Highlights
-
•
Categorization of DLBCL into four clusters based on survival- and ferroptosis-related factors.
-
•
Establishment of an efficient risk scoring model is established for patients with DLBCL.
-
•
Ferroptosis-based risk scoring model reveals immune infiltration correlation in DLBCL.
Keywords: DLBCL, Risk scoring model, Ferroptosis, Immune infiltration
Abbreviation: DLBCL, diffuse large b-cell lymphoma; TCGA, the cancer genome atlas; GEO, the gene expression omnibus; AUC, area under the receiver operating characteristic curve; LNCRNA, long non-coding RNAs; miRNA, microrna; UCSC, The University of California Santa Cruz; LASSO, least absolute shrinkage and selection operator; GO, gene ontology; BP, biological process; MF, molecular function; CC, cellular component; KEGG, Kyoto encyclopedia of genes and genomes; LDH, lactic dehydrogenase; NK, natural killer cells; ROC, the receiver operating characteristic
Abstract
Backgrounds
The prognostic significance of ferroptosis-related genes is well known. However, survival- and ferroptosis-related genes are not currently considered in risk scoring models for diffuse large B-cell lymphoma (DLBCL).
Materials and methods
Ferroptosis regulators and markers were downloaded from the FerrDb database. The transcriptome profiling data were collected from the cancer genome atlas (TCGA). Transcriptome data and corresponding clinical information of DLBCL were downloaded from the gene expression omnibus (GEO). The validation data were downloaded using the UCSC Xena browser. ConsensusClusterPlus was used to categorize DLBCL samples according to gene expression profiles. The survival function was plotted with the Kaplan-Meier plots. The nomogram was built using multivariate logistic regression analysis and the Cox proportional hazards regression model.
Results
Based on the GSE11318 dataset of 203 samples and 267 ferroptosis-related gene expression profiles, we identified four clusters. A total of 19 survival-related genes were found associated with ferroptosis. The prognostic risk scoring model was constructed based on the regression coefficients. The obtained area under the receiver operating characteristic curve (AUC) values were 0.769, 0.801, and 0.791 for 1-, 3-, and 5-year survival, respectively. DLBCL samples with cluster 2 or cancer stage IV have shorter survival. Correlations between the immune infiltration and risk scores of the 12 immune cells were demonstrated. The response of DLBCL to doxorubicin was effectively validated by the risk scoring model.
Conclusions
In this study, a ferroptosis-based risk scoring model for patients with DLBCL was constructed and validated in an independent dataset. This risk score model has a better efficacy in predicting survival compared to clinical characteristics.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive lymphoma that accounts for approximately 30% of malignant lymphomas [1]. As a clinical and genetic heterogeneous lymphoid malignancy, DLBCL exhibits a wide range of clinical outcomes and molecular signatures [2]. Patients typically display rapidly expanding lymphadenopathy and constitutional symptoms that necessitate immediate treatment [3]. More than half of DLBCL patients can be cured by current multidrug chemotherapy, radiation and immunotherapy regimens combined with autologous stem cell transplantation, which is one of the successes of modern cancer treatment [4,5]. Recently, several whole-genome and whole-exome sequencing studies have reported more than 200 DLBCL genes with recurrent mutation [6], [7], [8], [9], likely located in genes functionally associated with DLBCL tumors [10,11]. Ferroptosis has been recently revealed correlated with resistance to chemotherapeutic drugs and associated with the anti-tumor efficacy of immunotherapy [12,13]. Hence, therapeutic exploitation based on ferroptosis has shown to be of interest for cancer treatment.
Ferroptosis is an iron-dependent and oxidative form of cell death, distinct from autophagy, classic necrosis, apoptosis and other forms of cell death [14]. The bluntness of cellular glutathione-dependent antioxidant defenses initiates ferroptosis process, causing the enrichment of toxic lipid reactive oxidative species, membrane damage and then cell lysis [15]. Different degrees of ferroptosis sensitivity have been observed in cancer cells from different tissues [16,17], suggesting a roles of ferroptosis in cancer development. Recently, experimental data showed that cholesterol uptake-addicted lymphoma cells decrease glutathione peroxidase expression, leading to an accumulation of membrane-oxidized lipids and cell death by a mechanism consistent with ferroptosis [18]. Studies confirmed that imidazole ketone erastin used as ferroptosis inducers, causes glutathione depletion and lipid peroxidation, exerting an antitumor effect in DLBCL [19]. In Burkitt's lymphoma, artesunate was demonstrated to induce ferroptosis, leading to an endoplasmic reticulum stress response [20]. In addition, the prognostic significance of ferroptosis-related genes is becoming increasingly recognized [21,22]. Of particular interest, survival-related ferroptosis factors can be used for DLBCL scoring. Moreover, ferroptosis has been recently associated with resistance to chemotherapeutic drugs and associated with the anti-tumor efficacy of immunotherapy. Therefore, using ferroptosis as a therapeutic strategy is of interest for treatment of cancers, including DLBCL.
