Abstract
Objective
Uncoupling protein 1 (UCP1) is a mitochondrial protein critical for adaptive thermogenesis in adipose tissues, and it is typically believed to be restricted to thermogenic adipose tissues. UCP1-Cre transgenic mice are utilized in numerous studies to provide “brown adipose-specific” conditional gene targeting. Here, we examined the distribution of Cre and UCP1 throughout the body in UCP1-Cre reporter mice.
Methods
UCP1-Cre mice crossed to Ai14-tdTomato and Ai9-tdTomato reporter mice were used to explore the tissue distribution of Cre recombinase and Ucp1 mRNA in various tissues. UCP1-Cre mice were independently infected with either a Cre-dependent PHP.eB-tdTomato virus or a Cre-dependent AAV-tdTomato virus to determine whether and where UCP1 is actively expressed in the adult central nervous system. In situ analysis of the deposited single cell RNA sequencing data was used to evaluate Ucp1 expression in the hypothalamus.
Results
As expected, Ucp1 expression was detected in both brown and inguinal adipose tissues. Ucp1 expression was also detected in the kidney, adrenal glands, thymus, and hypothalamus. Consistent with detectable Ucp1 expression, tdTomato expression was also observed in brown adipose tissue, inguinal white adipose tissue, kidney, adrenal glands, and hypothalamus of both male and female UCP1-Cre; Ai14-tdTomato and UCP1-Cre; Ai9-tdTomato mice by fluorescent imaging and qPCR. Critically, expression of tdTomato, and thus UCP1, within the central nervous system was observed in regions of the brain critical for the regulation of energy homeostasis, including the ventromedial hypothalamus (VMH).
Conclusions
TdTomato expression in UCP1-Cre; tdTomato mice is not restricted to thermogenic adipose tissues. TdTomato was also expressed in the kidneys, adrenal glands, and throughout the brain, including brain regions and cell types that are critical for multiple aspects of central regulation of energy homeostasis. Collectively, these data have important implications for the utility of UCP1-Cre mice as genetic tools to investigate gene function specifically in brown adipose tissue.
Keywords: Adipose, Brain, VMH, Brown adipose tissue, Brown, Beige
Highlights
-
•
UCP1-Cre transgenic mice express Cre in tissues beyond thermogenic adipose tissues.
-
•
UCP1-Cre; tdTomato reporter mice show that Cre is also expressed throughout the brain, kidney, and adrenal glands.
-
•
Ucp1 mRNA is actively expressed in the central nervous system of adult mice.
1. Introduction
Adipose tissues maintain systemic energy homeostasis through the storage and release of lipids and adipokines and contribute to whole-body metabolic rate [[1], [2], [3]]. Within adipose tissues, there are at least three different types of adipocytes: white, beige/brite, and brown [4]. White adipocytes have a unilocular appearance and function to store energy. In contrast, brown adipocytes dissipate energy as heat through a process known as adaptive thermogenesis and histologically have a multilocular phenotype [4]. Brown adipocytes produce heat by utilizing glucose and lipids, which results in increased energy expenditure. This is accomplished by uncoupling protein 1 (UCP1), a protein localized to the mitochondrial inner membrane which uncouples proton transport across the inner mitochondrial membrane from the electron transport chain, leading to release of chemical energy as heat [[5], [6], [7]]. Beige or brite (brown in white) adipocytes phenotypically appear like white adipocytes, but when activated, they have the capacity to increase UCP1 expression and thermogenesis and obtain a multilocular appearance [8,9].
The role of UCP1 in the regulation of thermogenesis by brown adipose tissue (BAT) is well established in small mammals that require non-shivering thermogenesis for survival. Traditionally, UCP1 was believed to be restricted to BAT or thermogenic adipocytes, and numerous publications have used Cre recombinase driven by the UCP1 promoter to genetically alter expression of target genes in a BAT-specific manner [10]. In addition to adipose tissue, there have been multiple reports of Ucp1 mRNA expression in other tissues, including the kidney [11], thymus [12,13], and brain [[13], [14], [15], [16]]. Multiple studies have revealed neuronal UCP1 expression in animals, from amphibians [15] to mammals [14]. While the potential role of UCP1 in these tissues remains unclear, it is imperative to determine whether UCP1 is expressed outside of adipose tissue depots, as it would have major implications for the past and future use of UCP1 as a genetic driver to manipulate gene expression in an adipose-specific manner. Using multiple independent approaches, we show here that Cre expression in UCP1-Cre mice is not restricted to BAT. We report that Cre expression is observed in multiple areas throughout the central nervous system (CNS) of UCP1-Cre mice. Single-cell RNA sequencing analyses reveal that Ucp1 is indeed expressed in the hypothalamus during development and in the adult brain. Cre expression is not restricted to thermogenic adipocytes in UCP1-Cre mice, but is also expressed in regions of the CNS which regulate metabolism in a bottom-up fashion. Phenotypes attributed to genetic deletions using this model must consider potential direct effects on the CNS.
2. Materials and methods
2.1. Animals
B6.FVB-Tg (UCP1-Cre)1Evdr/J, referred to as UCP1-Cre (stock #024670), Ai14-tdTomato (stock #007914) and Ai9-tdTomato (stock #007909) mice were obtained from Jackson laboratory. According to Jackson Labs, the UCP1-Cre mice were backcrossed to C57BL/6J for at least 13 generations. Mice were housed at 22–23°C in a 12-h light–dark cycle with ad libitum access to food and water. The animals were 8–14 weeks old and not used for any other experiments. Health status was normal for all animals. All experiments presented in this study were conducted according to the animal research guidelines from NIH and were approved by the University of Iowa IACUC.
