Abstract
The development of organs-on-chip (OoC) has revolutionized in vitro cell-culture experiments by allowing a better mimicry of human physiology and pathophysiology that has consequently led researchers to gain more meaningful insights into disease mechanisms. Several models of hearts-on-chips and vessels-on-chips have been demonstrated to recapitulate fundamental aspects of the human cardiovascular system in the recent past. These 2D and 3D systems include synchronized beating cardiomyocytes in hearts-on-chips and vessels-on-chips with layer-based structures and the inclusion of physiological and pathological shear stress conditions. The opportunities to discover novel targets and to perform drug testing with chip-based platforms have substantially enhanced, thanks to the utilization of patient-derived cells and precise control of their microenvironment. These organ models will provide an important asset for future approaches to personalized cardiovascular medicine and improved patient care. However, certain technical and biological challenges remain, making the global utilization of OoCs to tackle unanswered questions in cardiovascular science still rather challenging. This review article aims to introduce and summarize published work on hearts- and vessels-on chips but also to provide an outlook and perspective on how these advanced in vitro systems can be used to tailor disease models with patient-specific characteristics.
Keywords: Organs-on-chips, Cell culture, Cardiovascular, Heart, Personalized medicine
1. Introduction
Cardiovascular diseases (CVDs) are a group of disorders affecting the heart and the vasculature that represent the number one cause of mortality globally.1 Only in Europe, CVDs causes over 4 million deaths each year,2 accounting for 47% of all deaths in Europe. The most common underlying pathology in CVDs is atherosclerosis, which causes an ischaemia in the heart and in peripheral arteries. Atherosclerosis is defined as a chronic disease of the vasculature, whose architecture is slowly remodelled over time. This disease and remodelling process involves the interplay of numerous cell subtypes, including endothelial cells (ECs) (becoming dysfunctional), leukocytes and macrophages (triggering inflammation), and smooth muscle cells (which dedifferentiate or undergo apoptosis).3,4
Unstable atherosclerotic plaques can rupture, which results in arterial thrombosis. Thrombosis, the formation of blood clots, prevents blood flow, and triggers life-threatening clinical conditions in the arterial system, such as myocardial infarction (MI) and ischaemic forms of stroke (IS). Although MI and IS are usually acute events resulting from chronic atherosclerotic processes affecting coronary and carotid arteries, respectively, venous thromboembolism (VTE) is mainly caused by haemostatic or coagulation abnormalities.5
Another severe result of tissue ischaemia is heart failure (HF), which is most commonly associated with coronary artery disease (CAD).6 Other complications of ischaemia include arrhythmias caused by discontinued oxygen supply to the cardiac conduction system. In particular long-term complications after an acute MI that triggers pathological myocardial remodelling remain unsolved and are a major cause for high re-hospitalization rates.7 Hallmark features of this cardiac remodelling process8 are excessive deposition of extracellular matrix (ECM) leading to cardiac fibrosis, chamber dilation (dilated cardiomyopathy), and cardiomyocyte hypertrophy. Apart from ischaemia being the key inducer leading to HF, several non-cardiac therapies can cause adverse reactions that induce a similar disease phenotype.9 In particular, anti-cancer treatment strategies (radiation as well as chemotherapies) are particularly known for their cardiotoxic potential. Here, the new field of cardio-oncology aims at improving our understanding of molecular and clinical alterations that cancer therapies generate in the cardiovascular system.10
Our knowledge about etiopathogenetic mechanisms in CVD has dramatically benefited from advances in ‘-omics’ technologies. A typical pipeline to tackle unanswered disease research questions exploits disciplines such as genomics and transcriptomics to identify novel targets in human cohorts as well as in vivo models to validate these findings. Genome-wide association studies (GWAS) to investigate CAD,11 VTE,12,13 IS,14 and HF15 have evaluated hundreds to thousands of individuals and led to the identification of multiple genetic loci associated with the respective disease. With the availability of new technologies and the combination of genomic data utilizing publicly available expression datasets, the translation from genomic loci to the discovery of causal genes is starting to become a reality. Functional assessments using in vitro modulation in cultured cells or in vivo animal models are considered irreplaceable to identify the actual genes or variants that are relevant for causing the associations while trying to validate biological relevance. However, the translation of discoveries across species remains challenging: the poor sequence conservation of most non-coding genes (unlike protein-coding genes) substantially limits the experimental studies of exiting candidates, such as long non-coding RNAs or circular RNAs.16 Moreover, the human circulatory system, including its mechanical, electrical, biochemical, and cellular complexity, is hard to mimic. Human in vitro models that consist of single types of cells cultured under static conditions on a plastic surface in two dimensions poorly represent our physiological constitution.17 These simplistic models lack the three-dimensional complexity of the tissue, the effect of flow, the cell–cell interaction between blood cells and the endothelium as well as the involvement of the ECM that characterizes vascular tissue in vivo.
In recent years, organs-on-chips (OoCs) have emerged as powerful new tools to fill the translational gap from animal models to human disease, with a particular potential to even replace animal testing in the future.18 OoC technology will improve the modelling of organs or organ systems for healthcare research while immensely impacting the precision medicine approach.18 OoCs comprise systems integrating either 2D cell cultures on permeable membranes or cells cultured in 3D hydrogel scaffolds. In this current review article, we are referring to OoCs as defined by the EU project ORCHID.19 Within the scale of novel physiologically relevant in vitro models, organoids need to be mentioned at this point. They will however not be covered in greater detail in this present review. Organoids are self-organized three-dimensional tissue cultures deriving from stem cells. They differ from OoCs especially in biological complexity, displaying multi-cellular self-assembled constructs,20 whereas OoCs are typically multi-structural engineered systems for on-chip cell cultures.
In the following pages, we introduce the general concept and scientific potential of OoC systems in CVD research. We further discuss the opportunities and challenges of utilizing OoCs in preclinical drug testing and target discovery.
2. Methods
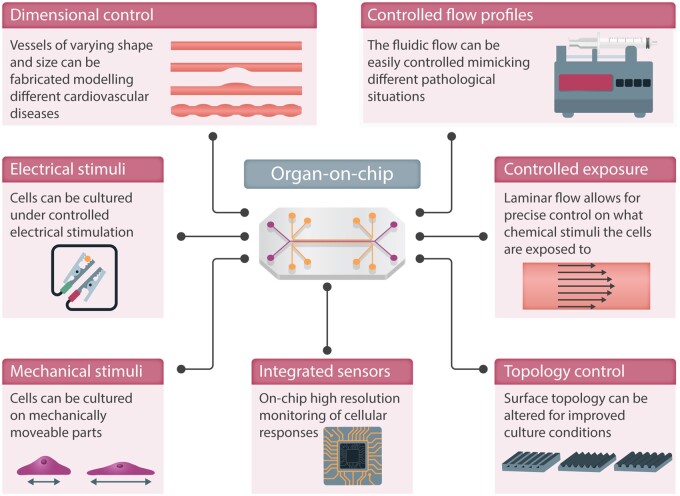
OoCs are micro-engineered in vitro models that recapitulate aspects of human physiology and pathology. They can be used in drug discovery as well as for efficacy and toxicology testing.21 OoCs are defined as microfluidic cell-culture devices that contain continuously perfused chambers being inhabited by living human cells arranged in a three-dimensional organization that preserves and mimics the tissue geometry.22,23 The high level of control, enabling customised cell-culture environment in OoCs, is illustrated in Figure 1. This includes custom ECM topology, the integration of sensors and actuators for monitoring and electrical/mechanical stimuli, control of microfluidic channel dimensions, and temporal and spatial flow profiles for pulsatile flow and chemical stimuli. Moreover, the precise microfluidic flow control enables an optimal growing environment (influx of nutrients and efflux of cell-waste) as well as the circulation of drugs, signalling molecules, or immune cells. The fabrication methods used to realize OoC systems were originally developed for the microelectronics industry, providing methods to define microfluidic channels and compartments with dimensions from a few micrometres (µm) to several millimetres (mm), thus matching the dimensions of real arteries, veins, and functional units of organs.
Figure 1.
Fabricating heart-on-chip and vessel-on-chip models using micromachining allows for integration of several advanced features.
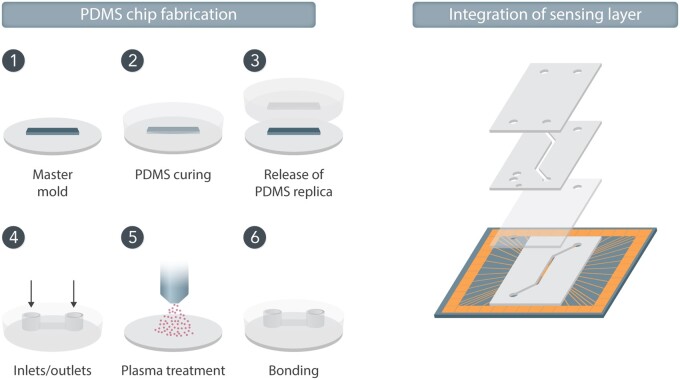
In order to fabricate vessel models, it is imperative to consider the geometry as this affects the flow profile, wall shear stress, culture area, and the total number of cells used in the specific model. The material most commonly used for proof-of-concept models is poly(dimethylsiloxane) (PDMS), a polymer developed in the late 1990s 24 to fabricate microfluidic channels. PDMS has several advantages for miniaturized cell cultures, such as simple fabrication, gas permeability, and optical transparency. Using PDMS, supportive microfluidic channels can be moulded off a master that has the reverse topographical features. By removing the PDMS from the mould and bonding the structure onto a glass slide, a sealed channel structure can be created (Figure 2). Inlet and outlet holes for connecting tubing for perfusion can easily be punched into the PDMS.
Figure 2.
Schematic drawing showing the six basic steps of PDMS moulding to form a microfluidic channel that can be used in organs-on-chip. 1—A master is prepared having the inverse topography of the final channel structures, 2—PDMS pre-polymer is poured onto the master and polymerised upon heat treatment, 3—The moulded channels are released from the master, 4—Holes for connecting tubing for media perfusion are prepared in the PDMS by punching, 5—The PDMS surface is activated for bonding via plasma treatment, 6—The microfluidic channels are sealed by bonding the PMDS slab onto a glass microscope slide which may include patterned electrodes.