Risk score models are statistical tools used to estimate the absolute risk of a disease, for instance, DLBCL, in the future for currently healthy individuals with specific risk factors [23], [24], [25]. With risk assessment models, it is possible to predict the distribution of absolute risk in a given population and ascertain individuals at high risk, contributing to intensive surveillance and chemoprevention [25], [26], [27]. Hence, it is necessary to improve the accuracy of risk evaluation and prognosis for DLBCL. Currently, a number of risk scoring models are available for DLBCL. For example, Saez et al. built a biological predictive model using eight protein markers to assign patients with DLBCL to different risk categories [25]. Schmitz et al. established the risk model for central nervous system DLBCL consisting of the international prognostic index factors to estimate the risk of relapse, and this model showed a high reproduction [23]. In addition, other predictive factors such as immune and inflammatory genes and non-coding RNA like long non-coding RNAs (lncRNAs) and microRNA (miRNA) have also been used to establish risk scoring models [28], [29], [30]. Recently, Chen et al. established a personalized risk assessment model based on ferroptosis-related gene signature for patients with DLBCL [31]. However, whether these ferroptosis-related genes are associated with survival remains to be investigated.
Studies have shown that ferroptosis has great potential in the treatment of cancer, and ferroptosis inducers such as sorafenib are currently used in the clinic. Studies have shown that ferroptosis shows great potential in the treatment of cancer [32,33]. In this study, we first obtained disease-related ferroptosis and performed clustering analysis. Then, we screened the prognosis-related ferroptosis factors and constructed a risk scoring model based on ferroptosis factors, followed by validation for its efficacy. The differences in prognosis between high and low risk were also analyzed.
Materials and methods
Data collection of ferroptosis regulators and markers
Ferroptosis regulators and markers were downloaded from the FerrDb database (URL: http://www.zhounan.org/ferrdb). In summary, 259 regulators, including 150 drivers (http://www.zhounan.org/ferrdb/fordownload/1_info_driver.csv) and 109 suppressors (http://www.zhounan.org/ferrdb/fordownload/1_info_suppressor.csv) were gathered, and 123 makers (http://www.zhounan.org/ferrdb/fordownload/1_info_marker.csv) were collected.
Collection of transcriptome data for DLBCL
The transcriptome profiling data were collected from the cancer genome atlas (TCGA), and the transcriptome data was saved as HiSeqV2 (https://tcga-xena-hub.s3.us-east-1.amazonaws.com/latest/TCGA.DLBC.sampleMap%2FHiSeqV2.gz), and the corresponding clinical data were stored in DLBC_survival.text file (https://tcga-xena-hub.s3.us-east-1.amazonaws.com/latest/survival%2FDLBC_survival.txt.gz). Meanwhile, transcriptome data and corresponding clinical information of DLBCL were downloaded from the gene expression omnibus (GEO) using the University of California Santa Cruz (UCSC) Xena browser. The GEO accession NO. was GSE11318, and the clinical information was saved in clinical_GPL570.csv. When a gene has multiple expressions in the same sample, the expression of the one with the largest mean expression is retained.
Validation data and medicine information for DLBCL
The validation data were downloaded using the UCSC Xena browser. The transcriptome data with a GEO accession NO. GSE10846 and clinical information saved in clinical_GSE10846.csv and clinical_GSE10846_stage.txt were derived from CEO database. The treatment and medicine information for DLBCL (saved in TCGA_DLBC_drug_clinical.txt) was downloaded using the TCGAbiolinks package [34].
Bioinformatics analysis
Clustering analysis
ConsensusClusterPlus (available through the Bioconductor project: http://www.bioconductor.org/) was used to categorize DLBCL samples according to gene expression profile [35]. The samples were clustered by the pam algorithm generating consensus in unsupervised class discovery. Ultimately, samples can be differentiated into subtypes based on transcriptomic data sets, allowing for the discovery of new disease subtypes or the comparative analysis of different subtypes.
Survival analysis
The relationship between the clustered subtypes and survival time and outcome of DLBCL was investigated. The survival function was plotted with the Kaplan-Meier plots. Between-group differences in survival functions were assessed with a weighted log-rank test was done for comparing between-group differences in survival functions. Additionally, the association between clinical indicators of DLBCL, including stage, lactate dehydrogenase ratio, and risk score and survival outcome, was investigated, and the risk and protective factors were filtered with p < 0.05.
Screening of differentially expressed genes
Differentially expressed genes between the clustering samples were screened using analysis of variance, and p < 0.05 was accepted. T-test was used for comparisons between the two groups, and p < 0.05 indicated the significance.
