Abstract
Cancer cells are dependent upon an abundance of ribosomes to maintain rapid cell growth and proliferation. The rate-limiting step of ribosome biogenesis is ribosomal RNA (rRNA) synthesis by RNA polymerase I (Pol I). Therefore, a goal of the cancer therapeutic field is to develop and characterize Pol I inhibitors. Here, we elucidate the mechanism of Pol I inhibition by a first-in-class small-molecule BMH-21. To characterize the effects of BMH-21 on Pol I transcription, we leveraged high-resolution in vitro transcription assays and in vivo native elongating transcript sequencing (NET-seq). We find that Pol I transcription initiation, promoter escape, and elongation are all inhibited by BMH-21 in vitro. In particular, the transcription elongation phase is highly sensitive to BMH-21 treatment, as it causes a decrease in transcription elongation rate and an increase in paused Pols on the ribosomal DNA (rDNA) template. In vivo NET-seq experiments complement these findings by revealing a reduction in Pol I occupancy on the template and an increase in sequence-specific pausing upstream of G-rich rDNA sequences after BMH-21 treatment. Collectively, these data reveal the mechanism of action of BMH-21, which is a critical step forward in the development of this compound and its derivatives for clinical use.
Keywords: cancer therapeutics, RNA polymerase I, transcription, rRNA, NET-seq
Abbreviations: CF, core factor; ETS1, external transcribed spacer 1; ITS, internal transcribed spacer; K-S test, Kolmogorov–Smirnov test; mRNA, messenger RNA; NET-seq, native elongating transcript sequencing; Pol I, RNA polymerase I; rDNA, ribosomal DNA; rRNA, ribosomal RNA; TBP, TATA-binding protein
RNA polymerase I (Pol I) activity accounts for 60% of the transcriptional activity of a growing eukaryotic cell (1). Pol I is responsible for synthesizing the majority of the ribosomal RNA (rRNA) that assembles with ribosomal proteins to form the eukaryotic ribosome. Since Pol I transcription of the ribosomal DNA (rDNA) is the first, rate-limiting step of ribosome biogenesis (2, 3, 4), Pol I activity is directly proportional to the rates of cell growth and proliferation (5). In Saccharomyces cerevisiae (yeast) cells, Pol I transcribes the rDNA that is organized in approximately 200 tandem repeats on chromosome XII. Each 9.1 kb repeat contains the 35S gene, transcribed by Pol I, and the 5S gene, transcribed by Pol III. Transcription of the 35S gene by Pol I yields the precursor 35S rRNA that is co- and post-transcriptionally modified (6) to generate mature 18S, 5.8S, and 25S rRNAs. The rRNA synthesized by Pols I and III is processed and complexed with ribosomal proteins in the nucleolus. Finally, ribosomes are exported to the cytoplasm to execute protein synthesis.
In most cancers, Pol I transcription is upregulated and drives ribosome biogenesis and subsequent protein synthesis (7, 8, 9). High levels of protein synthesis are required to maintain robust cancer cell growth and proliferation (9, 10, 11). While ribosome biogenesis has been targeted to reduce cancer growth, most therapeutics fail to directly influence Pol I transcription (12, 13, 14). It is critical to discover direct inhibitors of Pol I transcription to minimize DNA damage (15) and to reduce interference with messenger RNA (mRNA) synthesis and translation (16). Therefore, Pol I transcription is an excellent target for selective inhibition of cancer cell growth (8, 17, 18, 19). Several Pol I transcription inhibitors have been investigated as anticancer therapeutics (20), including ML-246 (21) and CX-5461 (22, 23). CX-5461 was the first Pol I inhibitor to complete Phase I clinical trials. It is thought to inhibit Pol I transcription by competing with the preinitiation complex protein, SL1, for the rDNA promoter (22, 24). CX-5461 treatment resulted in a decrease of rDNA transcription in patients (25) and a reduction in cancer cell proliferation and tumor volume in mammalian cancer cell lines (26). In light of the progress of Pol I transcription-targeting anticancer therapeutics, there is a demand to discover and characterize highly specific Pol I-inhibiting therapeutic compounds.
A high-throughput cell-based screen revealed a novel p53-activating compound, BMH-21 (27), whose derivatives are currently in preclinical development. BMH-21 intercalates into GC-rich regions of the rDNA (28) to inhibit Pol I transcription without inducing DNA damage (29). BMH-21 treatment destabilizes the largest subunit of Pol I, A190 in yeast and RPA194 in mammals, triggering its degradation (29, 30, 31). Early studies found that BMH-21 displayed antiproliferative effects in tumor cell lines, ex vivo prostate tissue culture, and mouse models (27). The mechanism of BMH-21 inhibition of rRNA synthesis is not fully understood. It was previously demonstrated that BMH-21 inhibits Pol I transcription elongation (31) and reduces Pol I occupancy on the rDNA in vivo (30). The effects of BMH-21 on Pol I transcription and the degradation of A190/RPA194 are conserved between yeast and humans, making the yeast system an exceptional tool for defining BMH-21’s mechanism of action. Our aim was to provide a comprehensive analysis of BMH-21’s influence on Pol I catalyzed transcription.
In this study, we leveraged high-resolution in vitro and in vivo techniques to elucidate BMH-21’s mechanism of inhibition of Pol I using the yeast model system. We discovered that Pol I transcription initiation, promoter escape, elongation, and occupancy of the rDNA were inhibited by BMH-21. BMH-21 reduced Pol I’s processivity in vitro, evidenced by reduced full-length RNA product synthesis. Additionally, we found that Pol I transcription elongation is the most sensitive step to BMH-21 treatment. Our in vitro conclusions were complemented in vivo with native elongating transcript sequencing (NET-seq). Pol I occupancy of the rDNA was reduced after acute BMH-21 treatment and Pol I elongation complexes preferentially paused upstream of G-rich sequences. This study reveals the mechanism of action for this small-molecule inhibitor that is supported both in vitro and in vivo.
Results
Promoter-dependent in vitro transcription experiments probe the sensitivity of PoI I transcription steps to BMH-21 treatment
To identify the impact of BMH-21 treatment on transcription catalyzed by Pol I, we utilized a fully reconstituted transcription assay (32) that includes all the required purified components for Pol I transcription: Pol I, core factor (CF), TATA-binding protein (TBP), and Rrn3 (Fig. 1). The template DNA includes a modified yeast rDNA template, mutated to not encode for C until the +56 position (with respect to the transcription start site). Once all the purified protein components are assembled, the DNA template and NTP substrates (ATP, GTP, α-32P-UTP) are added to allow the polymerase to initiate transcription and transcribe to the +55 position. Heparin is added to sequester unbound Pols to ensure we observe a single round of transcription. Finally, CTP and unlabeled UTP are added, releasing the polymerases from the +55 position to transcribe the remainder of the 800 base pair template. Reactions are run on polyacrylamide gels to resolve resultant RNA products for further analysis.