In order to form vessel mimics, the microfluidic channels can then be coated with human ECs, forming artificial intima layers of vessels in which blood, plasma, or other cells of interest (e.g. monocytes and platelets) can be perfused.25 Specific culture media with different added stimuli or drugs can be added to the system, and real-time observation of the system under a microscope can be used to evaluate the effect of certain stimuli. At the same time, variations in flow and shear stress can provide more information than static cultivation in well plates. To study disease onset, systems with higher complexity, including 3D lumen-chips that incorporate the media layer populated by SMCs, are required. A recent example of a perfusable artery-on-chip was published by Cho and Park,26 where SMCs and human umbilical vein ECs (HUVECs) were co-cultured in a PDMS channel. In an attempt to induce the proper morphology and orientation of SMCs, wrinkles were formed on the circular PDMS channel surface during the moulding as contact guidance, and the HUVECs were aligned by medial perfusion.
One major disadvantage of using PDMS is that the material is porous, thus absorbing especially hydrophobic compounds, which can lead to false-negative read-outs from drug-screening studies.27 Also, the polymer is not inert, meaning that silicon will leach from the structure into the cell culture environment.28 Developments of more inert yet biocompatible materials are therefore currently a major focus within the field.29
In an effort to make the vascular structure more biomimetic, one can design the system to include multiple microfluidic compartments where some can be filled with a biomimetic cell-culture scaffold to recreate the physiological environment of the vasculature.30–33 Alternatively, one can mould the complete device in a biological material such as collagen.34,35 Cells can further be introduced into the biomimetic scaffolds. This set-up allows investigation of the tissue-blood interface in a controlled environment that cannot be monitored in animals or patients. It can also be utilized to study the interaction with adjacent cells on disease sub-phenotypes, such as endothelial permeability, communication with blood cells, or platelet aggregation. Another approach to fabricate vessel models in a biomimetic material is to utilize the method of viscous fingering,36 thus forming co-centred channels instead of adjacent ones. The approach also has the advantage of generating circular vessel structures, which cannot be obtained via conventional PDMS moulding. The method has been used to study cell–cell interactions in 600 µm wide37 and 250 µm38 diameter vessel models. Although very interesting from a biological perspective, there are several technical challenges with integrating biomimetic scaffolds into OoC systems, which have been discussed in more detail elsewhere.39
One option to exploit the angiogenic potential of the cells themselves is to increase the biological relevance of the vasculature models even further. In such systems, vascular cells are seeded in the biomimetic scaffold and exposed to mechanical stimuli via slow perfusion40 or a chemical gradient of growth factors.41 This results in a vascular bed formation after 2–3 weeks, including both larger macro-vessels and dense capillary microvascular networks. Taking a completely different approach to investigating small and large artery diseases, one can utilize the advantages of microfluidics by developing advanced ex vivo culture platforms. They can be used if a higher complexity is needed to investigate, for example, cell–cell interactions and their relevance for the onset of a certain disease. An initial attempt to investigate structural changes occurring within the vessel wall was presented by Günther et al.42 They used a microfluidic platform for immobilizing small arteries obtained ex vivo from mice and long-term culturing under physiological conditions (37°C, 45 mmHg transmural pressure). Live imaging allowed them to determine the arteries’ inner and outer diameter in real time while assessing the effects of heterogeneous environmental changes on the microvascular structure and function.
Cardiac models are often realized using the multichannel approach described above, as it provides the possibility to compartmentalize the different domains of the cardiovascular system. This unprecedented modularity has led to the development of different heart-on-chip models that have focused on specific cardiovascular subdomains of great interest to cardiovascular researchers. The heart models can be fabricated to include multiple channels that are aligned in parallel either along the horizontal axis 43–45 or the vertical axis.46–48 The choice of layout is often related to the assays used, where horizontally structured channels, for example, allow for easy optical access of the separate compartments.
There are also several reports of 2D-based cardiac models,46,47 for example developed with an electrophysiological focus due to the increased simplicity of integrating planar electrodes with cell cultures. These systems are often fabricated in more robust materials, such as silicon- or glass-coated, with fibronectin, that enhance cellular adhesion.
Three-dimensional models, on the other hand, are more commonly found in models that mimic multi-organ systems or three-dimensional aspects that 2D models fail to recapitulate, such as the force of contraction measurements and maturation of the co-culture micro-tissues. Such models include mechanically moveable parts, such as suspended membranes49,50 or cantilevers structures51 that expose the encapsulated cardiac cells to mechanical stimulation during the culture.
A fabrication method that is rapidly gaining attention, both for realizing vessels and heart constructs, is ‘bioprinting’, a type of additive manufacturing or ‘3D printing’. In bio-printing, hydrogel cell suspensions are directly extruded onto a substrate, building up the final biological structures layer-by-layer. Advantage of bioprinting is that the final device can include a high level of topographical complexity and that multiple cell and material combinations can be realized. Bio-printed models of vascular networks52 have been reported, and recently, proof-of-principle of a drug toxicity screening heart-on-chip model using bioprinted cardiac cells was demonstrated.53 Although bioprinting is a rapidly developing technology, the challenge with interfacing bioprinted heart models with vascular models for controlled media perfusion still remains.
2.1. Integrated electrodes for cellular stimulation and read-out
The inclusion of electrodes in cell-culture systems assist in producing highly controlled environments and allow for continuous read-out of parameters essential to identifying cell behaviour. Various materials can be used for the fabrication of the electrodes, including bio-friendly metals such as gold44 and platinum,46,54 and organic conducting materials, such as carbon55 or poly (3,4-ethylenedioxythiophene) polystyrene sulfonate (PEDOT: PSS).56
For the realization of reliable heart-on-chip models, it is very important to support the development of cells with a mature electrophysiological conduction system, such as synchronized beating. Although some cultures of cardiomyocytes show spontaneous synchronized beating already after a few days of culture even without external interactions,44,54,57 an effective method to induce synchronised beating in immature cultures is to assist the synchronization of the beating by using external electrical stimulation58,59 (Figure 2). Typically, the cells are exposed to trains of electrical pulses similar to the electrical signalling of native cardiomyocytes. Often, this is achieved via large external electrodes, but more user-friendly custom systems have been developed using integrated electrodes on-chip.44,60 It has been shown that maturation of cells may be improved by pacing the cells as proved by expression of α-actinin, connexin,43 and cardiac troponin-T.60 In addition to external electrical stimulation, maturation schemes that rely on external mechanical43,61 and biochemical62 cues—or a combination of the aforementioned50,57—have also been presented.
The main use for microfabricated integrated electrodes in heart-on-chip systems is, however, for on-chip read-out of electrophysiology. Here, the ion currents of the cells are measured extracellularly as changes in the field potential. The extracellular field potential is closely linked to the QT interval through the corrected field potential duration.63 Multiple localized measurements of single cells enable assessment of the synchronization of beating cells and wavefront propagation. As demonstrated in open-format cell cultures, cells can be cultured on top of high-density electrodes, so-called microelectrode arrays (MEAs) for read-out of cellular electrophysiology.64–66 Customized MEAs can fit in virtually any microfluidic system and have been reported for several OoC systems.46,54,67
Normally, very close contact between the cells and the MEA is desired for optimal resolution and signal strength, resulting in cell culture on hard flat surfaces and 2D cell models. To make the culture environment more biomimetic, Kujala et al.64 cultured cardiomyocytes on a ∼100-µm-thick micro-grooved gelatin layer attached to the MEA and showed that the electrophysiology still could be mapped, although no longer with single-cell resolution. Other approaches that address the issue of 2D culture on hard surfaces are non-contact MEA measurements as explored by Sharf et al.68 and patterning of MEAs on a soft PDMS substrate as demonstrated by Gaio et al.57
Electrical sensors can also be integrated to monitor cell contraction. Quin et al.65 have demonstrated this in an open-top structure, where interdigitated electrodes were pattered for cell-contraction measurements in combination with MEAs to monitor beating. Although highly correlated in normally functioning cardiac tissue, contraction and electrophysiology are two different mechanisms that are important to follow in heart-on-chip models. Alternative ways to map cell contraction is to integrate mechanical sensors into the heart model,54,66 or to utilize an external optical read-out with computational motion tracking,62 or mapping the transient intracellular calcium signal using cells modified to include GCaMP6.44
Integrated electrodes are also interesting for vessel models, as they can be used to assess the barrier integrity of the cultured cells via trans-endothelial electrical resistance (TEER) measurements. Most commonly, this method works by integrating electrodes on either side of a porous membrane on which the ECs are cultured. As the cells form tight junctions, it becomes increasingly more difficult for any electrical current to flow through the cell layer, and the measured electrical resistance increases. It is possible to combine electrophysiological measurements and TEER as demonstrated by Maoz et al.,46 using a dual-channel, endothelialized heart-on-chip model. Further, 3D tubular vessels may incorporate TEER sensing capability by insertion of electrodes inside the vessel and in a surrounding hydrogel matrix.69
It may be noted that most models with integrated electrical sensing capabilities are two-dimensional, which is explained by the well-established technology to form electrodes on hard 2D surfaces. However, electrodes can also interface more in vivo-like cardiac microtissue as in the case of Weng et al.44 An increase in the number of publications on this topic is expected. Further, the hydrogel scaffold may be topologically patterned on top of the electrodes,64 or the electrodes themselves can be structured into 3D formats.60,70 Alternatively, conducting and biocompatible scaffolds can be prepared, thus enabling electrical read-out via the porous scaffold itself.71
2.2. Heart-on-chip disease models
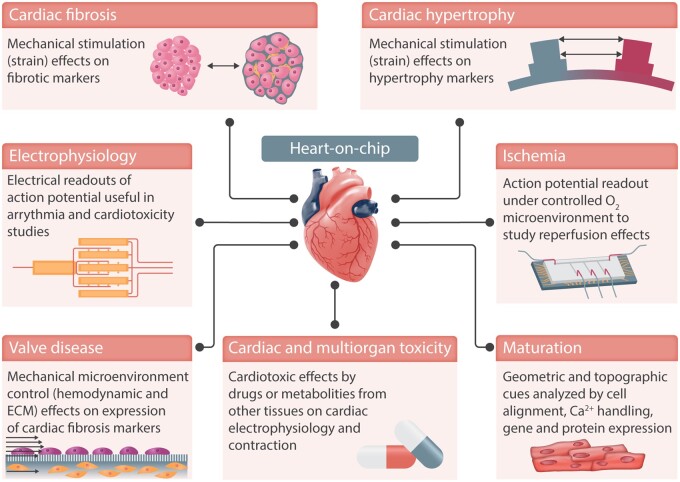
Heart-on-a-chip models developed so far have focused on establishing biomimetic, functional aspects of the heart, focusing in particular on the co-culture of multiple cell types, for example, cardiomyocytes and cardiac fibroblasts as well as on the electromechanical stimulation of the cells cultured on these systems (Figure 3). Heart-on-chip models aim to recapitulate the intricate conditions of the microenvironment that cells would experience in the heart. Nonetheless, the ability to include 3D cell cultures in these devices enables an increase in their biological complexity. Control over the microenvironment and the cultured cell types permits the recreation of relevant aspects of a specific disease (Table 1). Among the different reported heart-on-a-chip platforms, models to address cardiac ischaemia, cardiac fibrosis, and cardiotoxicity can be found in the literature (Figure 3), whereas many other exiting approaches have been developed and summarized in further review articles.62,72–77
Figure 3.
