, Ousset M, Dannau A, Blanpain C, Spatiotemporal regulation of multipotency during prostate development, Development 146(20) (2019) dev180224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shibata M, Epsi NJ, Xuan S, Mitrofanova A, Shen MM, Bipotent progenitors do not require androgen receptor for luminal specification during prostate organogenesis, Stem Cell Reports 15(5) (2020) 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McAuley E, Moline D, VanOpstall C, Lamperis S, Brown R, Vander Griend DJ, Sox2 expression marks castration-resistant progenitor cells in the adult murine prostate, Stem Cells 37(5) (2019) 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu HL, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM, A luminal epithelial stem cell that is a cell of origin for prostate cancer, Nature 461(7263) (2009) 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yoo YA, Roh M, Naseem AF, Lysy B, Desouki MM, Unno K, Abdulkadir SA, Bmi1 marks distinct castration-resistant luminal progenitor cells competent for prostate regeneration and tumour initiation, Nature Communications 7 (2016) 12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chua CW, Epsi NJ, Leung EY, Xuan S, Lei M, Li BI, Bergren SK, Hibshoosh H, Mitrofanova A, Shen MM, Differential requirements of androgen receptor in luminal progenitors during prostate regeneration and tumor initiation, eLife 7 (2018) e28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang BE, Wang X, Long JE, Eastham-Anderson J, Firestein R, Junttila MR, Castration-resistant Lgr5(+) cells are long-lived stem cells required for prostatic regeneration, Stem Cell Reports 4(5) (2015) 768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, Fan L, Dong B, Xue W, Wai Chua C, Wu D, Gao WQ, He Zhu H, Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate, Nature Communications 11(1) (2020) 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang D, Jeter C, Gong S, Tracz A, Lu Y, Shen J, Tang DG, Histone 2B-GFP label-retaining prostate luminal cells possess progenitor cell properties and are intrinsically resistant to castration, Stem Cell Reports 10(1) (2018) 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW, A cellular anatomy of the normal adult human prostate and prostatic urethra, Cell Reports 25(12) (2018) 3530–3542 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, Abler LL, Sandhu SK, Cadena MT, Malladi VS, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Roehrborn CG, Baker LA, Vezina CM, Strand DW, Urethral luminal epithelia are castration-insensitive cells of the proximal prostate, Prostate 80(11) (2020) 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Guo W, Li L, He J, Liu Z, Han M, Li F, Xia X, Zhang X, Zhu Y, Wei Y, Li Y, Aji R, Dai H, Wei H, Li C, Chen Y, Chen L, Gao D, Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips, Nat Genet 52(9) (2020) 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mevel R, Steiner I, Mason S, Galbraith LC, Patel R, Fadlullah MZ, Ahmad I, Leung HY, Oliveira P, Blyth K, Baena E, Lacaud G, RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development, eLife 9 (2020) a60225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Toivanen R, Mohan A, Shen MM, Basal progenitors contribute to repair of the prostate epithelium following induced luminal anoikis, Stem Cell Reports 6(5) (2016) 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kwon OJ, Zhang L, Ittmann MM, Xin L, Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin, Proc Natl Acad Sci USA 111(5) (2014) E592–E600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Xin L, Cells of origin for prostate cancer, Adv Exp Med Biol 1210 (2019) 67–86. [DOI] [PubMed] [Google Scholar]
- [62].Lee SH, Shen MM, Cell types of origin for prostate cancer, Curr Opin Cell Biol 37 (2015) 35–41. [DOI] [PubMed] [Google Scholar]
- [63].Wang ZA, Shen MM, Revisiting the concept of cancer stem cells in prostate cancer, Oncogene 30(11) (2011) 1261–1271. [DOI] [PubMed] [Google Scholar]
- [64].Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON, Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics, Proc Natl Acad Sci USA 105(52) (2008) 20882–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, Zhang H, Kohn DB, Huang J, Witte ON, Goldstein AS, Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells, Proc Natl Acad Sci USA 110(50) (2013) 20111–20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Taylor RA, Toivanen R, Frydenberg M, Pedersen J, Harewood L, Australian Prostate Cancer B, Collins AT, Maitland NJ, Risbridger GP, Human epithelial basal cells are cells of origin of prostate cancer, independent of CD133 status, Stem Cells 30(6) (2012) 1087–1096. [DOI] [PubMed] [Google Scholar]
