Abstract
We report here the potential role of a 4-strain probiotic suspension for use with patients with Parkinson's disease (PD). Stool samples from a group of three patients with diagnosed PD were used to create microbiotas in an in-vitro gut model. The effects of dosing with an oral probiotic suspension (Symprove) on bacterial composition and metabolic activity in the microbiotas was evaluated over 48 h and compared with healthy controls. Additionally, the effect of probiotic dosing on epithelial tight-junction integrity, production of inflammatory markers and wound healing were evaluated in cell culture models. In general, the relative proportions of the main bacterial phyla in the microbiotas of PD patients differed from those of healthy subjects, with levels of Firmicutes raised and levels of Bacteroidetes reduced. Dosing with probiotic resulted in a change in bacterial composition in the microbiotas over a 48 h period. Several other indicators of gut health changed upon dosing with the probiotic; production of short chain fatty acids (SCFAs) and lactate was stimulated, levels of anti-inflammatory cytokines (IL-6, IL-10) increased and levels of pro-inflammatory cytokines and chemokines (MCP-1 and IL-8) decreased. Tight junction integrity was seen to improve with probiotic dosing and wound healing was seen to occur faster than a control. The data suggest that if development and/or progression of PD is influenced by gut microbiota dysbiosis then supplementation of the diet with a properly formulated probiotic may be a useful adjunct to standard treatment in clinic.
Keywords: Probiotics, Gastrointestinal microbiome, Lactobacillus, Parkinson's disease, Epithelial gut wall integrity, Butyrate
Graphical abstract
1. Introduction
Parkinson's disease (PD) affects a significant proportion of the population over 65 years of age (ca. 3.7%,) and is the second most prevalent neurological disorder after Alzheimer's (Postuma and Berg, 2016). The condition is set to double in number by 2030 with a huge projected societal and medical burden (Dorsey et al., 2018). It is a progressive and chronic neurodegenerative disorder that primarily affects motor function but also includes a range of non-motor deficits. There is no cure and a definitive cause is not yet known. Disease progression affects multiple brain systems including but not limited to dopaminergic, noradrenergic, serotoninergic and cholinergic systems (Berg et al., 2014). Motor system impairments lead to a range of debilitating symptoms, including muscle stiffness, tremors, bradykinesia, impaired gait and postural instability. Non-motor symptoms include loss of smell and problems with gastrointestinal (GI), cardiovascular and urogenital systems (Chaudhuri et al., 2006).
The human gut hosts tens of trillions of microorganisms including more than 1000 species of bacteria, can be considered a separate enteric organ (Falony et al., 2016; Hill-Burns et al., 2017; Zhernakova et al., 2016) and is known to be affected by diet and lifestyle (Kau et al., 2011). James Parkinson identified gut problems as a diagnostic symptom of PD in his original classification of the shaking palsy (Hurwitz, 2014). Typically, patients suffer dry mouth and/or gastroparesis prior to the onset of motor symptoms (Cersosimo et al., 2013; Chiang and Lin, 2019; Fasano et al., 2015) and intractable constipation (Kaye et al., 2006) often accompanies or precedes (by many decades in some cases) motor dysfunction of PD (Elfil et al., 2020). Although there is no established microbial signature for constipation, microbiome changes are associated with gut dysbiosis (Carding et al., 2015) and are known to affect the absorption and metabolism of drugs (Hatton et al., 2019). This dysbiosis is thought to lead to a range of metabolic abnormalities including diminished production of neuroprotective factors, increased inflammation and neurotoxicity and adverse effects on the immune response to neuronal proteins (Elfil et al., 2020). PD is often characterised by a leaky gut denoting increased intestinal permeability, immune dysfunction (Bedarf et al., 2017; Dutta et al., 2019) and alpha synuclein (⍺Syn) accumulation (Perez-Pardoa et al., 2017). The latter are aggregates of insoluble fibrils found in several neuropathological conditions including PD. Because PD patients often have a degree of gut inflammation, faecal calprotectin is frequently raised (Weis et al., 2019) and has been suggested as an early biomarker for PD risk (Mulak et al., 2019). The GI abnormalities (particularly permeability issues and the finding of intestinal inflammation) found in PD are common to many other diseases, but their importance is that these changes may reflect pathophysiological processes that aggravate certain symptoms of PD (Tucker et al., 2020a, Tucker et al., 2020b).
The gut-brain axis has long been established as a pathway for reciprocal communication between the enteric system and the central nervous system and there is growing interest in its role in the pathogenesis of PD (Santos et al., 2019). One hypothesis is that, in susceptible individuals, ingested pathogens can trigger misfolding and aggregation of ⍺Syn deposits in the enteric nervous system which can then spread from the gut to the brain via the gut-brain axis (Braak et al., 2006; Houser and Tansey, 2017). In support of this theory, deposits of ⍺Syn were more frequently found in the gastrointestinal tract of PD patients compared with healthy controls, even in the prodromal phase of the condition (Bu et al., 2019), and several preclinical studies have supported the role of the vagus nerve in the bidirectional transmission of the synucleinopathy from the brain to the gut and vice versa (Breen et al., 2019).
Numerous studies (Bedarf et al., 2017; Bullich et al., 2019; Lin et al., 2019; Qian et al., 2018; Sampson, 2020; Unger et al., 2016) have categorised the bacterial composition of the gut microbiotas in PD patients compared with healthy subjects. Although these studies have not identified a particular microbiota composition that always accompanies PD, overall there is evidence that proinflammatory dysbiosis is usually present (Keshavarzian et al., 2015). In experimental animal work, Sampson et al. (2016) highlighted that signals from the gut microbiota are required for the development of neuroinflammation, gastrointestinal dysfunction and ⍺Syn-dependent motor deficits in mice overexpressing ⍺Syn. Choi et al. (2018) suggested that oral administration of Proteus mirabilis, whose abundance was found to be increased in PD mouse models, can trigger ⍺Syn aggregation both in the brain and in the colon and induce motor dysfunction, neurodegeneration as well as inflammation in the substantia nigra and striatum of mice models of PD. In addition, mouse-to-mouse transplants of healthy microbiomes protected mice from dopaminergic neuronal death, even following MPTP (a neurotoxin that induces Parkinsonian symptoms) (Sun et al., 2018). When MPTP was combined with exposure to P. mirabilis there was increased loss of dopaminergic neurons and worse motor impairment compared with MPTP treatment alone (Choi et al., 2018). Taken together, these results show that the gut microbiota could modulate the phenotypes associated with standard models of PD pathology.
In previous studies on probiotic use in human health and disease, we have shown that dosing with a 4-strain live probiotic suspension (Symprove™) had the potential to change the balance of species in the gut microbiotas of healthy human donors (Moens et al., 2019) and patients with ulcerative colitis (Ghyselinck et al., 2020) in an in-vitro model. Symprove showed excellent tolerance to gastric acid during in-vitro testing (Fredua-Agyeman and Gaisford, 2015), partly explaining its effectiveness in ameliorating the clinical symptom severity scores of a number of gut conditions in clinical studies. For instance, Symprove reduced clinical symptom severity scores in IBS (Sisson et al., 2014) and reduced abdominal pain scores and significantly reduced constipation, diarrhoea and mucorrhoea in diverticular disease (Kvasnovsky et al., 2017). Symprove has also shown anti-pathogenic activity against various common gut pathogens, including Clostridium difficile (Fredua-Agyeman et al., 2017), Escherichia coli, methicillin-resistant Staphylococcus aureus (MRSA) and Shigella sonnei (Dodoo et al., 2019).
Unsurprisingly, probiotic formulations have been mooted as a dietary supplement that might improve intestinal health and disease (Sanders et al., 2019) and alleviate PD symptoms (Gazerani, 2019), although reports of positive clinical effects with probiotic supplementation on PD progression are lacking. Confidence in the use of probiotic supplements in the management of PD will increase if there is evidence that probiotic bacteria can integrate and assimilate into an existing gut microbiota, improving overall gut health. Thus, the primary research objectives of this work were (i) to determine whether addition of the probiotic bacteria in Symprove could influence bacterial composition in the microbiotas of patients with PD and (ii) to determine whether other indicators of gut health in PD patients could be improved with probiotic supplementation. The results allowed a mechanism by which probiotic bacteria exert a positive effect to be established. The experimental difficulties of making such measurements in-vivo meant we employed an in-vitro dynamic, multi-compartment gastrointestinal model (the simulator of the human intestinal microbial ecosystem, equipped with a mucosal compartment, M-SHIME®).
2. Materials and methods
2.1. Symprove
Symprove, an aqueous suspension containing Lactobacillus acidophilus NCIMB 30175, Lactobacillus plantarum NCIMB 30173, Lactobacillus rhamnosus NCIMB 30174 and Enterococcus faecium NCIMB 30176, was obtained from Symprove Ltd. and used as received.
2.2. Donors
Faecal samples were obtained from six donors; three healthy control subjects and three diagnosed with PD. Each of the PD donors was on an established drug treatment regimen, details of which are shown in Table 1, and none had been on antibiotics in the three months prior to donation. Samples were collected according to the ethical approval of the University Hospital Ghent (reference number: B670201836585).
Table 1.