Heart-on-chip devices can recapitulate cardiac functions in vitro and integrate sensing units to monitor the cells in culture, e.g. action potential. Examples of cardiovascular diseases can be found in these devices, such as ischaemia and cardiac fibrosis. Integrated electrodes and mechanical actuation allow to monitor and stimulate the cells in culture, better recapitulating the cardiac microenvironment.
Table 1.
Summary of the heart-on-chip platforms.
| Aspects of human cardiac physiology and disease in organs-on-chips | ||
|---|---|---|
| Cardiac physiology | Defined 3D tissue organization | 43 , 48 , 61 , 62 , 64 , 135 |
| Force of contraction | 51 , 54 , 130 , 135 | |
| Electrophysiology | 43 , 46 , 57 , 62 , 64 , 65 , 67 , 135 | |
| Cardiac-vascular interactions | 44 , 46 , 62 , 135 | |
| Body-on-chip approach | 54 , 130 , 135 | |
| Cardiac disease and toxicity | Hypertrophy | 61 |
| Arrhythmia | 47 , 49 | |
| Ischaemia | 67 | |
| Fibrosis (e.g. fibroblast proliferation, collagen deposition, and valve calcification) | 47 , 49 | |
| Inflammation | 46 , 135 | |
| Cardiotoxicity & Pharmacology | 43 , 44 , 46 , 51 , 54 , 61 , 62 , 64 , 65 , 130 , 135 | |
The selection criteria employed in this table were that the devices used in the study could be considered a heart-on-a-chip device, i.e. a microfluidic device where the microenvironment of the cells or tissue in culture can be controlled and/or be stimulated mechanically and/or electrically. Platforms where constructs are cultured in well-plates or make use of spheroid technology were not considered due to the lack of their microfluidic character, which is seen as a requirement for organs-on-chips.
2.2.1. Ischaemia
The abrupt disruption of blood flow in ischaemia leads to the accumulation of metabolic by-products while reducing the oxygen supply to the tissue. This locally affects the contraction of cardiomyocytes. Liu et al.67 were able to replicate the hypoxic microenvironment and follow the action potential changes over time in the cell culture using patterned electrodes in a heart-on-a-chip device. The combination of the microenvironment cues, along with the insights gained from the electrophysiology of the cells depicts the solid control that can be attained with these devices.
2.2.2. Cardiac fibrosis
In the scope of cardiac fibrosis, Kong et al.49 were able to recreate the increased ECM stiffness using a photopolymerizable hydrogel while including the mechanical load similar to the stimulus that cardiac fibroblasts would experience under pro-fibrotic conditions. The cyclic mechanical loading, along with the exposure to a biochemical stimulus like transforming growth factor β (TGFβ) can even more closely mimic the fibrotic microenvironment of the heart. There have also been non-microfluidic devices that make use of 3D cardiac tissue models to mimic hallmarks of cardiac fibrosis using human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) along with fibroblasts. Mastikhina et al.78 reported a model where expression of collagen and brain natriuretic protein were upregulated when tissues were exposed to TGF-β, a pro-fibrotic agent, and downregulated when subsequently exposed to an anti-fibrotic drug. Wang et al.35 used cardiac fibroblast overpopulation of the tissues to mimic a fibrotic scenario, thus avoiding the pleiotropic effects of TGF-β. These two methods could be combined into microfluidic devices in additional adjacent compartments where, ECs can be integrated, and more complex, integrative models be made.
2.2.3. Cardiotoxicity
Interestingly, the vast majority of heart-on-chip devices reported in literature state cardiotoxicity as their primary goal. Although cardiotoxicity encompasses a myriad of aetiologies, two common trends can be found in these devices: (i) an investigation of the toxic effects that specific drugs have on cardiac cells and (ii) the toxic effect produced by the co-cultured cells from other organs (e.g. liver). The highly controlled environment of the heart-on-chip devices makes it a highly appealing platform for drug toxicity studies, as evidenced by a large number of devices with this as its motivation of the design.
The highly controlled microenvironment in heart-on-chip models makes them very useful for modelling diseases. There is plenty of room to study disease mechanisms that cannot be dissected in common platforms such as cell culture and animal models. However, one advantage of animal models compared with heart-on-a-chip models is the cross-talk between different organs, which are typically involved in the chronic diseases that lead to CVD.79 A major challenge in developing heart-on-a-chip disease models would be the difficulty of integrating multi-organ co-cultures for longer periods of time, typically around 1 week–7 days—after tissue formation. Nonetheless, aspects of complex chronic diseases can still be modelled with OoCs, e.g., cardiac fibrosis, as mentioned above.
Naturally, the aforementioned models make use of cardiac cells, a scarce resource since primary cardiomyocytes proliferate poorly in vitro. With the advent of human iPSC-CMs, this major cell source bottleneck is gradually being overcome. However, hurdles still remain to improve their immaturity, which ultimately affects the pharmacological response, with oncotherapy effects80 and the biomimetic aspects of the OoCs that employ human iPSC-CMs. Cardiac cell maturity can be defined in several aspects such as electrophysiology, metabolism, and morphology, to name a few. Unveiling what mechanisms underlie iPSC-derived cardiomyocytes maturity is an active field of research and is reviewed elsewhere.81 The models created with these cells naturally inherit their limitations. There are examples of initiatives that aim to characterize iPSC-CMs and their response to pharmacological agents with a known response as an attempt to benchmark different cell sources, such as the CiPA initiative82 and the Pulse CRACK-IT83 project. In the future development of heart-on-chip models, the drugs used for benchmarking in the mentioned initiatives can be used as a reference for new devices. With the aim of generating a model where the response of cardiac tissues exposed to libraries of compounds could determine the drug used, Lee et al.84 used a machine-learning model for the drug-response analysis. This clearly demonstrates that heart-on-chip development is a multidisciplinary field, where pharmacologists, engineers, biologists, regulators, and clinicians, among others, play a key role in the development of these models and their respective validation.
2.3. Studying atherosclerosis using chip-based systems
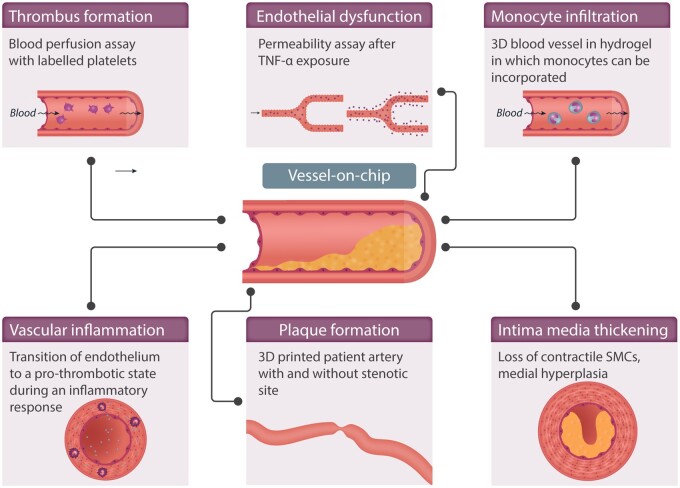
Modelling the different stages of the advancement of atherosclerosis is considered crucial in the development of vascular OoCs (summarized in Table 2 and Figure 4). Qiu et al.25 developed a 3D microvasculature-on-chip that is able to display physiological endothelial barrier function for several weeks, therefore allowing to study how chronic endothelial dysfunction develops. To model atherosclerosis progression, several devices have been created with occlusion or stenosis of the lumen, which recreates the higher shear region commonly found in developing atherosclerotic plaques.38
Table 2.
Summary of vessel-on-chip platforms.
| Aspects of human vascular physiology and disease in organs-on-chips | ||
|---|---|---|
| Vascular physiology | Perfusable 3D blood vessels with defined geometries | 37 , 38 , 87 , 93 , 97 , 139 , 140 |
| Perfusable microvasculature with self-organized geometries | 31 , 141 | |
| Angiogenesis | 31 , 34 | |
| Endothelial-mural interactions | 31 , 34 , 37 , 139 | |
| Blood perfusion | 34 , 85 , 87 , 94 , 98 , 99 | |
| Vascular disease modelling | Thrombosis | 34 , 85 , 87 , 93 , 94 , 97–99 |
| Inflammation (e.g. permeability and adhesion molecules) | 31 , 34 , 37 , 85 , 93 , 94 , 97 , 98 , 139 | |
| Immune cell recruitment | 31 , 34 | |
Vessel-on-chip devices included in this table consist of different types of perfusable blood vessels modelling human vascular physiology as well as pathophysiology.
Figure 4.
Vessels-on-chips devices are useful tools to study pathological mechanisms occurring within the vessel wall in early and later stages of atherosclerosis. Preliminary research can be performed in easier fabricated straight PDMS channels, whereas for more extensive and complicated research questions, a more elaborate model can be used by producing a 3D lumen in a hydrogel.
Westein et al.,85 Tovar-Lopez et al.,86 and Costa et al.87 have provided excellent examples of how to investigate atherosclerosis using chip-based systems. These three models differ regarding their biological and technical complexity. The chips developed by Westein et al. and Tovar-Lopez et al. consist of a square channel in which an artificial atherosclerotic plaque is already embedded (Figure 4). In order to study 3D vessel geometry in a more physiologic way, Costa et al.87 created different 3D vascular structures by using 3D-printed anatomical models based on observations generated by computed tomography angiography. They were, therefore, able to closely mimic architectures found in both healthy and stenotic blood vessels.
