Abstract
Various plants use antimicrobial proteins/peptides to resist phytopathogens. In the potato, Solanum tuberosum, the plant-specific insert (PSI) domain of an aspartic protease performs this role by disrupting phytopathogen plasma membranes. However, the mechanism by which PSI selects target membranes has not been elucidated. Here, we studied PSI-induced membrane fusion, focusing on the effects of lipid composition on fusion efficiency. Membrane fusion by the PSI involves an intermediate state whereby adjacent liposomes share their bilayers. We found that increasing the concentration of negatively charged phosphatidylserine (PS) phospholipids substantially accelerated PSI-mediated membrane fusion. NMR data demonstrated that PS did not affect the binding between the PSI and liposomes but had seminal effects on the dynamics of PSI interaction with liposomes. In PS-free liposomes, the PSI underwent significant motion, which was suppressed on PS-contained liposomes. Molecular dynamics simulations showed that the PSI binds to PS-containing membranes with a dominant angle ranging from −31° to 30°, with respect to the bilayer, and is closer to the membrane surfaces. In contrast, PSI is mobile and exhibits multiple topological states on the surface of PS-free membranes. Taken together, our data suggested that PS lipids limit the motion of the anchored PSI, bringing it closer to the membrane surface and efficiently bridging different liposomes to accelerate fusion. As most phytopathogens have a higher content of negatively charged lipids as compared with host cells, these results indicate that the PSI selectively targets negatively charged lipids, which likely represents a way of distinguishing the pathogen from the host.
Keywords: membrane fusion mechanism, plant-specific insert, NMR spectroscopy, molecular dynamic simulation
Abbreviations: AP, aspartic proteases; DLS, dynamic light scattering; HSQC, heteronuclear single quantum coherence; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific insert; SAPLIPs, saposin-like proteins
Aspartic proteases (APs) are ubiquitous in various life domains and have diverse functions (1, 2). Some APs in plants are involved in the innate immune systems, which inhibit to phytopathogens (3, 4). Like other APs that are synthesized as inactive proenzymes with prosegments, many plant APs have an additional segment known as the plant-specific insert (PSI) sequence consisting of approximately 100 amino acids between the N- and C-terminal domains of the proenzyme (5). The PSI domain exhibits activities against plant pathogens both as a part of AP enzyme (6, 7) and as an independent protein (8, 9). In vitro studies have shown that the AP from potato (Solanum tuberosum) inhibits germination of Phytophthora infestans and Fusarium solani, thus limiting their growth (6, 7). As an independent protein, a PSI from a S. tuberosum AP also acts as a defensive protein in response to attack by plant pathogens and is able to inhibit the plant fungal pathogen Botrytis cinerea in a dose-dependent manner (8, 9). Transgenic expression of a S. tuberosum PSI in Arabidopsis thaliana improved the resistance to B. cinerea attack by inducing jasmonic acid and salicylic pathway genes, thus the defensive activity of the PSI in vivo (8).
As a potential antimicrobial protein, the PSI's mechanism against the pathogen invasion has attracted much attention (5, 8, 9, 10, 11), although much remains unknown. The PSI forms dimers under acidic conditions (10) and adopts a fold similar to that of saposin-like proteins (SAPLIPs) in mammals (12, 13), which generally demonstrate the ability to bind and disrupt membranes (14, 15, 16). Similar to other SAPLIPs, the PSI interacts with lipid bilayers (10, 15, 17), inducing membrane permeability (18, 19) and membrane fusion (5, 11). Using vesicles with different lipid compositions, the PSI's ability to induce membrane fusion was pH dependent (10, 11).
Previous studies have suggested that the activity of PSI and its “parent” protein AP is organism dependent. Although being toxic to human and plant pathogens (including yeasts and prokaryotes) and cancer cells, it shows no toxicity to plant cells, human T-cells, or red blood cells (6, 7, 9, 18, 20, 21, 22). A common difference for cells that the PSI is toxic toward as compared with those which are not is the content of negatively charged phospholipids in their cell membranes. Human and plant pathogens are characterized by a higher content (20–25 %) of negatively charged phospholipids, whereas neutral phospholipids are dominant in the plasma membranes of human and plant cells, with only 5 to 10 % negatively charged phospholipids in their cell membranes (23, 24, 25, 26, 27, 28). As the PSI's activity is related to its interactions with membranes, it is possibly more inclined to interact with negatively charged liposomes in order to identify potential targets (6, 21, 29, 30, 31).
To date, the mechanistic details regarding the dependency of PSI activities on lipid composition, however, remains unclear. Therefore, this work attempts to yield further insights into the functions of the PSI. In this study, various complimentary biophysical techniques, that is, cryo-EM, Dynamic Light Scattering (DLS), FRET, NMR and Molecular Dynamics (MD) Simulations, were used to study the fusion activity dependence of the PSI as a function of brain-phosphatidylserine (PS) content, a negatively charged phospholipid, in liposomes. The cryo-EM studies trapped a stalk-like intermediate state, indicating that PSI-induced membrane fusion occurs via a stalk model within the hemi-fusion pathway. The rate of membrane fusion and the PSI's dynamics on liposomes were highly dependent on the content of PS. Increasing the PS content substantially accelerated PSI-induced membrane fusion. The NMR data showed that the PS concentration did not affect the ability of the PSI to bind vesicles but affected the dynamics of the PSI on liposomes. In PS-free liposomes, the PSI experienced pronounced motion, whereas this motion was suppressed when associated with PS-containing liposomes. Consistent with NMR, the MD data suggested that the PSI align a dominant angle ranging from −31° to 30°, on the surface of PS-containing membranes, driven by strong charge-charge interactions. Conversely, in PS-free membranes, the PSI experiences multiple topological states on or away from the surface of the membranes due to a lack of similar type of interactions. Taking into account the hemi-fusion pathway, the stable topology of the PSI on PS-containing liposomes could be more efficient in bridging different liposomes and bringing them into closer proximity, thereby accelerating the fusion process. Because pathogens have much higher amount of negatively charged lipids than host cells, these results suggest that target pathogens are identified through membrane composition.
Results
The hemi-fusion state of PSI-induced membrane fusion
Physiologically, PSI is involved in vacuole sorting; the environment of the vacuole being acidic. Our earlier work also demonstrated that the PSI forms dimers under acidic conditions and induces membrane fusion effectively below pH 4.5 (11). However, the fusion-intermediate states have not been observed. Membrane fusion possibly occurs via the lipid-stalk model or the fusion pore model (32), both of which are observed within the hemi-fusion pathway.