2.2. Histology
Mice were transcardially perfused with saline followed by 4% paraformaldehyde (PFA). Brain, heart, liver, pancreas, kidney, skeletal muscle, and testes were post-fixed in 4% PFA overnight followed by 30% sucrose for 48 h. Adipose tissues, adrenal glands, and pituitary glands were flash frozen in embedding media immediately following dissection. Coronal brain sections (40 μm) were collected using a cryostat (Leica) and all other tissues were sectioned (10 μm) utilizing a cryostat with cryo-jane system (Leica). For NeuN staining, free-floating brain sections from UCP1-Cre; Ai14-tdTomato mice were incubated with blocking solution (5% normal goat serum) for 1 h at room temperature (RT), followed by primary antibody (Abcam, ab104225) overnight at 4 °C. The following day, sections were incubated in goat anti-rabbit 488 secondary antibody (Life Technologies) for 1 h at RT and mounted onto slides. All tissue sections were mounted with VECTASHIELD antifade mounting media with DAPI (Vector Laboratories) and imaged using an Olympus BX61 Light Microscope.
2.3. Viral injections
Four-week-old UCP1-Cre mice were anesthetized with isoflurane and retro-orbitally injected with 10 μl of PHP.eB-pAAV-FLEX-tdTomato (8 × 1012 vg/ml). Following 4 weeks of recovery to allow for viral spread, brains were collected for fluorescent imaging. 9-Week-old UCP1-Cre mice were anesthetized with isoflurane and administered AAV1-CAG-FLEX-tdTomato through bilateral stereotactic injection into the VMH (AP -1.46; ML +/− 0.6; DV -5.3). Next, 1 μl of virus was injected with an injection speed of 0.02 μl/min. Following 3 weeks of recovery to allow for viral spread, brains were collected for fluorescent imaging.
2.4. Gene expression
Gene expression analyses were performed as described [17]. RNA was isolated from the indicated tissues following Trizol (Invitrogen) protocol. 2 μg RNA from each sample were used to generate cDNA (High-Capacity cDNA Reverse Transcription Kit; Life Technologies), and QPCR was conducted using SYBR green (Invitrogen). QPCR primer sequences are as follows: Ucp1: 5′-AAGCTGTGCGATGTCCATGT-3′, 5′-AAGCCACAAACCCTTTGAAAA-3′, tdTomato: 5′-GGGAAGGACAGCTTCTTGTAAT-3′, 5′-CGAGGAGGTCATCAAAGAGTTC-3′, U36B4: 5′-CGTCCTCGTTGGAGTGACA-3′, 5′CGGTGCGTCAGGGATTG-3′
2.5. In silico analysis of deposited single-cell RNA sequencing
To evaluate the expression of Ucp1 in the adult ventromedial hypothalamus (VMH), a publicly available dataset [18] (Mendelay Data, SMART-seq_VMH) was downloaded and loaded into the R package Seurat (v4.0.1). Within the VMH dataset, Ucp1-expressing cells were extracted and SCTransform was performed. Expressions of different marker genes were then evaluated in Ucp1-expressing cells. Developmental expression of Ucp1 in the hypothalamus was evaluated using the whole dataset from a study exploring the developmental gene expression at the single cell level in the hypothalamus ([19], GEO: GSE132730), again using the R package Seurat to visualize Ucp1 expression during development. The raw choroid plexus single cell and single nucleus RNA sequencing dataset [20] was downloaded from Gene Expression Omnibus GSE168704. After loading in the dataset, we followed the Seurat SCTransform pipeline to normalize, scale, and find variable features.
2.6. Tissue clearing, imaging, and processing
An augmented DISCO protocol was used to clear whole brain tissue of UCP1-Cre; Ai14-tdTomato and Cre negative control mice [21,22]. Mice were anesthetized and transcardially perfused with pH 9.5 saline followed by pH 9.5 PFA. After post-fixing in PFA overnight, the brains were washed 3 times in saline and dehydrated with 50% tetrahydrofuran (THF) in dH2O for 24 h. Every 24 h the brains were transferred to a higher concentration of THF (50%, 70%, 80%), all at pH 9.5. Brains were incubated in 100% THF for 48 h, and the solution was replaced every 12 h to maximize tissue THF concentration. After the final incubation in 100% THF, brains were transferred to dichloromethane (DCM) for 3 h followed by BABB-D15 for 4 h [23]. All steps were performed at 4 °C in the dark. After transferring brains to a new solution, argon gas was flowed over the sample to minimize oxygen interaction with the sample. Cleared brains were imaged in BABB-D15 using a light sheet fluorescence microscope (LaVision BioTec Ultramicroscope II) interfaced with an Andor Neo sCMOS camera, an Olympus MVX10 zoom microscope body, and MVPLAPO 2x dipping cap. Whole brain three-dimensional images were achieved using a 3 × 3 mosaic tile scan of a 2x optical zoom and a z-stack with step size of 2.5 μm. The raw lossless TIFF files of the collected tile scans were stitched using the ImspectorPro software and reconstructed into three dimensions with Imaris File Converter. Images and movies of brain samples were captured with Imaris.
2.7. Data analysis
Data were analyzed using Excel or GraphPad Prism and statistical differences between groups were determined by Student's t-test or two-way ANOVA using Sidak's correction for multiple comparisons.