A disadvantage of these models is the lack of the ECM, as PDMS is responsible for creating the lumen that directly surrounds the cells. If the occlusion mechanism and pathophysiology of the development of atherosclerosis are to be studied, a more flexible model in which monocytes and macrophages can be included would be highly appreciated.
2.4. Mimicking thrombosis on-a-chip
Thrombosis is a complex process influenced by genetic and environmental factors, involving three main components: abnormalities in the vessel wall (endothelium), abnormalities in components of the blood (coagulation proteins and platelets), or abnormalities in fluid dynamics (turbulence flow and shear stress).88,89 Unlike static in vitro cell-culture models, vessels-on-a-chip can mimic the effect of flow and its interaction with the vessel wall while overcoming inter-species differences of animal models. Microfluidic technology has therefore been used extensively to create new in vitro models to study thrombotic diseases.90–94
Although previous work has been published studying thrombosis on microfluidic devices lacking ECs (reviewed in Westein et al.91 and Zhu et al.92), we here review OoC models of thrombosis typically consisting of a tubular or rectangular mould that is covered by components of the ECM (typically fibrin or collagen). ECs form a monolayer, therefore recreating endothelial geometry and function. In normal healthy conditions, the endothelium has anticoagulant and anti-inflammatory properties, therefore allowing blood flow through the lumen and preventing platelet activation and fibrin clot formation.95 Upon endothelial damage or activation, coagulation and platelet aggregation are triggered to promote the formation of a thrombus.96 Besides endothelial damage, several stimuli and the effect of blood flow are known to activate the endothelium.
Several examples of OoC have been published that study the different triggers of clot formation through direct monitoring of platelet aggregation under a microscope. In this direction, Zheng et al.34 engineered microvascular networks by seeding HUVECs intro microfluidic circuits coated with collagen and demonstrated that upon perfusion with human blood, rolling and adhesion of platelets occurred only at sites of damage—or in stimulated endothelium where long fibres of von Willebrand factor (VWF) covered the surface of the activated endothelium. These structures had not been reported in previous planar cultures or mouse models of thrombosis. Similarly, a recent study by Brouns et al.94 explored the anticoagulant effect of intact endothelium (created by a microfluidic model coated with HUVECs) through natural anticoagulants while comparing with artificially damaged endothelium covering a highly thrombogenic surface created with collagen and tissue factor.
One of the key advantages of OoCs is their great capacity to tightly control vessel geometry and flow, which has contributed significantly to the understanding of flow-based changes on platelet activation and the risk of thrombosis. To better understand the contribution of atherosclerosis and vessel stenosis to thrombosis, stenotic vessels perfused with whole human blood have been developed. The results indicate that platelets aggregate at the outlet zone of constriction, therefore concluding that the shear rate is crucial for platelet adhesion and aggregation.85,87,90,97
Although these examples clearly contribute to advancing research and understanding of the coagulation process, most of these models are not practical to enhance clinical diagnosis. Several commercial devices are emerging with the idea to provide simple tools that can be used for clinical testing. Towards this aim, Mannino et al.97 proposed a commercial simple endothelial-coated cylindrical microchannel to test the effect of local vascular geometries on blood cell-endothelium interactions. One year later, Jain et al.98 demonstrated that a microfluidic device coated with ECs could be fixed and still retain their ability to modulate haemostasis under flow. The device was able to detect differences in patients taking antiplatelet medication and therefore providing a new tool for clinical laboratories for coagulation testing in more robust and practical diagnostic assays.
Mathur et al.99 created a model for thrombo-inflammation where they first isolated blood outgrowth endothelial cells (BOEC) from healthy subjects and then introduced these cells to microfluidic channels. These cells were then stimulated with TNFα and exposed to re-calcified human whole blood to compare the inflammatory response of these cells to models using HUVECs. Due to the easy isolation method of these BOECs, and a different response of healthy and unhealthy patients, this model could be used as a tool in personalized medicine approaches for certain pathologies, in which thrombo-inflammation is an essential contributor to disease exacerbation.
Sepsis is a disease often coinciding with thrombus formation in smaller vessels and arteries, which can lead to ischaemic events of the heart and subsequent HF.100 Many of the models described above can be used to model thrombotic sepsis in a microfluidic chip. In sepsis, high levels of inflammation in blood vessels are observed and accompanied by an elevation of TNF-α, interleukin-1, and interferons. These factors activate the endothelium to a pro-thrombotic state and strongly induce thrombus formation.101,102 After incubation with these factors, or other known contributors to sepsis, a blood perfusion assay can be performed to better understand more mechanisms involved in sepsis-associated thrombosis.
Finally, clinical applicability has also been demonstrated by using vessels-on-chips to predict thrombotic side effects in drug candidates prior to human clinical trials. For example, Tsai and collaborators used a vessel-on-chip device to perfuse blood samples from patients with sickle cell disease and test the effect of certain drugs in microvascular occlusion and thrombosis.93 Another study by Barrile et al.103 used a vascular channel coated with ECs and perfused whole human blood to study the potential for different drugs to promote blood clots. These studies serve as interesting examples for the potential of vessels-on-chips to evaluate thrombotic side effects that would otherwise be missed in prior animal or static cell culture studies. They are further proof of how OoCs can enhance drug safety during the process of developing novel treatment strategies.
2.5. Future importance of organs-on-a-chip in personalized cardiovascular medicine
Animal and genetic studies have implicated specific genes conferring increased susceptibility to many human diseases. Together with other emerging techniques (e.g. genome editing), OoCs have the potential to significantly enhance our understanding of how certain genes and epigenetic regulators are capable of influencing our personal CVD risk.
The novel genome editing CRISPR/Cas9 technology enables the introduction of targeted mutations or specific gene knock-outs104 into human cells. This methodology has allowed the generation of stable cell lines carrying the desired genetic mutation/knock-out while eliminating the effect of inter-individual variations due to the genetic background.
Although primary cells from specific individuals can be difficult to obtain and cannot be cultured indefinitely, stable cell lines with the desired genomic background can be generated from human iPSCs. Human iPSCs are obtained from a somatic cell and can be differentiated into all cell types.110,111 They can therefore be derived from accessible adult tissues of any patient, such as the blood or the skin. There are current protocols available for the generation of cardiomyocytes,112 defined atrial and ventricular cardiomyocyte subtypes,113 ECs as well as pericytes and vascular SMCs.114,115 An important limitation of iPSC-CMs relates to their immaturity. In fact, they share more similarities with foetal than adult human CMs. Although human iPSC-CMs express high levels of cardiac-specific genes and display a striated pattern for α-actinin and myosin light chain similar to the adult ventricular myocardium, their shape is typically roundish (and not elongated), whereas their cell body is smaller.111 In addition, the intrinsic immaturity of iPSC-CMs is reflected by electrophysiological impairments,111 which constitute a valid criticism when employing these cells for the investigation of arrhythmias. Various methods to enhance maturity are constantly developed and refined, including exposure to electrical stimulation, application of mechanical strain, and culturing human iPSC-CMs in three-dimensional tissue configuration.112 A great advantage of the iPSC differentiation protocols is that they allow for patient-specific iPSC-derived systems that can be conveniently edited to test the effect of specific disease-related mutations.113,114 The aforementioned gene-editing can be used to restore the effect of specific genes or mutations while exploring genotypic effects on specific phenotypes.
The opportunity to build OoC with disease-relevant cells can minimize the current challenges of most genomic studies. For example, vessel- and heart-on-chip designs could be used to test whether genetic associations emerging from recent CAD, VTE, HF, or IS GWAS will increase the disease risk through an effect in the cardiovascular system. At the same time, blood from individuals with a known genetic background can be perfused into different microchannels to study interactions between blood cells and the endothelium. Overall, the advanced possibilities of OoCs using novel available technologies offer great potential to finally elucidate the effect of genetic factors on CVD phenotypes, therefore enabling us to more rapidly and efficiently move towards the era of personalized medicine and pharmacogenomics. OoCs have already demonstrated their potential to support clinical trials of certain drug candidates.120 The opportunity to create patient-specific organ-on-chips could therefore pave the way for the development of precise and individualized therapies.
It is essential to mention that the majority of advances in personalized medicine stem from organoids-based research. In this context, Atchinson et al.116 have developed micro-engineered blood vessels based on SMCs derived from human iPSC of Hutchinson–Gilford progeria, a rare accelerating aging disorder that causes early onset of atherosclerosis. The work highlighted the possibility of the in vitro system to recapitulate the key features of the disease and how to utilize it as a patient-specific drug testing device. More recently, self-assembled 3D blood vessel organoids have been generated from human iPSCs. Blood vessel organoids are made of endothelial networks and pericytes that are genetically identical and can recapitulate the formation of a vascular lumen and basement membrane deposition.117 However, it is not yet possible to mimic blood flow perfusion in these models and therefore to obtain functional parameters, such as permeability or immune-cell adhesion/extravasation under defined in vitro conditions. This, as aforementioned, is one of the major advantages of OoC systems.
Moreover, several studies showing the integration of gene-editing technology and organoids culture systems have been published.118,119 An example was presented in a cystic fibrosis organoid model, where mutations in CFTR gene were repaired in intestinal stem cells by CRISPR/Cas9 system.120 Genome editing in iPSC-derived organoids have also been proved useful for drug testing,121 or to study the effect of virus infections.122,123 Very recent work in organoids made from iPSCs have shown that SARS-CoV-2 can infect engineered human blood vessel organoids and leak out into the bloodstream124 and that the infection could be inhibited by human recombinant soluble angiotensin-converting enzyme 2 (ACE2).
Few examples of personalized medicine accompanied by OoCs have been published in the CVD field.125,126 Wang et al.127 created a heart-on-a-chip using genetically engineered iPSC to proof that contractile deficiencies in cardiomyocytes associated with Barth syndrome were caused by a mutation in tafazzin gene, thus elucidating the biological mechanism and providing potential therapeutic targets to treat the disease. Another interesting approach is the integration of clinical data with the fabrication of OoC devices was used by Costa et al.,87 as described in detail in the atherosclerosis section above. Here, CTA data of a coronary artery formed the basis to construct personalized chips with the measured grade of stenosis. This method allowed the researchers to reproduce in vitro the unique (personalized) flow profile of individual patients and to observe the formation of thrombi dynamically. Similarly, blood vessel-on-chips can be perfused with human blood from individuals treated with different anticoagulant therapies to assess drug response of specific patients in a personalized manner.128
3. Conclusion and outlook
OoCs have advanced the drug development process by stimulating scientists to increase the level of complexity of better mimicking human biology and physiology on a more systemic level. Further, the possibility to integrate more than one organ in the same model is an important and on-going effort in the field.129–131 Microfluidic devices that contain the function of different organs have been connected with each other via vessels-on-chips to obtain the so-called ‘body-on-a-chip’. This concept captures the potential efficacy and toxicity of a drug in different organs.129,132 An important advancement in the validation of multi-organs-on-chip usage in drug screening has been the integration of sensors to ensure continuous assessment of the microenvironment parameters (pH, oxygen, and temperature).133 Kamei et al.134 have developed an integrated system composed by a healthy heart-on-chip connected to a liver cancer-on-chip and recapitulated the cardiotoxic effect of the anti-cancer drug doxorubicin. The side effect of the doxorubicin treatment on cardiac cells was due to toxic metabolites produced in the liver cancer cells.