Details of the drug treatment regimens of the three PD donors (all doses per day).
| Donor number | Drug treatment regimen |
|---|---|
| 1 | Artane |
| 2 | Azilect 1 mg, Requip 8 mg, Cymbalta, Silodyx, Melatonine 5 mg, Rilatine 2 × 10 mg |
| 3 | Azilect 1 mg, Requip 8 mg, Efexor 75 mg |
2.3. M-SHIME® testing
The M-SHIME® system was configured for short-term batch experiments (full details of the experimental arrangement of the M-SHIME® system are described elsewhere; Van den Abbeele et al., 2010; Van den Abbeele et al., 2012; Van den Abbeele et al., 2013). Briefly, this involved fermentation of faecal samples in single vessels in the presence or absence of the probiotic bacteria in Symprove. A sugar-depleted basal nutritional medium (56 mL) buffered at pH 6.5, containing the nutrients present in the colon (5.9 g/L K2HPO4, 18.3 g/L KH2PO4, 2.3 g/L NaHCO3, 2.3 g/L yeast extract, 2.3 g/L peptone, 0.6 g/L cysteine and 2.3 mL/L Tween 80) was co-administered with Symprove (7 mL) at the start of fermentation. A corresponding series of blank experiments were conducted by adding the basal nutritional medium to distilled water (7 mL, instead of Symprove). Subtraction of the blank data from the Symprove data allowed the effects of the probiotic formulation to be determined. A 7.5% (w/v) faecal suspension was prepared from each PD donor in anaerobic phosphate buffer (K2HPO4 8.8 g/L; KH2PO4 6.8 g/L; sodium thioglycolate 0.1 g/L; sodium dithionite 0.015 g/L) and was inoculated (7 mL) into the reactors, bringing the total volume to 70 mL. Finally, five mucin-covered microcosms were added to all colonic vessels, enabling maintenance of not only a luminal microbiota but also a specific mucosal microbiota in the colonic vessels. Each incubation was performed in triplicate, resulting in 36 independent incubations. The headspace of the vessels was flushed with nitrogen to ensure anaerobic conditions and incubations were performed for 48 h at 37 °C under continuous shaking.
2.4. Measurement of microbial metabolites and protein metabolism
SCFA levels, including acetate, propionate, butyrate and the branched-chain fatty acids (bCFA) isobutyrate, isovalerate and isocaproate, were monitored with gas chromatography (GC) coupled with flame ionization detection (FID). After addition of 2-methyl hexanoic acid as an internal standard, a sample (2.0 mL) was extracted with diethyl ether. The extracts were analyzed using a GC-2014 gas chromatograph (Shimadzu, ‘s-Hertogenbosch, the Netherlands), equipped with a GC SGE capillary column, 30 m × 0.32 mm ID-BP21x 0.25 μm (Achrom, Machelen, Belgium), a flame ionization detector and a split injector. The injection volume was 1 μL and the column temperature profile was set from 110 to 160 °C, rising at 6 °C min−1. The carrier gas was nitrogen and the temperatures of the injector and detector were both 200 °C.
Lactate quantification was performed using a commercially available enzymatic assay kit (R-Biopharm,Darmstadt, Germany) according to the manufacturer's instructions. Ammonium analysis was performed as previously described (Van de Wiele et al., 2004). Briefly, the ammonium in the liquid samples was quantified by initially performing a steam distillation. Subsequently, the ammonium in the distillate was determined titrimetrically with HCl.
2.5. Microbial community analysis
Illumina sequencing was performed at the beginning and end (48 h) of fermentation. Because the Illumina sequencing method is polymerase-chain reaction (PCR) based, microbial gene sequences are amplified until saturation is reached, meaning results at the phylogenetic level (phylum, family, genus and operational taxonomic unit, OTU) are expressed as proportional values. Two primers, 341F (5’-CCT ACG GGN GGC WGC AG −3′) and 785Rmod (5’-GAC TAC HVG GGT ATC TAA KCC-3′) (Klindworth et al., 2013), were used that spanned the hypervariable regions V3 and V4 of the 16 s rDNA. PCR products were run along DNA extracts on a 2% agarose gel for 30 min at 100 V. A sample (10 μL) of the original genomic DNA extract was sent for library preparation and sequencing on an Illumina Miseq platform with v3 chemistry with the primers mentioned above (LGC genomics GmbH, Germany).
2.6. Caco-2/THP1-blue™ co-culture model
Caco-2 cells (HTB-37, American Type Culture Collection) were seeded in semi-permeable inserts (24-well, 0.4 μm pore size) at a density of 1 × 105 cells/insert. The cells were cultured for 14 days, with three changes of medium per week, until a functional monolayer with a transepithelial electrical resistance (TEER) of more than 300 Ω cm2 was obtained. TEER was measured with a Millicell ERS-2 epithelial volt-ohm meter (Millipore). Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) containing glucose (25 mM) and glutamine (4 mM), supplemented with HEPES (10 mM) and heat-inactivated foetal bovine serum (HI-FBS, 20% v/v).
THP1-Blue™ cells (InvivoGen) were seeded in 24-well plates at a density of 5 × 105 cells/well and treated with phorbol 12-myristate 13-acetate (PMA) for 48 h. Roswell Park Memorial Institute (RPMI) 1640 medium containing glucose (11 mM) and glutamine (2 mM), supplemented with HEPES (10 mM), sodium pyruvate (1 mM) and HI-FBS (10% v/v), was used to maintain the cells.
To set up the co-culture, Caco-2 bearing inserts were placed on top of the PMA-differentiated THP1-blue™ cells. Sterile-filtered (0.22 μm) colonic SHIME media (diluted 1:5 v/v in Caco-2 complete medium) was placed in the apical (Caco-2) compartment. The basolateral (THP1-blue™) compartment was filled with Caco-2 complete medium. Sodium butyrate (12 mM, Sigma-Aldrich) was added to the apical compartment as a positive control. Cells were treated for 24 h, after which the TEER was measured. The basolateral compartment was then emptied, and cells were stimulated on the basolateral side with either (i) Caco-2 complete medium containing ultrapure lipopolysaccharide (LPS, E. coli K12, InvivoGen), (ii) Caco-2 complete medium with LPS and hydrocortisone (HC, Sigma-Aldrich) or (iii) Caco-2 complete medium without LPS. After LPS stimulation (6 h) the basolateral supernatants were collected for cytokine measurement (human TNF-α, IL-6, IL-8, IL-10, and CXCL10) by Luminex® multiplex (Affymetrix-eBioscience). The THP1-Blue™ cells contain a stably transfected reporter construct allowing the expression of a secreted alkaline phosphatase (SEAP) under the control of an NF-κB inducible promoter. Hence, upon LPS stimulation, NF-κB is activated, leading to the secretion of SEAP in the basolateral medium. Then, SEAP activity was measured in the basolateral medium using the QUANTI-Blue™ reagent (Invivogen). All measurements were performed in triplicate and cells were incubated at 37 °C in a humidified atmosphere of air/CO2 (95,5 v/v).
2.7. Scratch wound healing assay
T84 cells (Sigma-Aldrich) were seeded in 24-well plates and cultured for 7 days, with three changes of medium per week, until a complete, confluent, cell monolayer was formed. Cells were maintained in a basal medium (DMEM/F-12) containing l-glutamine and HEPES supplemented with antibiotic/antimycotic (Gibco, Life Technologies) and 5% HI-FBS. Cells were incubated at 37 °C in a humidified atmosphere of air/CO2 (95:5 v/v).
After 7 days a scratch was created in the monolayer, followed by treatment with 1/10 diluted colonic suspensions in serum-free T84 culture medium. Images were captured with a Cytation 5 Cell Imaging Multi-mode Reader at the initial time point (0 h) and after incubation (24 h). Images were compared in order to quantify the migration rate of the cells while the wound area was measured using ImageJ®. Serum-free culture medium and 5 mM NaB (Sigma-Aldrich) were used as negative and positive controls respectively. All measurements were conducted in triplicate.
2.8. Statistical tests for the cell assays
To evaluate differences in TEER and immune markers, colonic batch treatment samples were compared with their respective control samples using a two-way ANOVA with Sidak's multiple comparisons test. For the pure product, statistical significance for the TEER was calculated by using one-way ANOVA with Dunnett's multiple comparisons test and the different concentrations of Symprove were compared with CM. For the wound healing assay, statistical significance between CM and sodium butyrate was calculated with an unpaired, two-tailed t-test with Welch's correction. To evaluate differences in wound area, colonic batch treatment samples were compared with their respective control samples using a two-way ANOVA with Sidak's multiple comparisons test. For pure product, statistical significance was calculated by using one-way ANOVA with Dunnett's multiple comparisons test against CM. Statistically significant differences are represented by (*). (*), (**), (***) and (****) represent p < 0.05, p < 0.01, p < 0.001 and p < 0.0001, respectively.
All samples were taken as biological replicates in the cell assays (n = 3/donor) and statistics were performed on the average of the replicates.
3. Results and discussion
Table 2, Table 3 show the predominant phyla in the microbiotas of the healthy subjects and the PD patients before dosing with Symprove. The predominant phyla were Firmicutes, Bacteroidetes and Actinobacteria, although the PD patients showed lower levels of Firmicutes (70.3 ± 3.5%) and higher levels of Bacteroidetes (19.0 ± 15.1%) than healthy subjects (81.5 ± 10.8% and 10.5 ± 4.4% respectively). Table 4, Table 5 show the predominant phyla in the microbiotas of the healthy subjects and the PD patients after dosing with Symprove (familial detail of OTUs within phyla are given in Tables S1-S4). Table 6, Table 7 show the composition of the gut microbiota at bacterial family level.
Table 2.
Composition (%) at the phyla level of the microbiotas of the three healthy donors before dosing with probiotic.
| Phyla | Donor 1 | Donor 2 | Donor 3 |
|---|---|---|---|
| Firmicutes | 86.7 | 69.1 | 88.7 |
| Bacteroidetes | 8.9 | 15.4 | 7.1 |
| Actinobacteria | 2.5 | 15.2 | 3.9 |
| Proteobacteria | 1.1 | 0.3 | 0.3 |
| Verrucomicrobia | 0.9 | 0.0 | 0.0 |
Table 3.
Composition at the phyla level of the microbiotas of the three PD donors before dosing with probiotic.
| Phyla | Donor 1 | Donor 2 | Donor 3 |
|---|---|---|---|
| Firmicutes | 70.9 | 73.4 | 66.5 |
| Bacteroidetes | 13.2 | 22.2 | 21.7 |
| Actinobacteria | 14.1 | 3.0 | 6.1 |
| Proteobacteria | 0.9 | 0.8 | 5.2 |
| Verrucomicrobia | 0.8 | 0.3 | 0.2 |
Table 4.