COX-LASSO regression analysis
Univariable Cox regression analysis was carried out to screen the survival- and ferroptosis- related factors. A least absolute shrinkage and selection operator (LASSO)-type regularization method was used to penalize the factor data. The obtained model shrank the coefficients for non-informative factors toward zero, the reduced variance was acquired, and clustering performance was improved. Prognostic risk scoring models associated with ferroptosis-related factors in DLBCL were constructed based on the regression coefficients of these factors.
Immune infiltration analysis
To study the immune cell profile of each sample, the cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT) algorithm was used to analyze the abundance of member cell types based on gene expression data, and the CIBERSORT R package was downloaded for local use (available: http://cibersort.stanford.edu/) [36], and the solution of the kernel function is obtained by deconvolution of the SVM algorithm as the immune cell infiltration label.
Nomogram establishment for cancer prognosis
The nomogram was built using multivariate logistic regression analysis and the Cox proportional hazards regression model. Each influencing factor in the model was assigned a score at different value intervals according to the degree of contribution of the influencing factor to the outcome, i.e., the magnitude of the regression coefficient. The individual scores were then summed and obtained the total score. A functional transformation relationship between the total and the probability of the outcome was used to calculate the predictive value of the outcome for that individual. Ultimately, the complex regression equations were transformed into a visual graph, making the results of the predictive models more readable and to facilitate disease evaluation.
Results
Clustering and gene function analysis
Based on the GSE11318 dataset of 203 samples and the expression profile of 267 ferroptosis-related genes, the consensus clustering analysis identified four clusters were obtained by the consensus clustering analysis (Fig. 1A). The Kaplan-Meier survival analysis indicated that cluster 1, cluster 2, and cluster 3 had a significantly different in survival probability. Cluster 3 samples had an improved prognosis compared to cluster 1, and cluster 1 had a better prognosis compared to cluster 2 (Fig. 1B). In addition, there was a more significant difference in gene expression was found between cluster 1 and cluster 2, with a total of 115 genes were identified showing interclass differences using ANOVA (p < 0.05) (Fig. 1C). Next, we performed a functional analysis of the 115 differentially expressed genes among the four clusters was next performed. The top 9 biological processes corresponded to 33 genes, and these processes were related to the ferroptosis procedure, including responses to oxidative stress, chemical stress, extracellular stimulus, external stimulus, nutrient levels, and starvation (Fig. 2A). The 17 ferroptosis-associated genes mainly possess molecular functions like ubiquitin-protein ligase binding, tetrapyrrole binding, ubiquitin-like ligase binding, and heme-binding (Fig. 2B). We observed that 8 protein-coding proteins were linked to cellular components like mitochondrial outer membrane, organelle outer membrane, and outer cell membrane (Fig. 2C). KEGG pathway enrichment analysis indicated that these differentially expressed genes mediated the autophagy process, ferroptosis, necroptosis, neurodegeneration-multiple diseases, shigellosis infection, and Kaposi sarcoma-associated herpesvirus infection, etc. (Fig. 2D). Overall, survival-related ferroptosis factors show biological roles like responses to oxidative stress.
Fig. 1.
Consensus clustering analysis obtained 4 clusters of DLBCL based on ferroptosis-related genes expression signature. (A) The consensus clustering heatmap of 267 ferroptosis-related genes from 203 samples (GSE11318 from GEO); (B) The Kaplan-Meier survival probability of 204 patients with DLBCL categorized into 4 clusters; (C) The heatmap for the 267 ferroptosis-related gene expression and 203 DLBCL samples. DLBCL, diffuse large B-cell lymphoma.
Fig. 2.
Function and pathway analysis for differentially expressed genes among the four clusters. GO annotation of the enrichment of differentially expressed genes in (A) biological process, (B) molecular function, and (C) cellular component; KEGG analysis for the enrichment of differentially expressed genes among the four clusters. GO, gene ontology; BP, biological process; MF, molecular function; CC, cellular component; KEGG, Kyoto encyclopedia of genes and genomes.
Identification of ferroptosis and survival-related genes and construction of prognostic risk scoring model
Survival-related genes were identified based on ferroptosis-related genes with different expression profiles using one-way Cox regression analysis. Here, 19 survival-related genes were found associated with ferroptosis (p < 0.05) (Fig. 3A). The 19 ferroptosis factors were subjected to LASSO regression analysis for dimensionality reduction. The optimal penalty value lambda in the LASSO regression was first obtained using cross-validation (Fig. 3B). The regression coefficients of the 16 ferroptosis factors were obtained based on this value (Fig. 3C). The expression differences of the genes among cluster 1, cluster 2, cluster 3, and cluster 4 were compared by t-test. The relationship between gene expression levels and survival probability was analyzed by the Kaplan-Meier survival curves. A total of 16 genes associated with ferroptosis and survival in DLBCL (Fig. 3D). Patients with high survival probability showed abundance in UBC, CAPG, EPAS1, ACVR1B, GABARAPL1, TFAP2C, and AKR1C3. However, MT1G, IREB2, TRIB3, ATF4, ATG3, CHAC1, and ULK2 were low expressed in patients with DLBCL, which was associated with high survival probability. The prognostic risk scoring model was constructed based on the regression coefficients and calculated as follows, risk Score = -0.006906*DRD4-0.1427*TFAP2C-0.03086*AKR1C3+0.08295*CHAC1+0.2237*ULK2+0.1026*CXCL2-0.2478*GABARAPL1+0.07055*TRIB3-0.01005*CYBB+0.4483*IREB2-0.02139*EPAS1+0.1521*MT1G+0.2654*ATG3-0.06966*CAPG+0.4052*ATF4+0.03599*UBC.