In the present study, cryo-EM was used to observe fusion intermediates at pH 4.5 (Fig. 1). Figure 1B shows an example of an intermediate state of vesicle fusion induced by the PSI, in which the deformation of round vesicles was separated by a single bilayer. In contrast, the vesicles without the PSI did not show shared bilayers, even though some vesicles were in contact with one another (Fig. 1A). This observation agreed with the lipid-stalk model within the hemi-fusion pathway, which includes a diaphragm formed between the inner leaflets of the two membranes (32).
Figure 1.
Representative Cryo-electron micrographs of liposome with PSI.A, liposomes docking and two bilayers between two liposomes (white arrow); B, liposomes in intermediate state of the hemi-fusion liposomes. The white arrows point to the single bilayer between the fused liposome. The liposome composition was PC/PE/PS in a molar ratio of 1:1:1. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
Lipid composition dependence of PSI-induced membrane fusion
FRET experiments were conducted to investigate the lipid composition dependence of PSI-induced membrane fusion. Lipid-mixing experiments were performed by mixing nonfluorescent liposomes and fluorescent liposomes, where the PSI was added to induce vesicle fusion. Nonfluorescent liposomes were composed of phosphatidylcholine (PC)/phosphatidylethanolamine (PE) at a molar ratio of 1:1 (termed PS-free liposome hereafter) or PC/PE/PS (in a molar ratio of 1:1:0.5 or 1:1:1). Hereafter, the PC/PE/PS with a molar ratio of 1:1:1 was termed PS-containing liposome. The fluorescent liposomes were prepared by incorporating both Pho-PE and NBD-PE as the fluorescently labeled lipids (preparation details in method section FRET experiments). The close proximity between the fluorophore groups in the fluorescent liposomes quenched emission at 535 nm when using excitation wavelength at 465 nm. Lipid mixing by membrane fusion with nonfluorescent liposomes was expected to recover the emission intensity due to dilution of the fluorophore lipids with the nonfluorophore lipids and therefore separation of the FRET pairs.
The fluorescent and nonfluorescent liposomes were mixed in a pH 4.5 buffer and combined at a ratio of 1:9. The PSI solution was added to this mixture. Figure 2 showed that the fusion efficiency depends on the content of PS in liposomes. Increasing the PS concentration substantially increased the fusion efficiency. For liposomes of PS-containing liposomes, 90% fusion efficiency was achieved 1200 s after adding the PSI, with respect to the maximum fluorescent signal obtained upon the addition of Triton X-100 (positive control). However, the fusion efficiency of only 33% was observed for the PS-free liposomes and 68% for 1:1:0.5 PC/PE/PS liposomes (Fig. 2). These results clearly demonstrated that the fusion rates of PSI-induced membrane fusion were substantially faster in PS-containing liposomes than in PS-free liposomes, proving the PS dependency of membrane fusion by the PSI.
Figure 2.
Lipid mixing experiments demonstrates that PS content influences PSI-induced fusion efficiency. The molar ratio of protein to liposome is 1:20. A, lipid mixing curves for the PSI. The mixing curves for liposomes with PC/PE/PS of 1:1:1, PC/PE/PS of 1:1:0.5, and PC/PE of 1:1 are shown in red, blue, and green, respectively. The curves for control experiments with buffer and with Triton X-100 are shown in purple and black, respectively. B, membrane fusion efficiencies at 1200 s plotted against content of PS. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
Size distribution of liposomes after PSI-induced vesicle fusion
DLS can quantify the size distribution of the resultant fused liposomes. DLS data were collected before and after addition of the PSI to both types of liposomes (PS-free liposome and PS-containing liposomes). Before PSI addition, two peaks corresponding to vesicle radii of 10 and 100 nm were observed for both types of liposomes, with most vesicles being distributed around 100 nm (Fig. 3). After PSI addition, a broad peak appeared for both types of liposomes, corresponding to vesicles with different radii ranging from 100 to 1000 nm, thus indicating that PSI is able to fuse both PS-free and PS-containing liposomes with similar ranges of vesicle sizes, and that PS lipids did not affect the PSI fusion ability.
Figure 3.
Dynamic light-scattering data of liposomes after adding PSI. A, PS-containing liposomes and (B) PS-free liposomes. The size distribution of liposomes before (black) and after (red) the addition of PSI are shown. PS, phosphatidylserine; PSI, plant-specific interests.
Lipid compositions mediate PSI motions in the liposome
The lipid composition dependent fusion rate of PSI-induced membrane fusion may result from differences in the capability of the PSI to bind liposomes or differently affect the dynamics of the PSI in or on the liposomes. To clarify this point, NMR spectroscopy was used to study the interaction between PSI and different types of liposomes.
Solution NMR was used to study the binding capability between the PSI and different liposomes. As shown in Figure 4, the 1H-15N HSQC (heteronuclear single quantum coherence) of the PSI were recorded before and after adding different types of liposomes. Maximal PSI fusion activity has been shown to occur at approximately pH 4.5, which is near the PSI's PI. Owing to solubility issues at pH 4.5, all solution NMR experiments were conducted at 298 K and at pH 3.4. Combining PS-containing liposomes with the PSI at a protein:liposome molar ratio of 1:2 led to substantial reductions of the cross-peak intensities of the PSI. The addition of more liposomes (PSI:liposome molar ratio = 1:4) further reduced the spectral intensities. Only the cross-peaks corresponding to the highly flexible unstructured loop (i.e., 42–63) remained (Fig. 4, F and G). These results showed strong binding between the PSI and PS-containing liposomes (Fig. 4), consistent with earlier reports (11). The largely reduced signal intensities also indicated the limited motion of the PSI in liposomes.
Figure 4.
PSI-liposome interactions determined by the solution NMR. All spectra are 1H-15N HSQC spectra. A and E, the HSQC of spectra of free 15N-PSI at pH 3.4 and at 298 K. B and F, the HSQC of PSI after the addition of PS-free liposomes to a final protein-to-liposome molar ratio of 1:2 (B) and 1:4 (F). The boxes highlight the residues that are attenuated after the addition of liposomes. C and G, the HSQC of PSI after the addition of PS-containing liposomes to a final protein-to-liposome molar ratio of 1:2 (F) and 1:4 (G). D and H, the HSQC of spectra of the supernatant of the mixing solution from PSI + PS-free liposomes (D) and PSI + PS-containing liposomes (H). HSQC, heteronuclear single quantum coherence; PS, phosphatidylserine; PSI, plant-specific interests.
The same set of NMR titration experiments were also conducted between the PSI and the PS-free liposomes, where the 1H-15N HSQC spectra were collected accordingly (Fig. 4, A–C). Unlike the PS-containing liposomes, the cross-peak intensities for the PSI interacting with PS-free liposomes did not differ appreciably between the PSI alone, regardless of the samples containing protein:liposome in a molar ratio of 1:2 or 1:4 (Fig. 4, A–C).