Composition (%) in the lumen and in the mucosal layer at the phyla level of the microbiotas of the three healthy donors after 48 h in the M-SHIME® system (B, blank; P, dosing with probiotic).
| Phyla | Donor 1 |
Lumen Donor 2 |
Donor 3 |
Donor 1 |
Mucus Donor 2 |
Donor 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | P | B | P | B | P | B | P | B | P | B | P | |
| Firmicutes | 12 | 14 | 14 | 24 | 14 | 19 | 39 | 49 | 51 | 80 | 32 | 67 |
| Proteobacteria | 60 | 65 | 40 | 18 | 52 | 33 | 3 | 2 | 13 | 2 | 9 | 3 |
| Bacteroidetes | 22 | 18 | 45 | 42 | 32 | 41 | 56 | 47 | 35 | 18 | 55 | 21 |
| Verrucomicrobia | 6 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| Actinobacteria | 0 | 1 | 2 | 16 | 1 | 6 | 1 | 2 | 1 | 1 | 4 | 9 |
Table 5.
Composition (%) in the lumen and mucus at the phyla level of the microbiotas of the three PD donors after 48 h in the M-SHIME® system (B, blank; P, dosing with probiotic).
| Phyla | Lumen |
Mucus |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor 1 |
Donor 2 |
Donor 3 |
Donor 1 |
Donor 2 |
Donor 3 |
|||||||
| B | P | B | P | B | P | B | P | B | P | B | P | |
| Firmicutes | 43 | 47 | 19 | 46 | 25 | 42 | 30 | 41 | 41 | 50 | 31 | 30 |
| Proteobacteria | 26 | 26 | 29 | 25 | 36 | 34 | 13 | 13 | 20 | 11 | 10 | 21 |
| Bacteroidetes | 20 | 17 | 25 | 27 | 38 | 20 | 40 | 22 | 33 | 34 | 56 | 37 |
| Verrucomicrobia | 8 | 4 | 26 | 1 | 0 | 0 | 9 | 4 | 4 | 0 | 0 | 0 |
| Actinobacteria | 1 | 6 | 0 | 0 | 0 | 4 | 8 | 21 | 1 | 5 | 2 | 12 |
Table 6.
Composition (%) in the original inocula, the lumen after 48 h and the mucosal layer after 48 h at the family level of the microbiotas of the three healthy donors in the M-SHIME® system (B, blank; P, treatment with probiotic).
| Phylum | Family | Donor 1 |
Donor 2 |
Donor 3 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inoculum | Lumen |
Mucus |
Inoculum | Lumen |
Mucus |
Inoculum | Lumen |
Mucus |
||||||||
| B | P | B | P | B | P | B | P | B | P | B | P | |||||
| Actinobacteria | Bifidobacteriaceae | 1.0 | 0.1 | 0.5 | 0.3 | 0.8 | 14.3 | 1.4 | 15.5 | 0.6 | 0.3 | 3.7 | 0.7 | 5.6 | 1.7 | 2.5 |
| Coriobacteriaceae | 1.3 | 0.0 | 0.0 | 0.4 | 0.8 | 0.9 | 0.0 | 0.1 | 0.2 | 0.2 | 0.1 | 0.0 | 0.0 | 2.2 | 6.5 | |
| Eggerthellaceae | 0.2 | 0.2 | 0.2 | 0.0 | 0.0 | 0.0 | 0.1 | 0.2 | 0.0 | 0.0 | 0.2 | 0.5 | 0.6 | 0.1 | 0.1 | |
| Bacteroidetes | Bacteroidaceae | 5.8 | 20.2 | 17.2 | 55.8 | 46.6 | 15.1 | 42.5 | 36.7 | 33.9 | 17.8 | 6.3 | 26.2 | 23.6 | 45.1 | 14.6 |
| Bacteroidales_unclassified | 0.1 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 | 0.2 | 0.0 | 0.0 | 0.0 | 0.1 | 0.9 | 0.0 | 0.0 | |
| Muribaculaceae | 0.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Prevotellaceae | 1.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Rikenellaceae | 0.3 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.2 | 0.3 | 0.4 | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | |
| Tannerellaceae | 0.1 | 1.3 | 1.1 | 0.3 | 0.5 | 0.2 | 1.8 | 4.3 | 0.9 | 0.4 | 0.5 | 5.8 | 16.5 | 9.8 | 6.1 | |
| Firmicutes | Acidaminococcaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 | 0.7 | 1.0 | 1.4 | 0.8 |
| Anaerovoracaceae | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Butyricicoccaceae | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.1 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.1 | |
| Christensenellaceae | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Clostridia_UCG-014_fa | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Clostridiaceae | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 1.6 | 1.1 | 21.8 | 3.6 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | |
| Enterococcaceae | 0.0 | 1.6 | 0.1 | 0.5 | 0.1 | 0.0 | 0.6 | 0.0 | 0.2 | 0.0 | 0.0 | 0.9 | 0.0 | 2.7 | 0.2 | |
| Erysipelatoclostridiaceae | 0.6 | 0.0 | 0.0 | 0.1 | 0.1 | 0.7 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.2 | 0.0 | 1.1 | 1.4 | |
| Erysipelotrichaceae | 0.1 | 0.0 | 0.0 | 0.3 | 0.2 | 1.6 | 0.0 | 0.0 | 0.1 | 0.1 | 0.4 | 0.0 | 0.0 | 1.7 | 2.8 | |
| Lachnospiraceae | 54.4 | 9.7 | 8.9 | 35.7 | 47.3 | 43.5 | 11.1 | 13.2 | 26.9 | 75.3 | 81.3 | 12.5 | 13.4 | 24.1 | 58.7 | |
| Lactobacillaceae | 0.1 | 0.0 | 3.9 | 0.0 | 0.4 | 0.0 | 0.0 | 7.5 | 0.0 | 0.1 | 0.0 | 0.0 | 4.7 | 0.0 | 0.8 | |
| Monoglobaceae | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Oscillospiraceae | 2.0 | 0.4 | 0.3 | 1.3 | 0.2 | 0.3 | 0.1 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.2 | |
| Oscillospirales_fa | 0.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Peptococcaceae | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Peptostreptococcaceae | 6.7 | 0.0 | 0.0 | 0.0 | 0.1 | 4.4 | 0.1 | 0.0 | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.4 | |
| Ruminococcaceae | 13.8 | 0.1 | 0.1 | 0.9 | 0.2 | 15.5 | 0.0 | 0.0 | 0.7 | 0.2 | 5.7 | 0.0 | 0.0 | 0.5 | 1.3 | |
| Selenomonadaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.2 | 0.1 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Streptococcaceae | 0.3 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.2 | 0.5 | |
| Veillonellaceae | 5.1 | 0.4 | 0.3 | 0.2 | 0.2 | 0.1 | 0.3 | 0.5 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Proteobacteria | Desulfovibrionaceae | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.2 | 0.1 | 0.1 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 |
| Enterobacterales_unclassified | 0.0 | 0.1 | 0.2 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | 0.0 | |
| Enterobacteriaceae | 0.5 | 60.0 | 65.0 | 3.4 | 1.5 | 0.0 | 39.5 | 18.1 | 13.4 | 1.6 | 0.0 | 52.1 | 33.2 | 8.7 | 2.9 | |
| Sutterellaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Verrucomicrobia | Akkermansiaceae | 0.9 | 5.5 | 1.8 | 0.8 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Unclassified | Bacteria_unclassified | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Table 7.
Composition (%) in the original inocula, the lumen after 48 h and the mucosal layer after 48 h at the family level of the microbiotas of the three PD donors in the M-SHIME® system (B, blank; P, dosing with probiotic).
