Summary
Micropeptides are emerging as important regulators of various cellular processes. Long non-coding RNAs (lncRNAs) serve as a source of micropeptide-encoding small reading frames. The techniques to detect micropeptides or translating lncRNAs, such as mass spectrometry and ribosome profiling, are sophisticated and expensive. Here, we present an easy and cost-effective protocol to screen for potential micropeptide-encoding lncRNAs by polysome profiling in suspension cell lines. When combined with quantitative PCR, this protocol facilitates the identification of a number of translating lncRNAs simultaneously.
For complete details on the use and execution of this protocol, please refer to Sun et al. (2021).
Subject areas: Cell separation/fractionation, Molecular Biology
Graphical abstract

Highlights
-
•
A protocol for rapid detection of the potential micropeptide-encoding lncRNAs
-
•
Detail a complete schedule for polysome profiling
-
•
Provide a protocol to distinguish RNAs with low or high translation activity
Micropeptides are emerging as important regulators of various cellular processes. Long non-coding RNAs (lncRNAs) serve as a source of micropeptide-encoding small reading frames. The techniques to detect micropeptides or translating lncRNAs, such as mass spectrometry and ribosome profiling, are sophisticated and expensive. Here, we present an easy and cost-effective protocol to screen for potential micropeptide-encoding lncRNAs by polysome profiling in suspension cell lines. When combined with quantitative PCR, this protocol facilitates the identification of a number of translating lncRNAs simultaneously.
Before you begin
Make sure that all the required reagents and items (refer to “key resources table” and “materials and equipment” section) are ready to use. This protocol describes steps for MV4-11 cells. However, the adaptability of this method makes its application extended to other suspension cells or adherent cells with necessary modifications.
RNase-free environment is required for successfully executing this protocol. RNase ZAPTM could be applied for the work area, disposable gloves and other contacts to eliminate ubiquitous RNase contamination. Additional precautions, including the use of RNase-free consumables, baking of glass wares at 180°C for at least 4 h, operations and samples kept at 4°C, and the wearing of headgear, should also be taken.
MV4-11 cell culture and expansion
Timing: 1–2 weeks
-
1.
The minimum number of cells we used on day 1 of the protocol is 2 ∗ 107 MV4-11 cells per sample. The fewer number of cells than that may bring difficulties to recover sufficient RNAs for subsequent analysis. Expand the cells to the desired number ahead of time.
Preparation of buffer stock
Timing: 30 min
-
2.
The reagents required for this protocol are easy to get. The stock buffers are prepared as listed below. It is essential to prepare buffer stock and all the solutions mentioned below with DEPC-treated water.
Just in time preparation of sucrose buffer
Timing: 30 min
-
3.
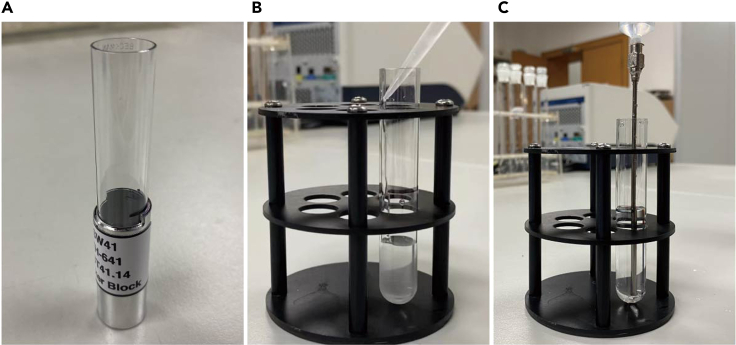
The day before starting the experiment (day 0), prepare the sucrose buffer as described below. Vortex to facilitate the dissolve of sucrose in the buffer and let it stand for overnight (14–16 h) to eliminate bubbles (Figure 1).
Note: The sucrose buffers could be filtered through 0.22 μm filters.
Note: For 10% and 45% sucrose buffer, preparation of about 7 mL buffer each is sufficient for one sample when using SW41 ultracentrifuge tubes. At least two samples (one experimental sample and one blank sample) are needed in the subsequent ultracentrifugation for balancing.
Figure 1.
Preparation of sucrose buffer
(A) Sucrose buffer with bubbles caused by vortexing to dissolve sucrose.
(B) Sucrose buffer without bubbles after standing overnight (14–16 h).
Just in time preparation of lysis buffer
Timing: 30 min
-
4.
