Abstract
As a component of N6-methyladenosine (m6A) “writers,” KIAA1429 was reported to promote breast cancer proliferation and growth in m6A-independent manners. However, the related mechanism of KIAA1429 in breast cancer metastasis has not been reported. In the present study, we found KIAA1429 could significantly promote the migration and invasion of breast cancer cells. Then we demonstrated that knockdown of KIAA1429 could impede breast cancer metastasis in nude mice in vivo. The level of SNAIL expression and epithelial-mesenchymal transition (EMT) progress was positively related with KIAA1429. Furthermore, we confirmed that the suppression of cell migration, invasion, and EMT progress by knockdown of KIAA1429 could be reversed by the upregulation of SNAIL. However, structural maintenance of chromosomes 1A (SMC1A), not KIAA1429, bound with the SNAIL promoter region directly and promoted the transcription of SNAIL. Then we confirmed that KIAA1429 could bind to the motif in the 3′ UTR of SMC1A mRNA directly and enhance SMC1A mRNA stability. In conclusion, our study revealed a novel mechanism of the KIAA1429/SMC1A/SNAIL axis in the regulation of metastasis of breast cancer. Moreover, it first provided detailed investigation of how KIAA1429 regulated the targeted gene expression at posttranscriptional levels as an RNA binding protein unrelated to its m6A modification.
Keywords: breast cancer, KIAA1429, SMC1A, SNAIL, metastasis
Graphical abstract
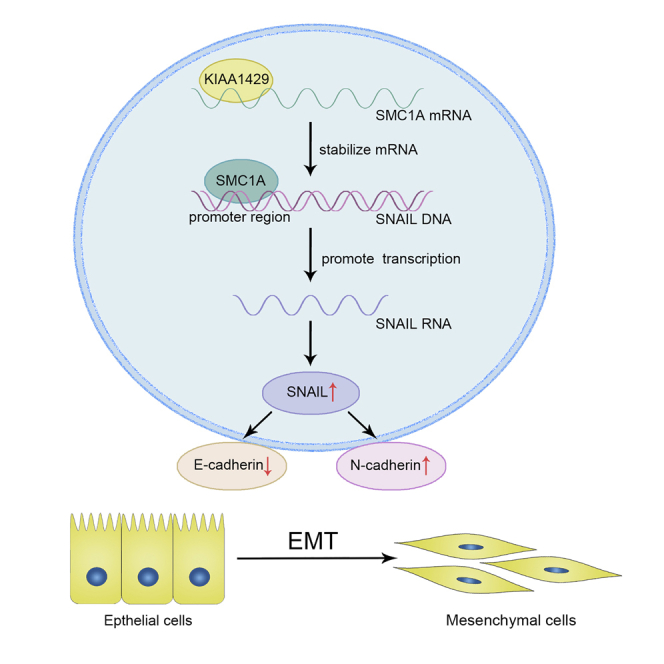
We provided a novel mechanism of the KIAA1429/SMC1A/SNAIL axis in the regulation of breast cancer invasion and metastasis, which may be a promising therapeutic target for human breast cancer.
Introduction
Breast cancer, originating from breast tissue, is the most commonly diagnosed malignant tumor and represents the leading cause of cancer-associated deaths among women worldwide.1,2 Although dramatic advancement has been made in the early diagnosis, and complex treatment such as surgical operation, radiotherapy, endocrine therapy, and immunotherapy in the past decades, the prognosis of breast cancer patients is still poor due to the high rate of lethal distant metastasis.3 Metastasis is a multi-step process by which cancer cells spread from the primary tumor to colonize distant sites. Epithelial-mesenchymal transition (EMT) is defined as a biologic procedure wherein cells lose their epithelial features and acquire mesenchymal characteristics, which enables them to migrate further and invade the underlying mesenchyme.4 Although recent studies have raised concerns about the true contribution of EMT to metastasis in pancreatic cancer5 and lung cancer,6 the experimental results acquired in breast cancer still support the idea that EMT plays a vital role in breast cancer metastasis.7, 8, 9, 10
Up to now, more than 100 kinds of RNA modification have been confirmed.11 Among them, N6-methyladenosine (m6A) has been considered as the most pervasive, abundant, and dynamic eukaryotic RNA modification, which affects RNA transcription, processing, translation, and metabolism.12 m6A RNA modification is regulated by methyltransferases (“writers”), demethylases (“erasers”), and RNA binding proteins (“readers”), and the m6A “writers” complex includes the methyltransferase-like 3 (METTL3), METTL14, WT1-associated protein (WTAP), RNA binding motif protein 15/15B (Rbm15/15B), and KIAA1429 (or VIRMA).13 It was found that the components in m6A “writers” complex were involved in many cancers’ metastasis and invasion via regulating the RNA fate of many oncogenes or tumor suppressor genes.14, 15, 16 In the m6A methyltransferase complex, KIAA1429 serves as a scaffold in bridging the catalytic core components METTL3 and METTL14 and acts as a positive regulator of oncogenesis. For instance, KIAA1429 facilitated the migration and invasion of hepatocellular carcinoma (HCC) through m6A modification of ID2 mRNA.17 It acts as an oncogenic factor in gastric cancer by stabilizing c-Jun mRNA in an m6A-independent manner.18 Furthermore, circ_KIAA1429, which came from KIAA1429, could accelerate HCC advancement and maintain the expression of Zeb1 through the mechanism of m6A-YTHDF3-Zeb1 in HCC.19 KIAA1429 also acted as an independent prognostic factor to classify lung cancers for patient stratification.20 In our previous studies, KIAA1429 could promote breast cancer cells proliferation in an m6A-independent manner as RNA binding protein.21 We also found that KIAA1429 could increase the metastasizing ability of breast cancers, but the detailed mechanism was still unknown.
In this study, we explored the deeper mechanisms that KIAA1429 participated in the invasion and metastasis of breast cancer. We first demonstrated that KIAA1429 was associated with breast cancer invasion and metastasis both in vivo and in vitro. Then we investigated the role of KIAA1429 in regulating cellular morphology and EMT markers, and we found that KIAA1429 could affect snail family transcriptional repressor 1 (SNAIL) expression obviously in the EMT regulator. However, we proved that KIAA1429 could not regulate SNAIL directly. So we found the structural maintenance of chromosomes 1A (SMC1A) was the potential targeting gene of KIAA1429 in breast cancer identified by RNA immunoprecipitation (RIP). Moreover, SMC1A could regulate SNAIL as a transcription factor, which promoted the process of EMT. We then confirmed that KIAA1429 could regulate SMC1A by an m6A-independent manner. Here we discovered a specific mechanism for KIAA1429 in the invasion and metastasis of breast cancer.