Especially relevant for cardiovascular research are models that include both vasculature and the heart. This can be achieved either by building a microfabricated vascular network that cardiac tissue can be shaped around135 or by generating vascularized cardiac microtissues via co-culture with ECs. The latter approach shows a higher biological relevance but comes with the challenge to interface the microtissue constructs with microfluidic circuits for controlled perfusion. To recapitulate complex, multi-layered, and interconnected tissue architectures remain impossible with the current engineering approaches utilised to build OoCs. However, the possibility to combine biological self-assembly capabilities of organoids with the controllable assembly of microfabricated OoCs represents an attractive advancement of both technologies. This has been demonstrated by a novel microphysiological model of the human retina derived from hiPSCs incorporated in a two-channel chip separated by a thin porous membrane mimicking the endothelial barrier and enabling the exchange of nutrients and metabolites.136
An important challenge for cardiovascular OoC platforms is the ability to reproduce the chronic aspect underlying the progression of CVDs. Animal models in this respect are still somewhat irreplaceable; however, investigators can benefit from OoCs to study a specific mechanism and subsequently refine the targets to be evaluated in further animal models.
The field of OoC is growing, and an increasing number of academic groups and companies are becoming active in the development of OoC systems.19 All these OoC systems have different layouts and interfaces, which prevents them from being easily connected, interchanged, or compared. This lack of common standards for OoC systems is considered to be one of the major challenges in their future development and implementation.137 Initiatives are currently emerging in the field to address this challenge, for example, the development of a Translational Organ-on-Chip Platform, which aims to be an ‘open’ platform based on standards that are defined and supported by the stakeholders (developers, manufacturers, users) in the field (https://top.hdmt.technology/).
Standardization and harmonization are not only of importance for the technical aspects of an OoC but also for the cells and tissues that are integrated into the chips. Human cells are highly variable in terms of growth, stability, and function, particularly when primary tissues or stem cells are used as a source. Mainly, users from the industry consider access to human cell material, including patient material, to be a major challenge.138 In the field of stem-cell technology, efforts are made to address this issue by setting up open databases of available human stem-cell lines, including their characteristics, sources, and restrictions to use (e.g. https://hpscreg.eu/).
Overall, the use of OoC is rapidly moving from basic science to translational research to validate results from genomic studies and provide better models for drug testing, paving the way for personalized medicine. The combination of organ-on-chips with gene editing and iPSC use for better control of genetic background is becoming an innovative, attractive alternative for the functional study of the cardiovascular system.
Acknowledgements
We thank Susan Peacock (Uppsala University) for manuscript editing and support with the literature review.
Conflict of interest: none declared.
Funding
MS-L is supported by a Miguel Servet contract from the ISCIII Spanish Health Institute (CP17/00142) and co-financed by the European Social Fund. MT and SJ have received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 757444) and the Knut and Alice Wallenberg Foundation (grant number WAF 2016.0112). HM and AvdM acknowledge financial support from the Dutch Research Council (NWO) under the Gravitation Grant ‘NOCI’ Program (Grant No. 024.003.001). AvdM acknowledges funding received from the Dutch Cardiovascular Alliance (CVON-2018-29) and the European Research Council (ERC) under the Advanced Grant ‘VESCEL’ Program (Grant no. 669768). Research in LM’s laboratories is supported by the European Research Council (ERC Starting Grant no. 679777), a DZHK Junior Research Group (JRG_LM_MRI), the SFB1123 and TRR267 of the German Research Council (DFG), the National Institutes of Health (NIH; 1R011HL150359-01), and the Bavarian State Ministry of Health and Care through the research project DigiMed Bayern.
References
- 1. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FGR, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo J-P, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KMV, Nasseri K, Norman P, O'Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De León FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh P-H, Yip P, Zabetian A, Zheng Z-J, Lopez AD, Murray CJL, AlMazroa MA, Memish ZA.. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Townsend N, Wilson L, Bhatnagar P, Wickramasinghe K, Rayner M, Nichols M.. Cardiovascular disease in Europe: epidemiological update 2016. Eur Heart J 2016;37:3232–3245. [DOI] [PubMed] [Google Scholar]
- 3. Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, Tokgözoğlu L, Lewis EF.. Atherosclerosis. Nat Rev Dis Primers 2019;5:56. [DOI] [PubMed] [Google Scholar]
- 4. Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z.. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol 2019;16:727–744. [DOI] [PubMed] [Google Scholar]
- 5. Phillippe HM. Overview of venous thromboembolism. Am J Manag Care 2017;23:S376–S382. [PubMed] [Google Scholar]
- 6. Bahit MC, Kochar A, Granger CB.. Post-myocardial infarction heart failure. JACC: Heart Failure 2018;6:179–186. [DOI] [PubMed] [Google Scholar]
- 7. Burchfield JS, Xie M, Hill JA.. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation 2013;128:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kehat I, Molkentin JD.. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation 2010;122:2727–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Page RL, O’Bryant CL, Cheng D, Dow TJ, Ky B, Stein CM, Spencer AP, Trupp RJ, Lindenfeld JAnn, Lindenfeld J, American Heart Association Clinical Pharmacology and Heart Failure and Transplantation Committees of the Council on Clinical Cardiology; Council on Cardiovascular Surgery and Anesthesia; Council on Cardiovascular and Stroke Nursing; and Council on Quality of Care and Outcomes Research. Drugs that may cause or exacerbate heart failure: a scientific statement from the American Heart Association. Circulation 2016;134:e32–e69. [DOI] [PubMed] [Google Scholar]
- 10. Albini A, Pennesi G, Donatelli F, Cammarota R, De Flora S, Noonan DM.. Cardiotoxicity of anticancer drugs: the need for cardio-oncology and cardio-oncological prevention. J Natl Cancer Inst 2010;102:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimäki T, Lu X, Lu Y, März W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Björkegren JLM, Schunkert H, Farrall M, Danesh J, Samani NJ, Watkins H, Deloukas P, EPIC-CVD Consortium. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385–1391. [DOI] [PubMed] [Google Scholar]
- 12. Lindström S, Wang L, Smith EN, Gordon W, Vlieg AH, de Andrade M, Brody JA, Pattee JW, Haessler J, Brumpton BM, Chasman DI, Suchon P, Chen M-H, Turman C, Germain M, Wiggins KL, MacDonald J, Braekkan SK, Armasu SM, Pankratz N, Jackson RD, Nielsen JB, Giulianini F, Puurunen MK, Ibrahim M, Heckbert SR, Damrauer SM, Natarajan P, Klarin D, de Vries PS, Sabater-Lleal M, Huffman JE, Bammler TK, Frazer KA, McCauley BM, Taylor K, Pankow JS, Reiner AP, Gabrielsen ME, Deleuze J-F, O'Donnell CJ, Kim J, McKnight B, Kraft P, Hansen J-B, Rosendaal FR, Heit JA, Psaty BM, Tang W, Kooperberg C, Hveem K, Ridker PM, Morange P-E, Johnson AD, Kabrhel C, Trégouët D-A, Smith NL, Million Veteran Program. Genomic and transcriptomic association studies identify 16 novel susceptibility loci for venous thromboembolism. Blood 2019;134:1645–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klarin D, Busenkell E, Judy R, Lynch J, Levin M, Haessler J, Aragam K, Chaffin M, Haas M, Lindström S, Assimes TL, Huang J, Min Lee K, Shao Q, Huffman JE, Kabrhel C, Huang Y, Sun YV, Vujkovic M, Saleheen D, Miller DR, Reaven P, DuVall S, Boden WE, Pyarajan S, Reiner AP, Trégouët D-A, Henke P, Kooperberg C, Gaziano JM, Concato J, Rader DJ, Cho K, Chang K-M, Wilson PWF, Smith NL, O’Donnell CJ, Tsao PS, Kathiresan S, Obi A, Damrauer SM, Natarajan P, Veterans Affairs’ Million Veteran Program. Genome-wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat Genet 2019;51:1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen W-M, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu F-C, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee J-M, Lemmens R, Leys D, Lewis CM, Lin W-Y, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O'Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S, Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Björkegren JLM, Codoni V, Civelek M, Smith NL, Trégouët DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD, Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT, Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M, MEGASTROKE Consortium. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, Hedman ÅK, Wilk JB, Morley MP, Chaffin MD, Helgadottir A, Verweij N, Dehghan A, Almgren P, Andersson C, Aragam KG, Ärnlöv J, Backman JD, Biggs ML, Bloom HL, Brandimarto J, Brown MR, Buckbinder L, Carey DJ, Chasman DI, Chen X, Chen X, Chung J, Chutkow W, Cook JP, Delgado GE, Denaxas S, Doney AS, Dörr M, Dudley SC, Dunn ME, Engström G, Esko T, Felix SB, Finan C, Ford I, Ghanbari M, Ghasemi S, Giedraitis V, Giulianini F, Gottdiener JS, Gross S, Guðbjartsson DF, Gutmann R, Haggerty CM, van der Harst P, Hyde CL, Ingelsson E, Jukema JW, Kavousi M, Khaw K-T, Kleber ME, Køber L, Koekemoer A, Langenberg C, Lind L, Lindgren CM, London B, Lotta LA, Lovering RC, Luan J, Magnusson P, Mahajan A, Margulies KB, März W, Melander O, Mordi IR, Morgan T, Morris AD, Morris AP, Morrison AC, Nagle MW, Nelson CP, Niessner A, Niiranen T, O'Donoghue ML, Owens AT, Palmer CNA, Parry HM, Perola M, Portilla-Fernandez E, Psaty BM, Rice KM, Ridker PM, Romaine SPR, Rotter JI, Salo P, Salomaa V, van Setten J, Shalaby AA, Smelser DT, Smith NL, Stender S, Stott DJ, Svensson P, Tammesoo M-L, Taylor KD, Teder-Laving M, Teumer A, Thorgeirsson G, Thorsteinsdottir U, Torp-Pedersen C, Trompet S, Tyl B, Uitterlinden AG, Veluchamy A, Völker U, Voors AA, Wang X, Wareham NJ, Waterworth D, Weeke PE, Weiss R, Wiggins KL, Xing H, Yerges-Armstrong LM, Yu B, Zannad F, Zhao JH, Hemingway H, Samani NJ, McMurray JJV, Yang J, Visscher PM, Newton-Cheh C, Malarstig A, Holm H, Lubitz SA, Sattar N, Holmes MV, Cappola TP, Asselbergs FW, Hingorani AD, Kuchenbaecker K, Ellinor PT, Lang CC, Stefansson K, Smith JG, Vasan RS, Swerdlow DI, Lumbers RT, Regeneron Genetics Center. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noviello TMR, Di Liddo A, Ventola GM, Spagnuolo A, D’Aniello S, Ceccarelli M, Cerulo L.. Detection of long non-coding RNA homology, a comparative study on alignment and alignment–free metrics. BMC Bioinformatics 2018;19:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pampaloni F, Reynaud EG, Stelzer EHK.. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 2007;8:839–845. [DOI] [PubMed] [Google Scholar]