On day 1, prepare fresh lysis buffers as described below. The RNase inhibitor, proteinase inhibitor or phenylmethylsulfonyl fluoride (PMSF), dithiothreitol (DTT), and cycloheximide are added before use.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Cycloheximide (CHX) | Sigma-Aldrich | Cat# 239764 |
| RNasin® Ribonuclease Inhibitor | Promega | Cat# N2115 |
| 1×complete ULTRA protease inhibitor | Roche | Cat# 5892970001 |
| Phenylmethylsulfonyl fluoride (PMSF) | Beyotime | Cat# ST506 |
| Dithiothreitol (DTT) | Sigma-Aldrich | Cat# D0632 |
| RNase ZAPTM | Thermo Fisher Scientific | Cat# AM9780 |
| Sodium deoxycholate | Sigma-Aldrich | Cat# D6750 |
| Sucrose | Sigma-Aldrich | Cat# V900116 |
| Magnesium chloride | Sigma-Aldrich | Cat# M8266 |
| IGEPAL | Sigma-Aldrich | Cat# 18896 |
| Tris | Asegene | Cat# AS430789 |
| IMDM modified medium with L-glutamine, HEPES | HyClone | Cat# SH30228.01 |
| Fetal Bovine Serum (FBS) | Gibco | Cat# 10270-106 |
| RNAiso Plus | Takara | Cat# 9109 |
| GlycoBlue™ Coprecipitant | Thermo Fisher Scientific | Cat# AM9516 |
| Ethanol | Sigma-Aldrich | Cat# 459844 |
| Chloroform | Sigma-Aldrich | Cat# C2432 |
| Isopropanol | Sigma-Aldrich | Cat# 563935 |
| DEPC Treated Water | Sangon Biotech | Cat# B5001005-0500 |
| Phosphate buffered saline (PBS) | Cellcook | Cat# CM2018 |
| Hydrochloric Acid | Sigma-Aldrich | Cat# 258148 |
| Critical commercial assays | ||
| PrimeScript RT reagent Kit | Takara | Cat# RR047A |
| TB Green Premix Ex Taq | Takara | Cat# RR420B |
| Experimental models: Cell lines | ||
| MV4-11 | ATCC | Cat# CRL-9591; RRID: CVCL_0064 |
| Oligonucleotides | ||
|
AL035446.1 Primer: Forward - GAAAAATGCGTGCCCTCTTGT Reverse - TCTTGGGCTCCATCATTCATCC |
Ruibiotech | N/A |
|
LINC01433 Primer: Forward - TTTGGAGCTTCCCTGCAGAC Reverse - CCGCTAGTGCATGACAGTGA |
Ruibiotech | N/A |
|
YTHDF3-AS1 Primer: Forward - CCAAACCCAGTTCAGCCGTA Reverse - TCAGAGCGGAGTCAAAAGCA |
Ruibiotech | N/A |
|
AC090515.2 Primer: Forward - TGGTGAGACATTCGCCCTAC Reverse - CCACTCCATGCAAAGTGAAGC |
Ruibiotech | N/A |
|
CASC2 Primer: Forward - AGCCATCTACGGTAACCCAGTAG Reverse - AAACCCACATCTGTCCCAAATT |
Ruibiotech | N/A |
| PROSER2-AS1 Primer: Forward - GCCACAGCCATTGTCTTCATC Reverse - ACCAGTGATGCCTCCCGTT |
Ruibiotech | N/A |
|
APPLE Primer: Forward - CGGTTGACCTGAGCCTACTTC Reverse - CTGACTAGGCGACCGGCA |
Ruibiotech | N/A |
|
AL713998.1 Primer: Forward - TCTTCTTGGGGAGACAGCCT Reverse - GGTTGTGTGGCTGGCAAATG |
Ruibiotech | N/A |
|
AL356356.1 Primer: Forward - CCAGCTCTTGCCAGCTATGT Reverse - GCAGATGAGCAAACTGCACC |
Ruibiotech | N/A |
|
LINC00649 Primer: Forward - TCTCTGGCACGAAAGTGACC Reverse - AATAGATGCAAGCCGGGACT |
Ruibiotech | N/A |
|
SATB2-AS2 Primer: Forward - GACCGGTAACACCAAGAGCC Reverse - GTCTCTCTCTGCGGCTTGTC |
Ruibiotech | N/A |
|
AC091057.1 Primer: Forward - AGACTTTGTTCGGTCCACGG Reverse - TCTGCTCCCGGTAAAGATGC |
Ruibiotech | N/A |
|
LINC00106 Primer: Forward - AGGACACCGTCTGTCTTACG Reverse - CCAGTGGTCACCTGAGATGG |
Ruibiotech | N/A |
|
LINC01215 Primer: Forward - CCCCATGTAGTCAGCTCGTT Reverse - GGAAATGCAACTGGACCCAC |
Ruibiotech | N/A |
|
hY1 Primer: Forward - GGCTGGTCCGAAGGTAGTGA Reverse - GCAGTAGTGAGAAGGGGGGA |
Ruibiotech | N/A |
|
GAPDH Primer: Forward - AGGTGAAGGTCGGAGTCAAC Reverse - AGTTGAGGTCAATGAAGGGG |
Ruibiotech | N/A |
| Software and algorithms | ||
| Triax™ FlowCell software | Biocomp Instruments | N/A |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/scientific- software/prism/ |
| QuantStudio Software v1.3 | Thermo Fisher Scientific | N/A |
| Excel | Microsoft | N/A |
| Other | ||
| Minisart® Syringe Filters | Sartorius | Cat# 16533 |
| Syringe | Jumin Bio-technologies | Cat# 1.2x35TWLB |
| Optima L-100 XP Ultracentrifuge | Beckman Coulter | N/A |
| SW 41 Ti Swinging-Bucket Rotor | Beckman Coulter | Cat# 331362 |
| Open-Top Thinwall Ultra-Clear Tube | Beckman Coulter | Cat# 344059 |
| Gradient MasterTM | Biocomp Instruments | N/A |
| Piston Gradient Fractionator™ | Biocomp Instruments | N/A |
| TriaxTM Flow Cell | Biocomp Instruments | N/A |
| Fraction Collector | Biocomp Instruments | N/A |
| QuantStudio 6 Flex Real-Time PCR System | Thermo Fisher Scientific | N/A |
| MicroAmp™ Optical Adhesive Film | Thermo Fisher Scientific | Cat# 4311971 |
| MicroAmp™ Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL | Thermo Fisher Scientific | Cat# 4346906 |
| NanoDrop 2000/2000c Spectrophotometer | Thermo Fisher Scientific | N/A |
| Centrifuge 5424 R | Eppendorf | N/A |
| Centrifuge 5810 R | Eppendorf | N/A |
| 50 mL Centrifuge Tubes | Corning | Cat# 430829 |
| Cell culture flask, 75 cm2 | Corning | Cat# 430641 |
| Axygen® 1.5 mL MaxyClear Snaplock Microcentrifuge Tube | Corning | Cat# MCT-150-C |
| Micro Centrifuge Tubes, 2 mL | Jet Bio-Filtration | Cat# CFT001020 |
| 0.2 mL Flat PCR Tube 8-Cap Strips, optical, ultraclear | Bio-Rad Laboratories | Cat# TCS0803 |
| Cellometer Auto T4 Bright Field Cell Counter | Nexcelom Bioscience | N/A |
| SD100 Slides | Nexcelom Bioscience | Cat# CHT4-SD100-002 |
| CO2 Incubator | Panasonic | Cat# MCO-170AICUVL-PC |
| Esco Biological Safety Cabinets | ESCO Technologies | N/A |
| Vortex Mixer V6 | ESSEN Scientific | N/A |
| SevenCompact pH meter S220 | METT.LER TOLEDO | Cat# 30019028 |
Materials and equipment
Stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 1 M | 6.057 g / 50 mL |
| NaCl | 4 M | 11.688 g / 50 mL |
| MgCl2 | 1 M | 4.761 g / 50 mL |
| Dithiothreitol (DTT) | 1 M | 1.55 g / 10 mL |
All the stock solutions can be stored at 4°C up to one year.