Figure 1.
Promoter-dependent in vitro transcription assay. A modified rDNA template, which does not encode for CTP until the +56 position, is incubated with proteins required for Pol I transcription. NTPs, limiting for CTP, are provided to allow transcription up to the +55 position for 5 min. Heparin is added to prevent multiple rounds of transcription. Finally, CTP and unlabeled UTP are added to release Pols from the +55 position, allowing them to transcribe the remainder of the 800 nt template. BMH-21 is added to different steps illustrated in the schematic (red stars) to test effects on transcription initiation, promoter escape, and elongation. Pol I, RNA polymerase I.
To determine the impact of BMH-21 on different steps in Pol I transcription, we added BMH-21 at different points in the reaction scheme. By varying the order of addition, these assays probe for the influence of BMH-21 on Pol I transcription initiation, promoter escape, and elongation (red stars in Fig. 1).
Transcription initiation by Pol I is reduced in the presence of BMH-21
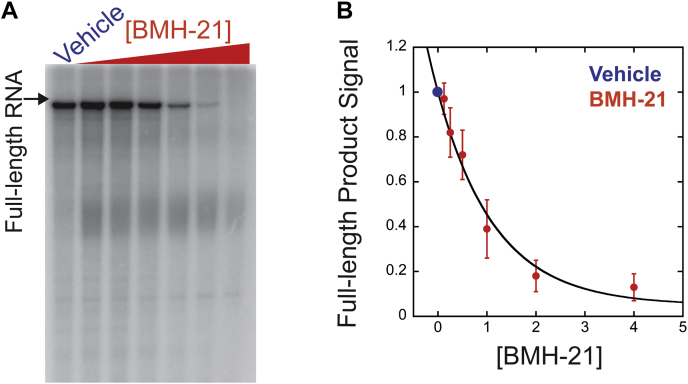
Transcription initiation involves many steps: first, transcription factors bind to the promoter region. Then, Pol I is recruited to form a preinitiation complex. DNA is unwound and initial RNA synthesis begins. The final stage of transcription initiation, promoter escape, occurs when Pol I clears the promoter region and advances on the template. To assay the impact of BMH-21 on transcription initiation, we added BMH-21 (125 nM–4 μM) or vehicle to the template prior to adding Pol I, TBP, CF, and Rrn3. ATP, GTP, and α-32P-UTP (20 μM) were provided for 5 min to allow Pol I to transcribe up to the +55 position. CTP and unlabeled UTP (20 μM) were added to the synchronized polymerases, releasing them from the +55 halt point. Each reaction proceeded for 10 min, allowing all complexes to synthesize full-length RNA products. Reactions were run on polyacrylamide gels and the abundance of full-length RNAs was analyzed (Fig. 2A). The amount of RNA synthesized by Pol I was reduced by increasing concentrations of BMH-21. Data were fit to a single exponential to calculate an IC50 value of 0.81 μM BMH-21 (Fig. 2B).
Figure 2.
BMH-21 inhibits early Pol I transcription initiation.A, vehicle or BMH-21 (125 nM–4 μM) was added to the rDNA template prior to preinitiation complex formation. Reactions proceeded for 10 min after CTP and unlabeled UTP were added. Full-length RNAs were resolved on polyacrylamide gels. B, plot of full-length RNA product signal over [BMH-21] treatment. Data were fit to a single exponential equation. Experiments were executed in triplicate and the mean is plotted with error bars corresponding to the standard deviation about the mean.
To control for the possibility that reduced full-length RNA products in the presence of BMH-21 (Fig. 2A) resulted from an inhibition of elongation rather than initiation, we treated Pol I transcription reactions during the elongation phase with vehicle or BMH-21 (1 μM) over a time course. Preinitiation complexes were incubated with ATP, GTP, and α-32P-UTP (20 μM) for 5 min. Simultaneously, BMH-21 (1 μM) or vehicle was added to the reactions with CTP and unlabeled UTP (20 μM). Reactions were stopped after 1, 4, and 10 min and run on polyacrylamide gels (Fig. S1). The time courses reveal that BMH-21 caused a delay in accumulation of full-length products, evident at the 4-min time points. However, the amplitude of the full-length products at the 10-min time points was identical between the vehicle- and BMH-21-treated samples (Fig. S1). Therefore, 10-min time points, as shown in Figure 2A, are not sensitive to transcription elongation rate effects. The simplest explanation for these data is that the reduction of full-length products in the presence of BMH-21, shown in Figure 2A, is due to inhibition of transcription initiation.
BMH-21 disrupts Pol I clearing of the rDNA promoter
We found a reduced amount of full-length RNA products when BMH-21 was added to the rDNA template, suggesting that BMH-21 inhibits transcription initiation. To determine if promoter escape was specifically inhibited by BMH-21, we formed the preinitiation complexes and added BMH-21 (300 nM) simultaneously with ATP, GTP, and α-32P-UTP (20 μM). Pol I complexes were allowed to synthesize through the promoter region up to the +55 position for 5 min. At t = 0, CTP and unlabeled UTP were added to the reactions and were stopped over time (30 s–4 min). The full-length RNAs were resolved on polyacrylamide gels (Fig. 3A).
Figure 3.
BMH-21 reduces promoter-escape efficiency.A, Pol I transcription reactions were treated with vehicle or BMH-21 (300 nM) during the addition of ATP, GTP, and α-32P-CTP. Reactions were stopped at 0.5, 1, 1.5, 2, and 4 min after CTP and unlabeled UTP addition. Polyacrylamide gels resolved full-length RNAs. B, plot of full-length RNA product signal over time for vehicle and BMH-21 conditions. Experiments were executed in triplicate and the mean is plotted with error bars corresponding to the standard deviation about the mean. Pol I, RNA polymerase I.
If BMH-21 interfered with Pol I’s ability to clear the promoter and move down the template, we would detect a reduction in full-length RNAs. We found that Pol I synthesized less full-length products in the presence of BMH-21 compared with the vehicle control. We observed a suppression of full-length products over the time course in the presence of BMH-21, maximally causing a 49% reduction at 4 min (Fig. 3B). We conclude that multiple steps in transcription initiation are vulnerable to BMH-21.
BMH-21 treatment robustly inhibits Pol I transcription elongation
Previously, we found that BMH-21 slows transcription elongation based on the observation that full-length RNA accumulation was slower in the presence of the compound (31). While strongly suggestive of direct inhibition of Pol I transcription elongation, this approach does not provide quantitative evaluation of transcription elongation kinetics. Here, we deploy a more quantitative assay (33) to directly measure Pol I transcription elongation rate in the presence of increasing concentrations of BMH-21.