The overall spectral changes of the PSI upon adding PS-free liposome were less pronounced than those when adding PS-containing liposomes. Two scenarios can explain the difference in the titration results. One possibility is that the PSI has a dramatically different binding affinity for two types of liposomes, whereby it interacts transiently with PS-free liposome, with most of the PSI remaining in solution, thus producing sharp lines for the protein signals. Conversely, the PSI may form stable complexes with PS-containing liposomes, thus diminishing its signals. Another possibility is that the PSI strongly binds both types of liposomes, but with different dynamic characteristics. In this scenario, the PSI may bind to PS-free liposomes, but moves relatively freely on their surface, thus potentially leading to narrow line-width for most residues of the PSI in PS-free PSI-liposome complexes. In contrast, the PSI when interacting with PS-containing liposomes may be less mobile.
To clarify these two possibilities, both of the PSI-liposome solutions were centrifuged to remove the liposomes, after the collection of the 1H-15N HSQC spectra of the supernatant. As shown in Figure 4, D and H, protein signals were not observed in either of the supernatants. These results showed that the PSI strongly binds on both types of liposomes, which in turn suggested that the narrow signals observed for the PSI on PS-free liposomes was because of the large dynamic motions of the PSI on PS-free liposomes.
Solid-state NMR was further used to validate the dynamics of PSI on two types of liposomes. The centrifuged liposomes with 15N-labeled PSI were packed into solid-state NMR rotors, and temperature dependent 1H-15N cross-polarization solid-state NMR spectra were collected. Because cross-polarization transfer was achieved using dipolar interactions, the motion of the amide groups would average and cancel out the dipolar couplings, yielding a reduction in signal intensities (33). As shown in Figure 5, the signal intensity of the PSI in PS-free liposomes substantially decreased when the temperature was increased from 268 K to 298 K; the signal intensity of the PSI at 298 K was only 20% with respect to that at 268 K (Fig. 5D). In contrast, using PS-containing liposomes, the temperature dependent spectral changes were much smaller. In PS-containing liposomes, the signal intensity at 298 K was about 76% that at 268 K (Fig. 5H). The different dynamic behavior of the PSI with different types of liposomes further supports the conclusion that the presence of PS phospholipids affects the dynamics of the PSI in membranes. This result was consistent with the solution NMR data where the PSI had additional molecular motion in PS-free liposomes with respect to PS-containing liposomes. The PSI is known to form dimers and bridge the two liposomes in close proximity to initiate membrane fusion (11). The rigid PSI in PS-containing liposomes discovered in the present study would be more efficient in bridging different liposomes, thus accelerating the process.
Figure 5.
Temperature variable15N-ssNMR spectra of the PSI in different type of liposomes.A–D, 1D 15N-PSI in PS-free liposomes at (left and right) 268K, 278K, 288K, and 298K. E–H, 1D 15N-PSI in PS-contained liposomes at (left and right) 268 K, 278K, 288 K, and 298 K. I, the relative intensities of 15N-PSI plotted against temperatures. PS, phosphatidylserine; PSI, plant-specific interests.
Lipid compositions mediate the interactions of the PSI with the liposome
To investigate the dynamics of the PSI on different types of liposomes, all-atom MD simulation of the PSI on two membranes compositions—1:1:1 POPC/POPE/POPS (PS-containing) and 1:1 POPC/POPE (PS-free)—was performed. The stability of secondary and tertiary structure of PSI dimer on the membranes was evaluated by Cα-RMSD and secondary structure analysis (Figs. S1 and S2). The majority of residues are quite stable in secondary structure, especially those in helical regions (Fig. S3). Qualitatively, no meaningful differences in secondary structure were found between the PS-free and PS-containing systems (Fig. S4).
We further computed the electron density distributions of the PSI on both types of bilayers and for the phospholipids in each of the bilayer compositions along the z-axis (a direction perpendicular to the membrane surface). The two pure membranes share overlapping density distributions, whereas the PSI's density distribution showed a lipid-dependent profile (Fig. 6). The PSI on a PS-containing membrane generally tended to the more closely associate with the membrane's surface compared with the PSI on a PS-free membrane. There is also more overlap with the respective membrane curves for the PSI on a PS-containing membrane than when on a PS-free membrane surface, indicating that the PSI forms stronger interaction with the PS-containing membrane. This is reasonable considering that PS has a net negatively charged head group and that the majority of acidic residues on the PSI are protonated under the simulated acidic condition (pH 4.0). The electrostatic interactions maintain a closer interaction between the PSI and the PS-containing membranes.
Figure 6.
Electron density profiles of PC/PE/PS and PC/PE membranes and associated PSI along the normal z-axis. The center of the membrane was set as the coordinate origin. The last 300 ns trajectory data of all 15 simulations were used for calculation. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
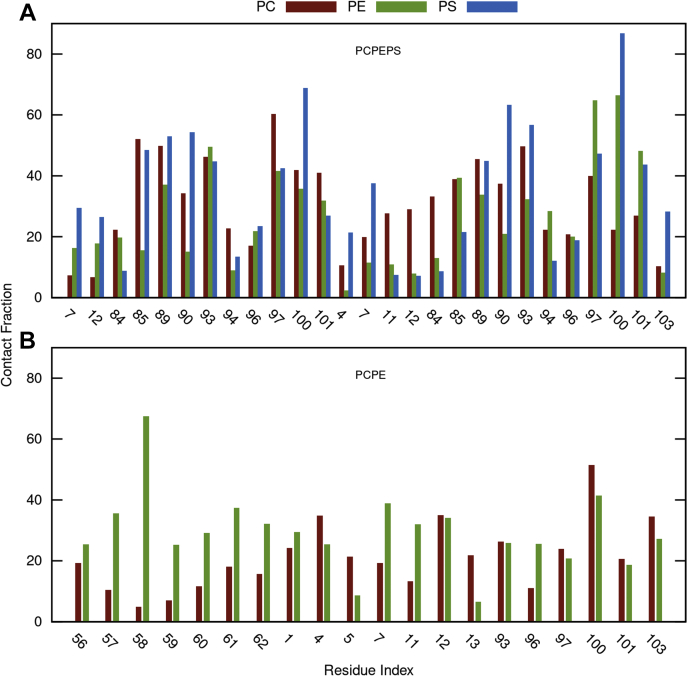
To identify the hotspot residues involved in the PSI-membrane interactions, we computed contact maps of the PSI residues relative to the head groups of each of the phospholipids in respective membranes (Fig. 7). Once the PS component is introduced, the overall contact fractions of PSI residues interacting with the membrane display a substantial increase. In the PS-containing system, the PSI residues preferentially form more contacts with PS molecules than with either PE or PC molecules (Fig. 7A). The interaction primarily mediated by residues (∗denotes protonated state of carboxylate group) of E100, K90, E93∗, Q97, E89∗, and E85. It is reasonable that the positively charged K90 and the (neutralized) polar residues E93∗, Q97, and E89∗ contribute highly to the interaction of PSI with the negatively charged PS lipids. However, of note are E100 and E85, which are deprotonated (negatively charged) and also form strong contacts with PS. A close inspection of the interaction details reveals the presence of strong hydrogen bonds (<2.1 Å, 140–160°) between the serine portion of the PS head group with the side chain and main chain oxygen atoms of the amino acids E100 and E85, respectively (Fig. 8). In the PS-free system, the hotspot residues are E58∗, E100, K7, and S61, with a similar contact fraction magnitude interacting with PC and PE to hotspots in the PS-containing system. Note that the hotspot residue E100 in the PS-containing system is also a major hotspot residue in the PS-free system, whereas the residues E58∗ and S61 are only among the top residues in the PS-free system.