3. Results
3.1. Cre expression is not restricted to thermogenic adipose tissue in UCP1-Cre mice
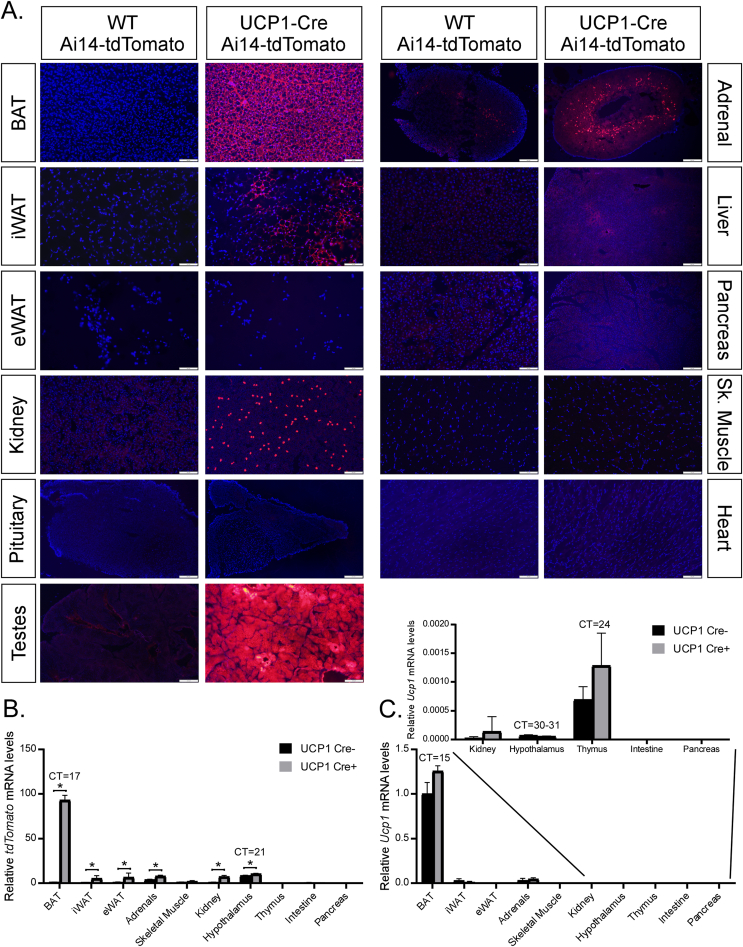
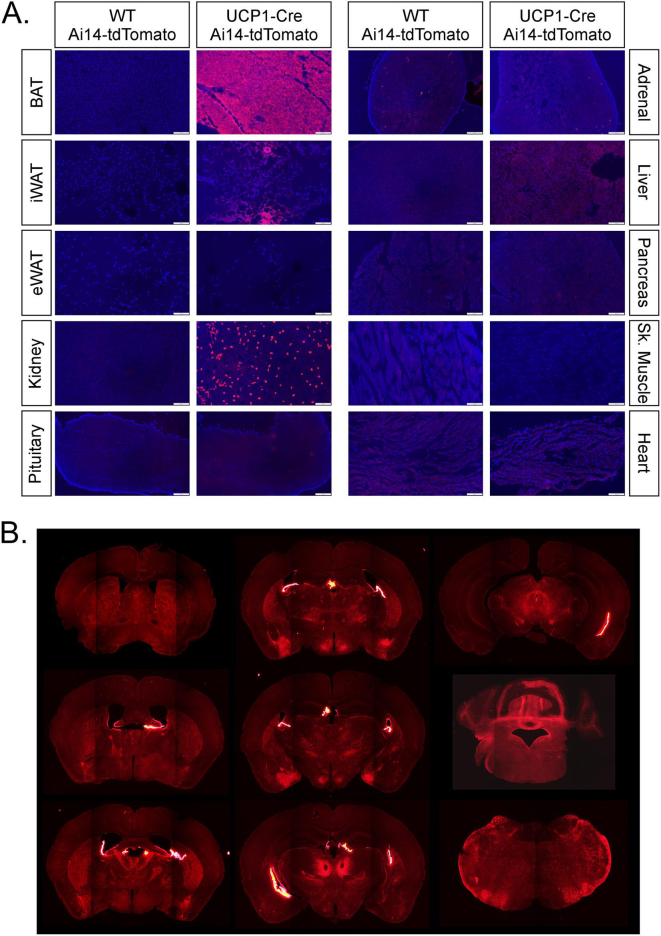
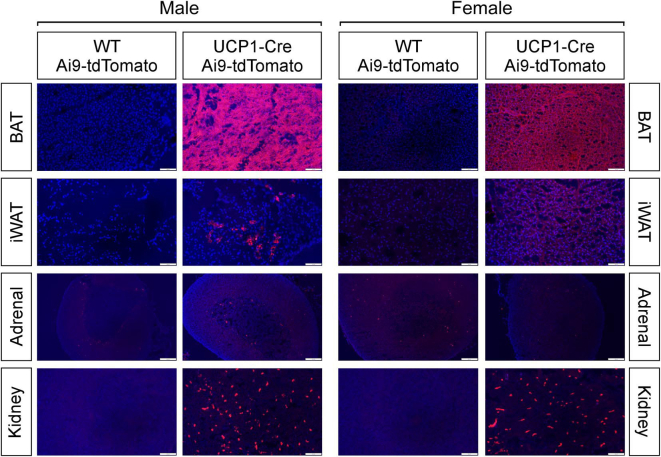
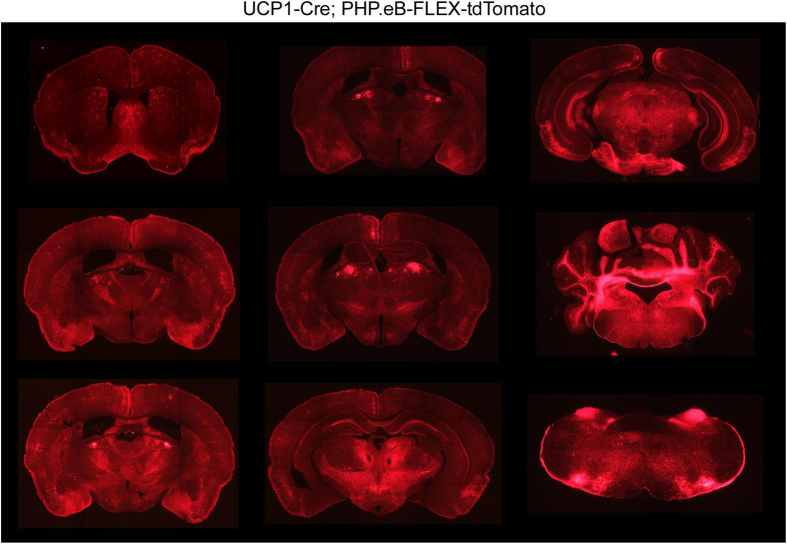
To examine the efficacy and specificity of Cre expression in UCP1-Cre mice, we crossed UCP1-Cre mice to either Ai14 Cre-dependent tdTomato reporter mice (UCP1-Cre; Ai14-tdTomato) or Ai9 Cre-dependent tdTomato reporter mice (UCP1-Cre; Ai9-tdTomato) to identify tissues that express Cre. As expected, both male and female UCP1-Cre; Ai14-tdtomato (Figure 1A, Supplemental Fig. 1A) and UCP1-Cre; Ai9-tdtomato mice (Supplemental Fig. 2) exhibited abundant tdTomato signal in the brown adipose tissue (BAT), and to a lesser extent in the inguinal white adipose tissue (iWAT). TdTomato signal was also detected in the kidney, adrenal glands, and brain in male and female UCP1-Cre; Ai14-tdTomato (Figure 1A, Figure 2A, Supplemental Fig. 1A) and UCP1-Cre; Ai9-tdTomato mice (Supplemental Figs. 2 and 3). Within the brain, tdTomato expression was abundantly expressed in the ventromedial hypothalamus (VMH), amygdala, and parafascicular thalamic nucleus (PF) (Figure 2A, Supplemental Fig. 3A). Whole-brain imaging following tissue clearing from UCP1-Cre; Ai14-tdTomato mice confirmed extensive expression of tdTomato throughout the brain (Supplemental Videos 1 and 2), including the choroid plexus, which also exhibits Ucp1 mRNA expression in a deposited single cell RNAseq dataset [20] (Supplemental Fig. 3B) and histologically (Supplemental Fig. 1B). Fluorescent tdTomato signal was not observed