| Phylum | Family | Donor 1 |
Donor 2 |
Donor 3 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inoculum | Lumen |
Mucus |
Inoculum | Lumen |
Mucus |
Inoculum | Lumen |
Mucus |
||||||||
| B | P | B | P | B | P | B | P | B | P | B | P | |||||
| Actinobacteria | Bifidobacteriaceae | 5.8 | 0.9 | 5.7 | 8.2 | 20.8 | 0.3 | 0.1 | 0.3 | 1.4 | 4.3 | 1.5 | 0.1 | 3.5 | 2.1 | 12.3 |
| Coriobacteriaceae | 6.6 | 0.3 | 0.4 | 0.1 | 0.3 | 2.2 | 0.0 | 0.0 | 0.0 | 0.6 | 2.7 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Coriobacteriales Incertae Sedis | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Eggerthellaceae | 1.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.3 | 0.1 | 0.0 | 0.0 | 0.0 | 1.1 | 0.1 | 0.0 | 0.0 | 0.0 | |
| Propionibacteriaceae | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Bacteroidetes | Bacteroidaceae | 1.3 | 15.0 | 13.7 | 27.9 | 14.2 | 6.2 | 20.8 | 22.3 | 24.7 | 25.8 | 9.1 | 34.1 | 17.0 | 55.0 | 35.8 |
| Bacteroidales_unclassified | 11.3 | 0.1 | 0.1 | 0.3 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Bacteroidia_unclassified | 0.0 | 0.2 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Barnesiellaceae | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Marinifilaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0,1 | 0.1 | 0.1 | 0.2 | 0.0 | 0.7 | 0.3 | 0.1 | 0.0 | 0.0 | |
| Muribaculaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 14.4 | 0.0 | 0.0 | 0.0 | 0.0 | 8.6 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Prevotellaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Rikenellaceae | 0.2 | 0.1 | 0.6 | 0.1 | 0.1 | 0.4 | 0.1 | 0.1 | 0.0 | 0.0 | 1.8 | 0.1 | 0.1 | 0.0 | 0.0 | |
| Tannerellaceae | 0.2 | 4.1 | 2.7 | 11.4 | 7.2 | 0.4 | 3.7 | 4.8 | 8.0 | 8.3 | 1.4 | 3.3 | 2.9 | 0.8 | 0.9 | |
| Veillonellaceae | 0.0 | 0.1 | 20.3 | 0.0 | 0.8 | 1.9 | 0.0 | 12.5 | 0.0 | 7.1 | 0.0 | 0.2 | 3.6 | 0.0 | 0.4 | |
| Firmicutes | Acidaminococcaceae | 0.4 | 0.6 | 0.5 | 0.3 | 0.2 | 0.2 | 2.4 | 1.2 | 1.6 | 0.9 | 3.0 | 5.3 | 2.4 | 3.6 | 1.5 |
| Christensenellaceae | 3.6 | 0.0 | 0.1 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.1 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Clostridiaceae 1 | 1.0 | 7.8 | 0.9 | 13.8 | 15.1 | 1.0 | 0.5 | 0.1 | 30.0 | 4.2 | 0.3 | 2.2 | 0.3 | 21.5 | 9.3 | |
| Clostridiales_unclassified | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.9 | 1.0 | 0.4 | 0.2 | |
| Clostridiales vadinBB60 group | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Enterococcaceae | 0.0 | 26.0 | 3.3 | 10.9 | 7.3 | 0.0 | 3.9 | 1.8 | 5.9 | 1.9 | 0.2 | 4.6 | 7.6 | 0.9 | 2.1 | |
| Erysipelotrichaceae | 4.0 | 0.1 | 0.9 | 0.2 | 5.7 | 1.4 | 0.0 | 0.0 | 0.0 | 0.4 | 1.6 | 0.0 | 0.1 | 0.0 | 0.3 | |
| Eubacteriaceae | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 | 2.8 | 0.3 | 0.5 | 0.1 | 4.4 | 6.9 | 0.4 | 0.5 | |
| Family_XI | 0.0 | 0.1 | 0.4 | 0.0 | 0.0 | 0.2 | 0.2 | 0.1 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | |
| Family_XIII | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.1 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Lachnospiraceae | 39.1 | 1.8 | 2.4 | 3.6 | 9.1 | 27.1 | 4.5 | 8.8 | 1.0 | 28.2 | 32.3 | 6.4 | 8.8 | 4.1 | 14.1 | |
| Lactobacillaceae | 0.0 | 0.0 | 7.9 | 0.0 | 0.6 | 0.0 | 0.0 | 7.7 | 0.0 | 0.9 | 0.1 | 0.0 | 10.9 | 0.0 | 0.7 | |
| Peptostreptococcaceae | 2.0 | 1.5 | 0.8 | 1.0 | 0.4 | 9.5 | 0.2 | 0.1 | 0.3 | 0.2 | 1.5 | 1.1 | 0.1 | 0.3 | 0.1 | |
| Ruminococcaceae | 20.0 | 0.4 | 0.7 | 0.1 | 0.3 | 28.6 | 0.1 | 0.1 | 0.0 | 0.1 | 23.9 | 0.2 | 0.2 | 0.0 | 0.2 | |
| Streptococcaceae | 0.4 | 4.6 | 8.5 | 0.3 | 1.2 | 0.0 | 6.5 | 10.8 | 2.3 | 5.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | |
| Fusobacteria | Fusobacteriaceae | 0.0 | 1.3 | 0.2 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Proteobacteria | Burkholderiaceae | 0.5 | 0.4 | 0.4 | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 3.8 | 0.3 | 0.0 | 0.1 | 0.0 |
| Desulfovibrionaceae | 0.3 | 0.1 | 0.0 | 0.0 | 0.0 | 0.4 | 0.1 | 0.2 | 0.0 | 0.0 | 1.1 | 0.1 | 0.1 | 0.0 | 0.0 | |
| Enterobacteriaceae | 0.0 | 25.8 | 25.2 | 12.5 | 12.7 | 0.0 | 28.9 | 25.3 | 20.3 | 11.0 | 0.2 | 35.1 | 34.2 | 10.3 | 21.2 | |
| Synergistetes | Synergistaceae | 0.0 | 0.7 | 0.1 | 0.0 | 0.0 | 0.1 | 1.0 | 0.2 | 0.2 | 0.1 | 0.1 | 0.9 | 0.0 | 0.0 | 0.0 |
| Tenericutes | Mollicutes_RF39_fa | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 |
| Uncultured | uncultured | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.8 | 0.0 | 0.0 | 0.0 | 0.0 |
| Verrucomicrobia | Akkermansiaceae | 0.4 | 7.8 | 3.9 | 8.9 | 3.6 | 0.1 | 25.8 | 0.6 | 3.7 | 0.2 | 0.2 | 0.3 | 0.1 | 0.0 | 0.0 |
| Puniceicoccaceae | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
Although the data cover only a 48 h period, some significant changes in bacterial composition are apparent. Considering the healthy subjects first, levels of Firmicutes, Actinobacteria and Bacteroidetes were raised. Two of the bacteria in Symprove (L. plantarum and L. rhamnosus) were significantly enriched in the luminal compartments of all three donors and in the mucosal compartments of two of the donors, contributing to the raised level of Firmicutes. Other contributors to the increase in Firmicutes included Lachnospiraceae, Ruminococcaceae, Selenomonadaceae, Streptococcaceae and Veillonellaceae. The proportion of Actinobacteria increased, principally by enrichment of Bifidobacteriaceae in the lumen of all donors. Coriobacteriaceae were enriched in the luminal compartments of two donors while Eggerthellaceae were enriched in the other donor. The proportion of Bacteroidetes was also raised, enriched by B. uniformis and Rikenellaceae in the lumen of donor 2. Tannerellaceae (principally Parabacteroides distasonis) was enriched in the luminal compartments of donors 2 and 3 and in the mucus of donor 1. The proportion of Proteobacteria reduced, principally through a lower abundance of Escherichia coli across the three donors (although it is noted that there may not have been an absolute reduction in Proteobacteria numbers, because the reduction in proportion could be caused by outgrowth in the other phyla).
In the PD patients the proportions of Actinobacteria and Firmicutes were raised. Enrichment of Firmicutes in the lumen of all donors and the mucus layer of donors 1 and 2 reflected partly the integration and proliferation of L. plantarum and L. rhamnosus from Symprove, but had contributions from other families, in particular Eubacteriaceae, Lachnospiracaea, Streptococcaceae and Veillonellaceae in the lumen and Erysipelotrichaceae, Lachnospiraceae and Veillonellaceae in the mucus. Bifidobacteriaceae stimulation (in particular Bifidobacterium longum) accounted for the elevation in Actinobacteria levels.
Many studies have shown the gut microbial profiles in those with PD differ from healthy subjects (Bedarf et al., 2017; Lin et al., 2019; Qian et al., 2018; Sampson, 2020; Unger et al., 2016). Species that have been found to be enriched in PD patients include Akkermansia, Alistipes shahii, Bifidobacteriaceae, Bilophila, Butyricimonas, Christensenellaceae, Enterococcus, Firmicutes, Lachnospiraceae, Lactobacillaceae, Mucispirillum, Odoribacter, Pasteurellaceae, Prevotellaceae, Porphyromonas, Tissierellaceae, Veillonella and Verrucomicrobiaceae. Species decreased in PD patients include Clostridium saccharolyticum, Erysipelotrichaceae (Eubacterium biforme), Faecalibacterium, members of the Lachnospiraceae family, including Blautia sp. and Roseburia sp., Lactobacillaceae and Prevotellaceae (Prevotella copri). These findings are not easily interpreted because of variations in methodology, sequencing, data collection, data analyses and variations across individuals in severity of condition, time since onset, medication, gender, ethnicity and geographical location (which is why machine learning may be a promising approach for identifying changes in microbial composition and linking them to disease progression, McCoubrey et al., 2021). It is clear from this study, however, that even a single dose of probiotic was able to effect a change in microbial diversity over 48 h, meaning that if development and/or progression of PD is influenced by gut microbiota dysbiosis then probiotic supplementation of the diet might be beneficial.
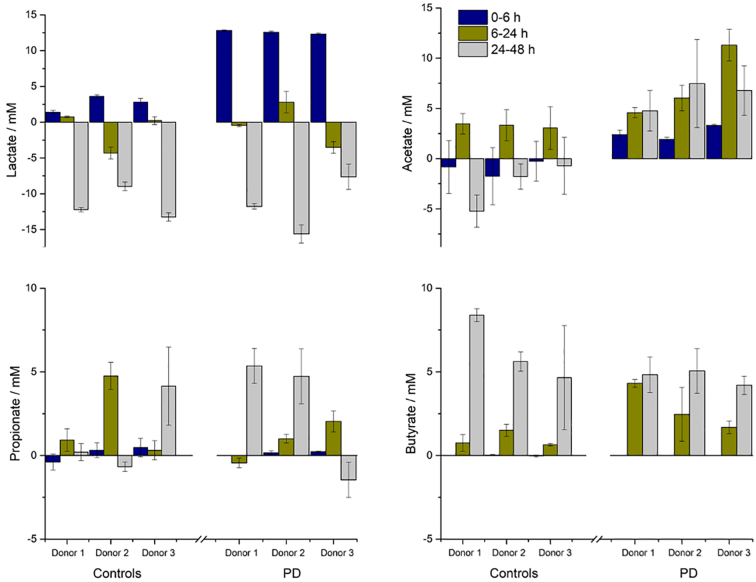
Fig. 1 shows the production and consumption of lactate and SCFA with time after dosing the donor microbiotas with probiotic. Lactate concentrations rose substantially in the first 6 h and then fell during the rest of the test as it was consumed. Acetate concentrations raised after 6 h but thereafter its utilisation in health and PD microbiomes differed; in healthy microbiomes there was considerable utilisation of acetate, while in PD microbiomes it continued to accumulate. Propionate and butyrate concentrations did not start to increase until 24 h. Propionate levels were fairly consistent in all microbiomes. Butyrate levels increased in all microbiomes, but the increase was greater in healthy subjects.
Fig. 1.
Effect of probiotic (Symprove) on the SCFA concentrations in the microbiotas of three PD donors compared with healthy control donors as a function of incubation time. Lactate (top left), acetate (top right), propionate (bottom left) and butyrate (bottom right). Data are shown as mean +/− standard deviation.