Figure 7.
Contact map of PSI residues with the lipid head groups PC, PE, and/or PS in PS-containing and PS-free membrane systems. The contact fraction was summed over the PSI residue and the lipid head groups and was averaged overall 15 MD simulations. Only the top residues interacting with the three lipid head groups with at least one fraction larger than 20 are present for clarity. The x-axis consists of the residues of chains A and B of PSI. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
Figure 8.
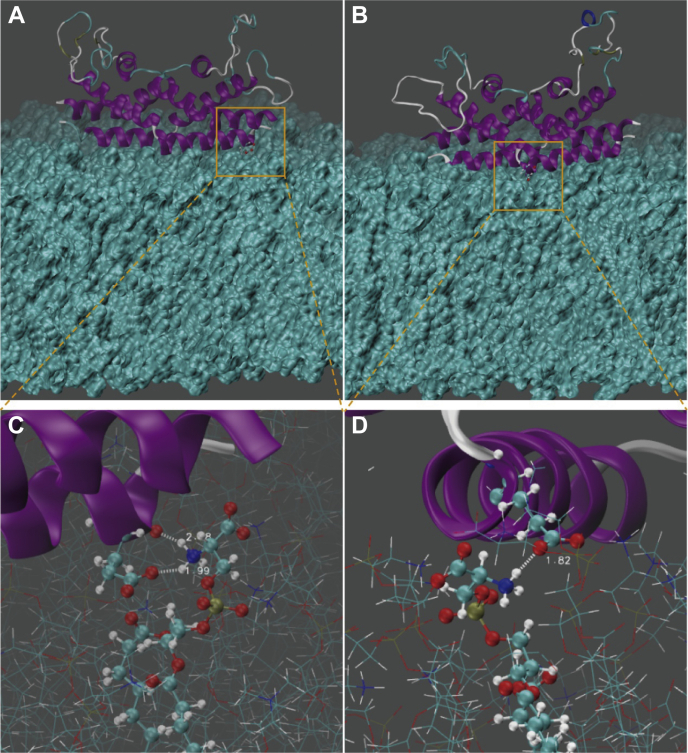
Snapshots of PSI with close contact with the membrane bilayer and the zoom-in details of interactions between PSI residues E100 and E85 and the phospholipid PS in the PC/PE/PS membrane. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
Lipid compositions regulate the dynamics of the PSI on bilayer surfaces
To examine the dynamics of the PSI on bilayer surfaces, we calculated the distance of the center of mass of the PSI to the central plane of the lipid bilayer and the angle between the principal vector of PSI with the XY-plane against time. As shown in Figure 9A, the curves are separated in distinct two groups. The PSI is quite stable in the PS-containing systems, whereas the PSI in the PS-free systems undergoes large fluctuations, indicating poor binding of the PSI in this system. This is also consistent with the aforementioned electron density profiles (Fig. 6), in which the PSI preferred to interact with a membrane containing PS. The principal vector of the PSI is defined as the vector from the Cα atom of the residue V2 to the Cα atom of the residue P66 to represent the principal direction (shown in red arrows in Fig. 10B). The angle between the principal vector of the PSI with the XY-plane also shows overall larger changes in the PSI's orientation in the PS-free system, suggesting that PSI often rotates during the simulation (Fig. 9B). Taken together, the dynamics of PSI are quite different in the membrane with or without the PS component, with more dynamics being observed in the PS-free system. These observations are consistent with our above solution NMR study of HSQC spectra (Fig. 4), which showed that the narrow signals of the PSI on PS-free liposomes, indicating large dynamic motions when associated with them.
Figure 9.
Time evolutions of the distance of the center of mass (COM) of PSI to the central plane of membrane bilayer and the angle θ between the principal vector of PSI and the XY-plane in PC/PE/PS and PC/PE systems. The curves of PS-containing systems are colored in red and PS-free systems in green. The last 300 ns trajectory data of all 15 simulations were used for calculation. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
Figure 10.
The range of the principle angles of PSI with the PS-free or PS-containing liposomes.A, probability density of the angle θ between the principal vector of PSI and the XY-plane using Freedman-Diaconis' binning rule (34) for bin width determination. The last 300 ns trajectory data of all 15 simulations were used for calculation. B, representative snapshots of typical angles for PSI in PC/PE/PS and PC/PE systems. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PSI, plant-specific interests.
To further characterize the typical states the PSI adopts in the interactions with the membrane, we plotted the normalized frequency histogram of the angle between the principal vector of PSI and the XY-plane using Freedman-Diaconis' binning rule (34) for bin width determination. As shown in Figure 10A, PSI predominantly adopts a flat orientation with the overall principal vector tilt angle ranging from −31° to 30° in the PS-containing system, whereas adopting diverse dominant angles with the overall angle spanning widely from −89° to 55° in the PS-free system. The snapshots of typical angles in those two systems are shown in Figure 10B. This demonstrates that the lipid compositions regulate not only the dynamics of the PSI on bilayer surfaces but also the dominant states which the PSI prefers to adopt in the interactions.
Discussion
Some plants use membrane fusion induced by membrane fusion proteins as a way to resist pathogen attack (35). Antimicrobial proteins have an important role in distinguishing host cells from pathogens, thus allowing for selective cytotoxicity (36). For antimicrobial proteins linked to membrane binding and disruption, the target is often negatively charged phospholipids as the content of negatively charged phospholipids varies widely between different organisms or different cell types in the same organisms (37). The plasma membranes of prokaryote or fungal pathogens contain 20 to 25% anionic phospholipids (38), whereas in eukaryotic cell membranes, they only compose 5 to 10% of total lipids. Furthermore, the distribution of phospholipids in mammalian cells is asymmetric. The outer leaflet is composed mainly of neutral lipids (39) and anionic phospholipids, such as PS lipids, are found in the inner leaflet of the plasma membrane, which only translocate to the outer leaflet during apoptosis (40). Thus, many antimicrobial proteins have evolved to preferentially bind and disrupt negatively charged phospholipids to selectively attack pathogen(s) (41).