in the epididymal white adipose tissue (eWAT), pituitary gland, liver, pancreas, skeletal muscle (Sk. Muscle), or heart from male or female UCP1-Cre; Ai14-tdTomato mice (Figure 1A, Supplemental Fig. 1A). To confirm and extend our fluorescent imaging from UCP1-Cre; Ai14-tdTomato tissue, we collected additional tissues from UCP1-Cre; Ai14-tdTomato male mice and Cre negative littermate controls to determine whole body mRNA expression of Ucp1 and tdTomato. Consistent with imaging data, tdTomato mRNA expression was detected in the BAT, iWAT, kidneys, adrenal glands, and hypothalamus in UCP1-Cre mice relative to Cre negative littermate controls (Figure 1B). Ucp1 mRNA expression was also detected in the BAT, iWAT, kidneys, adrenal glands, and hypothalamus (Figure 1C). Taken together, these data indicate that Ucp1 and Cre expression are not exclusive to thermogenic adipose tissues in UCP1-Cre mice.
Figure 1.
Peripheral and central expression of UCP1 in UCP1-Cre; Ai14-tdTomato male mice. (A) Representative fluorescence imaging for tdTomato expression in brown adipose tissue (BAT), inguinal white adipose tissue (iWAT), epididymal white adipose tissue (eWAT), kidney, pituitary gland, adrenal gland, liver, pancreas, skeletal muscle (Sk. Muscle), heart and testes from WT and UCP1-Cre positive male mice. (B–C) Relative tdTomato (B) and Ucp1 (C) mRNA levels in the BAT, iWAT, eWAT, adrenal gland, skeletal muscle, kidney, hypothalamus, thymus, intestine and pancreas from WT and UCP1-Cre; Ai14-tdTomato male mice (n = 6/group). Values are mean +/− SEM. ∗ = P < 0.05 compared with WT.
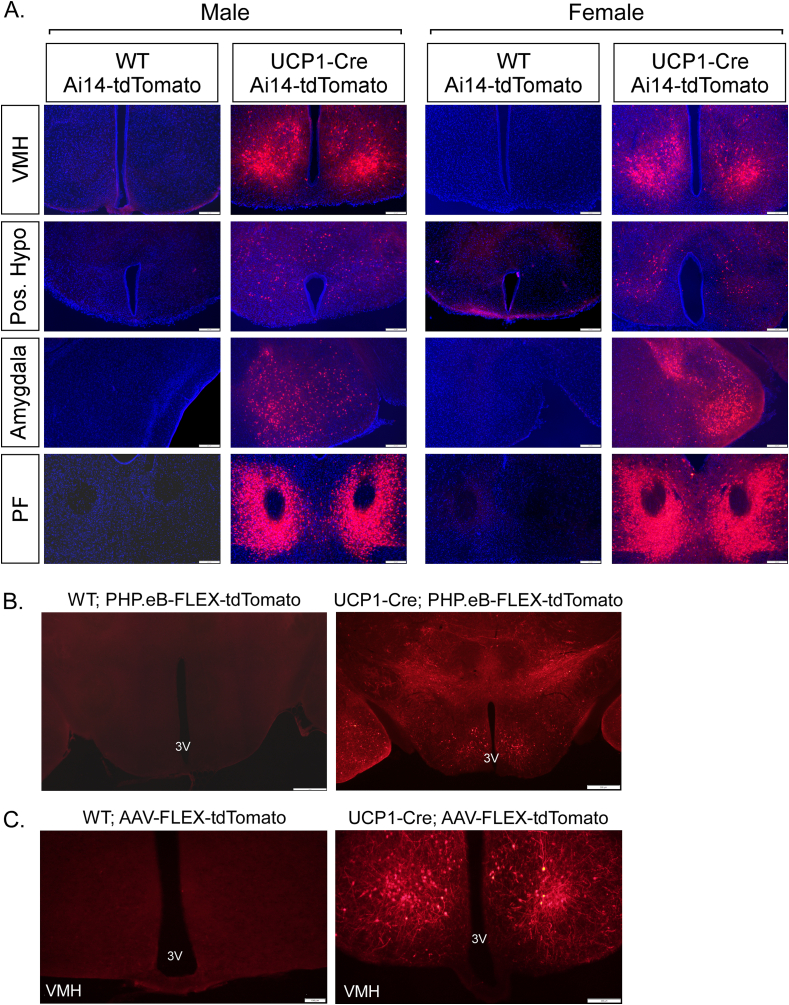
Figure 2.