Lysis buffer (pH 7.4)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M) | 25 mM | 1.25 mL |
| MgCl2 (1 M) | 5 mM | 0.25 mL |
| NaCl (4 M) | 100 mM | 1.25 mL |
| IGEPAL | 1 % | 0.5 mL |
| Sodium deoxycholate | 1 % | 0.5 g |
| DEPC H2O | n/a | up to 50 mL |
| Total | n/a | 50 mL |
This buffer can be stored at 4°C up to one year.
The 1 mM DTT, 100 μg/mL cycloheximide, 40 U/mL RNase inhibitor and 1 × protease inhibitor should be added before use. (Troubleshooting 1)
CRITICAL: HCl is highly volatile and is a high-risk inhalation hazard. It is toxic and causes severe skin burns and serious eye damage. Opening a concentrated HCl (36%) should be done in a fume hood. Wear personal protective equipment.
Sucrose buffer (pH 7.4)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M) | 25 mM | 1.25 mL |
| MgCl2 (1 M) | 5 mM | 0.25 mL |
| NaCl (4 M) | 100 mM | 1.25 mL |
| DEPC H2O | n/a | up to 50 mL |
| Total | n/a | 50 mL |
This buffer can be stored at 4°C up to one year.
The 100 μg/mL cycloheximide, 40 U/mL RNase inhibitor and 1 mM PMSF should be added before use.
Step-by-step method details
Whole cell lysate preparation – day 1
Timing: 1–1.5 h
Cells are harvested and lysed for subsequent ultracentrifugation on sucrose cushion.
-
1.
Grow MV4-11 cells with IMDM modified medium (10% fetal bovine serum) in the culture flask at 37°C and 5% CO2 to the desired number. Start culture at 1∗105–2∗105cells/mL and maintain between 1∗105 and 1∗106 cells/mL.
CRITICAL: If necessary, diluting the cells or passage the cells to a new flask to keep the cells on log phase at the day before starting the experiment. The polysome peak of over confluent cells that are less proliferative will not appear as robust because proliferation and translation rate are closely correlated (Johnson et al., 1976).
-
2.
Treat the cells with 100 μg/mL cycloheximide. After gentle shaking to mix thoroughly, incubate the cells in the culture flask at 37°C and 5% CO2 for 15 min.
Note: For suspension cells, it is recommended to count the desired number of cells before cycloheximide treatment. Too fewer cells may produce small peaks and weak signals below the detection threshold.
Note: Cycloheximide is a translation elongation inhibitor and used to prevent translating RNA from running off (Schneider-Poetsch et al., 2010). The cycloheximide treatment duration should be optimized for different cell lines. Preliminary experiments are needed to screen for the optimal cycloheximide concentration to generate robust polysome peaks.
-
3.Harvest and wash cells with ice-cold PBS supplemented with 100 μg/mL cycloheximide.
-
a.Collect cells in the culture with a 50 mL falcon tube and centrifuge the sample at 500 × g for 3 min at 4°C.
-
b.Discard the supernatant and wash the cells with ice-cold PBS supplemented with 100 μg/mL cycloheximide.
-
c.Centrifuge the cells at 500 × g for 3 min at 4°C. Repeat the wash step one more time.
-
d.Centrifuge the cells at 500 × g for 3 min at 4°C. Remove the supernatant as much as possible with precaution. Aspiration with a vacuum pump is preferred.
-
a.
Note: Keep the samples and solution on ice all the time. Make sure that all the items and reagents are ready to use so that the experiment can proceed quickly without haste. Leaving cells on ice for too long will result in suboptimal quality of polysome preparation.
Pause point: Cell pellets may be flash frozen in the liquid nitrogen and stored at −80°C until use. However, it is always recommended to use fresh cells for the subsequent analysis.
-
4.
Suspend the cell pellets by gently pipetting in 200–300 μL lysis buffer supplemented with 40 U/mL RNase inhibitor, 1 mM DTT, 1 mM PMSF, and 100 μg/mL cycloheximide. Immediately place the sample on ice for 30 min with occasional inverting to facilitate cell lysis.