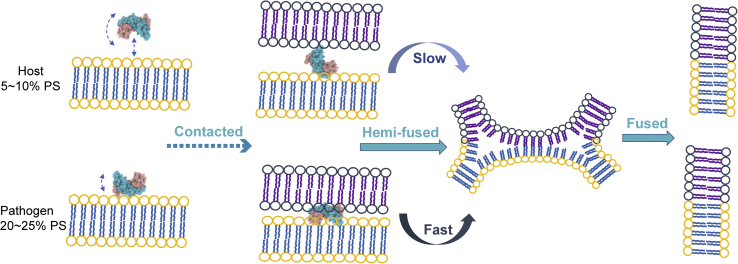
The PSI targets fungal and prokaryotic pathogens and shows lipid composition-dependent activity (15). The present study combines multiple biophysical techniques to identify the mechanism of PSI-membrane interactions as a function of PS content in the membrane fusion process. Each technique provides a unique aspect to determine the mechanism, whereby the combined techniques provide a more complete description. The results of the cryo-EM and DLS demonstrate, for the first time, that the PSI passes through a hemi-fusion intermediate state during the membrane fusion process. Although the PSI is capable of fusing both the PS-free and the PS-containing vesicle, FRET data showed that both the fusion rate and the fusion efficiency of PSI-induced membrane fusion are much higher for PS-containing liposomes. Lastly, NMR and MD experiments revealed the PS-dependent dynamics of the PSI when interacting with liposomes. In PS-containing liposomes, the PSI binds the phospholipid bilayer resulting in limited molecular motion. Conversely, in PS-free liposomes, PSI has a higher degree of motion at physiological temperature. Given that PSI-induced membrane fusion progresses through the hemi-fusion state, the rigid angle of the PSI when interacting with PS-containing liposomes (because of stronger binding) would be more effective in bringing two liposomes together in close proximity, yielding higher fusion efficiency and a faster fusion rate (Fig. 11).
Figure 11.
A schematic representation of PSI-induced fusion of liposomes. PSI, plant-specific interest.
Our results provide insights into its physiological function. In plants, the PSI localizes to the central vacuole, which plays a role in the defense against pathogen attack (5). Upon pathogen invasion-induced plant stress, two mechanisms of action involving the vacuole have been suggested; (i) fusing the vacuole's membrane with the plasma membrane of infected plant cells to induce apoptosis to prevent further infection and (ii) the release of toxic cargoes to attack the invading pathogens (17). For both mechanisms, the presence of the pathogen's cell membrane would result in an increase in the amount of negatively charged phospholipids. Therefore, the PSI may responded to the increase of the PS membrane, leading to reduced motion and faster disruption on membrane. The PSI may also be an important factor in the attack of pathogen membranes, that is, inducing leakage of the pathogen cells and leading to their lysis. The results obtained here also explained the organism-dependent activity (6, 7, 9, 18, 20, 21, 22). As an independent protein, the PSI has been shown to be cytotoxic toward yeast and prokaryotic plant pathogens, but not to the host plant cells. The main difference between two types of cell membranes is the lipid composition. Thus, the PSI may anchor on a prokaryotic membrane with limited motion and thereby induce faster disruption. Finally, SAPLIPs have potential medical applications; the results obtained here could facilitate the design of the SAPLIPs to fulfill the latter.
Experimental procedures
Isotope-labeled compounds (15NH4Cl) were obtained from Cambridge Isotopes Laboratories. The lipids were purchased from Avanti Polar Lipids.
PSI expression and purification
Uniformly 15N-labeled PSI was expressed and purified, as previously described (10).
Liposome preparations
The liposomes used for lipids mixing experiments were prepared, as reported previously (42). Fluorescent and nonfluorescent liposomes were prepared. The nonfluorescent liposomes were composed of egg-PE, egg-PC and brain-PS at a molar ratio of 1:1:X (X was either 0.5 or 1) or egg-PE and egg-PC at a molar ratio of 1:1. The PC/PE/PS fluorescent liposomes were comprised of PE, PC, PS, Rho-PE (N-(lissamine rhodamine B sulfonyl)-PE) and NBD-PE (N-(7-nitro-2,1,3-benzoxadiazol- 4-yl)-PE) at a molar ratio of 1:1:X:0.018:0.018 (X was 0.5 or 1). The PC/PE fluorescent liposomes were comprised of PE, PC, Rho-PE (N-(lissamine rhodamine B sulfonyl)-PE), and NBD-PE (N-(7-nitro- 2,1,3-benzoxadiazol-4-yl)-PE) at a molar ratio of 1:1:0.018: 0.018. After 10 freeze-thaw cycles, the liposomes were extruded 20 times through two stacked polycarbonate membranes with a pore of 100 nm.
Cryo-electron microscopy
The PSI was dissolved in a pH 4.5 buffer (27 mM Na-H3PO4 and 140 mM NaCl). A 3-μL aliquot of 100 μM PSI solution was mixed with a 500 μL of liposome solution (5 μM). The liposomes used were 1:1:1 PC/PE/PS. After addition of the PSI, the mixture was incubated for 5 min, followed by quick-freezing in liquid nitrogen to quench the fusion process.
A 4-μl aliquot of the samples was placed on a freshly glow-discharged lacey carbon EM grid. After 2.5 s of blotting with filter paper, the grid was flash-plunged in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific Inc.) at 20 °C and 100% humidity. The samples were imaged in low-dose conditions using a Titan Krios (Thermo Fisher Scientific Inc.) operating at 300 kV. The images of all conditions were collected at a nominal magnification of ×47,000 and an under-focus of 2.0 to 3.0 μm on a Falcon 3EC camera (4096 pixels × 4096 pixels).
FRET experiments
FRET experiments were conducted by mixing nonfluorescent and fluorescent liposomes at a molar ratio of 9:1 and diluted to the final concentration of 300 μM. The experiments were conducted at room temperature. The PSI was added to the mixtures of liposome to a final concentration of 15 μM. The molar ratio of protein to liposome is 1:20 to make the reaction rates in a reasonable time scale to be observed. The buffer condition was pH 4.5, 27 mM Na-H3PO4, and 140 mM NaCl. The excitation wavelength was set to 465 nm and emission was detected at 535 nm. The fluorescence intensity of the liposomes without PSI was set as the baseline (0 fluorescence value, f0), and the fluorescence intensity that induced by a solution of Triton X-100 with a final concentration after mixing of 0.2% was set as 100 fluorescence value (f100). The fusion efficiency, ft, was calculated according to the equation: % ft = (ft − f0)/(f100 − f0) × 100.