Fig. 3.
Expression and survival analysis of ferroptosis-related genes in DLBCL. (A) p-values and hazard ratios of the 19 genes associated with ferroptosis and survival; (B) Penalty values obtained by LASSO cross-validation; (C) Ferroptosis factors and regression coefficients after dimensionality reduction of optimal penalty values; (D) Expression profiles of 14 genes were compared between cluster 1, cluster 2, cluster 3 and cluster 4 by T-test. Kaplan-Meier survival probabilities were compared between the two groups with high and low gene expression. DLBCL, diffuse large B-cell lymphoma.
Validity assessment of prognostic model
To assess the validity of the prognostic risk score model, we used the risk score predicted by the model to classify the 203 DLBCL samples into two categories based on the median score (Fig. 4A). We also analyzed the predicted results of risk score classification for 1, 3, and 5 -year survival, and the obtained area under the receiver operating characteristic curve (AUC) values were 0.769, 0.801, and 0.791, respectively (Fig. 4B). Comparing the predictive validity of clinical stages, lactate dehydrogenase ratio and risk score for 3-year survivals, the obtained AUC value was 0.765 (Fig. 4C). In contrast, the predictive validity of age, gender and risk score on 3-year survival of patients was 0.69 (Fig. 4D). The effect of clinical characteristics and risk score on patient survival was verified using univariate and multifactorial Cox regression analysis, respectively, and risk score was found to be a more important risk factor (Fig. 4E, F). The expression of 16 ferroptosis-related factors in samples from high and low risk groups (Fig. 4G), sample size distribution, and patient survival status (Fig. 4H) also demonstrated a consistent association between high risk and high mortality. In addition, we found significantly different risk scores between cluster 1 and cluster 2 and between cluster 2 and cluster 3. Among them, cluster 2 had a higher risk score than cluster 1 and cluster 3 (Fig. 4I). The risk score of cancer stage IV was also found to be significantly higher than that of stage I (Fig. 4J). These results suggest that DLBCL samples with cluster 2 or cancer stage IV are associated with shorter survival.
Fig. 4.
Validity assessment of prognostic models and risk scores of clinical characteristics for DLBCL. (A) Survival probability of patients with high and low risks; (B) The AUC of the model containing risk categories and scores for 1, 2, and 3 -year survival rate; The AUC of the model based on (C) clinical stage, LDH ratio, and risk score, as well as (D) age, gender and risk score; (E) Univariate and (F) multivariate cox regression analyses for validation of the effect of clinical characteristics and risk scores on patient survival; (G) The heatmap of 16 ferroptosis and survival -related genes; (H) Sample size and survival status of groups with high and low risks; (I) Risk scores of 4 clusters (cluster 1–4) and 4 stages (stage 1–4) of diffuse large B-cell lymphoma; (J) Effects of LDH ratio (high and low), age (old and young), and gender (female and male) on risk scores. AUC, the area under the receiver operating characteristic curve; LDH, lactic dehydrogenase; DLBCL, diffuse large B-cell lymphoma.
Validation of the prognostic scoring model for DLBCL and external confirmation of the nomogram
Gene expression profiles from the GSE10846 dataset and the corresponding clinical phenotypes and survival data were used to validate the constructed risk score model. The GSE10846 expression profile was input into the model, and the risk scores were obtained. Fig. 5A shows that DLBCL patients with low risk had a high survival probability. The predictive validity of risk scores for 1, 3, and 5 -year survival probability is presented in Fig. 5B, with obtained AUC values of 0.673, 0.708 and 0.716, respectively. Clinical characteristics (age, gender, stage and lactic dehydrogenase ratio) and risk score for the model were found to be better predictors of patient survival (Fig. 5C). Univariate and multivariate analysis also confirmed the significant effect of risk score on patient survival (Fig. 5D). To calculate predictive values for individual outcome events, we constructed nomograms based on the effects of risk score, age, and gender on patient survival over 1, 3, and 5 years, as well as calibration curves for the nomograms (Fig. 5E). Notably, no significant difference was observed between the observed probabilities and the predicted outcomes for 1-year, 3-year, and 5-year survivals (Fig. 5F). The obtained results presented that the prediction based on an independent dataset GSE10846 was valid, which meanwhile proved the exportability of the risk scoring model for DLBCL disease.