Note: The amount of lysis buffer used is adjusted according to the cell number. Prepare excessive amount of fresh lysis buffer in advance and settle the buffer in the refrigerator at 4°C for a period of time to reduce bubbles caused by detergent.
-
5.
Centrifuge the cell lysate at 13,000 × g for 10 min at 4°C. Transfer the supernatant to a new pre-chilled 1.5 mL tube. Place the tube on ice until use.
Pause point: Alternatively, the cell lysate may be flash frozen in liquid nitrogen and stored at −80°C for a month. However, fresh cell lysates are always recommended.
Note: Avoid the contamination by precipitations at the bottom when transferring supernatant. It is recommended to immediately load the lysis on sucrose cushion as this will improve the quality of the preparation.
Sucrose cushion preparation
Timing: 15–20 min
Prepare sucrose gradient for density gradient centrifugation.
Note: Prepare the sucrose gradient while the cell sample is lysed on ice as described above.
-
6.
Mark the half-full position with the marker casing tube provided by the gradient maker (Figure 2A).
-
7.
Add 10% sucrose buffer first with 1 mL pipette until it reaches the half-full position (Figure 2B).
Note: Touch the tip to the upper side wall of the tube and dispensing the buffer along the wall at a slow constant rate. Be careful to not generate bubbles that may interfere with following gradient making and polysome sedimentation.
-
8.
Use long blunt needle to reach the bottom of the ultracentrifuge tube and layer 45% sucrose buffer until the upper 10% sucrose buffer reaches the top of the tube (Figure 2C).
Note: This process should be slow and gentle at a constant rate to avoid disturbing the 10% sucrose buffer layer.
-
9.
Lid the tube with the rate zonal cap provided by the gradient maker.
Note: When lidding the tube, contact the sucrose buffer with one side of the cap at first, and then slowly drop the other side that has the air hole into the tube to avoid making bubbles. The air hole is used to vent the bubbles (Figure 3).
-
10.
Put the tube into the magnetic tube holder on the gradient maker.
Note: Make sure the plate that the magnetic tube holder is attached on is level (Figures 4A and 4B).
-
11.
Run the sucrose gradient maker by selecting “10%–45%”. It takes about 1 min and 30 s to finish (Figure 4C). The sucrose gradient is ready to use immediately.
Pause point: Alternatively, the prepared sucrose cushion could be stored at −80°C for up to 6 months until use.
Note: Slide the magnetic tube holder out slowly and gently upon the ending of gradient making. Take extra precautions when moving the tube to avoid disturbing the prepared sucrose gradient.
Figure 2.
10%–45% sucrose gradient loading
(A) Use the tube holder to mark the half-full point as shown by the black marker.
(B) Add 10% sucrose buffer by a 1 mL pipette.
(C) Add 45% sucrose buffer a long blunt needle.
Figure 3.
Secure the tube with rate zonal cap
(A) The opposite side of air hole contacts the sucrose cushion first.
(B) Drop the cap slowly and gently.
(C) Lid the tube without making bubbles.
Figure 4.
10%–45% sucrose gradient preparation by gradient maker
(A) Check for the levelness of the magnetic plate.
(B) Adjust levelness using the button indicated by red arrow if the plate is unlevel.
(C) Select a gradient program and run it. In this example, 10%–45% gradient is used.
Polysome sedimentation by ultracentrifugation
Timing: 4 h
Ultracentrifugation to isolate 40S subunit, 60S subunit, 80S monosome, and polysome-bound RNA with different densities.
-
12.
Lift up the cap of the tube and layer the cell lysate onto the sucrose cushion. The A260 and A280 of MV4-11 cell lysate is approximately 65 and 35 respectively as measured by the NanoDrop 2000. If applicable, load lysis buffer onto the sucrose cushion of blank sample group.
Note: Use a 200 μL pipette to layer the cell sample onto the sucrose cushion. Touch the pipette tip to the upper side of the tube and dispense the cell lysate slowly and gently to avoid disturbing the sucrose gradient (Figure 5).
-
13.
Place the sucrose cushion tube loaded with samples into the SW41 rotor bucket.
-
14.
Use an analytical scale to balance the opposing rotor buckets.
Note: Remember to put the cap together with the bucket on the analytical scale. If unbalanced, load lysis buffer onto the cushion to balance the buckets. It is acceptable that the weight differences between opposing buckets are under 0.003 g.
-
15.
Once the buckets are balanced, screw the cap into the bucket and hook the buckets onto the rotor hanger. The buckets must be arranged symmetrically and all buckets, whether loaded or empty, must be hooked to rotor.
Note: Screw the cap anticlockwise at first until you hear the click sound that indicates the cap fitted into the thread. Then screw the cap in the clockwise direction until there is metal-to metal contact.
-
16.
Carefully lift the rotor with steady hands and install it on the drive hub of the ultracentrifuge perpendicularly.
Optional: You can check for the proper installation by manually turning the rotor clockwise and observe the speed reading on the screen.
-
17.
Centrifuge at 36,000 rpm (222,227 × g) for 3 h at 4°C with SW41 Ti rotor.
CRITICAL: Do not choose max deceleration for ultracentrifugation. Choose speed “5” instead for optimal polysome sedimentation.
Note: Check for the proper functioning of the ultracentrifuge occasionally to avoid the occurrence of errors.
Figure 5.
Loading cell lysate onto sucrose cushion
(A) Use a 200 μL to layer the cell lysate without disturbing the sucrose gradient.