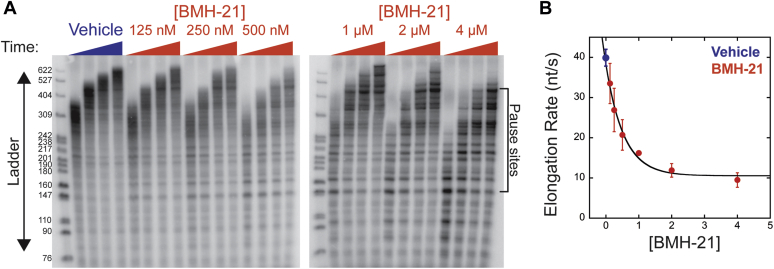
Preinitiation complexes were formed and incubated with ATP, GTP, and α-32P-UTP (1 mM) for 5 min. At t = 0, CTP and unlabeled UTP (1 mM) were added simultaneously with BMH-21 or vehicle to release synchronized Pols from the +55 position. For each treatment condition (vehicle or 125 nM–4 μM BMH-21), four time points were collected. Reactions were resolved on polyacrylamide gels alongside a reference nucleic acid ladder to calculate the RNA length (nt) of the leading population of synthesized RNAs at each time point (Fig. 4A). Transcription elongation rate was measured by plotting the RNA length as a function of time and calculating the slope of the resultant line (Fig. S2). Transcription elongation rate was reduced by BMH-21 in a concentration-dependent manner with an IC50 of 0.43 ± 0.06 μM BMH-21 (Fig. 4B). Additionally, we visualized increased Pol I pausing in the BMH-21-treated reactions, evidenced by shorter RNA bands observed throughout the time courses, but this pausing was absent in the vehicle-treated reactions (Fig. 4A). Interestingly, the locations of the pause sites were conserved across the BMH-21 treated reactions. These data suggest that BMH-21 causes sequence-specific pausing of Pol I in vitro. Taken together, these data show that BMH-21 inhibits transcription initiation, promoter escape, and elongation (Figure 2, Figure 3, Figure 4).
Figure 4.
BMH-21 decreases Pol I elongation rate and increases pausing.A, Pol I transcription reactions were treated with vehicle or BMH-21 (125 nM–4 μM) when CTP and unlabeled UTP were added. Reactions treated with vehicle or 125 to 500 nM BMH-21 were stopped at 4, 6, 8, and 10 s. Reactions treated with 1 to 4 μM BMH-21 were stopped at 5, 10, 15, and 20 s. Reactions were run on polyacrylamide gels alongside a nucleic acid ladder. B, elongation rate (nt/s) was plotted over [BMH-21] treatment. Data were fit to a single exponential equation. Experiments were executed in triplicate and the mean is plotted with error bars corresponding to the standard deviation about the mean. Pol I, RNA polymerase I.
Vehicle and BMH-21-treated NET-seq libraries are highly reproducible
NET-seq probes for polymerase occupancy, which we define here as the positioning of Pol I on the rDNA template, at single-nucleotide resolution in vivo. This technique was first developed to study Pol II (34), but we recently adapted and optimized it to detect Pol I occupancy with high reproducibility (35, 36, 37). NET-seq for both vehicle- and BMH-21-treated yeast was performed in biological triplicate. The vehicle-treated libraries were aligned to the yeast genome and the 5′ read ends (which correspond to the 3′ ends of nascent transcripts) were plotted with respect to their position on the 35S gene (Fig. S3A, left panel). At each position, the “counts” value indicates the number of Pol I complexes occupying that position on the rDNA at the time of harvest. A high count value represents a highly occupied position or peak, whereas a low count value can be interpreted as a lesser occupied position or trough. While transcription elongation kinetics cannot be determined from these experiments, it is reasonable to equate peaks with positions where Pol I is stalled on the template, whereas troughs may represent regions of rapid transcription elongation by Pol I. To assess whether the Pol I occupancy patterns displayed for the vehicle-treated replicates were statistically similar, the Spearman correlation test was executed (Fig. S3A, right panel). This test ranks the occupancy in two replicates by count (highest to lowest), compares the ranking between the replicates, and generates a coefficient value. This value indicates the correlation between the two replicates, where 1 represents 100% similarity. The histogram generation and Spearman correlation test analysis were repeated for the BMH-21-treated samples (Fig. S3B), and like the vehicle-treated data, these results also displayed high similarity. Therefore, we concluded that this technique is highly reproducible and is a powerful method to visualize Pol I occupancy under treatment conditions at high resolution in vivo.
BMH-21 reduces Pol I occupancy of the rDNA in vivo
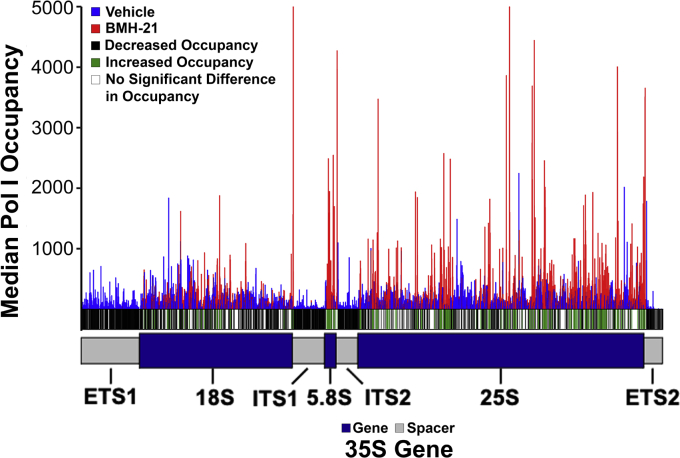
Using NET-seq, we tested the effect of acute BMH-21 treatment (50 μM for 2 min) on Pol I occupancy in vivo as compared with vehicle treatment. We plotted the median Pol I occupancy across the three replicates for each treatment (Fig. 5). At each position, a t-test was executed to determine the p-value between treatments. A p-value < 0.05 was deemed a significant difference in occupancy and was denoted by either a black (decreased) or green (increased) bar below the histogram, for the BMH-21-treated samples with respect to vehicle-treated. These data indicate that there is a significantly different occupancy pattern between the two treatment groups, as there is a depletion of Pol I especially in the spacer regions [external transcribed spacer 1 (ETS1), internal transcribed spacer 1 (ITS1), internal transcribed spacer 2 (ITS2), external transcribed spacer 2 (ETS2)].
Figure 5.
Pol I occupancy was decreased after BMH-21 treatment. The median Pol I occupancy for vehicle-treated (blue) and BMH-21-treated (red) libraries was plotted, and a t-test was executed at each position in the 35S gene. If the p-value was less than 0.05, the occupancy difference between treatment groups was deemed significant. Underneath the histogram, it was indicated whether occupancy was significantly increased (black), significantly decreased (green), or displayed no significant change (white) in the BMH-21-treated samples compared with the vehicle-treated. Pol I, RNA polymerase I.