The changes in SCFA concentrations result from the integration of the probiotic bacteria into the existing microbiota, followed by stimulation of various commensal bacterial groups through cross-feeding interactions (Moens et al., 2019). Fermentation of carbohydrate by the lactic-acid bacteria in Symprove means lactate is the first compound to increase in concentration; lactate does not, however, accumulate in the system because it is a substrate for propionate-producing species, such as Veillonella and Megasphaera (Reichardt et al., 2014), and butyrate-producing species, such as Anaerostipes caccae and E. hallii (Duncan et al., 2004a; Flint et al., 2015; Louis et al., 2014). Acetate results from saccharolytic fermentation by numerous bacterial groups including Bifidobacteria (De Vuyst et al., 2014), Bacteroidetes (Baxter et al., 2019: Macy et al., 1978) and acetogenic bacteria (Ragsdale and Pierce, 2008) and in human faeces it is the most abundant SCFA (accounting for more than 50% of the total, Louis et al., 2007). Finally, acetate is itself a substrate for many butyrate-producing species (such as Faecalibacterium prausnitzii and Roseburia spp., Duncan et al., 2004b) and is an essential co-substrate that must be consumed to enable butyrate synthesis from lactate or carbohydrate (Duncan et al., 2002).
It is well accepted that production of lactate and short-chain fatty acids (SCFA) by gut bacteria as a result of carbohydrate fermentation is key to good health. Of the SCFAs, acetate, propionate and butyrate are the most important in human health (Tremaroli and Bäckhed, 2012). These compounds are used as energy sources for commensal gut bacteria (acetate and propionate), peripheral tissues (acetate and propionate) and colonocytes (butyrate) (Hamer et al., 2008) but they also impact inflammation, vasodilation, gut motility and wound healing (Bergman, 1990). Lactate, although not an SCFA, is preferred to glucose as an energy source by neurons.
A recent review of the role of diet in the development of PD showed that general characteristics of the microbiota in patients with PD are a reduction in SCFA-producing bacteria and an increase in genes coding for lipopolysaccharides (Jackson et al., 2019). Despite evidence of reduced SCFA concentrations in the faeces of PD patients, Shin et al. (2020) found that acetate concentrations were significantly higher in the blood plasma of PD patients compared with controls after adjusting for covariates. They concluded that their findings indicated leakage of intestinal SCFAs because of subclinical inflammation induced by an altered microbiota in PD (i.e. the increased SCFA concentrations in blood plasma were not a marker of health in PD patients but rather a possible sign of altered metabolism/gut dysbiosis). Unger et al. (2016) quantitatively analyzed SCFA concentrations and microbiota composition in faecal samples of 34 PD patients and 34 age-matched controls. They found a significant absolute and relative reduction in faecal SCFA compared with matched and young healthy controls, consistent with the observed altered gut microbiota composition. Notably, they found a reduction in Lactobacillaceae in the PD group and increased abundance of Enterobacteriaceae.
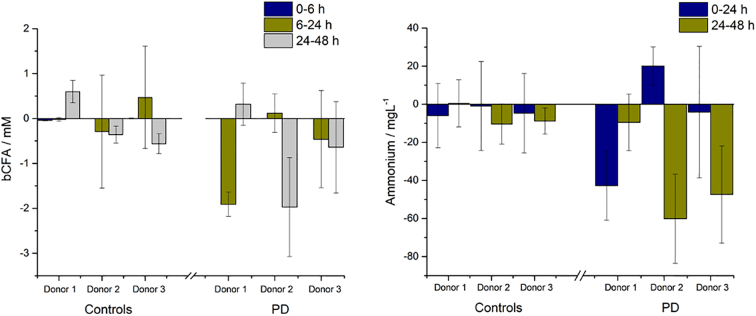
Once dietary carbohydrate has been exhausted, gut microbes will start to metabolise protein, producing various metabolic by-products including ammonium, numerous branched-chain fatty acids (bCFA), principally isobutyrate, isovalerate and isocaproate, and several amines, phenols/indoles and sulphides. Fig. 2 shows the bCFA and ammonium concentrations recorded in the microbiotas following dosing with Symprove; small concentrations of bCFAs and ammonium were seen after 6 h in all samples but were reduced after 48 h. Since these compounds are generally viewed as detrimental to human health (Scott et al., 2013) low levels are desirable and it is seen here that for all donors concentrations reduced after dosing with probiotic. In order to try and visualise the changes in SCFA, bCFA and ammonium levels following dosing with Symprove, principal component analysis plots (Fig. S1) are given in SI for both healthy and PD donors.
Fig. 2.
Effect of probiotic (Symprove) on the protein metabolite concentrations in the microbiotas of three PD donors compared with healthy control donors as a function of incubation time. bCFA (left) and ammonium (right). Data are shown as mean +/− standard deviation.
Table 8 shows the TEER values for Caco-2/THP1 cells treated with colonic media with and without dosing with Symprove for 24 h. For all donors TEER values without probiotic (healthy subjects, 83.3 ± 1.1; PD patients, 57.3 ± 1.7) were lower than the complete medium controls (healthy subjects, 88.8 ± 0.71; PD patients, 68.0 ± 1.8) while following dosing with probiotic TEER values were significantly higher for all donors (healthy subjects, 107.3 ± 3.3; PD patients, 76.3 ± 1.7, p < 0.001). Table 9 shows the TEER values measured when the apical side of the cell model was incubated with various concentrations of probiotic bacteria for 24 h; no increase in TEER was seen in these experiments.
Table 8.
Effect of colonic batch samples from healthy and PD donors on the transepithelial electrical resistance of the Caco-2/THP1 co-cultures. TEER was measured 24 h after dosing with probiotic (P). Samples without added probiotic were used as blanks (B). Δ shows the difference between control and probiotic.
| Healthy |
PD |
|||||
|---|---|---|---|---|---|---|
| B | P | Δ | B | P | Δ | |
| Donor 1 | 86 ± 0.7 | 113 ± 2.4 | 27 ± 2.5 | 61 ± 1 | 79 ± 2 | 18 ± 2.2 |
| Donor 2 | 82 ± 1.4 | 105 ± 4.1 | 23 ± 4.3 | 54 ± 1 | 73 ± 1 | 19 ± 1.4 |
| Donor 3 | 82 ± 1.3 | 104 ± 2.6 | 22 ± 2.9 | 57 ± 3 | 77 ± 2 | 20 ± 3.6 |
Table 9.
Effect of probiotic bacteria on the transepithelial electrical resistance of the Caco-2/THP1 co-cultures. TEER was measured 24 h after dosing.
| Concentration | TEER |
|---|---|
| 105 bacteria | 64 ± 2 |
| 106 bacteria | 62 ± 1 |
| 107 bacteria | 71 ± 2 |
Loss of epithelial tight-junction integrity and an increase in inflammatory markers are well-recognised factors in a number of disease states (Artis, 2008), and PD in particular (Dutta et al., 2019). The clinical issue is whether loss of the gut wall barrier function in PD patients is caused by dysbiosis in their gut microbiota and whether improvement is caused by simple addition of probiotic bacteria or by the influence of probiotic bacteria on the gut microbiota. Tight-junction integrity was quantified here with an in-vitro bilayer cell model comprising epithelial-like cells (Caco-2 cells) and immune cells of human origin (THP1 cells). The fact that following dosing with probiotic TEER values were significantly higher for all donors is not surprising, since many studies have shown positive effects of Lactobacillus and/or butyrate on TEER and wound healing (Gudadappanavar et al., 2017; Ma et al., 2012; Peng et al., 2009; Wang et al., 2012).
The question is whether these positive effects on TEER arise from simple addition of the probiotic bacteria or from stimulation of the microbiome as a whole. No increase in TEER was seen when the apical side of the cell model was incubated with various concentrations of probiotic bacteria for 24 h, indicating that the presence of the probiotic bacteria alone had no effect on gut barrier function. When exposed to sodium butyrate (12 mM) for 24 h an increase in TEER was observed (110.3 ± 1.3, p < 0.001). The implication from these results is thus that it is not the simple addition of probiotic bacteria to the gut that results in an improvement in epithelial tight-junction integrity, but the stimulatory effect of the probiotic bacteria on the commensal gut bacteria, and the resultant increase in SCFA concentration (butyrate in particular).
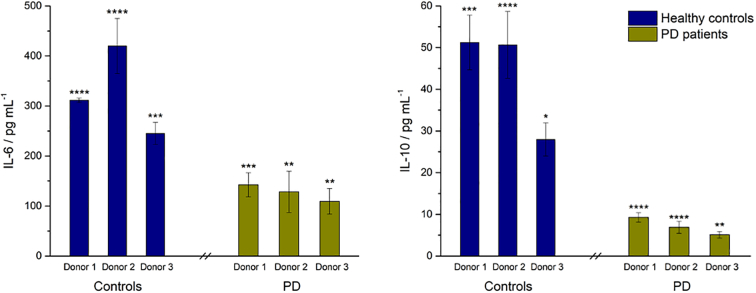
Fig. 3 shows the NF-κB activity, Fig. 4 shows the concentrations of anti-inflammatory cytokines IL-6 and IL-10 and Fig. 5 shows the concentrations of pro-inflammatory cytokines and chemokines TNFα, CXCL 10 and IL-8 following exposure of the cells to colonic media after dosing with Symprove for 24 h. For all donors, treatment with probiotic increased NF-κB activity and concomitant secretion of the anti-inflammatory cytokines compared with the untreated controls and IL-8 levels decreased. CXCL10 levels were increased while TNFα levels were highly dependent on the donor.
Fig. 3.