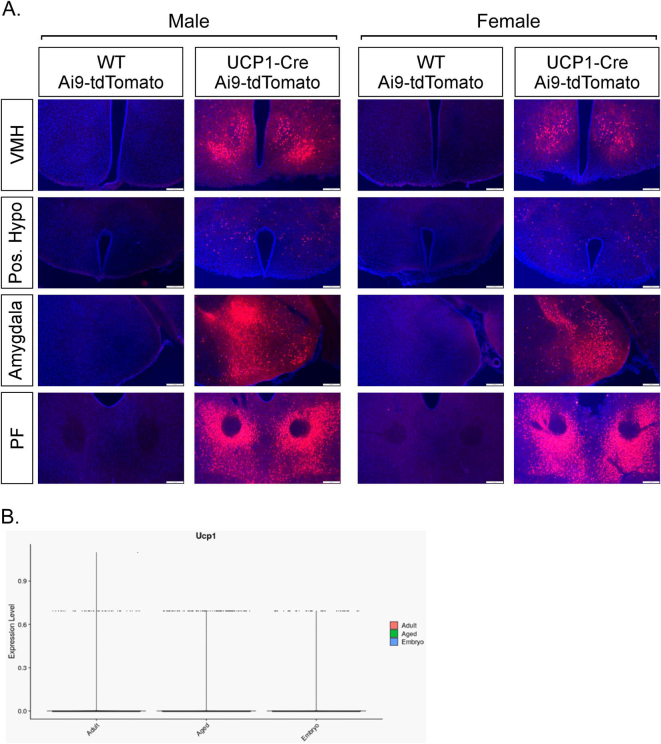
Central expression of UCP1 in adult mice. (A) Representative fluorescence imaging of tdTomato in the ventromedial hypothalamus (VMH), posterior hypothalamus, amygdala and parafascicular thalamic nucleus (PF) from WT and UCP1-Cre; Ai14-tdTomato male and female mice. (B) Representative fluorescence imaging of tdTomato in the VMH of 8-week-old male WT and UCP1-Cre male mice infected with a PHP.eB-FLEX-tdTomato virus. (C) Representative fluorescence imaging of tdTomato in the VMH of 12-week-old WT and UCP1-Cre male mice administered an AAV1-CAG-FLEX-tdTomato virus bilaterally into the VMH by stereotactic injection. 3V = third ventricle.
3.2. UCP1 is expressed in the adult central nervous system
To examine whether UCP1 is actively being expressed in the adult mouse brain, we injected UCP1-Cre male mice with either a PHP.eB pAAV-FLEX-tdTomato virus, which labels cells, predominantly neurons, throughout the CNS that actively express Cre recombinase [24], or a Cre-dependent AAV1-CAG-FLEX-tdTomato virus directly into the VMH. Consistent with UCP1-Cre; tdTomato reporter mice, UCP1-Cre mice, but not wild-type mice, infected with PHP.eB pAAV-FLEX-tdTomato virus exhibited robust tdTomato expression throughout numerous regions of the CNS including the VMH (Figure 2B and Supplemental Fig. 4). Robust tdTomato fluorescence was also observed in the VMH of UCP1-Cre mice bilaterally administered an AAV1-FLEX-tdTomato virus by stereotaxic injection (Figure 2C). These data indicate that UCP1 is actively expressed in the CNS.
3.3. UCP1 is expressed in the hypothalamus throughout development
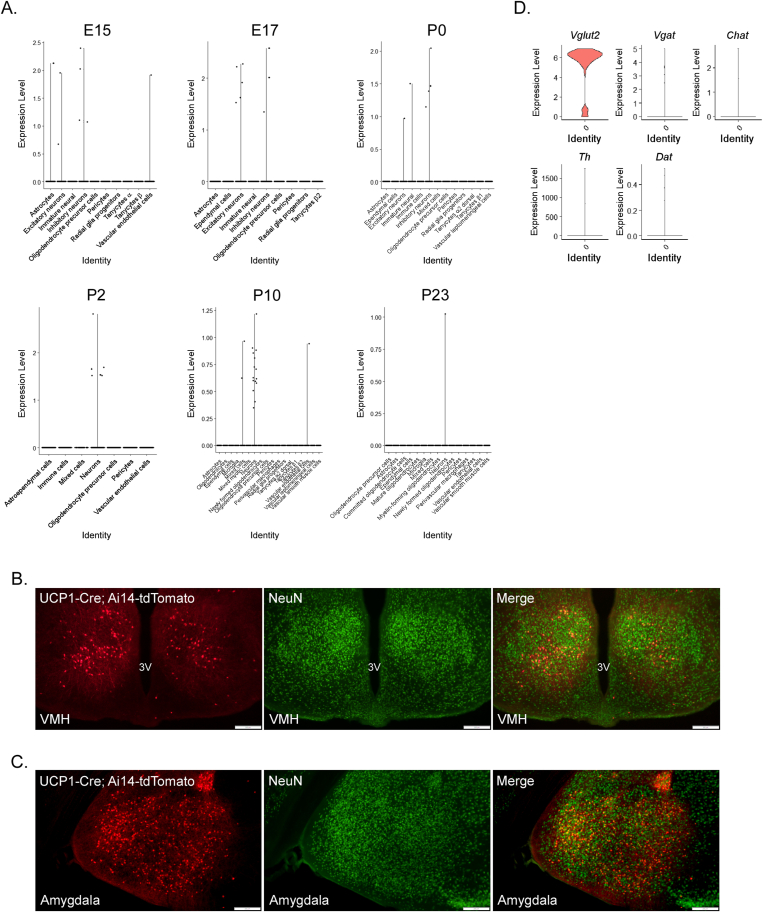
To further examine the expression of UCP1 within the CNS, we utilized deposited single-cell RNA sequencing (scRNAseq) datasets. To investigate developmental expression of Ucp1 in the hypothalamus, we assessed Ucp1 mRNA expression across different developmental stages of hypothalamic development at single cell resolution using a dataset derived via the Drop-seq methodology [19]. When assessing Ucp1 expression in the concatenated data from this study involving multiple developmental stages (E15-P23), it appears Ucp1 expression is primarily restricted to neurons (Figure 3A). To confirm that UCP1 is predominantly expressed in neurons, we immunostained UCP1-Cre; Ai14-tdTomato brain tissue with the neuronal marker, NeuN. As expected, a majority of tdTomato + cells within the VMH (Figure 3B) and amygdala (Figure 3C) colocalized to neurons. While Ucp1 is not widely expressed during embryonic development, Ucp1 expression appears to peak at post-natal day 10 (P10) in adolescent mice (Figure 3A). Within this dataset (of 51,199 cells), only ∼0.1% of cells expressed Ucp1. Ucp1 expression in the CNS is relatively low, and even sensitive scRNAseq protocols cannot often detect transcripts is some cells, which is termed dropout events [25].