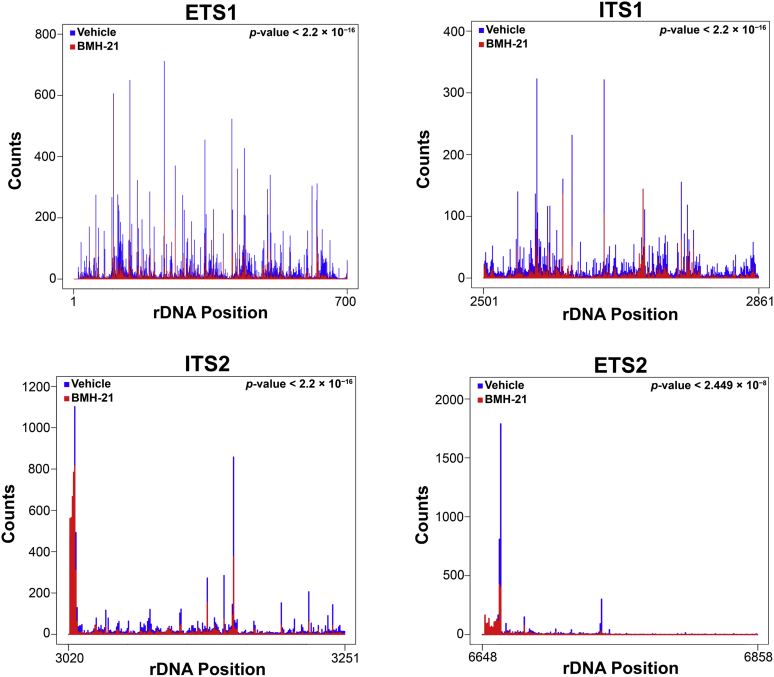
One limitation of the NET-seq technique is that there is unavoidable mature rRNA contamination in the libraries. This is especially evident in Figure 5, where there are still Pol I peaks present in the gene regions of the BMH-21-treated samples, despite the reduction in occupancy in the spacer regions. From previous literature, it has been established that processing/maturation of the rRNA begins to occur co-transcriptionally and results in removal of the spacer RNAs (32, 38, 39, 40). Furthermore, we performed NET-seq using a strain of yeast containing untagged Pol I to investigate the pattern of contamination in our samples. The data from this untagged Pol I control demonstrate that while there is high signal in the gene regions, reads mapping to the spacer regions were minimal (Fig. S4), confirming that the contamination in our sample is, indeed, mature rRNAs. Therefore, since it is impossible to remove mature rRNA contamination in the experiment, the spacer regions are critically important for observing nascent transcription by Pol I. Thus, we performed an in-depth analysis of Pol I occupancy in these regions (Fig. 6). In all four of the rDNA spacer regions, there was robust reduction in Pol I occupancy in the BMH-21-treated samples as compared with vehicle-treated. We employed the Kolmogorov–Smirnov test (K-S test) to evaluate the significance of this altered distribution between treatment groups within these regions and found these differences to be significant (indicated by the p-values in Fig. 6). These data are consistent with the patterns observed in Figure 5 and reveal that BMH-21 treatment significantly reduces Pol I occupancy on the rDNA in comparison to vehicle treatment even after only 2 min. In combination with the in vitro results shown in Figures 2 and 3, this observation suggests that BMH-21 inhibits transcription initiation by Pol I.
Figure 6.
Pol I occupancy patterns in spacer regions validate reduced occupancy after BMH-21 treatment gene-wide. The median Pol I occupancy was plotted for vehicle-treated (blue) and BMH-21-treated (red) libraries in each spacer region. The K-S test was performed to determine whether the overall distribution of occupancy in each spacer region was significantly different between the two treatment conditions. The p-value determined by that test is included in the top right corner of each graph. Pol I, RNA polymerase I.
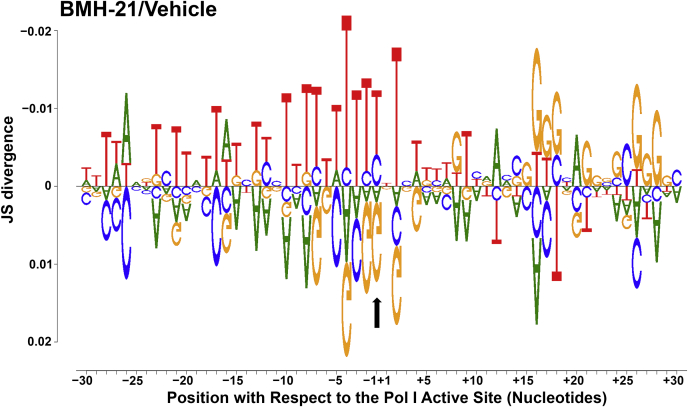
BMH-21 induces Pol I pausing upstream of G-rich rDNA sequences
We found that there was a reduction in overall occupancy of Pol I across the 35S gene, which suggests that BMH-21 treatment inhibits transcription initiation. To evaluate potential effects on transcription elongation, we compared in vivo pausing after either vehicle- or BMH-21-treatment by examining the sequence context of the top 2.5% most occupied positions (consistent with previously published findings (35, 36, 37)) in the spacer regions only. The DiffLogo plot in Figure 7 displays the BMH-21-treatment logo on top and the vehicle treatment logo underneath, with the black arrow indicating the position of the last incorporated nucleotide into the nascent transcript. We observed that in the BMH-21-treated samples, Pol I is frequently paused in T-rich regions of the rDNA that are directly upstream from G-rich regions. The opposite pattern is true in the vehicle-treated samples, where Pol I is stalled in G-rich regions of the template. This finding is consistent with previous literature suggesting that BMH-21 intercalates into GC-rich regions of DNA (30). We hypothesize that upon intercalation of BMH-21 directly downstream of the elongation complex, this could induce added torsional stress, causing Pol I to be halted on the template. Furthermore, these data are consistent with in vitro results (Fig. 4) that revealed sequence-specific effects by BMH-21 on transcriptional pausing. Thus, BMH-21 has direct negative effects on transcription initiation and elongation by Pol I in vitro and in vivo.
Figure 7.
Pol I is stalled upstream of particularly G-rich regions of the rDNA spacer regions. This figure was created using the DiffLogo program (version 2.14.0) and displays sequences both upstream and downstream of Pol I for the top 2.5% occupied positions in the spacer regions only (ETS1, ITS1, ITS2, and ETS2). The DiffLogo for the BMH-21-treated samples is plotted on top and displays the sequences that are enriched with comparison to the vehicle-treated libraries. In contrast, the DiffLogo for the vehicle-treated samples is displayed underneath and indicates the enriched sequences with comparison to the BMH-21-treated samples. The black arrow identifies the last incorporated nucleotide in the nascent transcript. ETS, external transcribed spacer; ITS, internal transcribed spacer; Pol I, RNA polymerase I.