Fig. 5.
Verification of the prognostic scoring model for DLBCL and external confirmation of the nomogram based on GSE10846 dataset. (A) Survival probability of patients with high and low risks from GSE10846 dataset; (B) The AUC of GSE10846 dataset for 1, 2, and 3 -year survival rate; (C) Clinical characteristics and risk score for validating survival status of patients from GSE10846; (D) Univariate and multivariate analysis for testing the validity of the predictive model using GSE10846 dataset; (E) Nomogram was used to predict 1-year, 3-year and 5-year survival probability of DLBCL patients according to risk score, age, gender, clinical stage, LDH ratio and total points; (F) The predicted percentages and the observed probabilities of 1-year, 3-year and 5-year survival. DLBCL, diffuse large B-cell lymphoma; LDH, lactic dehydrogenase.
Association between immune infiltration and risk score for DLBCL
Infiltration scores were assessed using CIBERSORT based on the expression profiles of all genes in 203 samples, and then the differences in immune infiltration of 22 immune cells (B cells naive, B cells memory, plasma cells, T cells CD8, T cells CD4 naive, T cells CD4 memory resting, T cells CD4 memory activated, T cells follicular helper, T cells regulatory Tregs, T cells gamma delta, NK cells resting, NK cells activated, monocytes, macrophages M0, macrophages M1, macrophages 2, dendritic cells resting, dendritic cells activated, mast cells resting, mast cells activated, eosinophils, neutrophils) were compared by t-test (Fig. 6A). It was suggested that activated CD4 memory T cells, follicular helper T cells, resting natural killer cells, activated natural killer cells, M0 and M2 macrophages, and neutrophils showed different proportions between high-risk and low-risk DLBCL. Additionally, we found that most of the ferroptosis-related and survival-related genes were positively correlated with tumor-filtrating immune cells, such as ACVR1B, CAPG, and CXCL2. A few were bidirectional, such as AKR1C3 and CAPG (Fig. 6B). We demonstrated correlations between the immune infiltration and risk scores of the 12 immune cells were demonstrated. In detail, all these 12 immune cells were associated with risk scores of DLBCL samples (Fig. 6C).
Fig. 6.
Association between the risk score and immune infiltration in DLBC. (A) Proportion of immune cells in DLBCL patients with high and low risk scores; (B) Pearson correlation between the 19 ferroptosis- and survival-related genes and the infiltrating immune cells; (C) Correlation analysis for the risk score and the infiltrating level of the significantly different immune cells in DLBCL. DLBCL, diffuse large B-cell lymphoma; NK, natural killer cells.
Risk score for predicting the response of chemotherapy dose
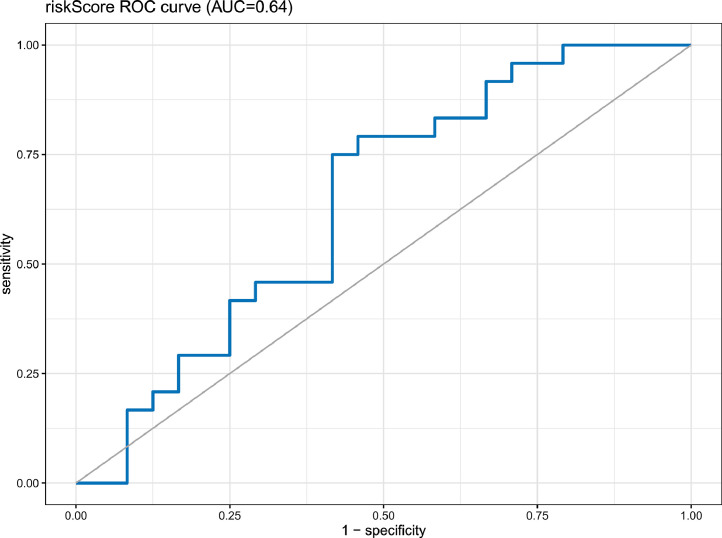
To further determine the clinical usefulness of the risk scores predicted by our model, clinical data on chemotherapy dosing for DLBCL were obtained using the TCGA biolinks tool to predict chemotherapy dosing outcomes. However, the data was not unavailable for progressive clinical disease, stable disease, and other dosing responses. Finally, we used the pRRopheticPredict R package to simulate dosing responses based on expression profiles. To simulate the actual dosing response as closely as possible, we used the drug most used in DLBCL presented by the TCGA database and the drug predicted by the pRRopheticPredict R package, i.e., doxorubicin. Then, we calculated the response of DLBCL to the drug and subsequently obtained the efficacy of the dosing response predicted by the sample risk score. The predicted AUC was found to be 0.64, indicating that our data are effective predictors of clinical drug application (Fig. 7).