Results
KIAA1429 could promote breast cancer migration and invasion both in vitro and in vivo
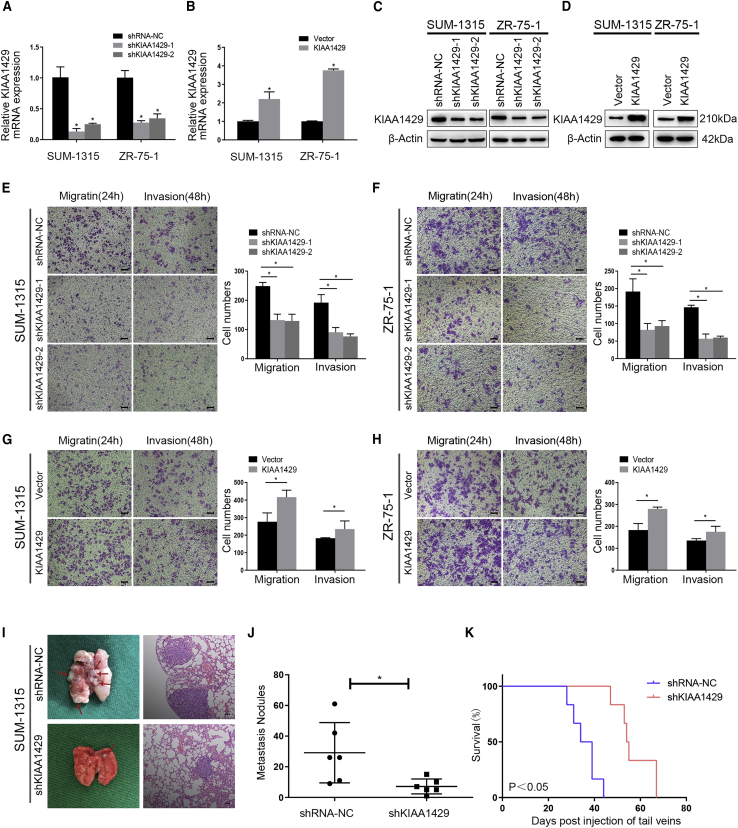
To investigate the effect of KIAA1429 on the metastasis of breast cancer, we transfected SUM-1315 and ZR-75-1 cells with lentivirus or adenovirus to repress or overexpress the expression of KIAA1429. KIAA1429 knockdown lentivirus constructs were generated and obtained as previously described.21 The mRNA and protein levels of KIAA1429 were verified by qRT-PCR (quantitative real-time PCR) (Figures 1A and 1B) and western blot (Figures 1C and 1D). As shown in Figures S1A and S1B, the migrated area of KIAA1429 knockdown increased, compared with the control cells (∗p < 0.05) in SUM-1315 and ZR-75-1 cells. On the contrary, the migrated area of KIAA1429 overexpression decreased, compared with the control in SUM-1315 and ZR-75-1 cells (Figures S1C and S1D, ∗p < 0.05). In addition, KIAA1429 knockdown showed significant decreasing ability of migration and invasion in SUM-1315 and ZR-75-1 cells (Figures 1E and 1F, ∗p < 0.05), while KIAA1429 overexpression showed significant increasing ability of migration and invasion in SUM-1315 and ZR-75-1 cells (Figures 1G and 1H, ∗p < 0.05).
Figure 1.
KIAA1429 promoted the migration and invasion of breast cancer cells in vitro and in vivo
(A–D) SUM-1315 and ZR-75-1 cell lines were respectively transfected with lentivirus and adenovirus to knock down (shKIAA1429-1, shKIAA1429-2) or overexpress KIAA1429 expression. qRT-PCR (A and B) and western blot (C and D) were applied to verify transfection efficiency. (E–H) Transwell experiment was performed to analyze the invasion and migration ability of SUM-1315 and ZR-75-1 cells. Data of migration and invasion are shown in the histogram on the right. Scale bars, 50 μm. Data were shown as mean ± SEM; ∗p < 0.05. (I) Representative lung images of the mice and H&E staining of lung section revealed the numbers and sizes of lung colonization in the control group and the KIAA1429 knockdown group. (J) Metastasis nodules plot was formed by the H&E staining of lung section of the mice (n = 6). Data were shown as mean ± SEM; ∗p < 0.05. (K) Kaplan-Meier survival curves of mice in the control group and the KIAA1429 knockdown group.
To investigate the effects of KIAA1429 on breast cancer metastasis in vivo, we injected the SUM-1315 cells into tail veins of nude mice. The tumor presence was validated by histological examination (Figure 1I). The results demonstrated that mice injected with sh-KIAA1429 cells produced less lung colonization compared with those with the control cells (Figure 1J). Moreover, we found that knockdown of KIAA1429 could prolong the survival time of mice compared with the control group (Figure 1K). These data strongly proved that KIAA1429 could promote breast cancer metastasis in vitro and in vivo.
KIAA1429 promoted EMT of breast cancer cells
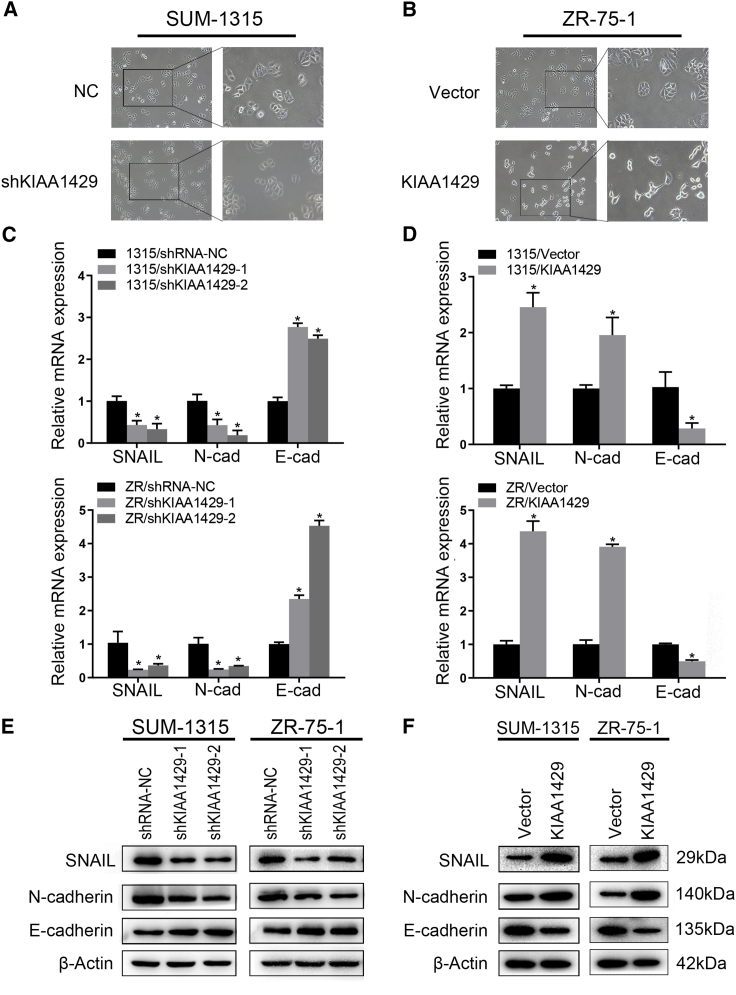
The morphological change in breast cancer cells was widely used for assessing EMT and was evaluated by microscopy. Significant morphological changes (from round-shaped to fibroblast-like cells) were observed in KIAA1429 overexpression cells compared with the control cells after 48 h of transfection in the SUM-1315 and ZR-75-1 cell lines (Figures 2A and 2B). We also revealed upregulation of the mesenchymal phenotype marker N-cadherin, as well as downregulation of the epithelial phenotype marker E-cadherin in KIAA1429 knockdown cells, when compared with the control cells. Reversed expression of E-cadherin and N-cadherin was observed in KIAA1429 overexpression cells compared with the control cells (Figures 2E and 2F). To explore the mechanism by which KIAA1429 regulates the EMT process in breast cancer cells, western blot and qRT-PCR were applied to show EMT regulator in KIAA1429 knockdown breast cancer cells, and we found SNAIL was mostly associated with KIAA1429 knockdown (Figure S4). SNAIL has a pivotal role in the regulation of EMT, which involves regulation of related biomarkers.22 These results imply that KIAA1429 might affect EMT progress by regulating SNAIL expression.
Figure 2.
KIAA1429 promoted the epithelial-mesenchymal transition of breast cancer cells
(A and B) KIAA1429 endowed breast cancer cells with a mesenchymal morphology. SUM-1315 (A) and ZR-75-1 (B) cells were observed under a microscope, and then we took pictures. The right panel shows the higher magnification of the boxed areas in the left panel. (C–F) qRT-PCR and western blot assays confirmed N-cadherin, E-cadherin, and SNAIL expression after knockdown (C and E) or overexpression (E and F) of KIAA1429 in SUM-1315 and ZR-75-1 cells. Results were representative of three independent experiments and presented as the mean ± SEM; ∗p < 0.05.