Figure 3.
In silico analysis of central UCP1 expression. (A) Expression of Ucp1 in the hypothalamus throughout development. (B–C) Representative fluorescence imaging of tdTomato and NeuN in the (B) ventromedial hypothalamus (VMH) and (C) amygdala of UCP1-Cre; Ai14-tdTomato mice. (D) Violin plot of single-cell RNA sequencing analysis of relative mRNA expression in UCP1-expressing neurons. 3V = third ventricle.
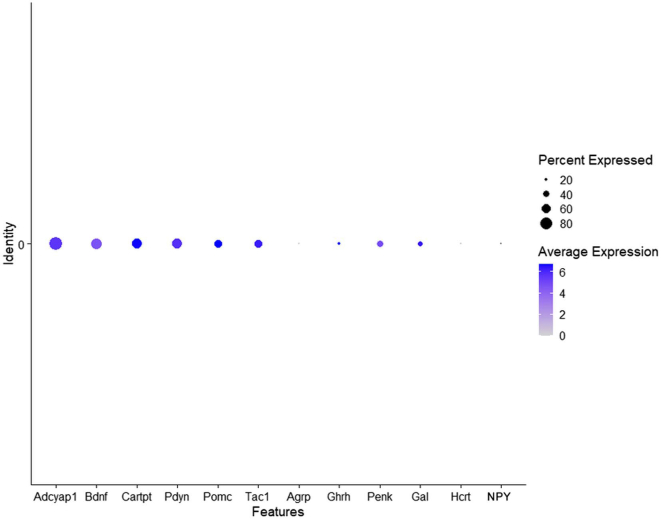
Given the abundant expression of tdTomato observed in the VMH (Figure 2A–C, Supplemental Fig. 3A, Supplemental Videos 1 and 2), we wanted to further investigate Ucp1 expression at the single cell level in the VMH of adult mice. We analyzed Ucp1 expression in a dataset (of 4,574 cells) which used the SMART-seq methodology to profile the transcriptomes of single cells of the VMH of adult mice [18]. Consistent with SMART-seq detecting a larger number of expressed genes compared to Drop-seq [26], 1.1% of Ucp1-expressing cells were identified in this dataset. This number reflects the percentage of cells expressing Ucp1 in the adult VMH and does not include the number of cells expressing Ucp1 or Cre recombinase (Figure 2A, Supplemental Fig. 3A, Supplemental Videos 1 and 2) throughout development (e.g., P10). In the adult VMH, Ucp1 expression is exclusive to Slc17a6+ (Vglut2) neurons (Figure 3D) which is consistent with previous work demonstrating that most neurons in the VMH are glutamatergic [27,28]. However, when profiling the expression of neuropeptides involved in regulation of appetite, we found that the majority of Ucp1+ neurons express transcripts for anorectic neuropeptides (Adcyap1, Bdnf, Cartpt, Pdyn, Pomc, Tac1) while exhibiting very little, if any, expression of transcripts for orexigenic peptides (Agrp, Ghrh, Penk, Gal, Hcrt, Npy) (Supplemental Fig. 5).
4. Discussion
The identification of thermogenic adipocytes in adult humans [[29], [30], [31]] has spawned numerous studies to evaluate the origins, regulation, and therapeutic potential of these exciting regulators of energy balance. Although classical brown adipocytes and beige/brite adipocytes do not share a common cellular progenitor [[32], [33], [34]], they both express the thermogenic protein UCP1 which is a critical regulator of adipocyte thermogenesis. With the development of the UCP1-Cre transgenic line in 2014 [10], investigators began exploring the role of various factors in thermogenic adipocyte function in vivo. In some cases, the phenotype of these animal models was compared to mice lacking a gene of interest from all adipose tissues using the Adiponectin-Cre transgenic mice. However, this has not occured in some other studies. Our finding that the UCP1-Cre line drives recombination of conditional alleles in non-thermogenic adipocyte cells, including central brain regions that regulate metabolism, has important implications for past and future studies to explore brown adipocyte function.
The expression of UCP1 in the VMH represents a potential confound of important consideration when attempting to interpret the metabolic phenotypes observed when UCP1-Cre mice are used to drive “tissue-specific” expression. The VMH is an important site for the regulation of energy homeostasis through modulation of both energy expenditure and food intake [35]. While the hypothesis that the VMH can function as a satiety center has been long proposed, albeit not without contention, the identification of a specific neuron population within the VMH which mediates this function has yet to be demonstrated. In this study, we observe that Ucp1+ cells in the VMH appear to favor expression of transcripts encoding anorectic hormones. Neuronal UCP1 expression in other animals, including carp [15] and ground squirrels [14], is critical for the regulation of thermogenesis when environmental temperatures are low. It has been suggested that the VMH is important for thermoregulation in response to cold in rabbits [36]. Together, this data suggests that neuronal UCP1 expression within the VMH may also regulate thermogenesis in response to cold exposure. However, while our in situ analysis and immunostaining suggests that UCP1 is predominantly restricted to neurons, we cannot exclude the possibility that UCP1 may also be expressed in glial cells. Although Ucp1 mRNA expression is observed in neurons, the level of Ucp1 in the CNS is markedly lower than what is observed in thermogenic adipose tissues. It is interesting to speculate that UCP1 may function in neurons to regulate reactive oxygen species as opposed to regulating heat production as it does in brown adipose tissue [37]. Future studies using specific targeting of UCP1+ neurons in the VMH will be critical to determine the contribution of this population to regulation of metabolism.