DLS assay
DLS was used to determine the size distribution of fused liposomes using a BI-200SM goniometer. The liposomes used for the DLS measurements were PS-free or PS-containing liposomes. The liposome solution was diluted to a final phospholipid concentration of 4 μM and mixed with the PSI at a molar ratio of 30:1. The liposomes before and after PSI addition were subjected to DLS measurement. The scattering angle (90°) and temperature (25 °C) were kept constant. The results were processed in the WinContin software, and the distribution of size was calculated with the regularization algorithm provided by the software.
Solution NMR spectroscopy
All solution NMR experiments were carried on a 500 MHz or 600 MHz Bruker Avance III spectrometer. The 1H-15N HSQC spectra of 15N-labeled PSI were collected at pH 3.4 and at a temperature of 298 K. The titration experiments between liposome and PSI were carried at 0.1 mM 15N-PSI at pH 3.4 (27 mM Na-H3PO4, 140 mM NaCl).
Solid-state NMR spectroscopy
The samples used for studying PSI-liposome interaction by ssNMR used 0.3 mM 15N-PSI at pH 3.4 in buffer (27 mM Na-H3PO4 and 140 mM NaCl). The 15N-PSI was mixed with liposomes at a molar ratio of 1:30 with gentle agitation at room temperature for 1 h followed by ultracentrifugation at 900,000 g at 4 °C for 3 h to collect the 15N-PSI-liposome complexes. The collected PSI-liposome complexes were packed into a 4.0 mm rotor for ssNMR data collection.
All solid-state NMR spectra were collected on a 600 MHz Bruker Avance III spectrometer equipped with a 4.0 mm E-free 1H/13C/15N probe. All the ssNMR experiments were conducted at a magic-angle spinning frequency of 10.0 kHz. The temperature was calibrated externally by the T1 relaxation time of 79Br in KBr powder (43). Typical π/2 pulses used were 2.5 μs for 1H, 4.0 μs for 13C, and 5.0 μs for 15N. All the experiments used the SPINAL-64 decoupling scheme at 100 kHz for 1H decoupling during the acquisition. The program TOPSPIN 3.2 was used to process NMR data. The 13C chemical shifts were calibrated externally to DSS (3-(trimethylsilyl) propane-1-sulfonic acid) and analyze the data using the program CARA.
Molecular dynamics simulation study
The PSI monomer structure was retrieved from the Protein Data Bank (PDB ID: 3RFI) (10), and the dimer recreated using the contained crystallographic symmetry information. Missing residues were fixed using the MODELLER package (44). The complete PSI dimer was then relaxed with a 40 ns MD simulation using the AMBER 19 package (45) at 300 K. Next, the relaxed PSI structure was submitted to the H++ web server (46) to determine the protonation states to approximate a simulation pH of 4.0. As a result, on the PSI, the following amino acids were simulated as protonated: E5, D19, D40, H44, E54, E56, E58, E64, E89, and E93. The residues E85 and E100 were simulated as deprotonated according to a previous constant pH simulation (47), which reported much lower pKa values for those two residues. Next, two membrane systems, POPC: POPE: POPS (“PS-containing”) and POPC: POPE (“PS-free”), with respective lipid ratios of 1:1:1 and 1:1 were generated using the CHARMM-GUI server (48). The TIP3P water model (49) was used, and different water thicknesses of 40 Å and 42 Å were specified to construct two PSI conformations and to ensure that there was sufficient space in the Z dimension for simulation. The X and Y dimensions were set as 79 Å, which led to a membrane bilayer patch of 34:34:34 or 50:50 for PC:PE:PS or PC:PE phospholipid components in one leaflet, respectively. The PSI structure and the membrane were combined together with the PSI positioned ∼6 Å above the surface of the membrane. Two simulation conformations (PSI lying down (“lie-down”) and PSI standing up (“stand-up”)) were constructed. The overlapped water molecules were deleted using visual molecular dynamics (50) after a reported criterion that the minimum distance of a water oxygen atom from any of the solute atoms was 2.6 Å, and the minimum distance of a water hydrogen atom was 2.0 Å (51).
The final system was neutralized with sodium or chloride ions as needed, and extra NaCl was added to model the experimental salt concentration of 0.14 M. All MD runs were performed using the AMBER 19 package (45) with the ff14SB protein force field (52) and the LIPID17 lipid force field (53). The van de Walls cutoff was set to 10 Å, and explicit solvent particle mesh Ewald was used for long-range electrostatic energy calculation. Minimization, heating, and holding processes were performed before production runs. All production simulations were performed at 303 K under the NPT ensemble with the integration step of 2 fs and an anisotropic pressure coupling. The temperature of 303 K was set to be higher enough than the phase transition temperatures of lipids. Five independent runs for each membrane system (PS-containing or PS-free) and conformation (lie-down/stand-up) were carried out to improve sampling, which led to a total of 20 simulations. Production trajectories of 700 ns were generated for all simulations. Two PS-containing trajectories and two PS-free trajectories with the stand-up conformation and one PS-free trajectory with the lie-down conformation were dropped for analysis due to the PSI drifting under the bottom of the membrane bilayer. Water molecules and ions of all trajectories were removed first before analysis. All the figures were prepared using gnuplot or visual molecular dynamics (50).
The equilibration of simulations was monitored by tracking the z-depth of the PSI center of mass and the PSI principal vector tilt angle and the rmsd of Cα atom (Cα-RMSD) of PSI excluding the loop regions (residues 40–63) during the simulation. As shown in Figures 9 and S1, the systems were equilibrated with some simulations of the PC/PE system undergoing fluctuations considering the typical dynamic properties of the simulated systems. Based on the observation, the last 300 ns trajectory data were used in the subsequent analysis to ensure converged calculations.
Data availability
All data relating to this manuscript is contained in the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare no conflicts of interest with the contents of this article.
Acknowledgments
All the NMR experiments were carried out at Beijing NMR Centre or the NMR facility at the National Centre for Protein Sciences at Peking University (Beijing, People's Republic of China). The Cryo-EM studies were carried out at Cryo-EM Center at Southern University of Science and Technology (Shenzhen, People's Republic of China). This work was supported by the National Key Research and Development Program of the Ministry of Science and Technology of the People's Republic of China (2016YFA0501203), the National Natural Science Foundation of China (Grant number 91953104 and 21874004), the State Key Laboratory of Medicinal Chemical Biology (2020031), the Fundamental Research Funds for the Central Universities, and the Beijing National Laboratory for Molecular Sciences. The authors thank Dr Hongwei Li and Dr Xiaogang Niu in the Beijing NMR Center of Peking University for assistance in collecting NMR data.