SNAIL reversed the suppression of migration and metastasis induced by KIAA1429 knockdown in vitro and in vivo
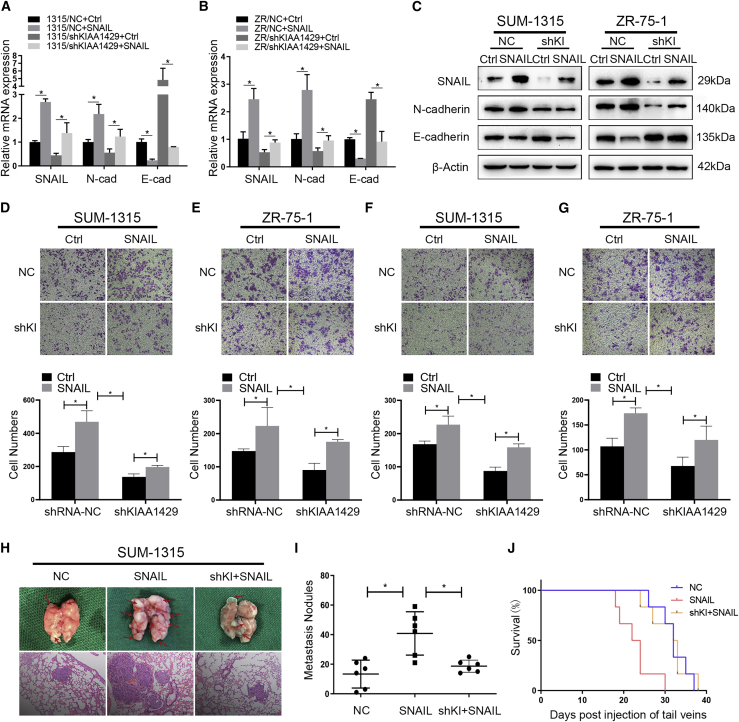
SNAIL is known as a significant transcription factor that can promote the EMT progress in breast cancer cells. To explore the effect of KIAA1429 on SNAIL-inducing EMT progress, we transfected KIAA1429 knockdown and the control groups of SUM-1315 and ZR-75-1 cells to overexpress SNAIL. The transfection efficiency was confirmed by qRT-PCR and western blot (Figures 3A–3C). In transwell migration and invasion assay, the SNAIL overexpression group exhibited a stronger ability of metastasis in SUM-1315 cells, while cell migration and invasion were significantly inhibited by the knockdown of KIAA1429. Similar results were also found in ZR-75-1 cells (Figures 3D–3G). Furthermore, NC (Negative countrol), shKIAA1429, and shKIAA1429+SNAIL groups of SUM-1315 cell lines were injected into tail veins of nude mice. Figures 3H–3J indicated that overexpression of SNAIL obviously increased lung metastases formed in numbers and shortened the survival time of mice, while knockdown of KIAA1429 strongly decreased the formation of metastases and prolonged the survival time. All the results indicated that KIAA1429 could promote SNAIL-inducing EMT progress both in vitro and in vivo.
Figure 3.
Snail reversed the suppression of migration and metastasis induced by KIAA1429 knockdown in vitro and in vivo
(A–C) KIAA1429 knockdown and the control groups of SUM-1315 and ZR-75-1 cells were transfected to overexpress SNAIL, followed by qRT-PCR and western blots examination. (D–G) Transwell experiment was performed to analyze the invasion and migration ability of SUM-1315 and ZR-75-1 cells. The lower panel of each picture showed the data of migration and invasion. Scale bars, 50 μm. Data were shown as mean ± SEM; ∗p < 0.05. (H and I) Representative images showed the sizes and numbers of lung metastasis. Metastasis nodules plot was formed by the H&E staining of lung section of the mice (n = 6). Data were shown as mean ± SEM; ∗p < 0.05. (J) Kaplan-Meier survival curves of mice in the control group, SNAIL group, and the shKIAA1429+SNAIL group(shKI+SNAIL).
SMC1A might be the potential target that was regulated by KIAA1429 and regulated the expression of SNAIL
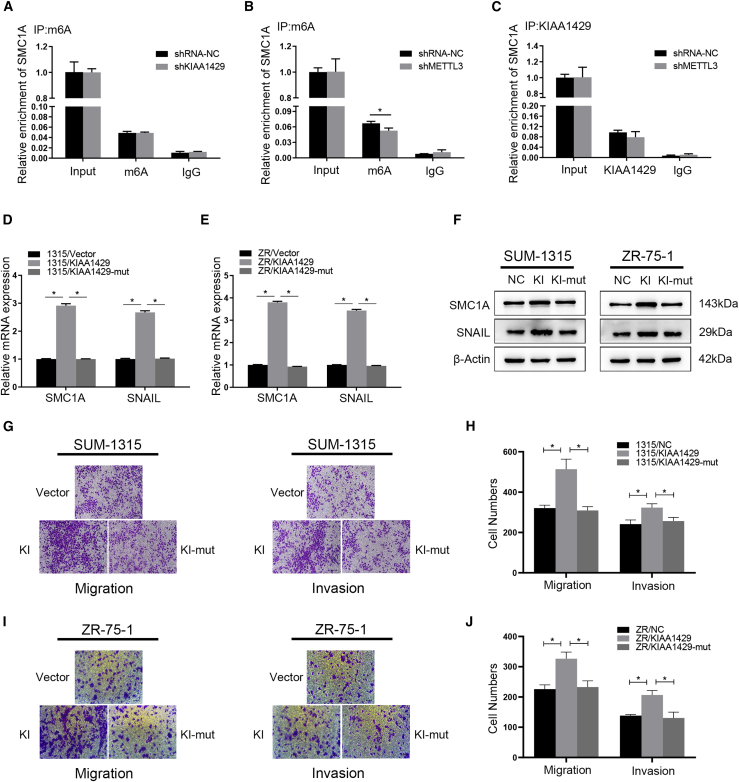
Figure S2A also demonstrated a positive correlation between the expression of KIAA1429 and SNAIL in 54 breast cancer patients in our hospital (∗p < 0.05). According to the previous study, the m6A level of total RNAs reduced obviously in the sh-KIAA1429 and sh-METTL3 cells, compared with the control groups in breast cancer cells.21 To explore the mechanism whether KIAA1429 would regulate SNAIL expression directly by an m6A-dependent manner, we immunoprecipitated RNAs of control, KIAA1429 knockdown, and METTL3 knockdown SUM-1315 cells by using the anti-m6A antibody. The results indicated that KIAA1429 knockdown did not decrease the m6A level of SNAIL mRNA (Figure S2B). Meanwhile METTL3 knockdown decreased the m6A level of SNAIL mRNA obviously (Figure S2C). These results indicated that KIAA1429 could not regulate SNAIL by an m6A-dependent manner. To investigate whether KIAA1429 regulated SNAIL in an m6A-independent manner, we treated KIAA1429 knockdown and control cells with actinomycin D (Act D, 5 mg/mL) at different time points in both SUM-1315 and ZR-75-1 cells lines. However, the half-life of the SUM-1315 and ZR-75-1 cells showed no difference (Figures S2D and S2E). Another RIP assay followed by RT-PCR was used, and we found that SNAIL mRNA transcript was not present in KIAA1429 (Figure S2F). Our former RIP sequencing (RIP-seq) of KIAA1429 indeed found SNAIL or other EMT markers were not bound with KIAA1429.21 According to the above results, KIAA1429 might not regulate SNAIL mRNA directly. So we speculated that there were intermediate links between KIAA1429 and SNAIL.