- [67].Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON, Identification of a cell of origin for human prostate cancer, Science 329(5991) (2010) 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu X, Grogan TR, Hieronymus H, Hashimoto T, Mottahedeh J, Cheng D, Zhang L, Huang K, Stoyanova T, Park JW, Shkhyan RO, Nowroozizadeh B, Rettig MB, Sawyers CL, Elashoff D, Horvath S, Huang J, Witte ON, Goldstein AS, Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome, Cell Reports 17(10) (2016) 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, Shen MM, Single luminal epithelial progenitors can generate prostate organoids in culture, Nat Cell Biol 16(10) (2014) 951–961, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Park JW, Lee JK, Phillips JW, Huang P, Cheng D, Huang J, Witte ON, Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay, Proc Natl Acad Sci USA 113(16) (2016) 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ratnacaram CK, Teletin M, Jiang M, Meng X, Chambon P, Metzger D, Temporally controlled ablation of PTEN in adult mouse prostate epithelium generates a model of invasive prostatic adenocarcinoma, Proc Natl Acad Sci USA 105(7) (2008) 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gao D, Zhan Y, Di W, Moore AR, Sher JJ, Guan Y, Wang S, Zhang Z, Murphy DA, Sawyers CL, Chi P, Chen Y, A Tmprss2-CreERT2 knock-in mouse model for cancer genetic studies on prostate and colon, PLoS One 11(8) (2016) e0161084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang ZA, Toivanen R, Bergren SK, Chambon P, Shen MM, Luminal cells are favored as the cell of origin for prostate cancer, Cell Reports 8(5) (2014) 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, Speers C, Cooperberg MR, Kim W, Ryan CJ, Den RB, Freedland SJ, Posadas E, Sandler H, Klein EA, Black P, Seiler R, Tomlins SA, Chinnaiyan AM, Jenkins RB, Davicioni E, Ross AE, Schaeffer EM, Nguyen PL, Carroll PR, Karnes RJ, Spratt DE, Feng FY, Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy, JAMA Oncol 3(12) (2017) 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Le Magnen C, Virk RK, Dutta A, Kim JY, Panja S, Lopez-Bujanda ZA, Califano A, Drake CG, Mitrofanova A, Abate-Shen C, Cooperation of loss of NKX3.1 and inflammation in prostate cancer initiation, Disease Models & Mechanisms 11(11) (2018) dmm035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Rybak AP, Bristow RG, Kapoor A, Prostate cancer stem cells: deciphering the origins and pathways involved in prostate tumorigenesis and aggression, Oncotarget 6(4) (2015) 1900–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mulholland DJ, Xin L, Morim A, Lawson D, Witte O, Wu H, Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model, Cancer Res 69(22) (2009) 8555–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Jiao J, Hindoyan A, Wang S, Tran LM, Goldstein AS, Lawson D, Chen D, Li Y, Guo C, Zhang B, Fazli L, Gleave M, Witte ON, Garraway IP, Wu H, Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells, PLoS One 7(8) (2012) e42564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sackmann Sala L, Boutillon F, Menara G, De Goyon-Pelard A, Leprevost M, Codzamanian J, Lister N, Pencik J, Clark A, Cagnard N, Bole-Feysot C, Moriggl R, Risbridger GP, Taylor RA, Kenner L, Guidotti JE, Goffin V, A rare castration-resistant progenitor cell population is highly enriched in Pten-null prostate tumours, J Pathol 243(1) (2017) 51–64. [DOI] [PubMed] [Google Scholar]
- [80].Agarwal S, Hynes PG, Tillman HS, Lake R, Abou-Kheir WG, Fang L, Casey OM, Ameri AH, Martin PL, Yin JJ, Iaquinta PJ, Karthaus WR, Clevers HC, Sawyers CL, Kelly K, Identification of different classes of luminal progenitor cells within prostate tumors, Cell Reports 13(10) (2015) 2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Shen MM, Illuminating the properties of prostate luminal progenitors, Cell Stem Cell 17(6) (2015) 644–646. [DOI] [PubMed] [Google Scholar]
- [82].Le Magnen C, Shen MM, Abate-Shen C, Lineage plasticity in cancer progression and treatment, Annu Rev Cancer Biol 2 (2018) 271–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Carceles-Cordon M, Kelly WK, Gomella L, Knudsen KE, Rodriguez-Bravo V, Domingo-Domenech J, Cellular rewiring in lethal prostate cancer: the architect of drug resistance, Nat Rev Urol 17(5) (2020) 292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Li W, Li L, Hui L, Cell plasticity in liver regeneration, Trends Cell Biol 30(4) (2020) 329–338. [DOI] [PubMed] [Google Scholar]