Author contributions
X. Z., X. M., J. J. T., R. Q., Z. Q., P. W., R. Y. Y., and S. W. methodology; X. Z., X. M., J. H. D., R. Q., J. J. T., J. C., X. O., Z. Q., D. L., P. W., R. Y. Y., and S. W. writing–review and editing; X. Z., X. M., Z. Q., D. L., P. W., R. Y. Y., and S. W. conceptualization; X. Z., X. M., J. H. D., R. Q., J. J. T., J. C., X. O., Z. Q., and D. L. data curation; X. Z., X. M., R. Q., J. J. T., J. C., X. O., Z. Q., and D. L. formal analysis; X. Z., X. M., P. W., R. Y. Y., and S. W. funding acquisition; X. Z., R. Q., Z. Q., D. L., P. W., and S. W. validation; X. Z., X. M., J. H. D., R. Q., J. J. T., J. C., X. O., Z. Q., D. L., P. W., R. Y. Y., and S. W. investigation; X. Z., X. M., J. H. D., R. Q., P. W., R. Y. Y., and S. W. writing–original draft; X. Z., P. W., R. Y. Y., and S. W. project administration; X. M., and R. Q. software; Z. Q., P. W., and R. Y. Y. supervision.
Funding and additional information
S. W. was supported by the National Key Research and Development Program of the Ministry of Science and Technology of the People's Republic of China (2016YFA0501203), the National Natural Science Foundation of China (Grant number 91953104 and 21874004), the State Key Laboratory of Medicinal Chemical Biology (2020031), the Fundamental Research Funds for the Central Universities, and the Beijing National Laboratory for Molecular Sciences. R. Q. was supported by the Guangdong Basic and Applied Basic Research Foundation (Grant no. 2019A1515110090). R. Y. Y. is supported by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (Grant no. RGPIN 04598). Financial support for J. H. D. and J. T. T. were through the MITACS Globalink program and via by NSERC.
Edited by Joseph Jez
Footnotes
Dedicated to the 100th anniversary of Chemistry at Nankai University.
Contributor Information
Peiyi Wang, Email: wangpy@sutech.edu.cn.
Rickey Y. Yada, Email: r.yada@ubc.ca.
Shenlin Wang, Email: wangshenlin@pku.edu.cn.
Supporting information
References
- 1.Davies D.R. The structure and function of the aspartic proteinases. Annu. Rev. Biophys. Biophys. Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- 2.Dunn B.M. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A., Janssen A., Bruin J., Posthumus M.A., Sabelis M.W. An ecological cost of plant defence: Attractiveness of bitter cucumber plants to natural enemies of herbivores. Ecol. Lett. 2002;5:377–385. [Google Scholar]
- 4.Shimada T., Takagi J., Ichino T., Shirakawa M., Hara-Nishimura I. Plant vacuoles. Annu. Rev. Plant Biol. 2018;69:123–145. doi: 10.1146/annurev-arplant-042817-040508. [DOI] [PubMed] [Google Scholar]
- 5.Cheung L.K.Y., Dupuis J.H., Dee D.R., Bryksa B.C., Yada R.Y. Roles of plant-specific inserts in plant defense. Trends Plant Sci. 2020;25:682–694. doi: 10.1016/j.tplants.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Guevara M., Oliva C., Huarte M., Daleo G. An aspartic protease with antimicrobial activity is induced after infection and wounding in intercellular fluids of potato tubers. Eur. J. Plant Pathol. 2002;108:131–137. [Google Scholar]
- 7.Guevara M.G., Almeida C., Mendieta J.R., Faro C.J., Verissimo P., Pires E.V., Daleo G.R. Molecular cloning of a potato leaf cDNA encoding an aspartic protease (StAsp) and its expression after P. infestans infection. Plant Physiol. Biochem. 2005;43:882–889. doi: 10.1016/j.plaphy.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Frey M.E., D'Ippolito S., Pepe A., Daleo G.R., Guevara M.G. Transgenic expression of plant-specific insert of potato aspartic proteases (StAP-PSI) confers enhanced resistance to Botrytis cinerea in Arabidopsis thaliana. Phytochemistry. 2018;149:1–11. doi: 10.1016/j.phytochem.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Munoz F.F., Mendieta J.R., Pagano M.R., Paggi R.A., Daleo G.R., Guevara M.G. The swaposin-like domain of potato aspartic protease (StAsp-PSI) exerts antimicrobial activity on plant and human pathogens. Peptides. 2010;31:777–785. doi: 10.1016/j.peptides.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Bryksa B.C., Bhaumik P., Magracheva E., De Moura D.C., Kurylowicz M., Zdanov A., Dutcher J.R., Wlodawer A., Yada R.Y. Structure and mechanism of the saposin-like domain of a plant aspartic protease. J. Biol. Chem. 2011;286:28265–28275. doi: 10.1074/jbc.M111.252619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X., Tian J., Yu H., Bryksa B.C., Dupuis J.H., Ou X., Qian Z., Song C., Wang S., Yada R.Y. Insights into the mechanism of membrane fusion induced by the plant defense element, Plant Specific Insert. J. Biol. Chem. 2020;295:14548–14562. doi: 10.1074/jbc.RA120.014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guruprasad K., Törmäkangas K., Kervinen J., Blundell T.L. Comparative modelling of barley-grain aspartic proteinase: A structural rationale for observed hydrolytic specificity. FEBS Lett. 1994;352:131–136. doi: 10.1016/0014-5793(94)00935-x. [DOI] [PubMed] [Google Scholar]
- 13.Ponting C.P., Russell R.B. Swaposins: Circular permutations within genes encoding saposin homologues. Trends Biochem. Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]
- 14.Bruhn H. A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 2005;389:249–257. doi: 10.1042/BJ20050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egas C., Lavoura N., Resende R., Brito R.M., Pires E., de Lima M.C., Faro C. The saposin-like domain of the plant aspartic proteinase precursor is a potent inducer of vesicle leakage. J. Biol. Chem. 2000;275:38190–38196. doi: 10.1074/jbc.M006093200. [DOI] [PubMed] [Google Scholar]
- 16.Kolter T., Sandhoff K. Principles of lysosomal membrane digestion: Stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu. Rev. Cell Dev. Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 17.Munoz F., Palomares-Jerez M.F., Daleo G., Villalain J., Guevara M.G. Cholesterol and membrane phospholipid compositions modulate the leakage capacity of the swaposin domain from a potato aspartic protease (StAsp-PSI) Biochim. Biophys. Acta. 2011;1811:1038–1044. doi: 10.1016/j.bbalip.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Mendieta J.R., Pagano M.R., Munoz F.F., Daleo G.R., Guevara M.G. Antimicrobial activity of potato aspartic proteases (StAPs) involves membrane permeabilization. Microbiology. 2006;152:2039–2047. doi: 10.1099/mic.0.28816-0. [DOI] [PubMed] [Google Scholar]