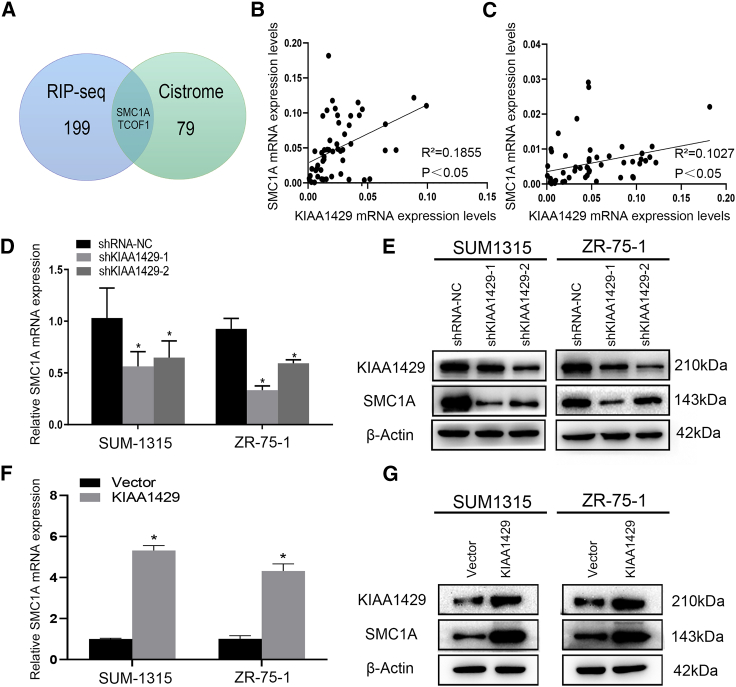
To identify potential mRNAs that were associated with KIAA1429 protein, we selected 201 genes that had different expressions between KIAA1429 and IgG group from former RIP-seq of KIAA1429 as described previously.21 It is known from the above that knockdown or overexpression of KIAA1429 would affect the EMT progress. Thereinto, SNAIL was together increased or decreased with KIAA1429, which was the best-characterized EMT effector. Then we predicted the potential target that was regulated by KIAA1429 and regulated the expression of SNAIL. Then we found about 125 targeted genes from the Cistrome that would promote the transcription of SNAIL. We took the intersection from these two groups, and we found SMC1A and TCOF1 met this criterion (Figure 4A). Then western blot and qRT-PCR analyses revealed that SMC1A might be the potential target (Figures 4D–4G), because TCOF1 did not change after KIAA1429 knockdown (Figure S3).
Figure 4.
Identification of KIAA1429-associated mRNAs by RIP
(A) Venn diagram showing SMC1A and TCOF1 were selected between former RIP-seq of KIAA1429 and target genes from the Cistrome, which would promote the transcription of SNAIL. (B and C) Correlation analysis between SMC1A and KIAA1429 or SNAIL expression in breast cancer tissues from patients (n = 54). (D–G) Expression of SMC1A was significantly decreased or increased following KIAA1429 knockdown or overexpression in SUM-1315 and ZR-75-1 cells at both the mRNA (D and F) and protein levels (E and G).
SMC1A could reverse the promotion of migration and metastasis induced by KIAA1429 overexpression
KIAA1429 overexpression and the control groups of SUM-1315 and ZR-75-1 cells were transfected into small interfering RNA (siRNA) to suppress SMC1A. The transfection efficiency of EMT-related proteins was confirmed by qRT-PCR and western blot (Figures S5A–S5C). In transwell migration and invasion assay, the SMC1A suppression group showed a weaker ability of metastasis in SUM-1315 cells and ZR-75-1 cells, while cell migration and invasion were significantly promoted by the overexpression of KIAA1429 (Figures S5D–S5G). These results indicated that SMC1A could reverse the promotion of metastasis induced by KIAA1429 overexpression.
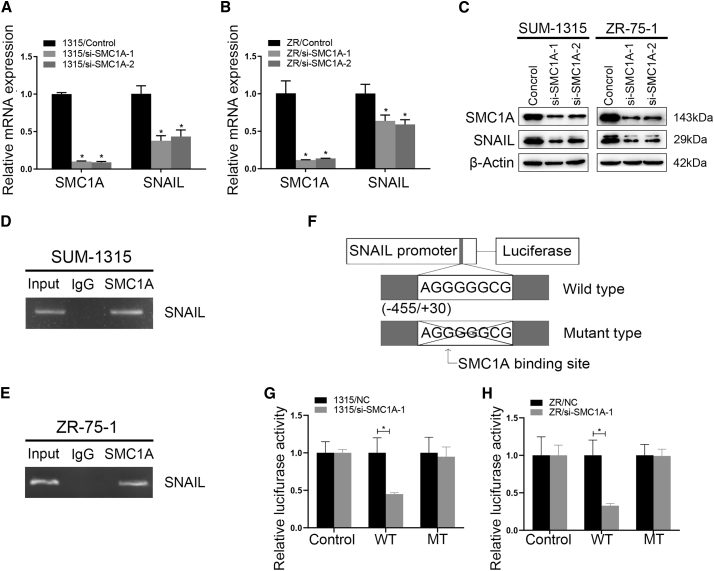
SMC1A regulated SNAIL expression by binding to the DNA recognition sequence in the promoter region of SNAIL gene in breast cancer cells
To investigate whether SMC1A would regulate SNAIL, we transfected siRNA or control into SUM-1315 and ZR-75-1 wild-type cells. The transfection efficiency was confirmed, and the expression of SNAIL was decreased at the same time by qRT-PCR and western blot (Figures 5A–5C). Subsequent analysis of the SNAIL gene revealed the presence of DNA recognition sequence of SMC1A23 (sequence: 5′-AGGGGGGC-3′) with the potential sites to be bound specifically by SNAIL in its promoter region. Chromatin immunoprecipitation (ChIP) assays showed that SMC1A bound to this recognition sequence of SNAIL gene in SUM-1315 and ZR-75-1 cells (Figures 5D and 5E). Luciferase activity assays found that luciferase activity for a reporter carrying the SMC1A recognition sequence in SNAIL promoter region was prominently increased in the absence of SMC1A in SUM-1315 (Figure 5G) and ZR-75-1 (Figure 5H) cells. These indicated that SMC1A could regulate SNAIL expression by binding to the recognition sequence in the promoter region of the SNAIL gene in breast cancer cells.
Figure 5.
SMC1A regulated SNAIL expression by binding to the DNA recognition sequence in the promoter region of SNAIL gene in breast cancer cells
(A–C) SUM-1315 and ZR-75-1 cell lines were transfected with small interfering SMC1A to knock down (siSMC1A-1, siSMC1A-2) SMC1A expression. qRT-PCR and western blot were used to confirm the transfection efficiency, and the SNAIL expression decreased obviously after downregulating SMC1A expression at both mRNA and protein levels. (D and E) SUM-1315 and ZR-75-1 cell lysates were collected and immunoprecipitated with SMC1A antibody or control IgG followed by RT-PCR and qRT-PCR to detect SNAIL mRNA levels. (F) Schematic diagram of the dual-luciferase reporter containing the motif in the promoter region of SNAIL gene. (G and H) The reporter containing SNAIL promoter region decreased by the knockdown of SMC1A in SUM-1315 and ZR-75-1 cells. Data were shown as mean ± SEM. ∗p < 0.05.
KIAA1429 regulated SMC1A mRNA expression in an m6A-independent manner
To explore the mechanism by which KIAA1429 regulates SMC1A and whether KIAA1429 would influence the m6A level of SMC1A RNAs directly, we conducted MeRIP (methylated RNA Immunoprecipitation), and the results indicated that KIAA1429 knockdown did not decrease the m6A level of SMC1A mRNA (Figure 6A). On the country, METTL3 knockdown decreased the m6A level of SMC1A mRNA obviously (Figure 6B). Moreover, the SMC1A mRNA that interacted with KIAA1429 did not change, while METTL3 knockdown changed (Figure 6C). Moreover, knockdown of METTL3 could not regulate SMC1A expression in breast cancer cells (Figure S6). These results showed that KIAA1429 could not disturb the m6A level of SMC1A mRNA, implying the m6A modification could not affect the interaction between KIAA1429 and SMC1A mRNA. All the results indicated that KIAA1429 regulated the SMC1A mRNA expression in an m6A-independent manner. To explore how KIAA1429 promotes breast cancer migration and invasion in an m6A-independent manner, we generated a KIAA1429 without RNA binding domain (KIAA1429-mut).24 SUM-1315 and ZR-75-1 cells were transfected with adenovirus containing KIAA1429 and KIAA1429-mut plasmids. We found the mRNA and protein levels of SMC1A and SNAIL were higher in the KIAA1429 overexpression group than the KIAA1429-mut overexpression group (Figures 6D–6F). In transwell migration and invasion assay, the KIAA1429 overexpression group exhibited a stronger ability of metastasis in SUM-1315 and ZR-75-1 cells, while the KIAA1429-mut group did not (Figures 6G–6J). These data proved that KIAA1429 could promote breast cancer metastasis in an m6A-independent manner.