- [85].Yuan S, Norgard RJ, Stanger BZ, Cellular plasticity in cancer, Cancer Discovery 9(7) (2019) 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, Foye A, Kothari V, Perry MD, Bailey AM, Playdle D, Barnard TJ, Zhang L, Zhang J, Youngren JF, Cieslik MP, Parolia A, Beer TM, Thomas G, Chi KN, Gleave M, Lack NA, Zoubeidi A, Reiter RE, Rettig MB, Witte O, Ryan CJ, Fong L, Kim W, Friedlander T, Chou J, Li H, Das R, Li H, Moussavi-Baygi R, Goodarzi H, Gilbert LA, Lara PN Jr., Evans CP, Goldstein TC, Stuart JM, Tomlins SA, Spratt DE, Cheetham RK, Cheng DT, Farh K, Gehring JS, Hakenberg J, Liao A, Febbo PG, Shon J, Sickler B, Batzoglou S, Knudsen KE, He HH, Huang J, Wyatt AW, Dehm SM, Ashworth A, Chinnaiyan AM, Maher CA, Small EJ, Feng FY, Genomic hallmarks and structural variation in metastatic prostate cancer, Cell 174(3) (2018) 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, Dhir R, Nelson JB, de la Taille A, Allory Y, Gerstein MB, Perner S, Pienta KJ, Chinnaiyan AM, Wang Y, Collins CC, Gleave ME, Demichelis F, Nanus DM, Rubin MA, Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets, Cancer Discovery 1(6) (2011) 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Li Q, Deng Q, Chao HP, Liu X, Lu Y, Lin K, Liu B, Tang GW, Zhang D, Tracz A, Jeter C, Rycaj K, Calhoun-Davis T, Huang J, Rubin MA, Beltran H, Shen J, Chatta G, Puzanov I, Mohler JL, Wang J, Zhao R, Kirk J, Chen X, Tang DG, Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses, Nat Commun 9(1) (2018) 3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Beltran H, Tomlins S, Aparicio A, Arora V, Rickman D, Ayala G, Huang J, True L, Gleave ME, Soule H, Logothetis C, Rubin MA, Aggressive variants of castration-resistant prostate cancer, Clin Cancer Res 20(11) (2014) 2846–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera JM, Reuter VE, Robinson BD, Troncoso P, Rubin MA, Proposed morphologic classification of prostate cancer with neuroendocrine differentiation, Am J Surg Pathol 38(6) (2014) 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Labrecque MP, Coleman IM, Brown LG, True LD, Kollath L, Lakely B, Nguyen HM, Yang YC, da Costa RMG, Kaipainen A, Coleman R, Higano CS, Yu EY, Cheng HH, Mostaghel EA, Montgomery B, Schweizer MT, Hsieh AC, Lin DW, Corey E, Nelson PS, Morrissey C, Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer, J Clin Invest 130 (2019) 4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, Bianchi-Frias D, Dumpit RF, Kaipainen A, Corella AN, Yang YC, Nyquist MD, Mostaghel E, Hsieh AC, Zhang X, Corey E, Brown LG, Nguyen HM, Pienta K, Ittmann M, Schweizer M, True LD, Wise D, Rennie PS, Vessella RL, Morrissey C, Nelson PS, Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling, Cancer Cell 32(4) (2017) 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Su W, Han HH, Wang Y, Zhang B, Zhou B, Cheng YK, Rumandia A, Gurrapu S, Chakraborty G, Su J, Yang G, Liang X, Wang G, Rosen N, Scher HI, Ouerfelli O, Giancotti F, The Polycomb Repressor Complex 1 drives double negative prostate cancer metastasis by coordinating stemness and immune suppression, Cancer Cell 36(2) (2019) 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Davies AH, Beltran H, Zoubeidi A, Cellular plasticity and the neuroendocrine phenotype in prostate cancer, Nat Rev Urol 15(5) (2018) 271–286. [DOI] [PubMed] [Google Scholar]
- [95].Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, Weinstein AS, Friedl V, Zhang C, Witte ON, Lloyd P, Gleave M, Evans CP, Youngren J, Beer TM, Rettig M, Wong CK, True L, Foye A, Playdle D, Ryan CJ, Lara P, Chi KN, Uzunangelov V, Sokolov A, Newton Y, Beltran H, Demichelis F, Rubin MA, Stuart JM, Small EJ, Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study, J Clin Oncol 36(24) (2018) 2492–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Laudato S, Aparicio A, Giancotti FG, Clonal evolution and epithelial plasticity in the emergence of AR-independent prostate carcinoma, Trends Cancer 5(7) (2019) 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Szczyrba J, Niesen A, Wagner M, Wandernoth PM, Aumuller G, Wennemuth G, Neuroendocrine cells of the prostate derive from the neural crest, J Biol Chem 292(5) (2017) 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, Baertsch R, Sokolov A, Meyerowitz JG, Mathis