(B) The cell lysate loaded onto the sucrose gradient as indicated by the red arrow.
Polysome fractionation
Collect each ribosome-bound RNA fractions for subsequent analysis.
Timing: 1 h
-
18.
Turn on the fractionation system 30 min ahead of the ending of ultracentrifugation.
Note: Switch on pump, UV detector, fractionation collector, and computer. Press “SCAN” on the control panel first, otherwise the computer may not be successfully connected (Figures 6A and 6B).
Note: Check whether the fractionation collector is at the initial position; if not, press “END” on the control panel of the fractionation station (Figure 6C).
-
19.
Open the software Triax™ FlowCell on the computer and set the parameters for fractionation.
Note: The parameters set for the fractionation station is shown in Figure 7.
-
20.
Fill the syringe attached to the fractionation station with DEPC-treated water to extensively rinse the system.
-
21.
Press “air” on the control panel to remove any residual water. Repeat the rinse and air operations for two more times.
Optional: A text run with DEPC-treated water is recommended to check whether the system works properly.
-
22.
Prepare 2 mL tube for collecting. Place them into tube rack on the fractionation collector.
Note: The tubes are placed into the rack to match the order of the sequentially collected fractions (Figure 8).
-
23.
Once the ultracentrifugation for polysome sedimentation ends, take out the rotor and place it onto the adapter.
-
24.
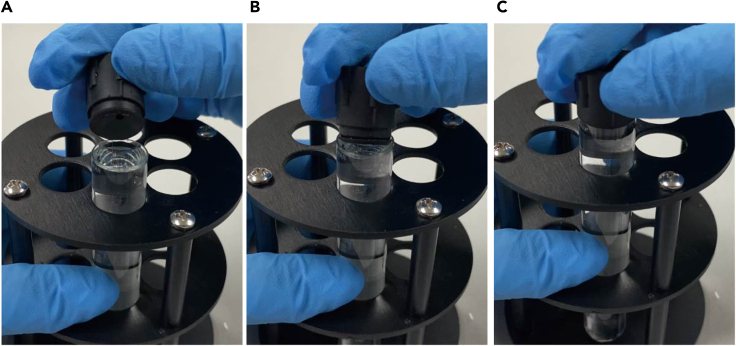
Use a screwdriver to remove the cap of the bucket, and take out the centrifuge tube with tweezers.
Note: Take extra care to ensure not to disturb the gradient at this stage. Be careful to not make the bucket sway on the hanger when taking it out from the ultracentrifuge.
-
25.
Put the cap of the SW41 tube holder onto the centrifuge tube of cell sample (Figures 9A–9C).
-
26.
Place the capped centrifuge tube into the SW41 tube holder provided by the fractionation station. Gently screw the cap clockwise to secure the centrifuge tube (Figure 9D).
-
27.
Transfer the loaded tube holder to let it slide into the fractionation station and turn it clockwise to be ready for collection (Figures 9E and 9F).
-
28.
Start the run by clicking the “START SCAN” in the software program. This will start the collection of each fraction by a piston tip from top to bottom, and at the same time the UV absorbance at 260 nm is recorded to plot the tracing. Make sure that every drop of each fraction is collected into its corresponding 2 mL tube (Troubleshooting 2, 3, and 4).
Note: Do not turn on the light from the piston tip for a prolonged period of time. The heat generated from the light may cause RNA degradation in the sucrose gradient.
Note: Press “Air” when the run ends to blow the residual drops out into the last tube for collection.
-
29.
Transfer the 2 mL tube on the ice and add equal volume RNAiso Plus solution immediately. Vigorously vortex each tube to mix thoroughly.
-
30.
Flash freeze each fraction with RNAiso Plus solution in liquid nitrogen and store them at −80°C until use.
Figure 6.
The fractionation station
(A) Overview of the fractionation station. It includes the piston tip and UV detector to collect and detect sucrose gradients (upper), the control panel (lower), and the fractionation collector to dispense each fraction into the tubes on the rack (left upper).
(B) The close view of control panel. Red arrows indicate the controls for Air and Rinse.
(C) The panel for fractionation collector. The red arrow indicates the “END” button. Press “END” could reset the fractionation collector to the initial position.
Figure 7.
The parameters set in the software Triax™ FlowCell for the fractionation station
(A) The parameters for UV detector and sucrose gradients.
(B) The parameters for fractionation collector.
Figure 8.
The order for the arrangement of collecting tubes
Figure 9.
Load the sample tube with the tube holder for collection
(A–C) Fit the cap of the tube holder onto the sample tube.
(D–F) Install the loaded tube holder onto the fractionation station.
Caution: RNAiso Plus solution is toxic and harmful for the researcher and surrounding environment. Precautionary measures should be taken, including working in a fume hood, wearing disposable latex gloves, coat, and goggles.
Note: Clearly mark the fraction number on the tube. Be careful not to spill RNAiso Plus solution which may erase the maker.
-
31.
Rinse and air the fractionation system three times before another run. After all the runs finish, launch the last run with DEPC-treated water and extensively rinse the system to remove any residual sucrose.
RNA extraction-day 2
This step can isolate RNA from sucrose buffer by RNAiso Plus solution.
Timing: 4–6 h
-
32.
Take samples from −80°C and quickly thaw them at room temperature (20°C–25°C).
-
33.
Add 0.2 mL of chloroform per 1 mL of RNAiso Plus solution.
-
34.
Vortex vigorously for 30 s to mix the chloroform thoroughly.