Figure 6.
KIAA1429 regulated SMC1A mRNA expression in breast cancer by an m6A-independent manner
(A) The detection of SMC1A m6A modification level by immunoprecipitation of m6A modified RNAs in control or KIAA1429 knockdown cells followed by qRT-PCR. (B) The detection of SMC1A m6A modification level by immunoprecipitation of m6A modified RNAs in control or METTL3 knockdown cells followed by qRT-PCR. ∗p < 0.05. (C) The detection of SMC1A binding to KIAA1429 by immunoprecipitation of KIAA1429-associated RNA in control and METTL3 knockdown cells followed by qRT-PCR. (D–F) KIAA1429 overexpression (KI), KIAA1429-mut overexpression (KI-mut), and the control groups of SUM-1315 and ZR-75-1 cells were transfected, followed by qRT-PCR and western blots examination. (G–J) Transwell experiment was performed to analyze the invasion and migration ability of SUM-1315 and ZR-75-1 cells. The right panel of each picture showed the data of migration and invasion. Scale bars, 50 μm. Data were shown as mean ± SEM; ∗p < 0.05.
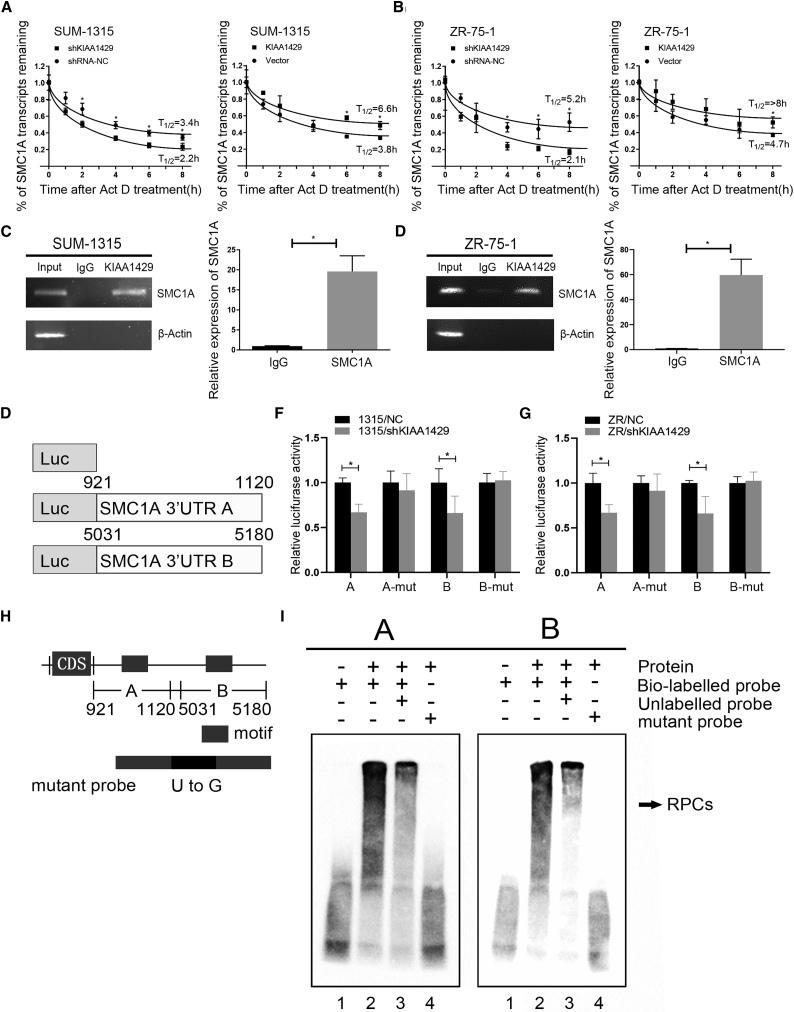
To investigate whether KIAA1429 regulates the expression of SMC1A in an m6A-independent manner, we treated KIAA1429 knockdown and the control breast cancer cells with 5 mg/mL Act D at different time points. Figure 7A indicated that downregulation of KIAA1429 decreased the half-life of SMC1A mRNA from 3.4 to 2.2 h. Overexpression of KIAA1429 increased the half-life of SMC1A mRNA from 3.8 to 6.6 h in SUM-1315 cells. Similar results were confirmed in ZR-75-1 cells (Figure 7B). These results suggested that KIAA1429 could increase SMC1A expression by regulating its mRNA stability. Furthermore, we investigated whether KIAA1429 bound to the SMC1A mRNA directly; RIP assay was performed in SUM-1315 (Figure 7C) and ZR-75-1 (Figure 7D) cell lines. The results showed that SMC1A mRNA was detected in KIAA1429 and the Input group, whereas it was not in the IgG group. It suggested that KIAA1429 could bind physically to SMC1A mRNA. To further explore if KIAA1429 could specifically bind to the motif in the 3′ UTR of SMC1A mRNA, we carried out luciferase reporter assay with pGL3 reporter containing A and B regions of the 3′ UTR. As the schematic diagram suggested (Figure 7E), 3′ UTR-A and -B contained the motif, while 3′ UTR-A-mut and -B-mut did not. The results indicated that the luciferase activity of a reporter carrying 3′ UTR-A and -B was greatly repressed in KIAA1429 knockdown of SUM-1315 and ZR-75-1 cells compared with that of 3′ UTR-A-mut and -B-mut (Figures 7F and 7G). RNA electrophoretic mobility shift assay (REMSA) was performed to demonstrate the direct binding sites of KIAA1429 in SMC1A mRNA. Using bio-UTP-labeled probes (probes A and B), we performed REMSA containing various fragments of the 3′ UTR of SMC1A mRNA (Figure 7H) to find out potential KIAA1429 binding sites in SMC1A mRNA. The KIAA1429 protein was able to form RNA-protein complexes (RPCs) with probes A and B (Figure 7I). These results proved that KIAA1429 could directly bind to the motif in the 3′ UTR of SMC1A mRNA to stabilize SMC1A expression.
Figure 7.
KIAA1429 bound to regions in the 3′ UTR of SMC1A mRNA directly
(A and B) In SUM-1315 and ZR-75-1 cell lines, KIAA1429 knockdown shortened the half-life of SMC1A mRNA, while overexpression of KIAA1429 prolonged the half-life of SMC1A mRNA. Cells were treated with 5 μg/mL Act D for 0, 1, 2, 4, 6, and 8 h, followed by qRT-PCR analysis. (C and D) SUM-1315 and ZR-75-1 cell lysates were collected and immunoprecipitated with KIAA1429 antibody or control IgG followed by RT-PCR and qRT-PCR to detect SMC1A mRNA levels, and β-actin was a negative control, which was not binding to KIAA1429. (E) Schematic of regions in the 3′ UTR of SMC1A mRNA. (F and G) The luciferase activity for the reporter containing SMC1A 3′ UTR-A and -B was repressed by knockdown of KIAA1429 in SUM-1315 and ZR-75-1 cells. Data were shown as mean ± SEM; ∗p < 0.05. (H) Schematic presentation of SMC1A mRNA and the location of probes used for REMSA. (I) REMSA was performed by mixing probes A and B with crude extracted protein, respectively. The bracket indicated RNA-protein complexes (RPCs). Lane 1, only bio-labeled probe; lane 2, mixing bio-labeled probes with protein; lane 3, mixing bio-labeled probes and unlabeled probes with protein; lane 4, mixing mutant probes with protein.
Discussion
Recent studies have demonstrated that KIAA1429 is associated with tumorigenesis and development in breast cancer, gastric cancer,18 HCC,25 and osteosarcoma.26 However, reports about the effect of KIAA1429 on breast cancer metastasis are rare. In this study, we first provided compelling evidence that KIAA1429 could promote breast cancer metastasis and invasion both in vitro and in vivo. We found that KIAA1429 could enhance SMC1A mRNA stability by targeting the motif of SMC1A mRNA directly and increased the SMC1A protein expression. Moreover, SMC1A promoted SNAIL expression by binding the promoter region of the SNAIL gene directly, and therefore promoted breast cancer cell migration and invasion.