Discussion
BMH-21 directly inhibits Pol I transcription and causes a decrease in rRNA synthesis by reducing Pol I occupancy of the rDNA
The data presented here, resulting from high-resolution in vitro and in vivo experiments, elucidate BMH-21’s mechanism of inhibition of transcription by Pol I. We showed that BMH-21 added during three transcription steps inhibited Pol I transcription through a decrease in full-length RNAs and elongation rate and an increase in amplitude of short, incomplete RNAs. Despite sensitivity to BMH-21 during all of the transcription steps tested, we conclude that the transcription elongation phase is most vulnerable to BMH-21 inhibition based on the IC50 values calculated (Figs. 2 and 4). Altogether, these in vitro observations suggest that BMH-21 treatment reduces the amount of Pol I transcription complexes that are capable of efficient and processive transcription. These findings were complemented by our in vivo NET-seq experiments. We found that Pol I occupancy of the rDNA was reduced across the 35S gene (Fig. 5). Specifically, there was a significant decrease in occupancy in the spacer regions of the 35S gene, which serve as a reporter for nascent transcripts, since these regions are removed prior to maturation of the rRNAs (Fig. 6). In the BMH-21-treated samples, Pol I was preferentially paused upstream of G-rich rDNA regions (Fig. 7). These findings suggest that the intercalation of BMH-21 into the GC-rich regions of the rDNA introduces a barrier to Pol I elongation complexes. In contrast, T-rich regions of the rDNA were enriched within the RNA:DNA hybrid and upstream of the polymerase. Furthermore, NET-seq was repeated to test a longer BMH-21 treatment time of 16 min (Figs. S5–S8), and these results were consistent with the 2-min treatment findings (Figure 5, Figure 6, Figure 7). Taken together, our in vitro and in vivo analyses are complementary, providing new insight into BMH-21’s mechanism of action.
BMH-21 influences Pol I activity similarly to other DNA intercalators
Our BMH-21 in vitro and in vivo results are consistent with the mechanism of action of other GC-intercalators including actinomycin, ethidium bromide, and proflavine (41, 42, 43). The Panov research group executed an extensive study of DNA intercalators using in vitro and cell-based techniques (41). They used a plasmid DNA template containing the rDNA promoter, very similar to our in vitro system. Consistent with our findings in Figures 2 and 3, early transcriptional events catalyzed by Pol I (initiation, promoter escape) are sensitive to DNA intercalators. Our in vitro IC50 concentration for transcription initiation by BMH-21, 810 nM, was within the range of IC50 values of proflavine, 9HE, Acridine Orange, and Thionine (41). Additionally, they found that Pol I transcription elongation is sensitive to DNA intercalators by measuring rRNA synthesis in human cells. Our IC50 measurement in Figure 4 revealed that BMH-21 is comparatively potent for inhibition of transcription elongation. Finally, DNA intercalators have been found to reduce Pol I occupancy similar to our results in Figures 5 and 6 (41). Taken together, our data suggest that BMH-21 acts similarly to other DNA intercalators by inhibiting early Pol I transcriptional events and Pol I occupancy of the rDNA.
BMH-21 and CX-5461 inhibit ribosome biogenesis through different mechanisms
The first Pol I inhibitor to complete Phase I clinical trials was CX-5461 (22, 23). It was found to inhibit Pol I transcription by interfering with transcription initiation. It binds SL1, comprised of TBP and TBP-associated factors, to disrupt the release of the Pol I-Rrn3 complex from the rDNA promoter (22, 44). In vivo studies showed promising anticancer effects as treatment with CX-5461 decreased cancer cell and tumor viability in human cancer cell lines and murine xenografts (45, 46, 47, 48). Despite the establishment of CX-5461 as a direct and selective inhibitor of Pol I transcription, a recent study found that CX-5461 may stabilize G-quadruplex DNA and cause DNA damage (49). This finding cast doubt on the previous classification of CX-5461 as a direct Pol I inhibitor. Considering the controversy, an in-depth, multidimensional investigation of the mechanism of CX-5461 found that it did not induce cell death via interference with Pol I function, rather, it accomplished its cytotoxic effects via topoisomerase II (Top2) poisoning (50). Thus, the mechanism of action for CX-5461 remains somewhat controversial, despite its initial promise in the clinic. Clarity on this will be required to facilitate selection of patients most responsive to its therapeutic effects.
The mechanism of Pol I inhibition by the BMH-21 class of molecules is significantly different than that of CX-5461. BMH-21 intercalates into GC-rich DNA without causing DNA lesions (27, 29, 30). Unlike CX-5461, multiple Pol I transcription steps (initiation, promoter escape, and elongation) are inhibited by BMH-21. BMH-21 inhibits Pol I transcription through binding to the highly GC-rich content of the rDNA, presumably without directly associating with Pol I transcription machinery. Previous studies with BMH-21 suggest a direct negative effect on Pol I transcription (29, 30, 31). Here, we define the mechanism of action for the compound using biochemical and genomic approaches. While these results do not exclude the possibility that BMH-21 may affect other processes, these findings clearly reveal acute, direct, and potent effects of the compound on Pol I.
How is BMH-21 specific to Pol I?
BMH-21 inhibition may be specific to Pol I transcription based on the chromatin organization of the rDNA. Our in vitro transcription DNA template lacks nucleosomes; therefore, it may be similar to the rDNA transcribed by Pol I in vivo. Unlike chromatinized genes transcribed by Pol II (51, 52), active rDNA repeats transcribed by Pol I lack ordered nucleosomes (28, 53, 54, 55, 56). Thus, based on BMH-21’s affinity for open, GC-rich rDNA, it could selectively impact Pol I transcription. Alternatively, it is possible that BMH-21 may bind many genomic loci, including loci transcribed by Pols II and III. Perhaps Pols II and III exhibit enzymatic features that render them insensitive to inhibition by BMH-21. We showed that Pols I and II possess unique biochemical properties (57). Nucleotide addition catalyzed by Pol I is significantly faster than by Pol II. Coupled with Pol I’s rapid elongation kinetics, it forms a less stable transcription elongation complex compared with Pol II. In contrast, Pol II nucleotide addition kinetics are significantly slower, and Pol II forms a very stable elongation complex. Pol I’s fast nucleotide addition kinetics and less stable elongation complexes may render Pol I specifically vulnerable to BMH-21 intercalation sites compared with Pol II. Determining whether BMH-21 selectively inhibits Pol I, compared with the other RNA polymerases, is a topic of ongoing investigation.
In conclusion, complementary in vitro and in vivo experiments demonstrate that BMH-21 inhibits Pol I transcription by reducing the efficiency of Pol I transcription initiation and elongation. Mechanistic studies are ongoing to elucidate the impact of BMH-21 on the kinetic mechanism of nucleotide addition by Pol I in the presence of BMH-21.