In addition to the brain, Ucp1 and tdTomato were also detected in the kidney and adrenal gland of UCP1-Cre; tdTomato mice. This is consistent with previously published data suggesting expression of UCP1 in these tissues [11,38]. Within the kidney, it has been demonstrated that UCP1 is expressed in renal tubular epithelial cells and that it functions to inhibit oxidative stress [11]. In contrast, a potential role of UCP1 in the adrenal glands remains unclear, although its expression does not appear to be regulated by cold exposure [38]. Some of the tdTomato signal in the adrenal gland is due to auto-fluorescence as there is also observable tdTomato signal in Cre negative control adrenal gland tissue. The adrenal glands are imbedded in perirenal adipose tissue, which expresses UCP1; therefore, some of the Ucp1 mRNA signal detected by qPCR may be due to small amounts of residual adipose tissue. Nevertheless, as an organ responsible for producing essential metabolic hormones, including epinephrine, norepinephrine, cortisol, and aldosterone, it will be critical to confirm that target gene expression is unaltered in the adrenal gland following gene knockout utilizing UCP1-Cre mice, and to determine whether adrenal UCP1 expression has any physiologically relevant function.
Here we have characterized UCP1 expression in tissues from B6.FVB-Tg (UCP1-Cre)1Evdr/J mice. However, there have been several other mouse lines driven by the UCP1 promoter. The first UCP1-Cre mouse model was developed in 2001 with a transgene that consisted of Cre cDNA inserted near exon 1 of the UCP1 gene, as well a portion of the 5’ UTR, and part of exon 3 and exons 4–6 downstream of Cre [39]. It was later reported that this line had issues with specificity [10]. In contrast, the B6.FVB-Tg(Ucp1-cre)1Evdr/J line investigated here was engineered using a bacterial artificial chromosome (BAC) construct containing the entire UCP1 gene, including key regulatory elements, with Cre recombinase inserted into the translation initiation site of exon 1 [10]. An inducible Tg(UCP1-Cre/ER)426Biat mouse line has also been generated using a BAC transgenic approach. In these animals, the administration of tamoxifen removes a stop cassette, thereby allowing expression of Cre in any cell that is currently expressing UCP1 [40]. While this line may eliminate off target genetic manipulations due to the developmental expression of UCP1, future studies are needed to determine whether this model also expresses Cre in non-adipose tissue cells that actively express UCP1 in the adult mouse as we have shown to be the case in several tissues, including the brain.
We have used multiple independent approaches to demonstrate that Cre expression in UCP1-Cre mice is not exclusive to thermogenic adipose tissue. We observe both Ucp1 expression and tdTomato fluorescence in non-adipose tissues, including the brain, kidney, and adrenal glands. UCP1 expression in the brain is not restricted to development as post-natal administration of Cre-dependent reporter viruses confirmed central UCP1 expression in the adult mouse. Phenotypes attributed to genetic deletions utilizing UCP1-Cre mice must consider potential direct effects on the CNS.
Author contributions
K.E.C. and K.H.F. designed and performed experiments, interpreted data, and wrote the paper. K.E.C., A.I.S., M.C.N., B.Z., T.N., K.H.F. and S.O.J performed experiments and interpreted data. M.J.P. conceived of the project, designed experiments, interpreted data, wrote the paper, and is responsible for the integrity of its content.
Acknowledgements
This work was funded by the National Institutes of Health (NIH) R01DK106104 (M.J.P.), F32 DK117510 (K.E.C.), T32 DK112751 (K.H.F.), and F31 DK117515 (S.O.J.). Veterans Affairs Merit Review Program I01BX004634 (M.J.P.), the University of Iowa Carver College of Medicine (M.J.P.). The authors would like to acknowledge use of the University of Iowa Central Microscopy Research Facility and the Iowa Neuroscience Institute's Neural Circuits and Behavior Core.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101405.
Conflict of interest
The authors have no conflicts to declare.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
FigS1.
FigS2.
FigS3.
FigS4.
FigS5.