As the largest known component in the m6A methyltransferase complex, KIAA1429 is identified as a scaffold that orchestrates the core components, which consist of METTL3, METTL14, and WTAP. In recent years, more and more evidence indicated that these components were involved in many cancers’ metastasis and invasion via regulating many oncogenes and tumor suppressor genes.27, 28, 29 For instance, METTL3 has been identified as an oncogene that could promote HCC tumorigenicity and metastasis by repressing the expression of SOCS (suppressor of cytokine signalling) in an m6A-dependent manner.30 METTL14 is an anti-metastatic factor and serves as a favorable factor in HCC by regulating m6A-dependent miRNA processing.31 In the present study, we found overexpression of KIAA1429 could increase breast cancer cells’ migration ability, while knockdown of KIAA1429 repressed breast cancer cell migration and invasion in vitro and inhibited lung metastases in vivo. Moreover, we found breast cancer cells reveal an EMT-like morphological change after changing the expression of KIAA1429, which verified that KIAA1429 was involved in EMT progress. Accordingly, qRT-PCR proved that SNAIL regulated this process, instead of other EMT regulators. However, RIP and MeRIP assay proved that KIAA1429 could not regulate SNAIL directly. So we speculated that there were intermediate links between KIAA1429 and SNAIL. We then combined these 125 genes from the Cistrome, which would promote the transcription of SNAIL with our former RIP-seq of KIAA1429, and verified SMC1A as the primary potential target of KIAA1429 in breast cancer. SMC1A encodes a subunit of the cohesion-core complex that tethers sister chromatids together to ensure correct chromosome segregation in both mitosis and meiosis and takes part in gene transcription regulation and genome organization.32 We did find SMC1A could bind directly to the DNA recognition sequence in the promotor region of SNAIL gene in breast cancer by ChIP experiment.
In the previous studies, KIAA1429 exhibited its activities by an m6A-dependent or m6A-independent manner.17,21 In this study, MeRIP experiment found that KIAA1429 could not disturb the m6A level of SMC1A mRNA, implying that m6A modification could not affect the interaction between KIAA1429 and SMC1A mRNA. The KIAA1429-mut group, which lacked the RNA binding domain, could not promote breast cancer metastasis. It seemed that KIAA1429 exhibited its activities in breast cancer by an m6A-independent manner. RNA stability, RIP, and luciferase reporter assay suggested that KIAA1429 could stabilize SMC1A mRNA dependent on the motif sites in the 3′ UTR of SMC1A. REMSA is commonly employed in a primary characterization of protein-RNA interactions.33 In this study, REMSA containing two fragments of SMC1A mRNA 3′ UTR (probes A and B) was performed, and the results proved that KIAA1429 could bind to the motif in the 3′ UTR of SMC1A mRNA directly as an RNA binding protein and regulate SMC1A expression. It seemed that KIAA1429 could regulate the targeted gene expression at posttranscriptional levels, unrelated to its m6A modification. Overall, our study revealed that KIAA1429 could promote EMT progress by regulating SMC1A in an m6A-independent manner in breast cancer.
Conclusions
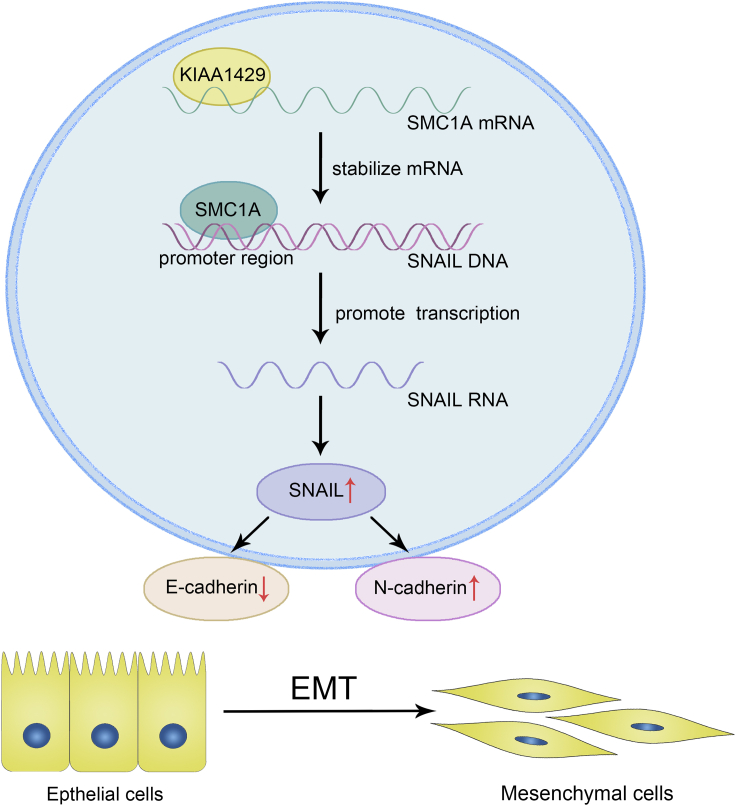
In total, we demonstrated that KIAA1429 promoted breast cancer migration and invasion in vitro and in vivo. We provided a novel mechanism of the KIAA1429/SMC1A/SNAIL axis in the regulation of breast cancer invasion and metastasis, which may be a promising therapeutic target for human breast cancer. Moreover, it first provided detailed investigation of how KIAA1429 regulated the targeted gene expression at posttranscriptional levels as an RNA binding protein, unrelated to its m6A modification (Figure 8).
Figure 8.
The graphic illustration of KIAA1429 modulating breast cancer invasion and metastasis via promoting EMT progress and detailed mechanism
Materials and methods
Cell cultures
The human breast cancer ZR-75-1 cell lines were obtained from the American Type Culture Collection (ATCC, USA), and the SUM-1315 cell lines were kindly provided by Dr. Stephen Ethier, University of Michigan. The cells were cultured in complete high-glucose DMEM (Wisent, China), supplemented with 10% fetal bovine serum, 100 μg/mL penicillin-streptomycin (Hyclone, USA). All cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
Lentivirus transfection and adenovirus transfection
The breast cancer cells were transfected with lentivirus (Obio, Shanghai, China) to repress KIAA1429 (ENSEMBL: ENSG00000164944 ENTREZ ID:8243) and METTL3 (ENSEMBL ID: ENSG00000165819 ENTREZ ID:56339) expression (termed as shKIAA1429-1, shKIAA1429-2, and shMETTL3). Also, pLKDCMV-G&PR-U6 negative control vectors were termed as shRNA-NC. Then we transfected the breast cancer cells with adenovirus (Obio, Shanghai, China) to overexpress KIAA1429 and KIAA1429 without RNA binding domain (KIAA1429-mut). The deleted domain in KIAA1429 protein was shown in Figure S7. Accordingly, pLenti-CMV-3FLAG negative control vectors were termed as Vector. Cells were plated in six-well dishes overnight at ∼30%–40% confluence and infected with the retroviruses. Meanwhile, we added the polybrene (5 μg/mL) to enhance the target cells’ infection efficiency. Then puromycin (3 μg/mL) was used to select the stable cells for 2 weeks.
To explore the further relationship between KIAA1429, SNAIL (ENSEMBL: ENSG00000072501, ENTREZ: 25962), and SMC1A (ENSEMBL: ENSG00000124216, ENTREZ: 6615) in breast cancer cells, these cells were seeded in six-well plates overnight and then transfected with SMC1A-siRNA (GenePharma, Shanghai, China) and the nonspecific siRNA control, using Lipofectamine 3000 transfection agent (Invitrogen, USA). The sequences of the siRNAs are as follows: si-SMC1A-1 sequences: 5′-GCAGCGAAAGGCAGAGAUATT-3′ and si-SMC1A-2 sequences: 5′- GCUAUGAGCCACCUCAUAUTT-3′.