Experimental procedures
Yeast strain used for NET-seq experiments
Wild-type (WT) yeast (ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11,15 can1-100 RPA135-(HA)3- (His)7::TRP1m × 6 rpa190Δ::HIS3M × 6 carrying pRS315-RPA190) was used for these experiments. This strain was used previously to generate NET-seq data for Pol I occupancy in other studies (35, 36). To perform NET-seq for this study, cells were cultured at 30 °C with nutation. A yeast strain containing untagged Pol I (ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11,15 can1-100) was used to generate the NET-seq data in Figure S4.
Purified proteins used within in vitro transcription assays
Pol I was purified from a protease-deficient yeast strain (58) with the largest subunit, A190, tagged with ten histidine residues. CF, Rrn3, and TBP were cloned into overexpression vectors and transformed into BL21(DE3)-pRosetta-2 E. coli cells. Purifications were performed as previously published (36, 59, 60).
Promoter-dependent in vitro transcription assay
In vitro transcription experiments were performed as previously described (59). The purification of the modified rDNA template (C-less until +56 position) and protein components were confirmed with agarose and SDS-PAGE gels, respectively. Template DNA was incubated with Pol I, CF, TBP, and Rrn3 to allow for preinitiation complex formation. ATP, GTP, and α-32P-UTP were added to allow transcription until the +55 halt point for 5 min. Heparin is added to ensure a single round of transcription by binding unbound Pol I’s. CTP and unlabeled UTP were added to release the Pols from the +55 position, allowing for the synthesis of full-length RNA products. Reactions were stopped with the addition of 1 M ammonium acetate in 100% ethanol. RNA was precipitated overnight at −20 °C and then centrifuged at 13,000g for 10 min. Supernatant was discarded and the RNA pellets were resuspended in RNA loading dye. Samples were boiled and loaded on a 5% polyacrylamide (29:1, acrylamide to bis), 1 X TBE, 7 M urea gel. Gels were vacuumed dry and exposed in a phosphorimager cassette overnight. The phosphorimager screen was imaged by an Amersham Typhoon scanner, and the image was analyzed using ImageQuant.
Determination of BMH-21 concentration for promoter-escape experiments
We originally treated Pol I transcription reactions during promoter escape with the IC50 calculated from transcription initiation experiments (Fig. 2). Due to low ratio of signal to background, we were unable to execute subsequent experiments at ∼800 nM BMH-21. To determine a BMH-21 concentration that would allow for quantifiable and reproducible signal, we tested different concentrations of BMH-21, 100 to 1600 nM. We selected 300 nM BMH-21 for experiments shown in Figure 3.
Elongation rate determination
Quantification of Pol I transcription elongation rate was executed similarly as previously published (33). Gel images from transcription elongation experiments were analyzed in ImageQuant. Lanes are boxed within columns, each encompassing one reaction collected at a certain time point. The leading population RNAs are boxed and the average intensity point within the box is calculated. The average intensity point is correlated to the ladder to calculate the RNA length for each time point. Elongation rate is calculated by plotting RNA length (nt) over time (s) per condition and fitting to a line (Fig. S2).
NET-seq experiments
NET-seq was performed based on an adaptation from previously described methods (35, 36, 37). Yeast was cultured the same as in previous literature (36), except that this adapted method required only 1 l of yeast per replicate, compared with the previous 3 l per replicate. Once cells reached an OD600 of 0.3, they were treated with either 50 μM of BMH-21 (for the BMH-21 samples) or an equivalent volume of the vehicle buffer (0.1 M NaH2PO4, pH 6; for the vehicle samples). BMH-21 (12H-Benzo[g]pyrido[2,1-b]quinazoline-4- carboxamide, N-[2(dimethylamino)ethyl]-12-oxo) was verified for purity using LC/MS mass spectrometry and 1H-NMR as previously published (31). Immediately following treatment, cells were placed back in the incubator to culture for an additional 1 min (for the 2-min experiment, Figure 5, Figure 6, Figure 7 and S3) or 15 min (for the 16-min experiment, Figs. S5–S8). Cells were harvested using an ∼1 min method and lysis was performed, both described previously (36).
To prepare for immunoprecipitation (IP), lysis buffer (20 mM Tris-HCl pH 7.9, 0.4% Triton X-100, 0.1% NP-40, 100 mM NH4Cl, 5 mM EDTA-Na pH 8.5, 1x HALT Protease Inhibitor, 25 U/ml SUPERase-in RNAse Inhibitor) (36) and blocking buffer (same as lysis buffer, with 10 mg/ml BSA added) were prepared on ice. Pierce anti-HA beads for each sample (equivalent to 4% of the lysis buffer required based on grindate weight of sample (36)) were washed three times at 4 °C with 1 ml lysis buffer and nutation. After the final wash, lysis buffer was discarded, and 500 μl blocking buffer was added to each bead-containing tubes. Beads were blocked overnight at 4 °C with nutation. IP, RNA extraction, and precipitation were performed as previously described (35, 36) except that 2 mg/ml BSA was added to the wash buffer, and bead washing was eliminated during the IP.
Following precipitation, samples were centrifuged, and linker ligation was performed the same as previously described (35, 36), except that RNA pellets were dissolved in 10 mM Tris-HCl, pH 6.9, and 20 U/μl SUPERase-in RNAse inhibitor was added the ligation mix. A DNA linker containing a unique molecular identifier was used (35). After linker ligation was complete, an enzymatic digestion of the excess linker was performed instead of a gel extraction using the following steps. First, 2 μl of 5′ deadenylase (NEB) was added to each sample and mixed thoroughly. Samples were incubated at 30 °C for 45 min. Next, samples were diluted 2.5X with water supplemented with 0.6X NEBuffer 2, and 2 μl of RecJf (NEB) was added and mixed well. Samples were incubated at 37 °C for 45 min. Then, 2.2 μl zinc fragmentation buffer (100 mM Tris-HCl, pH 7.0, 100 mM ZnCl2) (35) was added to each sample, and samples were incubated at 70 °C for 16 min. Finally, 2.5 μl 200 mM EDTA, pH 8.5, 1 μl glycoblue, and 360 μl ammonium acetate precipitation solution were added, and samples were precipitated for at least 2 h at −80 °C.
Samples were centrifuged, pellets were washed, and reverse transcription was performed as previously described (35, 36), except that reactions were incubated at 45 °C (instead of the previous 50 °C) for 30 min. A 10% polyacrylamide gel was prepared and pre-run, samples were loaded, and gel was run as previously described (36). Slices between approximately 120 and 600 nucleotides were excised from the gel and pulverized for each sample. Then, 500 μl water was added and gel slurries were incubated at −80 °C for 15 min and then at 70 °C for 15 min. Finally, slurries were placed at 30 °C with nutation overnight. The following day, each slurry was transferred to a Costar Spin-X Centrifuge Tube Filters (Corning), and tubes were centrifuged at 16,000g for 3 min at room temperature. The flow-through from each tube was transferred to a new 1.5 ml tube, and 32 μl 3 M NaCl, 940 μl 100% isopropanol, and 1 μl glycoblue were added to each sample. Samples were precipitated for at least 2 h at −80 °C.