- 18. Ronaldson-Bouchard K, Vunjak-Novakovic G.. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell 2018;22:310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mastrangeli M, Millet S, Eijnden-van Raaij JVD, The ORCHID partners. Organ-on-chip in development: towards a roadmap for organs-on-chip. Atlex 2019;650–668. [DOI] [PubMed] [Google Scholar]
- 20. Jackson EL, Lu H.. Three-dimensional models for studying development and disease: moving on from organisms to organs-on-a-chip and organoids. Integr Biol (Camb) 2016;8:672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huh D, Torisawa Y, Hamilton GA, Kim HJ, Ingber DE.. Microengineered physiological biomimicry: organs-on-chips. Lab Chip 2012;12:2156–2164. [DOI] [PubMed] [Google Scholar]
- 22. An F, Qu Y, Liu X, Zhong R, Luo Y.. Organ-on-a-chip: new platform for biological analysis. Anal Chem Insight 2015;10:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H, Wang Y, Cui K, Guo Y, Zhang X, Qin J.. Advances in hydrogels in organoids and organs‐on‐a‐chip. Adv Mater 2019;31:1902042. [DOI] [PubMed] [Google Scholar]
- 24. Duffy DC, McDonald JC, Schueller OJ, Whitesides GM.. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal Chem 1998;70:4974–4984. [DOI] [PubMed] [Google Scholar]
- 25. Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF, Lam WA.. Microvasculature-on-a-chip for the long-term study of endothelial barrier dysfunction and microvascular obstruction in disease. Nat Biomed Eng 2018;2:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cho M, Park J-K. Fabrication of a perfusable 3D in vitro artery-mimicking multichannel system for artery disease models. ACS Biomater Sci Eng2020; 6: 5326–5336. [DOI] [PubMed] [Google Scholar]
- 27. Meer BV, Vries HD, Firth KSA, Weerd JV, Tertoolen LGJ, Karperien HBJ, Jonkheijm P, Denning C, IJzerman AP, Mummery CL.. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun 2017;482:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carter S-SD, Atif A-R, Kadekar S, Lanekoff I, Engqvist H, Varghese OP, Tenje M, Mestres G.. PDMS leaching and its implications for on-chip studies focusing on bone regeneration applications. Organs-on-a-Chip 2020;2:100004. [Google Scholar]
- 29. Campbell S, Wu Q, Yazbeck J, Liu C, Okhovatian S, Radisic M.. Beyond polydimethylsiloxane: alternative materials for fabrication of organ on a chip devices and microphysiological systems. ACS Biomater Sci Eng. DOI: 10.1021/acsbiomaterials.0c00640. [DOI] [PubMed] [Google Scholar]
- 30. Shin Y, Han S, Jeon JS, Yamamoto K, Zervantonakis IK, Sudo R, Kamm RD, Chung S.. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat Protoc 2012;7:1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S, Lee H, Chung M, Jeon NL.. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip 2013;13:1489–1500. [DOI] [PubMed] [Google Scholar]
- 32. Lee D-H, Bae CY, Kwon S, Park J-K.. User-friendly 3D bioassays with cell-containing hydrogel modules: narrowing the gap between microfluidic bioassays and clinical end-users’ needs. Lab Chip 2015;15:2379–2387. [DOI] [PubMed] [Google Scholar]
- 33. Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, Lanz HL, Nicolas A, Ng CP, Joore J, Kustermann S, Roth A, Hankemeier T, Moisan A, Vulto P.. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat Commun 2017;8:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD.. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci USA 2012;109:9342–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X-Y, Jin Z-H, Gan B-W, Lv S-W, Xie M, Huang W-H.. Engineering interconnected 3D vascular networks in hydrogels using molded sodium alginate lattice as the sacrificial template. Lab Chip 2014;14:2709–2716. [DOI] [PubMed] [Google Scholar]
- 36. Bischel LL, Lee S-H, Beebe DJ.. A practical method for patterning lumens through ECM hydrogels via viscous finger patterning. J Lab Autom 2012;17:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herland A, Meer AVD, FitzGerald EA, Park T-E, Sleeboom JJF, Ingber DE.. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3d human blood-brain barrier on a chip. PLoS One 2016;11:e0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Graaf M. D, Cochrane A, Hil FVD, Buijsman W, Meer AVD, Berg AVD, Mummery CL, Orlova VV.. Scalable microphysiological system to model three-dimensional blood vessels. APL Bioeng 2019;3:026105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tenje M, Cantoni F, Porras Hernández AM, Searle SS, Johansson S, Barbe L, Antfolk M, Pohlit HA.. Practical guide to microfabrication and patterning of hydrogels for biomimetic cell culture scaffolds. Organs-on-a-Chip 2020;2:100003. [Google Scholar]
- 40. Hsu Y-H, Moya ML, Hughes CCW, George SC, Lee AP.. A microfluidic platform for generating large-scale nearly identical human microphysiological vascularized tissue arrays. Lab Chip 2013;13:2990–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shin Y, Jeon JS, Han S, Jung G-S, Shin S, Lee S-H, Sudo R, Kamm RD, Chung S.. In vitro 3D collective sprouting angiogenesis under orchestrated ANG-1 and VEGF gradients. Lab Chip 2011;11:2175–2181. [DOI] [PubMed] [Google Scholar]
- 42. Günther A, Yasotharan S, Vagaon A, Lochovsky C, Pinto S, Yang J, Lau C, Voigtlaender-Bolz J, Bolz S-S.. A microfluidic platform for probing small artery structure and function. Lab Chip 2010;10:2341–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Veldhuizen J, Cutts J, Brafman DA, Migrino RQ, Nikkhah M.. Engineering anisotropic human stem cell-derived three-dimensional cardiac tissue on-a-chip. Biomaterials 2020;256:120195. [DOI] [PubMed] [Google Scholar]
- 44. Weng K-C, Kurokawa YK, Hajek BS, Paladin JA, Shirure VS, George SC.. Human induced pluripotent stem-cardiac-endothelial-tumor-on-a-chip to assess anticancer efficacy and cardiotoxicity. Tissue Eng Part C Methods 2020;26:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellis BW, Acun A, Can UI, Zorlutuna P.. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics 2017;11:024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maoz BM, Herland A, Henry OYF, Leineweber WD, Yadid M, Doyle J, Mannix R, Kujala VJ, FitzGerald EA, Parker KK, Ingber DE.. Organs-on-chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017;17:2294–2302. [DOI] [PubMed] [Google Scholar]
- 47. Chen MB, Srigunapalan S, Wheeler AR, Simmons CA.. A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell–cell interactions. Lab Chip 2013;13:2591–2598. [DOI] [PubMed] [Google Scholar]
- 48. Schneider O, Zeifang L, Fuchs S, Sailer C, Loskill P.. User-friendly and parallelized generation of human induced pluripotent stem cell-derived microtissues in a centrifugal heart-on-a-chip. Tissue Eng Part A 2019;25:786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kong M, Lee J, Yazdi IK, Miri AK, Lin Y-D, Seo J, Zhang YS, Khademhosseini A, Shin SR.. Cardiac fibrotic remodeling on a chip with dynamic mechanical stimulation. Adv Healthcare Mater 2019;8:1801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marsano A, Conficconi C, Lemme M, Occhetta P, Gaudiello E, Votta E, Cerino G, Redaelli A, Rasponi M.. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016;16:599–610. [DOI] [PubMed] [Google Scholar]
- 51. Agarwal A, Goss JA, Cho A, McCain ML, Parker KK.. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013;13:3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA.. Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci USA 2016;113:3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mehrotra S, Melo BAG, Hirano M, Keung W, Li RA, Mandal BB, Shin SR.. Nonmulberry silk based ink for fabricating mechanically robust cardiac patches and endothelialized myocardium‐on‐a‐chip application. Adv Funct Mater 2020;30:1907436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pires de Mello CP, Carmona-Moran C, McAleer CW, Perez J, Coln EA, Long CJ, Oleaga C, Riu A, Note R, Teissier S, Langer J, Hickman JJ.. Microphysiological heart-liver body-on-a-chip system with a skin mimic for evaluating topical drug delivery. Lab Chip 2020;20:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoshida S, Sumomozawa K, Nagamine K, Nishizawa M.. Hydrogel microchambers integrated with organic electrodes for efficient electrical stimulation of human iPSC‐derived cardiomyocytes. Macromol Biosci 2019;19:1900060. [DOI] [PubMed] [Google Scholar]