Note: RNAiso Plus solution is caustic so that precautionary measures should be taken, including the wearing of eye protection. The tube cap should be tightly closed, and if necessary, use cap locks.
-
35.
Keep the samples at room temperature (20°C–25°C) for 5 min.
-
36.
Centrifuge the samples at 12,000 × g for 15 min at 4°C.
Note: The centrifuge should be pre-cooled to 4°C before use.
Note: The samples are separated into three layers: the bottom layer is a red organic phase containing protein; the middle layer is a white semi-solid containing DNA; the top layer is a colorless aqueous phase containing RNA.
-
37.
Transfer the top layer into new RNase-free 1.5 mL tubes.
CRITICAL: Be careful not to touch the middle layer as it could lead to DNA contamination.
Note: Two or more tubes should be used for each fraction to make sure that the volume of the aqueous phase is not over than 0.75 mL to allow the addition of isopropanol.
-
38.
Add equal volume of ice-cold isopropanol and 1 μL GlycoBlue™ Coprecipitant. Mix thoroughly.
CRITICAL: It is recommended to add carrier because the RNA recovered from polysome fractions may not be easily visible. GlycoBlue™ Coprecipitant is glycogen linked with a blue dye. It facilitates nucleic acid precipitation and increases the size and visibility of the pellet without interfering subsequent analysis.
-
39.
Keep the mixture at room temperature (20°C–25°C) for 10 min to precipitate.
Optional: Keep the mixture at −20°C for at least 1 hour to increase RNA yield.
-
40.
Centrifuge at 12,000 × g for 10 min at 4°C.
-
41.
Discard the supernatant carefully without touching the pellet.
Note: Centrifuge shortly and remove the remaining isopropanol completely.
-
42.
Add 1 mL ice-cold 75% ethanol to wash the RNA pellet.
-
43.
Vortex briefly and centrifuge at 7,500 × g for 5 min at 4°C.
Note: Do not disperse the pellet during vortexing.
-
44.
Discard the supernatant carefully and add 1 mL ice-cold 100% ethanol to wash the RNA pellet.
Pause point: The RNA can be stored in 100% ethanol at −20°C for several months.
-
45.
Vortex briefly and centrifuge at 7,500 × g for 5 min at 4°C.
-
46.
Centrifuge shortly and remove the remaining ethanol completely.
-
47.
Air dry the pellet for 5 min.
-
48.
Dissolve the pellet with 10–30 μL RNase-free water.
CRITICAL: The final volume of the RNA solution should be identical for each fraction of polysome profiling.
Note: The RNA solution should be kept on ice for immediate subsequent analyses to prevent degradation.
Pause point: Alternative, the RNA solution could be stored at −80°C. RNA is generally stable at −80°C for up to a year. Use aliquots to avoid freeze-thaw cycles.
-
49.
Measure the RNA concentrations by NanoDrop™ 2000 spectrophotometer (Troubleshooting 5).
Optional: The DNase I treatment should be applied if a genomic DNA elimination reaction is not included in the subsequent reverse-transcription step.
RNA detection by quantitative reverse transcription PCR (RT-qPCR)
RNA quantifications for each fraction to map the distribution of potentially translating lncRNAs in polysome profiles.
Timing: 2–3 h (depending on number of samples)
Note: RNAs are reversely transcribed to cDNA. The synthesized cDNA could serve as template for quantification of lncRNAs of interest by real-time PCR. The following reverse-transcription steps are performed using PrimeScript™ RT Reagent Kit with gDNA Eraser.
Alternatives: Other kits or methods could also be used for reverse transcription.
-
50.
Prepare a genomic DNA elimination reaction mix as described in the following table.
Dispense equal volume of RNA samples from different fractions into each reaction.
| gDNA elimination reaction | |
|---|---|
| Reagent | Volume |
| 5× gDNA Eraser Buffer | 2 μL |
| gDNA Eraser | 1 μL |
| RNA | up to 1 μg |
| RNase-Free dH2O | to 10 μL |
| Total | 10 μL |
Note: All reaction components should be added on ice. A mix except for RNA samples is recommended to save time and should be prepared with at least one extra reaction.
Note: To calculate the percent RNA of each fraction, the equal volume of RNA solution should be used for each fraction of polysome profiling.
-
51.
Gently mix the reagent and quickly spin the samples to collect all liquid to the bottom of tubes.
-
52.
Incubate the samples at 42°C for 2 min and place them at 4°C until use.
-
53.
Prepare a reverse-transcription reaction mix shown below. Add 10 μL of mix to each reaction solution from last step.
| Reverse-transcription reaction | |
|---|---|
| Reagent | Volume |
| Reaction solution from step 52 | 10 μL |
| 5× PrimeScript Buffer 2 | 4 μL |
| PrimeScript RT Enzyme Mix I |
1 μL |
| RT Primer Mix | 1 μL |
| RNase-Free dH2O | 40 μL |
| Total | 20 μL |
-
54.
Gently mix the reagent and quickly spin the samples to collect all liquid to the bottom of tubes.
-
55.
Incubate the samples at 37°C for 15 min, 85°C for 5 s and place them at 4°C until use.
Pause point: The cDNAs could be stored at −20°C for long term.
-
56.
The cDNAs should be diluted at least two times with sterilized double-distilled water before use.
-
57.