References
- 1.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Scheja L., Heeren J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nature Reviews Endocrinology. 2019;15(9):507–524. doi: 10.1038/s41574-019-0230-6. [DOI] [PubMed] [Google Scholar]
- 3.Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fedorenko A., Lishko P.V., Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151(2):400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heaton G.M., Wagenvoord R.J., Kemp A., Jr., Nicholls D.G. Brown-adipose-tissue mitochondria: photoaffinity labelling of the regulatory site of energy dissipation. European Journal of Biochemistry. 1978;82(2):515–521. doi: 10.1111/j.1432-1033.1978.tb12045.x. [DOI] [PubMed] [Google Scholar]
- 7.Klingenberg M., Huang S.G. Structure and function of the uncoupling protein from brown adipose tissue. Biochimica et Biophysica Acta. 1999;1415(2):271–296. doi: 10.1016/s0005-2736(98)00232-6. [DOI] [PubMed] [Google Scholar]
- 8.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J., et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. Journal of Clinical Investigation. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajimura S., Spiegelman B.M., Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metabolism. 2015;22(4):546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong X., Banks A., Liu T., Kazak L., Rao R.R., Cohen P., et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158(1):69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia P., Wu X., Pan T., Xu S., Hu J., Ding X. Uncoupling protein 1 inhibits mitochondrial reactive oxygen species generation and alleviates acute kidney injury. EBioMedicine. 2019;49:331–340. doi: 10.1016/j.ebiom.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frontini A., Rousset S., Cassard-Doulcier A.M., Zingaretti C., Ricquier D., Cinti S. Thymus uncoupling protein 1 is exclusive to typical brown adipocytes and is not found in thymocytes. Journal of Histochemistry and Cytochemistry. 2007;55(2):183–189. doi: 10.1369/jhc.6A7013.2006. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Willershauser M., Karlas A., Gorpas D., Reber J., Ntziachristos V., et al. A dual Ucp1 reporter mouse model for imaging and quantitation of brown and brite fat recruitment. Molecular Metabolism. 2019;20:14–27. doi: 10.1016/j.molmet.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laursen W.J., Mastrotto M., Pesta D., Funk O.H., Goodman J.B., Merriman D.K., et al. Neuronal UCP1 expression suggests a mechanism for local thermogenesis during hibernation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(5):1607–1612. doi: 10.1073/pnas.1421419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jastroch M., Buckingham J.A., Helwig M., Klingenspor M., Brand M.D. Functional characterisation of UCP1 in the common carp: uncoupling activity in liver mitochondria and cold-induced expression in the brain. Journal of Comparative Physiology B. 2007;177(7):743–752. doi: 10.1007/s00360-007-0171-6. [DOI] [PubMed] [Google Scholar]
- 16.Lengacher S., Magistretti P.J., Pellerin L. Quantitative rt-PCR analysis of uncoupling protein isoforms in mouse brain cortex: methodological optimization and comparison of expression with brown adipose tissue and skeletal muscle. Journal of Cerebral Blood Flow and Metabolism. 2004;24(7):780–788. doi: 10.1097/01.WCB.0000122743.72175.52. [DOI] [PubMed] [Google Scholar]
- 17.BonDurant L.D., Ameka M., Naber M.C., Markan K.R., Idiga S.O., Acevedo M.R., et al. FGF21 regulates metabolism through adipose-dependent and -independent mechanisms. Cell Metabolism. 2017;25(4):935–944. doi: 10.1016/j.cmet.2017.03.005. e934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D.W., Yao Z., Graybuck L.T., Kim T.K., Nguyen T.N., Smith K.A., et al. Multimodal analysis of cell types in a hypothalamic node controlling social behavior. Cell. 2019;179(3):713–728. doi: 10.1016/j.cell.2019.09.020. e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romanov R.A., Tretiakov E.O., Kastriti M.E., Zupancic M., Haring M., Korchynska S., et al. Molecular design of hypothalamus development. Nature. 2020;582(7811):246–252. doi: 10.1038/s41586-020-2266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dani N., Herbst R.H., McCabe C., Green G.S., Kaiser K., Head J.P., et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell. 2021;184(11):3056–3074 e3021. doi: 10.1016/j.cell.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Y., Yu T., Xu J., Wan P., Ma Y., Zhu J., et al. FDISCO: advanced solvent-based clearing method for imaging whole organs. Science Advances. 2019;5(1) doi: 10.1126/sciadv.aau8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Xu J., Wan P., Yu T., Zhu D. Optimization of GFP fluorescence preservation by a modified uDISCO clearing protocol. Frontiers in Neuroanatomy. 2018;12:67. doi: 10.3389/fnana.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan C., Cai R., Quacquarelli F.P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nature Methods. 2016;13(10):859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- 24.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nature Neuroscience. 2017;20(8):1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque A., Engel J., Teichmann S.A., Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Medicine. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., et al. Comparative analysis of single-cell RNA sequencing methods. Molecular Cell. 2017;65(4):631–643. doi: 10.1016/j.molcel.2017.01.023. e634. [DOI] [PubMed] [Google Scholar]
- 27.Shimazu T., Minokoshi Y. Systemic glucoregulation by glucose-sensing neurons in the ventromedial hypothalamic nucleus (VMH) Journal of Endocrine Society. 2017;1(5):449–459. doi: 10.1210/js.2016-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong Q., Ye C., McCrimmon R.J., Dhillon H., Choi B., Kramer M.D., et al. Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metabolism. 2007;5(5):383–393. doi: 10.1016/j.cmet.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B., et al. Identification and importance of brown adipose tissue in adult humans. New England Journal of Medicine. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D., et al. Cold-activated brown adipose tissue in healthy men. New England Journal of Medicine. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 31.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T., et al. Functional brown adipose tissue in healthy adults. New England Journal of Medicine. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 32.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raajendiran A., Ooi G., Bayliss J., O'Brien P.E., Schittenhelm R.B., Clark A.K., et al. Identification of metabolically distinct adipocyte progenitor cells in human adipose tissues. Cell Reports. 2019;27(5):1528–1540. doi: 10.1016/j.celrep.2019.04.010. e1527. [DOI] [PubMed] [Google Scholar]
- 35.Timper K., Bruning J.C. Hypothalamic circuits regulating appetite and energy homeostasis: pathways to obesity. Disease Models Mechanisms. 2017;10(6):679–689. doi: 10.1242/dmm.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morimoto A., Murakami N., Ono T., Watanabe T., Sakata Y. Stimulation of ventromedial hypothalamus induces cold defense responses in conscious rabbits. American Journal of Physiology. 1986;250(4 Pt 2):R560–R566. doi: 10.1152/ajpregu.1986.250.4.R560. [DOI] [PubMed] [Google Scholar]
- 37.Jastroch M. Uncoupling protein 1 controls reactive oxygen species in brown adipose tissue. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(30):7744–7746. doi: 10.1073/pnas.1709064114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita H., Habuta M., Hattori T., Kubota S., Kumon H., Ohuchi H. UCP1 expression in the mouse adrenal gland is not upregulated by thermogenic conditions. Biochemical and Biophysical Research Communications. 2021;566:184–189. doi: 10.1016/j.bbrc.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Guerra C., Navarro P., Valverde A.M., Arribas M., Bruning J., Kozak L.P., et al. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. Journal of Clinical Investigation. 2001;108(8):1205–1213. doi: 10.1172/JCI13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenwald M., Perdikari A., Rulicke T., Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nature Cell Biology. 2013;15(6):659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.