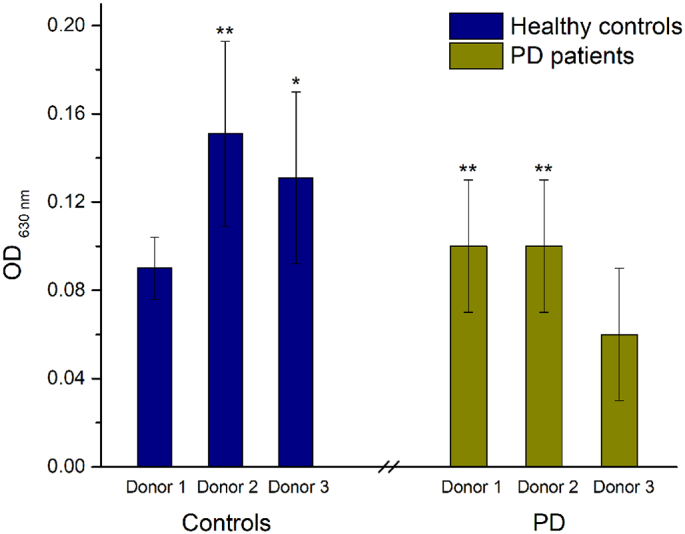
Effect of colonic batch samples from patients with PD, compared with colonic batch samples from healthy controls, on the NF-κB activity of THP1-Blue™ cells. (*) represents statistically significant differences between the control and probiotic-dosed, p < 0.1; (**) p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Effect of colonic batch samples from patients with PD, compared with colonic batch samples from healthy controls, on secretion of anti-inflammatory cytokines IL-6 (left) and IL-10 (right). (*) represents statistically significant differences between the control and probiotic-dosed samples, p < 0.1; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
Fig. 5.
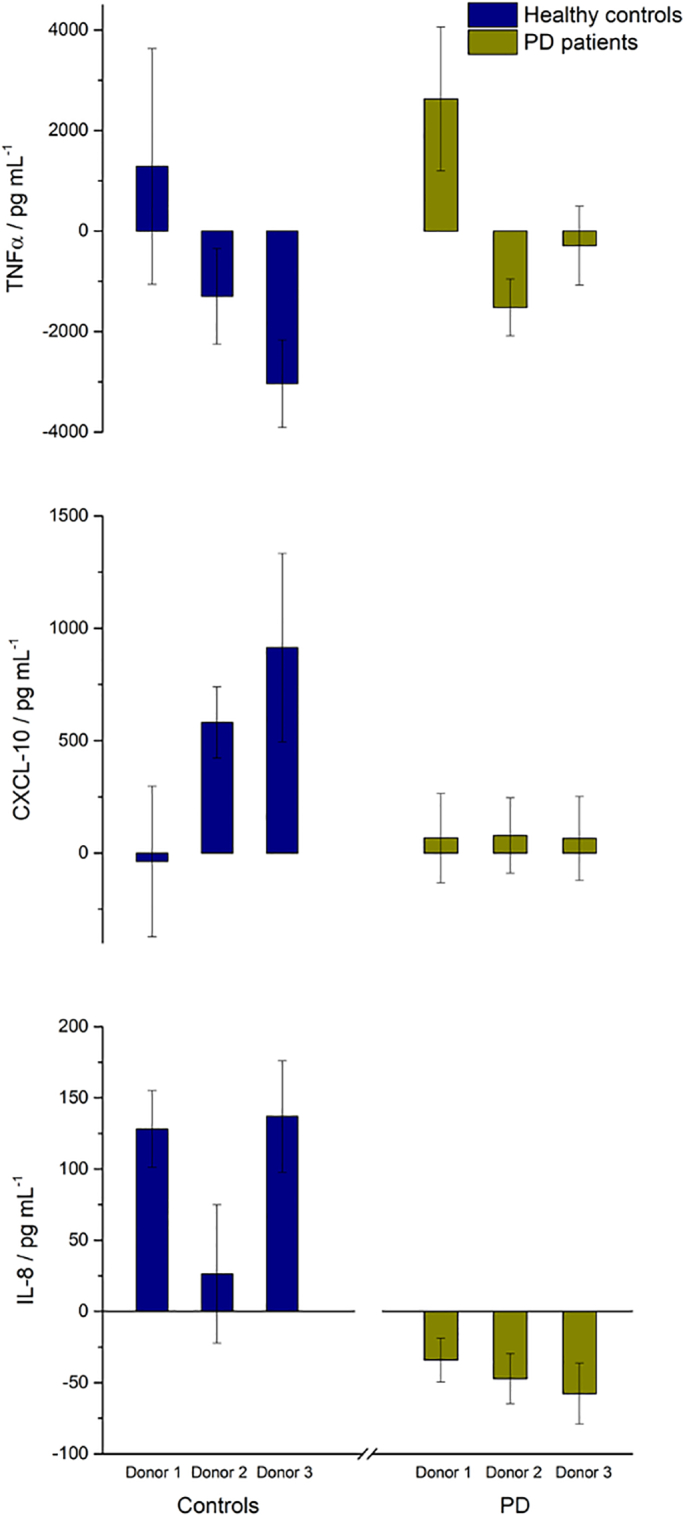
Effect of colonic batch samples from patients with PD, compared with colonic batch samples from healthy controls, on secretion of inflammatory cytokines TNFα (top), CXCL 10 (middle) and IL-8 (bottom).
For all donors, treatment with Symprove increased NF-κB activity and concomitant secretion of the anti-inflammatory cytokines compared with the untreated controls. While IL-10 is considered to be a prototypical anti-inflammatory cytokine, IL-6 can exert both pro- and anti-inflammatory effects (Scheller et al., 2011). Indeed, IL-6 was shown to inhibit the secretion of pro-inflammatory cytokines, such as IL-1β. Furthermore, in the gut, IL-6 demonstrated positive effects on epithelial regeneration and wound healing (Dann et al., 2008). Therefore the increased secretion of IL-6 upon treatment in this ‘leaky gut’ model could be seen as beneficial in terms of protection of the epithelial barrier. Likewise, NF-κB activation has been linked to both pro- and anti-inflammatory functions (Pires et al., 2018), including expression of IL-10 (Lyer and Cheng, 2012). Hence, NF-κB activation under inflammatory conditions leads additionally to the upregulation of anti-inflammatory cytokines, needed to resolve the inflammation and prevention of tissue disruption.
Although NF-κB inhibition has been proposed as a therapeutic intervention in PD to prevent microglia activation and neurodegeneration (Flood et al., 2011), butyrate-induced NF-κB activation in the inflamed gut might restore the balance to a rather anti-inflammatory state by differential modulation of cytokines due to its HDAC inhibitory potential (Vinolo et al., 2011).
Secretion of the pro-inflammatory cytokines and chemokines TNFα, CXCL 10 and IL-8 were modulated following exposure of the cells to colonic media with Symprove for 24 h. For all donors, IL-8 levels decreased upon probiotic dosing although CXCL10 levels were increased. TNFα levels were highly dependent on the donor. For PD microbiomes, upon probiotic dosing TNFα secretion was raised in donor 1, while the levels decreased in donors 2 and 3, but these changes were not statistically significant. Thus, treatment with Symprove demonstrated some positive effects on inflammation by increasing the secretion of anti-inflammatory cytokines, while decreasing some, but not all, pro-inflammatory cytokines and chemokines tested in the co-culture model used. As above, a principal component analysis plot (Fig. S2) is given in SI for both healthy and PD donors, showing the change in TEER and inflammatory markers following dosing with Symprove.
Fig. 6 shows images of the scratch wound as created and after 24 h exposure to complete medium (CM), sodium butyrate and healthy donor microbiotas (with and without probiotic). Fig. 7 shows similar images for PD donors (note that the data for donor 3 were excluded due to toxicity of the sample to the T84 cells). Calculated percentages of the wound closures are given in Table 10. In all cases, dosing with probiotic significantly (p < 0.001) increased wound healing compared with the control.
Fig. 6.
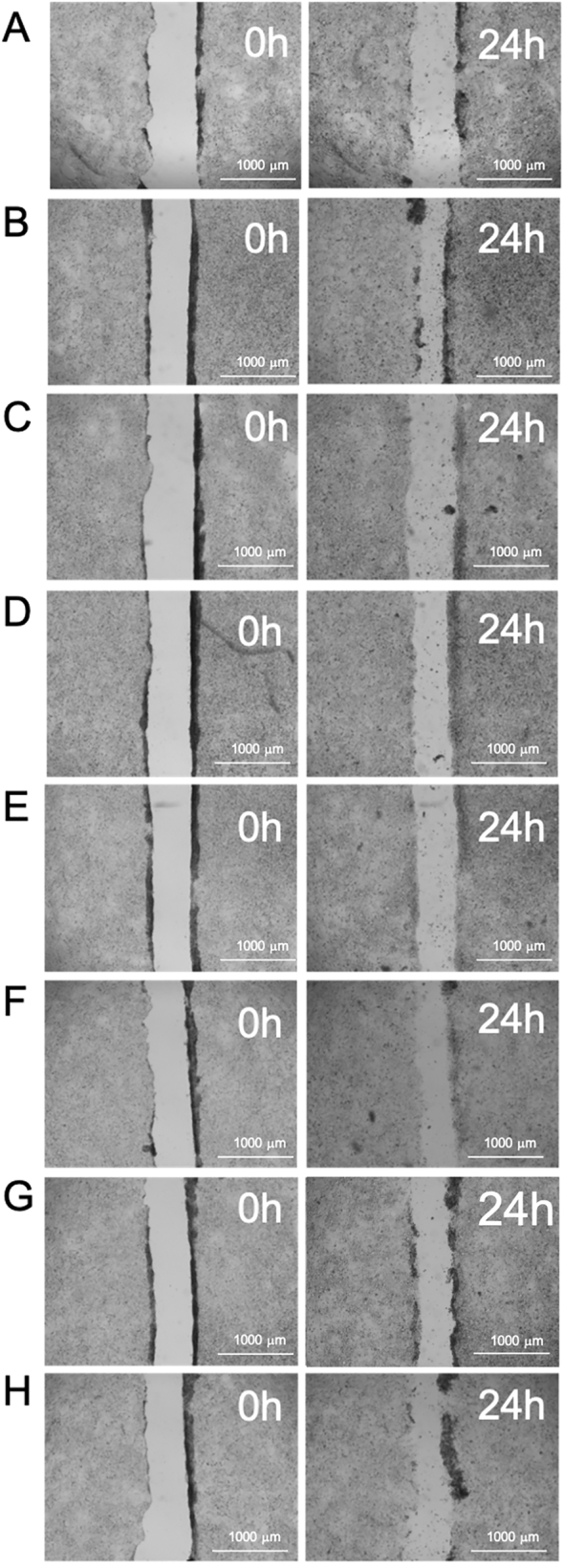
Images of the wound area of T84 cells at the start of treatment (0 h) and after incubation (24 h) for healthy control donors. Treated with (A) complete medium (B) sodium butyrate, (C) Donor 1 control, (D) Donor 1 dosed with probiotic, (E) Donor 2 control, (F) Donor 2 dosed with probiotic, (G) Donor 3 control and (H) Donor 3 dosed with probiotic.