Samples were centrifuged at 16,000g for 1 h at 4 °C and then washed as previously described (35, 36). Pellets were resuspended in 15 μl of 10 mM Tris-OAc, pH 7.9. Circularization, amplification, sample preparation/purification, and sequencing were performed as previously described (35, 36). Amplification primers for each sample are included in Table S1.
NET-seq data analysis and statistical analysis
The data analysis pipeline used was identical to a previous publication (35).
Software and the versions used for these studies are included in Table S2. Reads were aligned to the S. cerevisiae genome assembly R64-1-1. The data used to generate the figures in this study have been deposited into NCBI’s Gene Expression Omnibus (61) and are available through the series accession number GSE175553.
The Spearman correlation test was used to detect reproducibility between replicates, and these data are included in the right panel of Figures S3 and S5. For this test, n = 2, where n is the number of replicates being compared for similarity. The K-S test was used to determine any significant differences in occupancy distribution between treatment groups. The results from the K-S test are shown in the inset in the top right corner of the graphs in Figures 6 and S7.
Data availability
The datasets generated during this study are available at: GSE175553 on the NCBI GEO database. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Schneider (dschneid@uab.edu).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the present and past members of the Schneider lab for their critical evaluation of this work and useful discussion. The authors gratefully acknowledge the resources provided by the University of Alabama at Birmingham IT-Research Computing group for high performance computing (HPC) support and CPU time on the Cheaha computer cluster.
Author contributions
D. A. S., M. L., and R. Q. J. funding acquisition; R. Q. J, A. K. H., and D. A. S. investigation; R. Q. J, A. K. H., and D. A. S. methodology; D. A. S., M. L., and R. Q. J. supervision; R. Q. J., A. K. H., M. L., and D. A. S. writing–original draft.
Funding and additional information
NIH (NIGMS R01 GM121404 to D. A. S. and M. L. and R35 GM140710 to D. A. S.) and NIH (T32 GM109780 to R. Q. J.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Karin Musier-Forsyth
Supporting information
References
- 1.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Kopp K., Gasiorowski J.Z., Chen D., Gilmore R., Norton J.T., Wang C., Leary D.J., Chan E.K.L., Dean D.A., Huang S. Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells. Mol. Biol. Cell. 2007;18:394–403. doi: 10.1091/mbc.E06-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chédin S., Laferté A., Hoang T., Lafontaine D.L., Riva M., Carles C. Is ribosome synthesis controlled by pol I transcription? Cell Cycle. 2007;6:11–15. doi: 10.4161/cc.6.1.3649. [DOI] [PubMed] [Google Scholar]
- 4.Laferté A., Favry E., Sentenac A., Riva M., Carles C., Chédin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell J., Zomerdijk J.C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scull C.E., Schneider D.A. Coordinated control of rRNA processing by RNA polymerase I. Trends Genet. 2019;35:724–733. doi: 10.1016/j.tig.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 8.Bywater M.J., Poortinga G., Sanij E., Hein N., Peck A., Cullinane C., Wall M., Cluse L., Drygin D., Anderes K., Huser N., Proffitt C., Bliesath J., Haddach M., Schwaebe M.K., et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montanaro L., Treré D., Derenzini M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim. Biophys. Acta. 2012;1825:101–110. doi: 10.1016/j.bbcan.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Williamson D., Lu Y.J., Fang C., Pritchard-Jones K., Shipley J. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2006;45:839–845. doi: 10.1002/gcc.20347. [DOI] [PubMed] [Google Scholar]
- 11.van Riggelen J., Yetil A., Felsher D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 12.Burger K., Mühl B., Harasim T., Rohrmoser M., Malamoussi A., Orban M., Kellner M., Gruber-Eber A., Kremmer E., Hölzel M., Eick D. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 2010;285:12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awad D., Prattes M., Kofler L., Rössler I., Loibl M., Pertl M., Zisser G., Wolinski H., Pertschy B., Bergler H. Inhibiting eukaryotic ribosome biogenesis. BMC Biol. 2019;17:46. doi: 10.1186/s12915-019-0664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catez F., Dalla Venezia N., Marcel V., Zorbas C., Lafontaine D.L.J., Diaz J.J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019;159:74–81. doi: 10.1016/j.bcp.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Waldman T., Zhang Y., Dillehay L., Yu J., Kinzler K., Vogelstein B., Williams J. Cell-cycle arrest versus cell death in cancer therapy. Nat. Med. 1997;3:1034–1036. doi: 10.1038/nm0997-1034. [DOI] [PubMed] [Google Scholar]
- 16.Harris C.C. The carcinogenicity of anticancer drugs: A hazard in man. Cancer. 1976;37:1014–1023. doi: 10.1002/1097-0142(197602)37:2+<1014::aid-cncr2820370805>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 17.Hannan R.D., Drygin D., Pearson R.B. Targeting RNA polymerase I transcription and the nucleolus for cancer therapy. Expert Opin. Ther. Targets. 2013;17:873–878. doi: 10.1517/14728222.2013.818658. [DOI] [PubMed] [Google Scholar]
- 18.Drygin D., Rice W.G., Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira R., Schneekloth J.S., Jr., Panov K.I., Hannan K.M., Hannan R.D. Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells. 2020;9:266. doi: 10.3390/cells9020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laham-Karam N., Pinto G.P., Poso A., Kokkonen P. Transcription and translation inhibitors in cancer treatment. Front. Chem. 2020;8:276. doi: 10.3389/fchem.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankowski K.J., Wang C., Patnaik S., Schoenen F.J., Southall N., Li D., Teper Y., Sun W., Kandela I., Hu D., Dextras C., Knotts Z., Bian Y., Norton J., Titus S., et al. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drygin D., Lin A., Bliesath J., Ho C.B., Brien S.E., Proffitt C., Omori M., Haddach M., Schwaebe M.K., Siddiqui-Jain A., Streiner N., Quin J.E., Sanij E., Bywater M.J., Hannan R.D., et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 23.Haddach M., Schwaebe M.K., Michaux J., Nagasawa J., O'Brien S.E., Whitten J.P., Pierre F., Kerdoncuff P., Darjania L., Stansfield R., Drygin D., Anderes K., Proffitt C., Bliesath J., Siddiqui-Jain A., et al. Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 2012;3:602–606. doi: 10.1021/ml300110s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorski J.J., Pathak S., Panov K., Kasciukovic T., Panova T., Russell J., Zomerdijk J.C. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007;26:1560–1568. doi: 10.1038/sj.emboj.7601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khot A., Brajanovski N., Cameron D.P., Hein N., Maclachlan K.H., Sanij E., Lim J., Soong J., Link E., Blombery P., Thompson E.R., Fellowes A., Sheppard K.E., McArthur G.A., Pearson R.B., et al. First-in-human RNA polymerase I transcription inhibitor CX-5461 in patients with advanced hematologic cancers: Results of a phase I dose-escalation study. Cancer Discov. 2019;9:1036. doi: 10.1158/2159-8290.CD-18-1455. [DOI] [PubMed] [Google Scholar]