In addition, the KIAA1429 negative control and knockdown breast cancer cells were transfected stably with SNAIL negative control vectors and SNAIL overexpression vectors pLenti-CMV-SNAIL-2A-mCherry3FLAG-PGK-Zeo (termed as shRNA-NC + Vector, shKIAA1429 + Vector, shRNA-NC + SNAIL, and shKIAA1429 + SNAIL). All plasmids were verified by sequencing (GenePharma, Shanghai, China).
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted using TRIzol reagent (Takara, Japan), and about 1,000 ng RNA was reverse transcribed into cDNA using PrimeScript RT Reagent (Takara, Japan). The qRT-PCR were carried out as described previously.34 The following PCR primers were used: β-actin forward, 5′-TCACCCACACTGTGCCCATCTACGA-3′; β-actin reverse, 5′-CAGCGGAACCGCTCATTGCCAATGG-3′; KIAA1429 forward, 5′-CAACGATGGCACGAATTAC-3′; KIAA1429 reverse, 5′-TGTTGCTTCATATTCCGACA-3′; SMC1A forward, 5′-TGATGCTGCCTTGGATAACA-3′; SMC1A reverse, 5′-GGGTACTTGGTGAGGTCGAA-3′; SNAIL forward, 5′-GCTCCACAAGCACCAAGAGT-3′; SNAIL reverse, 5′-CAGGCAGAGGACACAGAACC-3′; E-cadherin forward, 5′-GAACGCATTGCCACATACAC-3′; E-cadherin reverse, 5′-GAGGATGGTGTAAGCGATGG-3′; N-cadherin forward, 5′-ATGGAAGGCAATCCCACATA-3′; N-cadherin reverse, 5′-CAGTAGGATCTCCGCCACTG-3′; METTL3 forward, 5′-GCTGACCATTCCAAGCTCTC-3′; and METTL3 reverse, 5′-ATTTCTTGGCTGGCTCCTTT-3′.
Western blot analysis
Western blot analysis was conducted as previously described.35 In brief, cells were ruptured with RIPA (P0013C; Beyotime, China) buffer containing 1% PMSF, 1% phosphatase inhibitor, and 0.1% protease inhibitor. Cell lysates were resolved by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). After incubation with the primary antibodies, the membranes were then incubated with secondary antibodies (CST, USA). The primary antibodies included anti-mouse β-actin (3700S; CST, USA), anti-rabbit KIAA1429 (88358S; CST, USA), SMC1A (ab243875; Abcam, UK), SNAIL (3879S; CST, USA), E-cadherin (3195S; CST, USA), N-cadherin (13116S; CST, USA), METTL3 (86132S; CST, USA), and TWIST (69366S; CST, USA). The dilutions of antibodies were according to the product usage information.
Wound healing assay
Breast cancer cells were seeded in a six-well plate and grown to full confluence. The monolayer was scratched with a sterile plastic tip and washed twice with PBS to remove loose cells, and the media were replaced with serum-free media. The wounded areas were photographed under a microscope. ImageJ was used to measure the area between the invading fronts of cells in three random microscopic fields (×100) for each condition and time point (0, 24 h).
Transwell migration and invasion assay
The migration and invasion assay of breast cancer cells were conducted as described previously.36 In brief, 5 × 104 cells were seeded into the upper chambers of each transwell, which was coated with or without Matrigel (BD Biosciences, USA) for the invasion and migration assays, and 500 μL of medium with 10% FBS was added to the lower chamber. After incubation at 37°C for 48 h, non-invading cells were wiped from the upper side of the membrane. Then cells adhered to the lower membrane were fixed in methanol and stained with 0.1% crystal violet. Images of three random fields (×100) were captured from each membrane, and the number of invading or migrating cells was counted.
Experimental metastasis assay
The BALB/c female nude mice (4 weeks old) were obtained from Animal Research Center of Nanjing Medical University (Nanjing, China), and the use of animals was approved by Institutional Animal Care and Use Committee of Nanjing Medical University. For the mouse lung metastasis model, SUM-1315 cells (2 × 106/0.2 mL) expressing NC, shKIAA1429, SNAIL, or shKIAA1429+SNAIL were injected into the nude mice through the tail vein. After the mice died, the lungs were excised, imaged, and examined for lung metastases using hematoxylin and eosin (H&E) staining.
RNA stability assays
KIAA1429-knockdown cell lines, KIAA1429-overexpressing cell lines, and their control cell lines were cultured in six-well plates. Then we added Act D 5 μg/mL at 0, 1, 2, 4, 6, and 8 h before cell scraping the collection. Total RNA was isolated using TRIzol reagent, and qRT-PCR was performed to quantify the relative levels of SMC1A.
Luciferase reporter assay
The targeted genes that promote the transcription of SNAIL were obtained from Cistrome (http://cistrome.org/db/#/). We clicked on what factors regulated the gene of interest and wrote “SNAIL.” Then we found about 125 targeted genes from the Cistrome. Luciferase reporter assay was performed as previously described.37 In brief, the breast cancer cells were seeded into a 24-well plate and transfected with Renilla luciferase vector and pGL3 reporter. After 2 days, the cells were harvested, and the luciferase activity was tested by the Dual-Luciferase reporter assay system (Promega, USA).
RIP
RIP assay was conducted as previously described.38 Cell lysates were prepared with RIP lysis buffer (Magna RIP Kit; Millipore, USA) and then incubated with 5 μg of anti-KIAA1429 or rabbit IgG at 4°C overnight. The RNA-protein immunocomplexes were collected by protein A/G magnetic beads. After elution and purification, the purified RNA was analyzed by RT-PCR and qRT-PCR.
The m6A RIP was performed as previously described with some modifications.27 Total RNAs were isolated from SUM-1315 stable KIAA1429/METTL3 knockdown and control cells and treated with DNase I (Sigma Aldrich, USA). RNAs were fragmented by RNA fragmentation reagents. Immunoprecipitations were performed using an anti-m6A antibody (1:1,000; Abcam, USA) previously bound to magnetic Dynabeads (Life Technologies, USA) in the RIP Immunoprecipitation Buffer (Magna RIP Kit; Millipore, MA, USA) and incubated with DNA-free fragmented RNAs. RNAs were extracted by miRNeasy Mini kit (QIAGEN, Germany) and subjected to qRT-PCR and normalized to input.
REMSA
To generate REMSA probes, we amplified various regions (A and B) in SMC1A mRNA by PCR with T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′). Biotin-labeled RNA probes were made from in vitro transcription with a MEGA shortscript Kit (Ambion, Waltham, MA, USA) in the presence of biotin-16-UTP (Roche) according to the manufacturer’s instructions. The primers for probes A and B are listed as follows: probe A, 5′-GGGGCTAACAATATTACCTACCTCATAGGATTTAATGATGTCAAGCTCCTCA CTGGAGGCCTTATCCCTTCGTGGAGCCCACTAGGTGCCGACCCCTCAGAATATAACCCTCATGCCTGGACCCCTGAGAGCTTCTGATCCCAGCTATTAGGGACAGAAGAAGCCTCCAAATCTGGAAGGTGCTGAATGCCCTGCTGACT-3′; and probe B, 5′-AGGACTTCACCTTACAGGGGTGGCATGTATCAAATGGCAAATGTATGAAACAACCAGATCTTTCAGGGAGGCAGAATGTGAGCTATTCAGAAGAAGTGAACGTTAATTAGAATTTAATGAGGCATTAGTGGTGGTGGATGAGGGGTGGCC-3′. The REMSA was performed with a LightShift Chemiluminescent RNA EMSA Kit (20158; Thermo, USA) following the manufacturer’s instruction. In brief, crude extracted protein from SUM-1315 cell lines and bio-labeled, unlabeled, or mutant RNA probe were mixed in REMSA binding buffer and incubated for 25 min at room temperature. The RNA-protein complexes were then electrophoresed by 4% native polyacrylamide gel and transferred to nylon membrane (77015; Thermo, USA). The RNA was UV crosslinked to the membrane, and the membrane was blocked in blocking buffer and then replaced for the blocking buffer with conjugate/blocking buffer. After washing with 1× wash buffer for three times, the membrane was incubated in substrate equilibration buffer. Then the membrane was incubated in working solution and exposed.