Prepare the real-time PCR reaction mix except for cDNAs as described in the following table.
| Real-time PCR | |
|---|---|
| Reagent | Volume |
| TB Green Premix Ex Taq II (Tli RNaseH Plus) (2×) |
5 μL |
| PCR Forward Primer (10 μM) | 0.2 μL |
| PCR Reverse Primer (10 μM) | 0.2 μL |
| ROX Reference Dye or Dye II (50×) | 0.2 μL |
| RT reaction solution (cDNA) from step 56 | 2 μL |
| Sterile purified water | 2.4 μL |
| Total | 10 μL |
Note: All reaction components should be added on ice.
-
58.
Mix the reaction solution and quickly spin the samples.
-
59.
Transfer 8 μL of the mix to 96/384-well plate.
Note: The 384-well plate is recommended to detect several lncRNAs for the same batch of fractions on the same plate.
-
60.
Add 2 μL of the diluted cDNA to the corresponding wells.
-
61.
Seal the 96/384-well plate by an optical adhesive film.
CRITICAL: Make sure that the wells were completely sealed to avoid sample evaporation during PCR reaction.
-
62.
Centrifuge the plate at 1,500 × g for 2 min at 4°C to collect all liquid to the bottom of the plate and to eliminate bubbles.
-
63.
Run the real-time PCR reaction with the appropriate program. An example is shown in the following table.
| PCR cycling conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial denaturation | 95°C | 30 s | 1 |
| PCR reaction | 95°C | 5 s | 40 cycles |
| 60°C | 30 s | ||
-
64.
The percent lncRNA of interest in each fraction is calculated by RT-qPCR data as described in quantification and statistical analysis section.
Expected outcomes
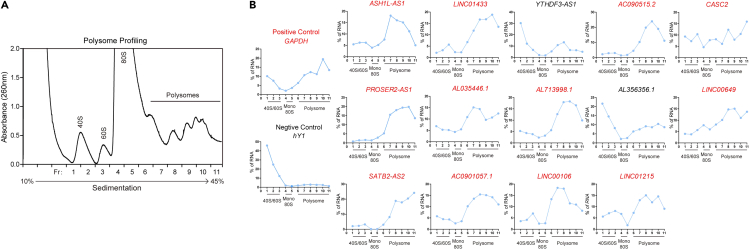
A successful polysome profiling should show clear 40S subunit, 60S subunit, 80S monosome, and several polysome peaks. The polysome peaks represent the RNAs bound to increasing number of ribosomes (>=2). The robustness of polysome peaks is dependent on the proliferation state of the cell samples and varies between different cell lines. Generally, at least three polysome peaks are observed for a good polysome profile.
It is important to include both positive and negative controls when screening for potentially translating lncRNAs by RT-qPCR following RNA extractions of each fraction. The mRNA of housekeeping genes, such as GAPDH, could serve as the positive control; the known cytoplasmic non-coding RNAs, such as hY1, could serve as the negative control. The peak of positive control, GAPDH, should be in the high polysome fraction, while the negative control, hY1, should be enriched in the submonosome fractions. The candidate lncRNAs that have prominent peaks in the polysome fractions may indicate activate translation in cells. An example is shown for polysome profiling in MV4-11 cells and RT-qPCR detection of the candidate lncRNA distributions in each fraction (Figure 10), as we previously described (Sun et al., 2021).
Figure 10.
An example of polysome profiling followed by lncRNA distribution calculations
(A) Polysome profile of MV4-11 cells.
(B) Distributions of lncRNAs of interest. Protein-coding mRNA GAPDH is used as a positive control. Non-coding RNA hY1 is used as a negative control.
Figure reprinted with permission from Sun et al. (2021).
Quantification and statistical analysis
The calculations of the percent RNA in each fraction are as follows:
X = the number of the fraction that is calculated; Y= the total number of fractions.
Limitations
This protocol requires a large number of cells to perform. Too few cells may result in low RNA yield and unreliable results. Therefore, this protocol may not apply to rare and precious cell samples.
The part of polysome profiling is tricky due to a couple of reasons as follows: 1. RNA prone to rapid degradation; 2. the requirement for preservation of both RNA and protein to detect the translation complex; 3. the need to keep the sucrose gradient undisturbed despite that the researcher has to move it, layer the lysate onto it, and collect fractions from it. Although manual collection of each fraction is feasible, it is uneasy to do it without disturbing the gradient. Therefore, it is recommended to use a fractionation station to collect each fraction, which may not be handy in some cases. It is also a prerequisite to carry on the experiment in an isolated and clean environment. The RNA and protein are prone to degradation in a crowded and contaminated workplace.
As no control could be used for controlling the potential differences in RNA extraction, it requires high demand for handling the samples of each fraction with identical operations. The misoperation for one sample may lead to incorrect results. The inclusion of positive control, such as coding-gene GAPDH, and negative control, such as non-coding hY1, is important to evaluate the robustness of the results.