- 26.Li L., Li Y., Zhao J., Fan S., Wang L., Li X. CX-5461 induces autophagy and inhibits tumor growth via mammalian target of rapamycin-related signaling pathways in osteosarcoma. Onco. Targets Ther. 2016;9:5985–5997. doi: 10.2147/OTT.S104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peltonen K., Colis L., Liu H., Jäämaa S., Moore H.M., Enbäck J., Laakkonen P., Vaahtokari A., Jones R.J., af Hällström T.M., Laiho M. Identification of novel p53 pathway activating small-molecule compounds reveals unexpected similarities with known therapeutic agents. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dammann R., Lucchini R., Koller T., Sogo J.M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colis L., Peltonen K., Sirajuddin P., Liu H., Sanders S., Ernst G., Barrow J.C., Laiho M. DNA intercalator BMH-21 inhibits RNA polymerase I independent of DNA damage response. Oncotarget. 2014;5:4361–4369. doi: 10.18632/oncotarget.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peltonen K., Colis L., Liu H., Trivedi R., Moubarek M.S., Moore H.M., Bai B., Rudek M.A., Bieberich C.J., Laiho M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014;25:77–90. doi: 10.1016/j.ccr.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei T., Najmi S.M., Liu H., Peltonen K., Kucerova A., Schneider D.A., Laiho M. Small-molecule targeting of RNA polymerase I activates a conserved transcription elongation checkpoint. Cell Rep. 2018;23:404–414. doi: 10.1016/j.celrep.2018.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider D.A., Michel A., Sikes M.L., Vu L., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scull C.E., Ingram Z.M., Lucius A.L., Schneider D.A. A novel assay for RNA polymerase I transcription elongation sheds light on the evolutionary divergence of eukaryotic RNA polymerases. Biochemistry. 2019;58:2116–2124. doi: 10.1021/acs.biochem.8b01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Churchman L.S., Weissman J.S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huffines A.K., Edwards Y.J.K., Schneider D.A. Spt4 promotes pol I processivity and transcription elongation. Genes. 2021;12:413. doi: 10.3390/genes12030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke A.M., Engel K.L., Giles K.E., Petit C.M., Schneider D.A. NETSeq reveals heterogeneous nucleotide incorporation by RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11633–E11641. doi: 10.1073/pnas.1809421115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scull C.E., Clarke A.M., Lucius A.L., Schneider D.A. Downstream sequence-dependent RNA cleavage and pausing by RNA polymerase I. J. Biol. Chem. 2020;295:1288–1299. doi: 10.1074/jbc.RA119.011354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venema J., Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 39.Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 40.Kos M., Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews W.J., Ray S., Panova T., Engel C., Panov K.I. DNA intercalators inhibit eukaryotic ribosomal RNA synthesis by impairing the initiation of transcription. Genes. 2021;12:1412. doi: 10.3390/genes12091412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pommier Y., Sun Y., Huang S.-y. N., Nitiss J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momparler R.L., Karon M., Siegel S.E., Avila F. Effect of adriamycin on DNA, RNA, and protein synthesis in cell-free systems and intact cells. Cancer Res. 1976;36:2891–2895. [PubMed] [Google Scholar]
- 44.Mars J.-C., Tremblay M.G., Valere M., Sibai D.S., Sabourin-Felix M., Lessard F., Moss T. The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. NAR Cancer. 2020;2 doi: 10.1093/narcan/zcaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanij E., Hannan K.M., Xuan J., Yan S., Ahern J.E., Trigos A.S., Brajanovski N., Son J., Chan K.T., Kondrashova O., Lieschke E., Wakefield M.J., Frank D., Ellis S., Cullinane C., et al. CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 2020;11:2641. doi: 10.1038/s41467-020-16393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duo Y., Yang M., Du Z., Feng C., Xing C., Wu Y., Xie Z., Zhang F., Huang L., Zeng X., Chen H. CX-5461-loaded nucleolus-targeting nanoplatform for cancer therapy through induction of pro-death autophagy. Acta Biomater. 2018;79:317–330. doi: 10.1016/j.actbio.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 47.El Hassouni B., Mantini G., Immordino B., Peters G.J., Giovannetti E. CX-5461 inhibits pancreatic ductal adenocarcinoma cell growth, migration and induces DNA damage. Molecules. 2019;24:4445. doi: 10.3390/molecules24244445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismael M., Webb R., Ajaz M., Kirkby K.J., Coley H.M. The targeting of RNA polymerase I transcription using CX-5461 in combination with radiation enhances tumour cell killing effects in human solid cancers. Cancers (Basel) 2019;11:1429. doi: 10.3390/cancers11101429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O'Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J., Kabeer F., Lai D., Soriano P., Banáth J., Chiu D.S., et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017;8:14432. doi: 10.1038/ncomms14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruno P.M., Lu M., Dennis K.A., Inam H., Moore C.J., Sheehe J., Elledge S.J., Hemann M.T., Pritchard J.R. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc. Natl. Acad. Sci. U. S. A. 2020;117:4053–4060. doi: 10.1073/pnas.1921649117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brogaard K., Xi L., Wang J.-P., Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwabish M.A., Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dutta D., Shatalin K., Epshtein V., Gottesman Max E., Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.French S.L., Osheim Y.N., Schneider D.A., Sikes M.L., Fernandez C.F., Copela L.A., Misra V.A., Nomura M., Wolin S.L., Beyer A.L. Visual analysis of the yeast 5S rRNA gene transcriptome: Regulation and role of La protein. Mol. Cell Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones H.S., Kawauchi J., Braglia P., Alen C.M., Kent N.A., Proudfoot N.J. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittner M., Hamperl S., Stöckl U., Seufert W., Tschochner H., Milkereit P., Griesenbeck J. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell. 2011;145:543–554. doi: 10.1016/j.cell.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs R.Q., Ingram Z.M., Lucius A.L., Schneider D.A. Defining the divergent enzymatic properties of RNA polymerases I and II. J. Biol. Chem. 2020;296:100051. doi: 10.1074/jbc.RA120.015904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engel C., Sainsbury S., Cheung A.C., Kostrewa D., Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 59.Schneider D.A. In: Transcriptional Regulation: Methods and Protocols. Vancura A., editor. Springer New York; New York, NY: 2012. Quantitative analysis of transcription elongation by RNA polymerase I in vitro; pp. 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bedwell G.J., Appling F.D., Anderson S.J., Schneider D.A. Efficient transcription by RNA polymerase I using recombinant core factor. Gene. 2012;492:94–99. doi: 10.1016/j.gene.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available at: GSE175553 on the NCBI GEO database. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Schneider (dschneid@uab.edu).