Fig. 7.
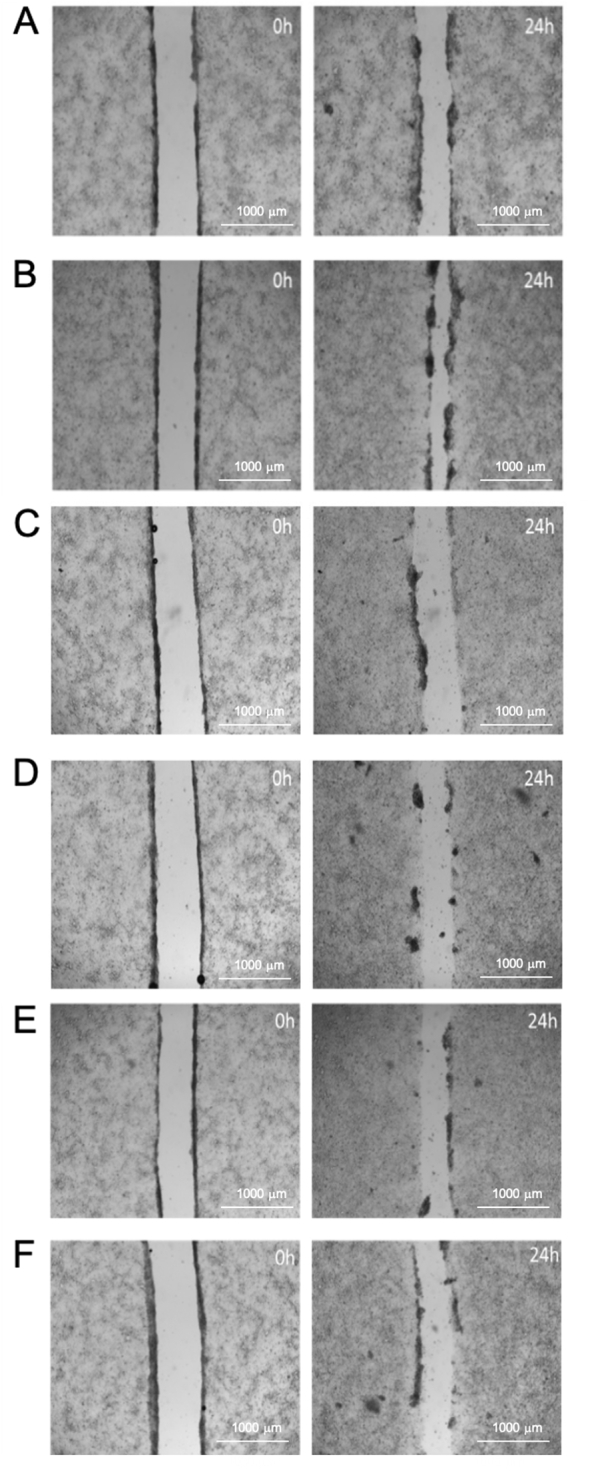
Images of the wound area of T84 cells at the start of treatment (0 h) and after incubation (24 h) for PD donors. Treated with (A) complete medium (B) sodium butyrate, (C) Donor 1 control, (D) Donor 1 dosed with probiotic, (E) Donor 2 control and (F) Donor 2 dosed with probiotic.
Table 10.
Improvement in wound area healing after 24 h following dosing with colonic samples from healthy and PD donors. Each value is expressed as a percentage change from its relevant blank.
| Healthy controls |
PD patients |
||
|---|---|---|---|
| Donor 1 | 9.0 ± 3.6 | Donor 1 | 16.1 ± 1.7 |
| Donor 2 | 15.5 ± 3.2 | Donor 2 | 15.2 ± 1.7 |
| Donor 3 | −3.5 ± 5.1 | Donor 3 | ND |
| Mean ± sd | 7.0 ± 7.0 | Mean ± sd | 15.7 ± 2.4 |
After 24 h, the CM sample wound had reduced to 75% of its initial area while the sodium butyrate wound reduced significantly (p < 0.0001) more, to 45% of its initial area. In all cases, dosing with probiotic significantly (p < 0.001) increased wound healing compared with the control. Again, the results suggest that it is not the presence of the probiotic species that positively influences wound healing; rather, butyrate is the key factor that encourages wound closure and the positive results from dosing with probiotic arise from increased SCFA concentrations from stimulation of the commensal gut bacteria.
4. Summary
This work suggests that probiotic supplementation might improve gut health in PD patients. It is not just the simple presence of the probiotic bacteria that causes these positive effects, but a cascade of events (proliferation of the probiotic bacteria generating lactate, lactate stimulating propionate- and butyrate-producing commensal species) leading to raised levels of butyrate. If development and/or progression of PD is influenced by gut microbiota dysbiosis then supplementation of the diet with a properly formulated probiotic may be a useful adjunct to standard treatment in clinic. Because of the small number of donors, results should be interpreted with caution and need replication, but combined with human and animal data they provide a compelling indication that probiotics may be a potential and cost-effective intervention in managing patients with Parkinson's Disease.
Funding
Symprove Ltd. funded the studies conducted by ProDigest BV.
Author contributions
Jonas Ghyselinck; Investigation/Formal analysis, Lynn Verstrepen; Investigation/Formal analysis, Frédéric Moens; Investigation/Formal analysis, Pieter Van den Abbeele; Investigation/Formal analysis, Arnout Bruggeman; Formal analysis, Jawal Said; Reviewing and editing, Barry Smith; Conceptualisation, Lynne Ann Barker; Reviewing and editing, Caroline Jordan; Reviewing and editing, Valentina Leta; Reviewing and editing, Ray Chaudhuri; Reviewing and editing, Abdul W. Basit; Reviewing and editing, Simon Gaisford; Original draft, formal analysis, project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Symprove Ltd. funded the M-SHIME studies conducted by ProDigest BV and the research team thanks the PD patient donors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpx.2021.100087.
Appendix A. Supplementary data
Supplementary material
References
- Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- Baxter N.T., et al. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibres. mBio. 2019;10 doi: 10.1128/mBio.02566-18. e02566–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedarf J.R., et al. Functional implications of microbial and viral gut metagenomic changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017;9:39. doi: 10.1186/s13073-017-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D., et al. Time to redefine PD? Introductory statement of the MDS Task Force on the definition of Parkinson’s disease. Mov. Disord. 2014;29:454–462. doi: 10.1002/mds.25844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- Braak H., de Vos R.A., Bohl J., Del Tredici K. Gastric alpha-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neurosci. Lett. 2006;396:67–72. doi: 10.1016/j.neulet.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Breen D.P., Halliday G.M., Lang A.E. Gut-brain axis and the spread of α-synuclein pathology: Vagal highway or dead end? Mov. Disord. 2019;34:307–316. doi: 10.1002/mds.27556. [DOI] [PubMed] [Google Scholar]
- Bu J., Liu J., Liu K., Wang Z. Diagnostic utility of gut α-synuclein in Parkinson’s disease: a systematic review and meta-analysis. Behav. Brain Res. 2019;364:340–347. doi: 10.1016/j.bbr.2019.02.039. [DOI] [PubMed] [Google Scholar]
- Bullich C., Keshavarzian A., Garssen J., Kraneveld A., Perez-Pardo P. Gut vibes in Parkinson’s disease: the microbiota-gut-brain axis. Mov. Disord. Clin. Prac. 2019;6:639–651. doi: 10.1002/mdc3.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carding S., Verbeke K., Vipond D.T., Corfe B.M., Owen L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cersosimo M.G., et al. Gastrointestinal manifestations in Parkinson’s disease: prevalence and occurrence before motor symptoms. J. Neurol. 2013;260:1332–1338. doi: 10.1007/s00415-012-6801-2. [DOI] [PubMed] [Google Scholar]
- Chaudhuri K.R., Healy D.G., Schapira A.H. Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol. 2006;5:235–245. doi: 10.1016/S1474-4422(06)70373-8. [DOI] [PubMed] [Google Scholar]
- Chiang H.L., Lin C.-H. Altered gut microbiome and intestinal pathology in Parkinson’s disease. J. Mov. Disord. 2019;12:67–83. doi: 10.14802/jmd.18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.G., et al. Oral administration of Proteus mirabilis damages dopaminergic neurons and motor functions in mice. Sci. Rep. 2018;8:1275. doi: 10.1038/s41598-018-19646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann S.M., et al. IL-6-dependent mucosal protection prevents establishment of a microbial niche for attaching/effacing lesion-forming enteric bacterial pathogens. J. Immunol. 2008;180:6816–6826. doi: 10.4049/jimmunol.180.10.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vuyst L., Moens F., Selak M., Rivière A., Leroy F. Summer meeting 2013: growth and physiology of bifidobacteria. J. Appl. Microbiol. 2014;116:477–491. doi: 10.1111/jam.12415. [DOI] [PubMed] [Google Scholar]
- Dodoo C.C., et al. Use of a water-based probiotic to treat common gut pathogens. Int. J. Pharm. 2019;556:136–141. doi: 10.1016/j.ijpharm.2018.11.075. [DOI] [PubMed] [Google Scholar]
- Dorsey E.R., Sherer T., Okun M.S., Bloem B.R. The emerging evidence of the Parkinson pandemic. J. Parkinsons Dis. 2018;8:S3–S8. doi: 10.3233/JPD-181474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Barcenilla A., Stewart C.S., Pryde S.E., Flint H.J. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Louis P., Flint H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., et al. Contribution of acetate to butyrate formation by human faecal bacteria. Br. J. Nutr. 2004;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- Dutta S.K., et al. Parkinson’s disease: the emerging role of gut dysbiosis, antibiotics, probiotics, and fecal microbiota transplantation. J. Neurogastroenterol. Motil. 2019;25:363–376. doi: 10.5056/jnm19044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfil M., Kamel S., Kandil M., Koo B.B., Schaefer S.M. Implications of the gut microbiome in Parkinson’s Disease. Mov. Disord. 2020;35:921–933. doi: 10.1002/mds.28004. [DOI] [PubMed] [Google Scholar]
- Falony G., et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- Fasano A., Visanji N.P., Liu L.W.C., Lang A.E., Pfeiffer R.F. Gastrointestinal dysfunction in Parkinson’s disease. Lancet. 2015;14:625–639. doi: 10.1016/S1474-4422(15)00007-1. [DOI] [PubMed] [Google Scholar]
- Flint H.J., Duncan S.H., Scott K.P., Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Sci. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- Flood P.M., et al. Transcriptional factor NF-κB as a Target for Therapy in Parkinson’s Disease. Parkinson’s Dis. 2011;2011(216298) doi: 10.4061/2011/216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredua-Agyeman M., Gaisford S. Comparative survival of commercial probiotic formulations: tests in biorelevant gastric fluids and real-time measurements using microcalorimetry. Benefic. Microbes. 2015;6:141–151. doi: 10.3920/BM2014.0051. [DOI] [PubMed] [Google Scholar]