The polysome profiling analysis described here is rapid and cost-effective. It allows the evaluation of overall translation status and the status of individual RNAs, which comes handy in screening for potential micropeptide-encoding lncRNAs. The major limit of this method is the lack of information regarding ribosome localization on lncRNA, therefore it is impossible to predict which potential small open reading frame in the lncRNA encodes micropeptide. In contrast, ribosome profiling followed by RNA sequencing (Ribo-seq) could determine the ribosome position at a single-nucleotide resolution. However, Ribo-seq is unable to determine the translation status of an lncRNA like polysome profiling analysis does. For example, in the scenario where two ribosomes show occupancy on two lncRNA As (A1 and A2) at position a and b; the Ribo-seq analysis cannot distinguish between that the two ribosomes occupy A1 or A2 at these two positions and that the two ribosomes occupy A1 and A2 at position a and b respectively. Furthermore, the information of regions that are unprotected by ribosomes is lost in Ribo-seq, which makes it impossible to distinguish the transcript variants that have different translation efficiencies (Ingolia, 2014). Therefore, the protocol described here and Ribo-seq are complementary methods to look for micropeptide-encoding lncRNAs. Moreover, polysome profiling could also combine with microarray and RNA-sequencing for high-throughput screening. In addition, ribosome affinity purification followed by RNA analysis is another method that can be used for characterizing ribosome-bound RNAs (Halbeisen et al., 2009; Heiman et al., 2014). This method could remove contaminations that might exist in sucrose gradients, such as lipid rafts, P-body components or pseudo-polysome (Halbeisen et al., 2009; Thermann and Hentze, 2007). Despite this advantage, it cannot determine the translation efficiency of lncRNAs except for their bindings to ribosome. In sum, these methods should be combined under different scenarios to screen for the potential translated lncRNAs.
Troubleshooting
Problem 1
The sodium deoxycholate could not be dissolved in the lysis buffer.
Potential solution
Reprepare the lysis buffer and adjust the addition order of sodium deoxycholate. Add all the reagents except sodium deoxycholate and adjust the pH under ∼7.8. Then add the sodium deoxycholate followed by vortexing for dissolution. Adjust the pH to the final 7.4.
Problem 2
The fractionation system is clogged.
Potential solution
Sucrose solutions should be filtered through 0.22 μm filters to prevent clogging. Once clogged, immediately use a large amount of DEPC-treated water to extensively rinse and air the system until the water flow smoothly.
Problem 3
No peaks or small peaks are detected and recorded by the fractionation system.
Potential solution
Increase the number of cells used. The RNA amount of each fraction yielded by too few cells may be under the threshhold of UV detector.
The RNA might be degraded due to RNase activity in reagents and environment. The dissolved cycloheximide solutions and buffers can be sterilized by filtering through 0.22 μm filters before adding RNase and protease inhibitors. Change the disposable latex gloves frequently, and apply RNase ZAPTM on it and other contacts. The material and equipment that make close contact with the sucrose gradient, such as the rate zonal cap for sucrose gradient making and the piston tip for collection, could also be cleaned by RNase ZAPTM followed by extensive washes with DEPC-treated water.
The sucrose may be disturbed. Remake the sucrose buffer and be careful not to make bubbles when layering the gradient. Keep steady hand when handling it and make moves with no rushes.
Problem 4
Non-robust or flat polysome peaks are observed.
Potential solution
Use fresh cycloheximide solution. It is better to use fresh-made or stored cycloheximide solution under one month in the freezer.
Try to adjust the MgCl2 concentration in the lysis and sucrose buffer. Mg2+ is important for complex formation and prevents polysomes from dissociating into subunits (Klein et al., 2004). Optimal Mg2+ concentration may be beneficial for obtaining robust polysome peaks.
Adjust the cell numbers cultured in the flask or dishes to avoid over confluency so that cells are in a proliferative state.
Harvest cells as quickly as possible and cool them immediately on ice. Shorten the time interval between the completion of cell lysis and loading onto the sucrose cushion.
Problem 5
Low RNA yield from the extraction of each fraction.
Potential solution
Increase the number of cells used. The amount of RNA retrieved from some fractions might be relatively low, so a large number of cells are needed to obtain sufficient RNA.
Use ethanol precipitation instead of isopropanol to increase the amount of RNA recovered. Additionally, precipitate the RNA in -80°C overnight (14–16 h) to improve RNA yield.
Pool the corresponding fractions from parallel samples together to increase the final RNA amount. If doing so, it is important to make the gradients of parallel samples as identical as possible.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yueqin Chen (lsscyq@mail.sysu.edu.cn).
Materials availability
This study did not generate new unique materials.
Acknowledgments
This research was supported by the Natural Science Foundation of Guangdong Province (2021B1515020002 and 2018A030310073), the China Postdoctoral Science Foundation (2021 T140771), and the National Natural Science Foundation of China (81770174 and 31700719).
Author contributions
Conceptualization and Protocol Optimization, C.H. and L.S.; Investigation and Writing, C.H. and L.S.; Review and Editing, Q.P., Y.S., W.W., and Y.C.; Funding Acquisition, Y.S., W.W., and Y.C.; Supervision, Y.C.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Cai Han, Email: hancai@mail2.sysu.edu.cn.
Yueqin Chen, Email: lsscyq@mail.sysu.edu.cn.
Data and code availability
This study did not generate any data or code
References
- Halbeisen R.E., Scherrer T., Gerber A.P. Affinity purification of ribosomes to access the translatome. Methods. 2009;48:306–310. doi: 10.1016/j.ymeth.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Heiman M., Kulicke R., Fenster R.J., Greengard P., Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat. Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- Johnson L.F., Levis R., Abelson H.T., Green H., Penman S. Changes in RNA in relation to growth of the fibroblast. IV. Alterations in theproduction and processing of mRNA and rRNA in resting and growing cells. J. Cell Biol. 1976;71:933–938. doi: 10.1083/jcb.71.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.J., Moore P.B., Steitz T.A. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wang W., Han C., Huang W., Sun Y., Fang K., Zeng Z., Yang Q., Pan Q., Chen T., et al. The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol. Cell. 2021;81:P4493–P4508.E9. doi: 10.1016/j.molcel.2021.08.033. [DOI] [PubMed] [Google Scholar]
- Thermann R., Hentze M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any data or code