- 56. Koutsouras DA, Perrier R, Villarroel Marquez A, Pirog A, Pedraza E, Cloutet E, Renaud S, Raoux M, Malliaras GG, Lang J.. Simultaneous monitoring of single cell and of micro-organ activity by PEDOT: PSS covered multi-electrode arrays. Mater Sci Eng C Mater Biol Appl 2017;81:84–89. [DOI] [PubMed] [Google Scholar]
- 57. Gaio N, Meer B. V, Quirós Solano W, Bergers L, Stolpe A. V D, Mummery C, Sarro P, Dekker R.. Cytostretch, an organ-on-chip platform. Micromachines 2016;7:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jacobson S, Piper H.. Cell cultures of adult cardiomyocytes as models of the myocardium. J Mol Cell Cardiol 1986;18:661–678. [DOI] [PubMed] [Google Scholar]
- 59. Liu J, Laksman Z, Backx PH.. The electrophysiological development of cardiomyocytes. Adv Drug Deliv Rev 2016;96:253–273. [DOI] [PubMed] [Google Scholar]
- 60. Zhang N, Stauffer F, Simona BR, Zhang F, Zhang Z-M, Huang N-P, Vörös J.. Multifunctional 3D electrode platform for real-time in situ monitoring and stimulation of cardiac tissues. Biosens Bioelectron 2018;112:149–155. [DOI] [PubMed] [Google Scholar]
- 61. Parsa H, Wang BZ, Vunjak-Novakovic G.. A microfluidic platform for the high-throughput study of pathological cardiac hypertrophy. Lab Chip 2017;17:3264–3271. [DOI] [PubMed] [Google Scholar]
- 62. Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Healy KE.. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep 2015;5:8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Caspi O, Itzhaki I, Kehat I, Gepstein A, Arbel G, Huber I, Satin J, Gepstein L.. In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev 2009;18:161–172. [DOI] [PubMed] [Google Scholar]
- 64. Kujala VJ, Pasqualini FS, Goss JA, Nawroth JC, Parker KK.. Laminar ventricular myocardium on a microelectrode array-based chip. J Mater Chem B 2016;4:3534–3543. [DOI] [PubMed] [Google Scholar]
- 65. Qian F, Huang C, Lin Y-D, Ivanovskaya AN, O'Hara TJ, Booth RH, Creek CJ, Enright HA, Soscia DA, Belle AM, Liao R, Lightstone FC, Kulp KS, Wheeler EK.. Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip 2017;17:1732–1739. [DOI] [PubMed] [Google Scholar]
- 66. Oyunbaatar N-E, Dai Y, Shanmugasundaram A, Lee B-K, Kim E-S, Lee D-W.. Development of a next-generation biosensing platform for simultaneous detection of mechano- and electrophysiology of the drug-induced cardiomyocytes. ACS Sens 2019;4:2623–2630. [DOI] [PubMed] [Google Scholar]
- 67. Liu H, Bolonduro OA, Hu N, Ju J, Rao AA, Duffy BM, Huang Z, Black LD, Timko BP.. Heart-on-a-chip model with integrated extra- and intracellular bioelectronics for monitoring cardiac electrophysiology under acute hypoxia. Nano Lett 2020;20:2585–2593. [DOI] [PubMed] [Google Scholar]
- 68. Sharf T, Hansma PK, Hari MA, Kosik KS.. Non-contact monitoring of extra-cellular field potentials with a multi-electrode array. Lab Chip 2019;19:1448–1457. [DOI] [PubMed] [Google Scholar]
- 69. Mori N, Morimoto Y, Takeuchi S, Transendothelial electrical resistance (TEER) measurement system of 3D tubular vascular channel. In: 2018 IEEE Micro Electro Mechanical Systems (MEMS). Belfast: IEEE, 2018. pp. 322–325.
- 70. Pan Y, Jiang D, Gu C, Qiu Y, Wan H, Wang P.. 3D microgroove electrical impedance sensing to examine 3D cell cultures for antineoplastic drug assessment. Microsyst Nanoeng 2020;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Inal S, Hama A, Ferro M, Pitsalidis C, Oziat J, Iandolo D, Pappa A-M, Hadida M, Huerta M, Marchat D, Mailley P, Owens RM.. Conducting polymer scaffolds for hosting and monitoring 3D cell culture. Adv Biosys 2017;1:1700052. [Google Scholar]
- 72. Weinberger F, Mannhardt I, Eschenhagen T.. Engineering cardiac muscle tissue: a maturating field of research. Circ Res 2017;120:1487–1500. [DOI] [PubMed] [Google Scholar]
- 73. Jastrzebska E, Tomecka E, Jesion I.. Heart-on-a-chip based on stem cell biology. Biosens Bioelectron 2016;75:67–81. [DOI] [PubMed] [Google Scholar]
- 74. Kitsara M, Kontziampasis D, Agbulut O, Chen Y.. Heart on a chip: micro-nanofabrication and microfluidics steering the future of cardiac tissue engineering. Microelectron Eng 2019;203-204:44–62. [Google Scholar]
- 75. Conant G, Lai BFL, Lu RXZ, Korolj A, Wang EY, Radisic M.. High-content assessment of cardiac function using heart-on-a-chip devices as drug screening model. Stem Cell Rev Rep 2017;13:335–346. [DOI] [PubMed] [Google Scholar]
- 76. Cho KW, Lee WH, Kim B-S, Kim D-H.. Sensors in heart-on-a-chip: a review on recent progress. Talanta 2020;219:121269. [DOI] [PubMed] [Google Scholar]
- 77. Zhao Y, Rafatian N, Wang EY, Wu Q, Lai BFL, Lu RX, Savoji H, Radisic M.. Towards chamber specific heart-on-a-chip for drug testing applications. Adv Drug Deliv Rev 2020;165–166:60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mastikhina O, Moon BU, Williams K, Hatkar R, Gustafson D, Mourad O, Sun X, Koo M, Lam AYL, Sun Y, Fish JE, Young EWK, Nunes SS.. Human cardiac fibrosis-on-a-chip model recapitulates disease hallmarks and can serve as a platform for drug testing. Biomaterials 2020;233:119741. [DOI] [PubMed] [Google Scholar]
- 79. Savoji H, Mohammadi MH, Rafatian N, Toroghi MK, Wang EY, Zhao Y, Korolj A, Ahadian S, Radisic M.. Cardiovascular disease models: a game changing paradigm in drug discovery and screening. Biomaterials 2019;198:3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gintant G, Burridge P, Gepstein L, Harding S, Herron T, Hong C, Jalife J, Wu JC, on behalf of the American Heart Association Council on Basic Cardiovascular Sciences. Use of human induced pluripotent stem cell–derived cardiomyocytes in preclinical cancer drug cardiotoxicity testing: a scientific statement from the American Heart Association. Circ Res 2019;125:75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Karbassi E, Fenix A, Marchiano S, Muraoka N, Nakamura K, Yang X, Murry CE.. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat Rev Cardiol 2020;17:341–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Strauss DG, Gintant G, Li Z, Wu W, Blinova K, Vicente J, Turner JR, Sager PT.. Comprehensive in vitro proarrhythmia assay (CiPA) update from a Cardiac Safety Research Consortium/Health and Environmental Sciences Institute/FDA meeting. Ther Innov Regul Sci 2019;53:519–525. [DOI] [PubMed] [Google Scholar]
- 83. Saleem U, Meer B. V, Katili PA, Mohd Yusof NAN, Mannhardt I, Garcia AK, Tertoolen L, Korte TD, Vlaming MLH, McGlynn K, Nebel J, Bahinski A, Harris K, Rossman E, Xu X, Burton FL, Smith GL, Clements P, Mummery CL, Eschenhagen T, Hansen A, Denning C.. Blinded, multicenter evaluation of drug-induced changes in contractility using human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol Sci 2020;176:103–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lee EK, Tran DD, Keung W, Chan P, Wong G, Chan CW, Costa KD, Li RA, Khine M.. Machine learning of human pluripotent stem cell-derived engineered cardiac tissue contractility for automated drug classification. Stem Cell Report 2017;9:1560–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Westein E, Meer AVD, Kuijpers MJE, Frimat J-P, Berg AVD, Heemskerk JWM.. Atherosclerotic geometries exacerbate pathological thrombus formation poststenosis in a von Willebrand factor-dependent manner. Proc Natl Acad Sci USA 2013;110:1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tovar-Lopez FJ, Rosengarten G, Westein E, Khoshmanesh K, Jackson SP, Mitchell A, Nesbitt WS.. A microfluidics device to monitor platelet aggregation dynamics in response to strain rate micro-gradients in flowing blood. Lab Chip 2010;10:291–302. [DOI] [PubMed] [Google Scholar]
- 87. Costa PF, Albers HJ, Linssen JEA, Middelkamp HHT, Hout LVD, Passier R, Berg AVD, Malda J, Meer AVD.. Mimicking arterial thrombosis in a 3D-printed microfluidic in vitro vascular model based on computed tomography angiography data. Lab Chip 2017;17:2785–2792. [DOI] [PubMed] [Google Scholar]
- 88. Blann AD, Lip GY.. Virchow’s triad revisited: the importance of soluble coagulation factors, the endothelium, and platelets. Thromb Res 2001;101:321–327. [DOI] [PubMed] [Google Scholar]
- 89. Lowe GDO. Virchow’s triad revisited: abnormal flow. Pathophysiol Haemost Thromb 2003;33:455–457. [DOI] [PubMed] [Google Scholar]
- 90. Nesbitt WS, Westein E, Tovar-Lopez FJ, Tolouei E, Mitchell A, Fu J, Carberry J, Fouras A, Jackson SP.. A shear gradient-dependent platelet aggregation mechanism drives thrombus formation. Nat Med 2009;15:665–673. [DOI] [PubMed] [Google Scholar]
- 91. Westein E, de Witt S, Lamers M, Cosemans JMEM, Heemskerk JWM.. Monitoring in vitro thrombus formation with novel microfluidic devices. Platelets 2012;23:501–509. [DOI] [PubMed] [Google Scholar]
- 92. Zhu S, Herbig BA, Li R, Colace TV, Muthard RW, Neeves KB, Diamond SL.. In microfluidico: recreating in vivo hemodynamics using miniaturized devices. Biorheology 2015;52:303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tsai M, Kita A, Leach J, Rounsevell R, Huang JN, Moake J, Ware RE, Fletcher DA, Lam WA.. In vitro modeling of the microvascular occlusion and thrombosis that occur in hematologic diseases using microfluidic technology. J Clin Invest 2012;122:408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Brouns SLN, Provenzale I, Geffen JV, Meijden PVD, Heemskerk JWM.. Localized endothelial-based control of platelet aggregation and coagulation under flow: a proof-of-principle vessel-on-a-chip study. J Thromb Haemost 2020;18:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hinsbergh VV. Endothelium – role in regulation of coagulation and inflammation. Semin Immunopathol 2012;34:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Versteeg HH, Heemskerk JWM, Levi M, Reitsma PH.. New fundamentals in hemostasis. Physiol Rev 2013;93:327–358. [DOI] [PubMed] [Google Scholar]