- 19.Munoz F., Palomares-Jerez M.F., Daleo G., Villalain J., Guevara M.G. Possible mechanism of structural transformations induced by StAsp-PSI in lipid membranes. Biochim. Biophys. Acta. 2014;1838:339–347. doi: 10.1016/j.bbamem.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Guevara M.G., Oliva C.R., Machinandiarena M., Daleo G.R. Purification and properties of an aspartic protease from potato tuber that is inhibited by a basic chitinase. Physiol. Plant. 1999;106:164–169. [Google Scholar]
- 21.Guevara M.G., Veríssimo P., Pires E., Faro C., Daleo G.R. Potato aspartic proteases: Induction, antimicrobial activity and substrate specificity. J. Plant Pathol. 2004;86:233–238. [Google Scholar]
- 22.Mendieta J.R., Fimognari C., Daleo G.R., Hrelia P., Guevara M.G. Cytotoxic effect of potato aspartic proteases (StAPs) on Jurkat T cells. Fitoterapia. 2010;81:329–335. doi: 10.1016/j.fitote.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Gidalevitz D., Ishitsuka Y., Muresan A.S., Konovalov O., Waring A.J., Lehrer R.I., Lee K.Y.C. Interaction of antimicrobial peptide protegrin with biomembranes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6302–6307. doi: 10.1073/pnas.0934731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huijbregts R.P.H., de Kroon A.I.P.M., de Kruijff B. Topology and transport of membrane lipids in bacteria. Biochim. Biophys. Acta. 2000;1469:43–61. doi: 10.1016/s0304-4157(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 25.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 26.Simons K., Vaz W.L. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 27.Slotte J.P. Sphingomyelin–cholesterol interactions in biological and model membranes. Chem. Phys. Lipids. 1999;102:13–27. doi: 10.1016/s0009-3084(99)00071-7. [DOI] [PubMed] [Google Scholar]
- 28.Tannert A., Pohl A., Pomorski T., Herrmann A. Protein-mediated transbilayer movement of lipids in eukaryotes and prokaryotes: The relevance of ABC transporters. Int. J. Antimicrob. Agents. 2003;22:177–187. doi: 10.1016/s0924-8579(03)00217-6. [DOI] [PubMed] [Google Scholar]
- 29.Chen F., Foolad M.R. Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol. Biol. 1997;35:821–831. doi: 10.1023/a:1005833207707. [DOI] [PubMed] [Google Scholar]
- 30.Faro C., Ramalho-Santos M., Veríssimo P., Pissarra J., Frazão C., Costa J., Lin X.-L., Tang J., Pires E. Structural and functional aspects of cardosins. Adv. Exp. Med. Biol. 1998;436:423–433. doi: 10.1007/978-1-4615-5373-1_58. [DOI] [PubMed] [Google Scholar]
- 31.Runeberg-Roos P., Saarma M. Phytepsin, a barley vacuolar aspartic proteinase, is highly expressed during autolysis of developing tracheary elements and sieve cells. Plant J. 1998;15:139–145. doi: 10.1046/j.1365-313x.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 32.Jackson M., Chapman E. The fusion pores of Ca2+-triggered exocytosis. Nat. Struct. Mol. Biol. 2008;15:684–689. doi: 10.1038/nsmb.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Aslimovska L., Glaubitz C. Molecular dynamics of proteorhodopsin in lipid bilayers by solid-state NMR. J. Am. Chem. Soc. 2011;133:4874–4881. doi: 10.1021/ja109766n. [DOI] [PubMed] [Google Scholar]
- 34.Freedman D., Diaconis P. On the histogram as a density estimator: L 2 theory. Z. Wahrscheinlichkeitstheorie verw Gebiete. 1981;57:453–476. [Google Scholar]
- 35.Hatsugai N., Iwasaki S., Tamura K., Kondo M., Fuji K., Ogasawara K., Nishimura M., Hara-Nishimura I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009;23:2496–2506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glukhov E., Stark M., Burrows L.L., Deber C.M. Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J. Biol. Chem. 2005;280:33960–33967. doi: 10.1074/jbc.M507042200. [DOI] [PubMed] [Google Scholar]
- 37.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 38.Glukhov E., Stark M., Burrows L.L., Deber C.M. Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J. Biol. Chem. 2005;280:33960–33967. doi: 10.1074/jbc.M507042200. [DOI] [PubMed] [Google Scholar]
- 39.Verkleij A.J., Zwaal R.F.A., Roelofsen B., Comfurius P., Kastelijn D., Vandeene Ll. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim. Biophys. Acta. 1973;323:178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]
- 40.Verhoven B., Schlegel R.A., Williamson P. Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J. Exp. Med. 1995;182:1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouny Y., Rapaport D., Mor A., Nicolas P., Shai Y. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogs with phospholipid membranes. Biochemistry. 1992;31:12416–12423. doi: 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- 42.Schuette C.G., Hatsuzawa K., Margittai M., Stein A., Riedel D., Küster P., König M., Seidel C., Jahn R., Südhof T.C. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thurber K.R., Robert T. Measurement of sample temperatures under magic-angle spinning from the chemical shift and spin-lattice relaxation rate of 79Br in KBr powder. J. Magn. Reson. 2009;196:84–87. doi: 10.1016/j.jmr.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 45.Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Giambasu G., Giese T.J., Gilson M.K., Gohlke H., Goetz A.W., Greene D., et al. University of California; San Francisco, CA: 2019. AMBER 2019. [Google Scholar]
- 46.Gordon J.C., Myers J.B., Folta T., Shoja V., Heath L.S., Onufriev A. H++: A server for estimating p Ka s and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupuis J.H., Yu H., Habibi M., Peng X., Plotkin S.S., Wang S., Song C., Yada R.Y. pH dependent membrane binding of the Solanum tuberosum plant specific insert: An in silico study. Biochim. Biophys. Acta Biomembr. 2018;1860:2608–2618. doi: 10.1016/j.bbamem.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Lee J., Patel D.S., Ståhle J., Park S.J., Kern N.R., Kim S., Lee J., Cheng X., Valvano M.A., Holst O., Knirel Y.A., Qi Y., Jo S., Klauda J.B., Widmalm G., et al. CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J. Chem. Theory Comput. 2019;15:775–786. doi: 10.1021/acs.jctc.8b01066. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 50.Humphrey W., Dalke A., Schulten K. Vmd: Visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Kandt C., Ash W.L., Peter Tieleman D. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson C.J., Madej B.D., Skjevik Å.A., Betz R.M., Teigen K., Gould I.R., Walker R.C. Lipid14: The amber lipid force field. J. Chem. Theor. Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relating to this manuscript is contained in the article.