Fig. 7.
External validation of the nomogram for predicting chemotherapy response of patients with DLBC. The obtained AUC is 0.64. AUC, the area under the receiver operating characteristic curve; ROC, the receiver operating characteristic; DLBCL, diffuse large B-cell lymphoma.
Discussion
One of the pivotal challenges for killing cancer cells is effectively triggering apoptotic cell death and controlling therapy-resistant cells [37]. As an iron-catalyzed form of regulated necrosis, ferroptosis is involved in the silencing or downregulation of genes initiating or executing necroptosis in cancers [38,39]. DLBCL outcome is the result of interactions between the genetic abnormalities of ferroptosis-related factors and the clinical status of the patients [31,40]. Identification of clinical risk is helpful to better design the therapeutic intervention in different DLBCL patients [41,42]. Here, we report a ferroptosis-based risk scoring model for patients with DLBCL, and tits validation in an independent dataset. This risk score model has a better efficacy in predicting survival compared to clinical characteristics. Additionally, the risk score suggested the consistency with immune cell infiltration in DLBCL. Of note, this model can predict the medical response of DLBCL patients to doxorubicin.
Ferroptosis is characterized by the excessive peroxidation of polyunsaturated fatty acids containing phospholipids catalyzed by iron in cell membranes [14]. Ferroptosis is being studied as preventive strategy in several cancers, including fibrosarcoma, melanoma, ovarian cancer [17], mesothelioma [16], gastric cancer [43] and liver cancer [44]. In terms of DLBCL, imidazole ketone erastin, artesunate, and cholesterol cause lipid peroxidation, induce ferroptosis biomarkers, and result in an endoplasmic reticulum stress response in vivo and in vitro, showing a therapeutic mechanism consistent with ferroptosis [45], [46], [47]. Recently, Zhou et al. linked the ferroptosis process to metabolites and biomolecules, annotating several regulators and markers of ferroptosis, which is meaningful to linking ferroptotic process to metabolites and biomolecules in cancers [48]. According to the ferroptotic signature of cancers, we performed the consensus clustering analysis and obtained 4 clusters with different survivalprobabilities. The results suggested that cluster 3 and cluster 4 showed an improved prognosis compared with cluster 1 and cluster 2. These ferroptosis-related and survival-related molecules participate in stimulus-response, membrane composition, lipid oxidation, regulation of autophagy, and mitophagy, as shown in previous studies [49,50].
Furthermore, we identified 16 ferroptosis- and survival-related genes after LASSO regression analysis. A risk scoring model was developed based on 16 genes, followed by validating the discriminatory accuracy of the model by the AUC method using an independent dataset. The discriminatory accuracy was obviously improved compared with the model that only included risk factors like the clinical stage, LDH ratio, age and gender. Beside, the metabolic prognostic model has been generated to assign patients with DLBCL into high-or low-risk clusters, which shows a superiority in aspect of a short-term [51]. Other prognostic scoring models for DLBCL have been reported based on infiltrating immune cells [52] and tumor microenvironment [53]. All these models have effectively and independently determined the prognosis of patients with DLBCL. Similarly, Chen et al. reported a risk assessment model for patients with DLBCL, and this personalized model was constructed based on ferroptosis-related gene signature [31]. They established a reliable prognostic prediction for subgroup analysis using the 8-gene associated with ferroptosis [31]. Here, our study integrated 16 ferroptosis-related gene signatures and clinical factors, which showing an improved prognostic value in classifying patients with DLBCL into high-or low-risk groups.
Malignant cells change the generation of cytokines that typically govern proliferation, and consequently participate as part of a dysregulated immune environment [54,55]. Hence, understanding the types and roles of immune cells is critical to develop strategies to target tumors and improve patient prognosis. The participation of ferroptosis in T cell immunity has been confirmed by Wang et al., [17]. They reported that immunotherapy induces the activation of CD8+ T cells, which fortifies ferroptosis-specific lipid peroxidation [17]. Furthermore, the genes associated with survival and ferroptosis were correlated with immune infiltration. It has been proved that ferroptosis-related genes affect immune infiltration and invasion in ovarian cancer [56] and breast cancer [57]. However, it remains unclear whether ferroptosis shows effects on immune infiltration in DLBCL. Here, we noticed that patients with high and low risk showed differences in the proportions of activated CD4 memory T cells, follicular helper T cells, resting natural killer cells, activated natural killer cells, M0 and M2 macrophages, and neutrophils. To estimate the risk of relapse and progression in patients with DLBCL treated with doxorubicin, the efficacy of the dosing response was predicted. The predicted AUC was 0.64, suggesting an effective predictor for clinical drug application.