Statistical analysis
All experiments were performed at least three times, unless otherwise specified. The data were analyzed using the GraphPad Prism 7.0 Software (GraphPad, La Jolla, CA, USA). Student’s t test was used to research the statistical significance of the differences between groups, and p < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81972486, 81802644, and 81802748), the ‘333’ High-level Talents Training Project of Jiangsu Province (BRA2016505), and the Key Medical Talents of Jiangsu Province (ZDRCA2016029). The work was also funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the International Cooperation Project of Jiangsu Provincial Science and Technology Department (BZ2018054).
Author contributions
Q.D., J.F.-W., and L.S. designed the study; X.Z., X.-Y.D., and J.Y.-Q. carried out the experiments; F.X. and Z.W.-W. performed the statistical analysis; T.X., X.J.-Z., and X.X.-L. participated in the clinical specimens detection; X.Z. and J.F.-W. wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.009.
Contributor Information
Liang Shi, Email: shiliang@njmu.edu.cn.
Ji-Fu Wei, Email: weijifu@hotmail.com.
Qiang Ding, Email: dingqiang@njmu.edu.cn.
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Desantis C.E., Ma J., Gaudet M.M., Newman L.A., Miller K.D., Goding Sauer A., Jemal A., Siegel R.L. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 3.Song Z., Zhang X., Lin Y., Wei Y., Liang S., Dong C. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J. Cell. Mol. Med. 2019;23:7554–7565. doi: 10.1111/jcmm.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Staalduinen J., Baker D., Ten Dijke P., van Dam H. Epithelial-mesenchymal-transition-inducing transcription factors: new targets for tackling chemoresistance in cancer? Oncogene. 2018;37:6195–6211. doi: 10.1038/s41388-018-0378-x. [DOI] [PubMed] [Google Scholar]
- 5.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T., Choi H., El Rayes T., Ryu S., Troeger J., et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao N., Fu Y., Chen L., Liu Z., He J., Zhu Y., Xia T., Wang S. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38:7216–7233. doi: 10.1038/s41388-019-0904-5. [DOI] [PubMed] [Google Scholar]
- 8.Tan X., Li Z., Ren S., Rezaei K., Pan Q., Goldstein A.T., Macri C.J., Cao D., Brem R.F., Fu S.W. Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer. Breast Cancer Res. 2019;21:89. doi: 10.1186/s13058-019-1173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou T., Yu L., Huang J., Zhao X., Li Y., Hu Y., Lei Y. GDF10 inhibits proliferation and epithelial-mesenchymal transition in triple-negative breast cancer via upregulation of Smad7. Aging (Albany NY) 2019;11:3298–3314. doi: 10.18632/aging.101983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia X., Shi L., Wang X., Luo L., Ling L., Yin J., Song Y., Zhang Z., Qiu N., Liu H., et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019;10:373. doi: 10.1038/s41419-019-1566-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Dunin-Horkawicz S., Czerwoniec A., Gajda M.J., Feder M., Grosjean H., Bujnicki J.M. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X., Wang X., Cao C., Gao Y., Zhang S., Yang Z., Liu Y., Zhang X., Zhang W., Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Yu H.L., Ma X.D., Tong J.F., Li J.Q., Guan X.J., Yang J.H. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. OncoTargets Ther. 2019;12:6191–6201. doi: 10.2147/OTT.S205730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Xu M., Xu X., Zeng K., Liu X., Pan B., Li C., Sun L., Qin J., Xu T., et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer. 2020;19:106. doi: 10.1186/s12943-020-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan T., Li H., Zhang D., Xu L., Liu H., Hao X., Yan X., Liao H., Chen X., Xie K., et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer. 2019;18:186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao R., Dai C.C., Mei L., Xu J., Sun S.W., Xing Y.L., Wu L.S., Wang M.H., Wei J.F. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J. Cell. Physiol. 2020;235:7420–7432. doi: 10.1002/jcp.29645. [DOI] [PubMed] [Google Scholar]
- 19.Wang M., Yang Y., Yang J., Yang J., Han S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m6A-YTHDF3-Zeb1. Life Sci. 2020;257:118082. doi: 10.1016/j.lfs.2020.118082. [DOI] [PubMed] [Google Scholar]
- 20.Li N., Zhan X. Identification of pathology-specific regulators of m6A RNA modification to optimize lung cancer management in the context of predictive, preventive, and personalized medicine. EPMA J. 2020;11:485–504. doi: 10.1007/s13167-020-00220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian J.Y., Gao J., Sun X., Cao M.D., Shi L., Xia T.S., Zhou W.B., Wang S., Ding Q., Wei J.F. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38:6123–6141. doi: 10.1038/s41388-019-0861-z. [DOI] [PubMed] [Google Scholar]
- 22.Kaufhold S., Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J. Exp. Clin. Cancer Res. 2014;33:62. doi: 10.1186/s13046-014-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z., Zhang P., Yan A., Guo Z., Ban Y., Li J., Chen S., Yang H., He Y., Li J., et al. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci. Adv. 2017;3:e1601602. doi: 10.1126/sciadv.1601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niessen M., Schneiter R., Nothiger R. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics. 2001;157:679–688. doi: 10.1093/genetics/157.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng X., Li M., Rao X., Zhang W., Li X., Wang L., Huang G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. OncoTargets Ther. 2019;12:3421–3428. doi: 10.2147/OTT.S180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Q., Yang J., Yang H., Li C., Li J., Cao Y. KIAA1429 promotes osteosarcoma progression by promoting stem cell properties and is regulated by miR-143-3p. Cell Cycle. 2020;19:1172–1185. doi: 10.1080/15384101.2020.1749465. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Han J., Wang J.Z., Yang X., Yu H., Zhou R., Lu H.C., Yuan W.B., Lu J.C., Zhou Z.J., Lu Q., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Peng C., Chen J., Chen D., Yang B., He B., Hu W., Zhang Y., Liu H., Dai L., et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Zhang S., He C., Xue P., Zhang L., He Z., Zang L., Feng B., Sun J., Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 31.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F., Wang T.T., Xu Q.G., Zhou W.P., Sun S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 32.Musio A. The multiple facets of the SMC1A gene. Gene. 2020;743:144612. doi: 10.1016/j.gene.2020.144612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramanathan M., Porter D.F., Khavari P.A. Methods to study RNA-protein interactions. Nat. Methods. 2019;16:225–234. doi: 10.1038/s41592-019-0330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L., Xia T.S., Wei X.L., Zhou W., Xue J., Cheng L., Lou P., Li C., Wang Y., Wei J.F., Ding Q. Estrogen receptor (ER) was regulated by RNPC1 stabilizing mRNA in ER positive breast cancer. Oncotarget. 2015;6:12264–12278. doi: 10.18632/oncotarget.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X.J., Wu J., Shi L., Li X.X., Zhu L., Sun X., Qian J.Y., Wang Y., Wei J.F., Ding Q. PTEN expression is upregulated by a RNA-binding protein RBM38 via enhancing its mRNA stability in breast cancer. J. Exp. Clin. Cancer Res. 2017;36:149. doi: 10.1186/s13046-017-0620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu L., Xi P.W., Li X.X., Sun X., Zhou W.B., Xia T.S., Shi L., Hu Y., Ding Q., Wei J.F. The RNA binding protein RBMS3 inhibits the metastasis of breast cancer by regulating Twist1 expression. J. Exp. Clin. Cancer Res. 2019;38:105. doi: 10.1186/s13046-019-1111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X., Hu Y., Wu J., Shi L., Zhu L., Xi P.W., Wei J.F., Ding Q. RBMS2 inhibits the proliferation by stabilizing P21 mRNA in breast cancer. J. Exp. Clin. Cancer Res. 2018;37:298. doi: 10.1186/s13046-018-0968-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X.X., Shi L., Zhou X.J., Wu J., Xia T.S., Zhou W.B., Sun X., Zhu L., Wei J.F., Ding Q. The role of c-Myc-RBM38 loop in the growth suppression in breast cancer. J. Exp. Clin. Cancer Res. 2017;36:49. doi: 10.1186/s13046-017-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.