- Fredua-Agyeman M., et al. In vitro inhibition of Clostridium difficile by commercial probiotics: a microcalorimetric study. Int. J. Pharm. 2017;517:96–103. doi: 10.1016/j.ijpharm.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Gazerani P. Probiotics for Parkinson’s disease. Int. J. Mol. Sci. 2019;20:4121. doi: 10.3390/ijms20174121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck J., et al. A 4-strain probiotic supplement influences gut microbiota composition and gut wall function in patients with ulcerative colitis. Int. J. Pharm. 2020;587:119648. doi: 10.1016/j.ijpharm.2020.119648. [DOI] [PubMed] [Google Scholar]
- Gudadappanavar A.M., Hombal P.R., Timashetti S.S., Javali S.B. Influence of Lactobacillus acidophilus and Lactobacillus plantarum on wound healing in male Wistar rats – an experimental study. Int. J. Appl. Basic Med. Res. 2017;7:233–238. doi: 10.4103/ijabmr.IJABMR_329_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer H.M., et al. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Hatton G.B., Madla C.M., Rabbie S.C., Basit A.W. Gut reaction: Impact of systemic diseases on gastrointestinal physiology and drug absorption. Drug Discov. Today. 2019;24:417–427. doi: 10.1016/j.drudis.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Hill-Burns E.M., et al. Parkinson’s disease and PD medications have distinct signatures of the gut microbiome. Mov. Disord. 2017;32:739–749. doi: 10.1002/mds.26942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser M.C., Tansey M.G. The gut-brain axis: is intestinal inflammation a silent driver of Parkinson’s disease pathogenesis? NPJ Parkinsons Dis. 2017;11:3. doi: 10.1038/s41531-016-0002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz B. Urban observation and sentiment in James Parkinson’s essay on the shaking palsy (1817) Lit. Med. 2014;32:74–104. doi: 10.1353/lm.2014.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A., et al. Diet in Parkinson’s disease: critical role for the microbiome. Front. Neurol. 2019;10:1245. doi: 10.3389/fneur.2019.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J., Gage H., Kimber A., Storey L., Trend P. Excess burden of constipation in Parkinson’s disease: a pilot study. Mov. Disord. 2006;21:1270–1273. doi: 10.1002/mds.20942. [DOI] [PubMed] [Google Scholar]
- Keshavarzian, et al. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015;30:1351–1360. doi: 10.1002/mds.26307. [DOI] [PubMed] [Google Scholar]
- Klindworth A., et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvasnovsky C.L., et al. A randomized double-blind placebo-controlled trial of a multi-strain probiotic in treatment of symptomatic uncomplicated diverticular disease. Inflammopharmacol. 2017;25:499–509. doi: 10.1007/s10787-017-0363-y. [DOI] [PubMed] [Google Scholar]
- Lin C.-H., et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflammation. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Scott K.P., Duncan S.H. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G.L., Flint H. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- Lyer S.S., Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012;32:23–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., et al. Butyrate promotes the recovering of intestinal wound healing through its positive effect on the tight junctions. J. Anim. Sci. 2012;90:266–268. doi: 10.2527/jas.50965. [DOI] [PubMed] [Google Scholar]
- Macy J.M., Ljungdahl L.G., Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J. Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoubrey L.E., Elbadawi M., Orlu M., Gaisford S., Basit A.W. Harnessing machine learning for development of microbiome therapeutics. Gut Microbes. 2021;13:1–20. doi: 10.1080/19490976.2021.1872323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens F., et al. A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int. J. Pharm. 2019;555:1–10. doi: 10.1016/j.ijpharm.2018.11.020. [DOI] [PubMed] [Google Scholar]
- Mulak A., Koszewicz M., Panek-Jeziorna M., Koziorowska-Gawron E., Budrewicz S. Fecal calprotectin as a marker of the gut immune system activation is elevated in Parkinson’s disease. Front. Neurol. 2019;13:992. doi: 10.3389/fnins.2019.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Li Z.-R., Green R.S., Holzman I.R., Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pardoa P., et al. The gut-brain axis in Parkinson's disease: Possibilities for food-based therapies. Eur. J. Pharmacol. 2017;817:86–95. doi: 10.1016/j.ejphar.2017.05.042. [DOI] [PubMed] [Google Scholar]
- Pires B.R.B., Silva R.C.M.C., Ferreira G.M., Abdelhay E. NF-kappaB: two Sides of the same Coin. Genes. 2018;9:24. doi: 10.3390/genes9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postuma R.B., Berg D. Advances in markers of prodromal Parkinson’s disease. Nat. Rev. Neurol. 2016;12:622–634. doi: 10.1038/nrneurol.2016.152. [DOI] [PubMed] [Google Scholar]
- Qian Y., et al. Alteration of fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 2018;70:194–202. doi: 10.1016/j.bbi.2018.02.016. [DOI] [PubMed] [Google Scholar]
- Ragsdale S.W., Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt N., et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T. The impact of indigenous microbes on Parkinson’s disease. Neurobiol. Dis. 2020;135:104426. doi: 10.1016/j.nbd.2019.03.014. [DOI] [PubMed] [Google Scholar]
- Sampson T.R., et al. Gut microbiota regulate motor deficits and Neuroinflammation in a model of Parkinson's disease. Cell. 2016;167:1469–1480. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M.E., Merenstein D.J., Reid G., Gibson G.R., Rastall R.A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019;16:605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- Santos S.F., de Oliveira H.L., Yamada E.S., Neves B.C., Pereira A., Jr. The gut and Parkinson’s disease – a bidirectional pathway. Front. Neurol. 2019;10:574. doi: 10.3389/fneur.2019.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- Scott K.P., Gratz S.W., Sheridan P.O., Flint H.J., Duncan S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013;69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Shin C., Lim Y., Lim H., Ahn T.-B. Plasma short-chain fatty acids in patients with Parkinson’s Disease. Mov. Disord. 2020;2020(28016) doi: 10.1002/mds.28016. [DOI] [PubMed] [Google Scholar]
- Sisson G., et al. Randomised clinical trial: A liquid multi-strain probiotic vs. placebo in the irritable bowel syndrome – a 12 week double-blind study. Aliment. Pharmacol. Ther. 2014;40:51–62. doi: 10.1111/apt.12787. [DOI] [PubMed] [Google Scholar]
- Sun M.F., et al. Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson's disease mice: gut microbiota, glial reaction and TLR4/TNF-alpha signaling pathway. Brain Behav. Immun. 2018;70:48–60. doi: 10.1016/j.bbi.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Tucker R.M., et al. Distinctive pathophysiology underlying constipation in Parkinson’s disease: Implications for cognitive inefficiency. J. Clin. Med. 2020;9:1916. doi: 10.3390/jcm9061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker R.M., et al. Role of Helicobacters in neuropsychiatric disease: a systematic review in idiopathic Parkinsonism. J. Clin. Med. 2020;9:2159. doi: 10.3390/jcm9072159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger M.M., et al. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 2016;32:66–72. doi: 10.1016/j.parkreldis.2016.08.019. [DOI] [PubMed] [Google Scholar]
- Van de Wiele T., Boon N., Possemiers S., Jacobs H., Verstraete W. Prebiotic effects of chicory inulin in the simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 2004;51:143–153. doi: 10.1016/j.femsec.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Van den Abbeele P., et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium Cluster IX. Appl. Environ. Microbiol. 2010;76:5237–5246. doi: 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 2012;5:106–115. doi: 10.1111/j.1751-7915.2011.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P., et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;6:335–340. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo M.A., Rodrigues H.G., Nachbar R.T., Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.B., Wang P.Y., Wang X., Wan Y.L., Liu Y.C. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig. Dis. Sci. 2012;57:3126–3135. doi: 10.1007/s10620-012-2259-4. [DOI] [PubMed] [Google Scholar]
- Weis S., et al. Effect of Parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. NPJ Parkinson’s Disease. 2019;5:28. doi: 10.1038/s41531-019-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material