Conclusions
In conclusion, our study established a ferroptosis-based risk scoring model for patients with DLBCL, and this model was validated in an independent dataset. The risk scoring model showed improved efficacy in predicting survival compared to clinical characteristics. In addition, the risk score showed consistency with immune cell infiltration in DLBCL. This model implemented stratification of DLBCL patients into low- and high-risk cohorts.
CRediT authorship contribution statement
Dan Xiong: Conceptualization, Supervision, Formal analysis, Writing – original draft, Writing – review & editing. Mojuan Li: Formal analysis, Writing – original draft, Writing – review & editing. Chong Zeng: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Availability of data and materials
Datasets used and/or analyzed in this study can be obtained from the corresponding author upon reasonable request.
Funding
This work was supported by grants from the Medical Scientific Research Foundation of Guangdong Province (grant no. B2021195), the Science and Technology Program of Foshan (grant no. 1920001000694), the key program of Shunde Hospital(grant no. SRSP2021003), the key specialty funding department of Hematology, Shunde Hospital (No. 0520).
Acknowledgments
Not applicable.
Contributor Information
Dan Xiong, Email: xiongdancn@aliyun.com.
Chong Zeng, Email: wonder10@126.com.
References
- 1.Li S., Young K.H., Medeiros L.J. Diffuse large B-cell lymphoma. Pathology. 2018;50(1):74–87. doi: 10.1016/j.pathol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Miyawaki, K., et al., A Germinal center-associated microenvironmental signature reflects malignant phenotype and outcome of diffuse large B-cell lymphoma. 2019: p. 833947. [DOI] [PMC free article] [PubMed]
- 3.Liu Y., Barta S.K. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am. J. Hematol. 2019;94(5):604–616. doi: 10.1002/ajh.25460. [DOI] [PubMed] [Google Scholar]
- 4.Skrabek P., et al. Emerging therapies for the treatment of relapsed or refractory diffuse large B cell lymphoma. Curr. Oncol. 2019;26(4):253–265. doi: 10.3747/co.26.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris L.J., Patel K., Martin M. Novel therapies for relapsed or refractory diffuse large B-cell lymphoma. Int. J. Mol. Sci. 2020;21(22):8553. doi: 10.3390/ijms21228553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasqualucci L., et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenz G., et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. U. S. A. 2008;105(36):13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin R.D., et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohr J.G., et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U. S. A. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan A., Dogan A. Prognostic and predictive biomarkers in diffuse large B-cell lymphoma. Surg. Pathol. Clin. 2019;12(3):699–707. doi: 10.1016/j.path.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobashi A. Molecular Pathogenesis of Diffuse Large B-Cell Lymphoma. J. Clin. Exp. Hematopathol. 2016;56(2):71–78. doi: 10.3960/jslrt.56.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia X., et al. The relationship between ferroptosis and tumors: a novel landscape for therapeutic approach. Curr. Gene Ther. 2019;19(2):117–124. doi: 10.2174/1566523219666190628152137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedmann Angeli J.P., Krysko D.V., Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 14.Dixon S.J., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J., et al. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572(7769):402–406. doi: 10.1038/s41586-019-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rink J.S., et al. Targeted reduction of cholesterol uptake in cholesterol-addicted lymphoma cells blocks turnover of oxidized lipids to cause ferroptosis. J. Biol. Chem. 2021:296. doi: 10.1074/jbc.RA120.014888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., et al. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 2019;26(5):623–633. doi: 10.1016/j.chembiol.2019.01.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N., et al. Artesunate activates the ATF4-CHOP-CHAC1 pathway and affects ferroptosis in Burkitt's Lymphoma. Biochem. Biophys. Res. Commun. 2019;519(3):533–539. doi: 10.1016/j.bbrc.2019.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y., et al. Ferroptosis-related genes are potential prognostic molecular markers for patients with colorectal cancer. Clin. Exp. Med. 2021;21(3):467–477. doi: 10.1007/s10238-021-00697-w. [DOI] [PubMed] [Google Scholar]
- 22.Zhou N., Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database. 2020;2020:baaa021. doi: 10.1093/database/baaa021. (Oxford) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz N., et al. CNS international prognostic index: a risk model for CNS relapse in patients with diffuse large B-cell lymphoma treated with R-CHOP. J. Clin. Oncol. 2016;34(26):3150–3156. doi: 10.1200/JCO.2015.65.6520. [DOI] [PubMed] [Google Scholar]