- 97. Mannino RG, Myers DR, Ahn B, Wang Y, Gole H, Lin AS, Guldberg RE, Giddens DP, Timmins LH, Lam WA.. Do-it-yourself in vitro vasculature that recapitulates in vivo geometries for investigating endothelial-blood cell interactions. Sci Rep 2015;5:12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jain A, Meer AVD, Papa A-L, Barrile R, Lai A, Schlechter BL, Otieno MA, Louden CS, Hamilton GA, Michelson AD, Frelinger AL, Ingber DE.. Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium. Biomed Microdev 2016;18:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mathur T, Singh KA, Pandian NKR, Tsai S-H, Hein TW, Gaharwar AK, Flanagan JM, Jain A.. Organ-on-chips made of blood: endothelial progenitor cells from blood reconstitute vascular thromboinflammation in vessel-chips. Lab Chip 2019;19:2500–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gami AS, Hayman SR, Grande JP, Garovic VD.. Incidence and prognosis of acute heart failure in the thrombotic microangiopathies. Am J Med 2005;118:544–547. [DOI] [PubMed] [Google Scholar]
- 101. Albers P, van den B, van der M.. Automated analysis of platelet aggregation on cultured endothelium in a microfluidic chip perfused with human whole blood. Micromachines 2019;10:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Belair DG, Whisler JA, Valdez J, Velazquez J, Molenda JA, Vickerman V, Lewis R, Daigh C, Hansen TD, Mann DA, Thomson JA, Griffith LG, Kamm RD, Schwartz MP, Murphy WL.. Human vascular tissue models formed from human induced pluripotent stem cell derived endothelial cells. Stem Cell Rev Rep 2015;11:511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Barrile R, Meer AVD, Park H, Fraser JP, Simic D, Teng F, Conegliano D, Nguyen J, Jain A, Zhou M, Karalis K, Ingber DE, Hamilton GA, Otieno MA.. Organ-on-chip recapitulates thrombosis induced by an anti-CD154 monoclonal antibody: translational potential of advanced microengineered systems. Clin Pharmacol Ther 2018;104:1240–1248. [DOI] [PubMed] [Google Scholar]
- 104. Hsu PD, Lander ES, Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stadtfeld M, Hochedlinger K.. Induced pluripotency: history, mechanisms, and applications. Genes Dev 2010;24:2239–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Takahashi K, Yamanaka S.. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol 2016;17:183–193. [DOI] [PubMed] [Google Scholar]
- 107. Batalov I, Feinberg AW.. Differentiation of cardiomyocytes from human pluripotent stem cells using monolayer culture. Biomark Insight 2015;10:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kleinsorge M, Cyganek L.. Subtype-directed differentiation of human iPSCs into atrial and ventricular cardiomyocytes. STAR Protoc 2020;1:100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sinha S, Iyer D, Granata A.. Embryonic origins of human vascular smooth muscle cells: implications for in vitro modeling and clinical application. Cell Mol Life Sci 2014;71:2271–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lin Y, Gil C-H, Yoder MC.. Differentiation, evaluation, and application of human induced pluripotent stem cell-derived endothelial cells. Arterioscler Thromb Vasc Biol 2017;37:2014–2025. [DOI] [PubMed] [Google Scholar]
- 111. Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ.. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circul Res 2009;104:30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Casini S, Verkerk AO, Remme CA.. Human iPSC-derived cardiomyocytes for investigation of disease mechanisms and therapeutic strategies in inherited arrhythmia syndromes: strengths and limitations. Cardiovasc Drug Ther 2017;31:325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Freiermuth JL, Powell-Castilla IJ, Gallicano GI.. Toward a CRISPR Picture: use of CRISPR/Cas9 to model diseases in human stem cells in vitro. J Cell Biochem 2018;119:62–68. [DOI] [PubMed] [Google Scholar]
- 114. Hockemeyer D, Jaenisch R.. Induced pluripotent stem cells meet genome editing. Cell Stem Cell 2016;18:573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Jang K-J, Otieno MA, Ronxhi J, Lim H-K, Ewart L, Kodella KR, Petropolis DB, Kulkarni G, Rubins JE, Conegliano D, Nawroth J, Simic D, Lam W, Singer M, Barale E, Singh B, Sonee M, Streeter AJ, Manthey C, Jones B, Srivastava A, Andersson LC, Williams D, Park H, Barrile R, Sliz J, Herland A, Haney S, Karalis K, Ingber DE, Hamilton GA.. Reproducing human and cross-species drug toxicities using a liver-chip. Sci Transl Med 2019;11:eaax5516. [DOI] [PubMed] [Google Scholar]
- 116. Atchison L, Zhang H, Cao K, Truskey GA.. A tissue engineered blood vessel model of hutchinson-gilford progeria syndrome using human iPSC-derived smooth muscle cells. Sci Rep 2017;7:8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wimmer RA, Leopoldi A, Aichinger M, Kerjaschki D, Penninger JM.. Generation of blood vessel organoids from human pluripotent stem cells. Nat Protoc 2019;14:3082–3100. [DOI] [PubMed] [Google Scholar]
- 118. Nie J, Hashino E.. Organoid technologies meet genome engineering. EMBO Rep 2017;18:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, Chuva de Sousa Lopes S, Zon J. V, Tans S, Clevers H.. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol 2020;22:321–331. [DOI] [PubMed] [Google Scholar]
- 120. Schwank G, Koo B-K, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, Ent CVD, Nieuwenhuis EES, Beekman JM, Clevers H.. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013;13:653–658. [DOI] [PubMed] [Google Scholar]
- 121. Corbett JL, Duncan SA.. iPSC-derived hepatocytes as a platform for disease modeling and drug discovery. Front Med (Lausanne) 2019;6:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, Kommineni S, Bilican B, Klim JR, Hill EJ, Kane LT, Ye C, Kaykas A, Eggan K.. Genetic ablation of AXL does not protect human neural progenitor cells and cerebral organoids from zika virus infection. Cell Stem Cell 2016;19:703–708. [DOI] [PubMed] [Google Scholar]
- 123. Kim J, Koo B-K, Yoon K-J.. Modeling host-virus interactions in viral infectious diseases using stem-cell-derived systems and CRISPR/Cas9 technology. Viruses 2019; 11:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM.. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020;181:905–913.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Berg A. V D, Mummery CL, Passier R, Meer A. V D.. Personalised organs-on-chips: functional testing for precision medicine. Lab Chip 2019;19:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ribas J, Sadeghi H, Manbachi A, Leijten J, Brinegar K, Zhang YS, Ferreira L, Khademhosseini A.. Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl in Vitro Toxicol 2016;2:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang D-Z, Li K, Wang J, Wanders RJA, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT.. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 2014;20:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jain A, Graveline A, Waterhouse A, Vernet A, Flaumenhaft R, Ingber DE.. A shear gradient-activated microfluidic device for automated monitoring of whole blood haemostasis and platelet function. Nat Commun 2016;7:10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, Lindner M, Drewell C, Bauer S, Thomas A, Sambo NS, Sonntag F, Lauster R, Marx U.. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015;15:2688–2699. [DOI] [PubMed] [Google Scholar]
- 130. Oleaga C, Bernabini C, Smith AST, Srinivasan B, Jackson M, McLamb W, Platt V, Bridges R, Cai Y, Santhanam N, Berry B, Najjar S, Akanda N, Guo X, Martin C, Ekman G, Esch MB, Langer J, Ouedraogo G, Cotovio J, Breton L, Shuler ML, Hickman JJ.. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep 2016;6:20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Satoh T, Sugiura S, Shin K, Onuki-Nagasaki R, Ishida S, Kikuchi K, Kakiki M, Kanamori T.. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab Chip 2017;18:115–125. [DOI] [PubMed] [Google Scholar]
- 132. Sung JH, Wang YI, Sriram NN, Jackson M, Long C, Hickman JJ, Shuler ML.. Recent advances in body-on-a-chip systems. Anal Chem 2019;91:330–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, Avci H, Zhang W, Silvestri A, Sanati Nezhad A, Manbohi A, De Ferrari F, Polini A, Calzone G, Shaikh N, Alerasool P, Budina E, Kang J, Bhise N, Ribas J, Pourmand A, Skardal A, Shupe T, Bishop CE, Dokmeci MR, Atala A, Khademhosseini A.. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci USA 2017;114:E2293–E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kamei K, Kato Y, Hirai Y, Ito S, Satoh J, Oka A, Tsuchiya T, Chen Y, Tabata O.. Integrated heart/cancer on a chip to reproduce the side effects of anti-cancer drugs in vitro. RSC Adv 2017;7:36777–36786. [Google Scholar]
- 135. Lai BFL, Huyer LD, Lu RXZ, Drecun S, Radisic M, Zhang B.. InVADE: integrated vasculature for assessing dynamic events. Adv Funct Mater 2017;27:1703524. [Google Scholar]
- 136. Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J, Nikolova M, Cora V, Antkowiak L, Haq W, Shen N, Schenke-Layland K, Ueffing M, Liebau S, Loskill P.. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife 2019;8:e46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mastrangeli M. Building blocks for a European organ-on-chip roadmap. Altex 2019;481–492. [DOI] [PubMed] [Google Scholar]
- 138. Middelkamp HHT, Meer AVD, Hummel JM, Stamatialis DF, Mummery CL, Passier R, IJzerman MJ.. Organs-on-chips in drug development: the importance of involving stakeholders in Early Health Technology Assessment. Appl In Vitro Toxicol 2016;2:74–81. [Google Scholar]
- 139. Hasan A, Paul A, Memic A, Khademhosseini A.. A multilayered microfluidic blood vessel-like structure. Biomed Microdev 2015;17:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jia L, Han F, Yang H, Turnbull G, Wang J, Clarke J, Shu W, Guo M, Li B.. Microfluidic fabrication of biomimetic helical hydrogel microfibers for blood‐vessel‐on‐a‐chip applications. Adv Healthcare Mater 2019;8:1900435. [DOI] [PubMed] [Google Scholar]
- 141. Loessberg-Zahl J, Meer AVD, Berg AVD, Eijkel JCT.. Flow focusing through gels as a tool to generate 3D concentration profiles in hydrogel-filled microfluidic chips. Lab Chip 2019;19:206–213. [DOI] [PubMed] [Google Scholar]