C, Cheng D, Stuart JM, Shokat KM, Gustafson WC, Huang J, Witte ON, N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells, Cancer Cell 29(4) (2016) 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, Smith BA, Cheng C, Tsai BL, Cheng D, Huang J, Kurdistani SK, Graeber TG, Witte ON, Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage, Science 362(6410) (2018) 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW, Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance, Science 355(6320) (2017) 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, Cyrta J, Sboner A, Noorzad Z, MacDonald T, Cheung C, Yuen KS, Gao D, Chen Y, Eilers M, Mosquera JM, Robinson BD, Elemento O, Rubin MA, Demichelis F, Rickman DS, N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer, Cancer Cell 30(4) (2016) 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, Nikitin AY, Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer, Cancer Res 66(16) (2006) 7889–7898. [DOI] [PubMed] [Google Scholar]
- [103].Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, Le Magnen C, Chester D, Mostaghel EA, Califano A, Rubin MA, Shen MM, Abate-Shen C, Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer, Cancer Discovery 7(7) (2017) 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Foster BA, Evangelou A, Gingrich JR, Kaplan PJ, DeMayo F, Greenberg NM, Enforced expression of FGF-7 promotes epithelial hyperplasia whereas a dominant negative FGFR2iiib promotes the emergence of neuroendocrine phenotype in prostate glands of transgenic mice, Differentiation 70(9–10) (2002) 624–632. [DOI] [PubMed] [Google Scholar]
- [105].Kaplan-Lefko PJ, Chen TM, Ittmann MM, Barrios RJ, Ayala GE, Huss WJ, Maddison LA, Foster BA, Greenberg NM, Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model, Prostate 55(3) (2003) 219–237. [DOI] [PubMed] [Google Scholar]
- [106].Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, Ward JM, Cardiff RD, Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee, Cancer Res 64(6) (2004) 2270–2305. [DOI] [PubMed] [Google Scholar]
- [107].Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A, Weissman JS, Molecular recording of mammalian embryogenesis, Nature 570(7759) (2019) 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bowling S, Sritharan D, Osorio FG, Nguyen M, Cheung P, Rodriguez-Fraticelli A, Patel S, Yuan WC, Fujiwara Y, Li BE, Orkin SH, Hormoz S, Camargo FD, An engineered CRISPR-Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells, Cell 181(7) (2020) 1693–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H, Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas, Science 337(6095) (2012) 730–735. [DOI] [PubMed] [Google Scholar]
- [110].Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C, Defining the mode of tumour growth by clonal analysis, Nature 488(7412) (2012) 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF, A restricted cell population propagates glioblastoma growth after chemotherapy, Nature 488(7412) (2012) 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Han X, Zhang Z, He L, Zhu H, Li Y, Pu W, Han M, Zhao H, Liu K, Li Y, Huang X, Zhang M, Jin H, Lv Z, Tang J, Wang J, Sun R, Fei J, Tian X, Duan S, Wang QD, Wang L, He B, Zhou B, A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting, Cell Stem Cell 28(6) (2021) 1160–1176. e7. [DOI] [PubMed] [Google Scholar]
- [113].He L, Li Y, Li Y, Pu W, Huang X, Tian X, Wang Y, Zhang H, Liu Q, Zhang L, Zhao H, Tang J, Ji H, Cai D, Han Z, Han Z, Nie Y, Hu S, Wang QD, Sun R, Fei J, Wang F, Chen T, Yan Y, Huang H, Pu WT, Zhou B, Enhancing the precision of genetic lineage tracing using dual recombinases, Nat Med 23(12) (2017) 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, Sato T, Visualization and targeting of LGR5(+) human colon cancer stem cells, Nature 545(7653) (2017) 187–192. [DOI] [PubMed] [Google Scholar]
- [115].Biehs B, Dijkgraaf GJP, Piskol R, Alicke B, Boumahdi S, Peale F, Gould SE, de Sauvage FJ, A cell identity switch allows residual BCC to survive Hedgehog pathway inhibition, Nature 562(7727) (2018) 429–433. [DOI] [PubMed] [Google Scholar]
- [116].de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, Dijkgraaf GJ, Piskol R, de Sauvage FJ, A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer, Nature 543(7647) (2017) 676–680. [DOI] [PubMed] [Google Scholar]
- [117].Shibata M, Shen MM, The roots of cancer: stem cells and the basis for tumor heterogeneity, Bioessays 35(3) (2013) 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]