- 24.Lossos I.S., et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Engl. J. Med. 2004;350(18):1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 25.Sáez A.I., et al. Building an outcome predictor model for diffuse large B-cell lymphoma. Am. J. Pathol. 2004;164(2):613–622. doi: 10.1016/S0002-9440(10)63150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dembrower K., et al. Comparison of a deep learning risk score and standard mammographic density score for breast cancer risk prediction. Radiology. 2020;294(2):265–272. doi: 10.1148/radiol.2019190872. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q., Fan C. A novel risk score system for assessment of ovarian cancer based on co-expression network analysis and expression level of five lncRNAs. BMC Med. Genet. 2019;20(1):103. doi: 10.1186/s12881-019-0832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habermann T.M., et al. Host immune gene polymorphisms in combination with clinical and demographic factors predict late survival in diffuse large B-cell lymphoma patients in the pre-rituximab era. Blood. 2008;112(7):2694–2702. doi: 10.1182/blood-2007-09-111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J., et al. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci. Rep. 2016;6(1):27842. doi: 10.1038/srep27842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R., et al. A novel prognostic model based on four circulating miRNA in diffuse large B-cell lymphoma: implications for the roles of MDSC and Th17 cells in lymphoma progression. Mol. Oncol. 2021;15(1):246–261. doi: 10.1002/1878-0261.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H., et al. Ferroptosis-related gene signature: a new method for personalized risk assessment in patients with diffuse large B-cell lymphoma. Pharmgenom. Pers. Med. 2021;14:609–619. doi: 10.2147/PGPM.S309846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H., et al. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J. Cell. Mol. Med. 2017;21(4):648–657. doi: 10.1111/jcmm.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu T., et al. Molecular mechanisms of ferroptosis and its role in cancer therapy. J. Cell. Mol. Med. 2019;23(8):4900–4912. doi: 10.1111/jcmm.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colaprico A., et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman A.M., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carneiro B.A., El-Deiry W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020;17(7):395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimada K., et al. Cell-line selectivity improves the predictive power of pharmacogenomic analyses and helps identify NADPH as biomarker for ferroptosis sensitivity. Cell Chem. Biol. 2016;23(2):225–235. doi: 10.1016/j.chembiol.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuo S., et al. Clinical and biological significances of a ferroptosis-related gene signature in glioma. Front. Oncol. 2020;10(2572):590861. doi: 10.3389/fonc.2020.590861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt A., et al. Dimethyl fumarate induces ferroptosis and impairs NF-κB/STAT3 signaling in DLBCL. Blood. 2021;138(10):871–884. doi: 10.1182/blood.2020009404. [DOI] [PubMed] [Google Scholar]
- 41.Abramson J.S., Shipp M.A. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106(4):1164–1174. doi: 10.1182/blood-2005-02-0687. [DOI] [PubMed] [Google Scholar]
- 42.Saez A.I., et al. Identification of biological markers of sensitivity to high-clinical-risk-adapted therapy for patients with diffuse large B-cell lymphoma. Leuk. Lymphoma. 2009;50(4):571–581. doi: 10.1080/10428190902785528. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H., et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer. 2020;19(1):43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., et al. Ferroptosis is governed by differential regulation of transcription in liver cancer. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., et al. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 2019;26(5):623–633. doi: 10.1016/j.chembiol.2019.01.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang N., et al. Artesunate activates the ATF4-CHOP-CHAC1 pathway and affects ferroptosis in Burkitt's Lymphoma. Biochem. Biophys. Res. Commun. 2019;519(3):533–539. doi: 10.1016/j.bbrc.2019.09.023. [DOI] [PubMed] [Google Scholar]
- 47.Rink J.S., et al. Targeted reduction of cholesterol uptake in cholesterol-addicted lymphoma cells blocks turnover of oxidized lipids to cause ferroptosis. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.014888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou N., Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database. 2020;2020:baaa021. doi: 10.1093/database/baaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 2016;73(11):2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y., et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H., et al. Identification of a prognostic metabolic gene signature in diffuse large B-cell lymphoma. J. Cell. Mol. Med. 2021;25(14):7066–7077. doi: 10.1111/jcmm.16720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma S.Y., et al. A prognostic immune risk score for diffuse large B-cell lymphoma. Br. J. Haematol. 2021;194(1):111–119. doi: 10.1111/bjh.17478. [DOI] [PubMed] [Google Scholar]
- 53.Wang G., et al. Construction of a DLBCL prognostic signature based on tumor microenvironment. Expert Rev. Hematol. 2021;14(7):1–8. doi: 10.1080/17474086.2021.1943349. [DOI] [PubMed] [Google Scholar]
- 54.Baba Y., et al. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci. 2020;111(9):3132–3141. doi: 10.1111/cas.14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petitprez F., et al. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front. Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.You Y., et al. Ferroptosis-related gene signature promotes ovarian cancer by influencing immune infiltration and invasion. J. Oncol. 2021;2021 doi: 10.1155/2021/9915312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K., et al. A ferroptosis-related lncRNAs signature predicts prognosis and immune microenvironment for breast cancer. Front. Mol. Biosci. 2021;7(8):678877. doi: 10.3389/fmolb.2021.678877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets used and/or analyzed in this study can be obtained from the corresponding author upon reasonable request.