Summary
Drosophila flight muscles are highly enriched with mitochondria and have emerged as a powerful genetic system for studying how oxidative phosphorylation (OXPHOS) complexes are assembled. Here, we describe a series of protocols for analyzing the integrity of OXPHOS complexes in Drosophila via blue native polyacrylamide gel electrophoresis (BN PAGE). We have also included protocols for the additional steps that are typically performed after OXPHOS complexes are separated by BN PAGE, such as Coomassie staining, silver staining, and in-gel OXPHOS activities.
For complete details on the use and execution of this protocol, please refer to Murari et al. (2020).
Subject areas: Cell Biology, Genetics, Metabolism, Model Organisms, Protein Biochemistry
Graphical abstract
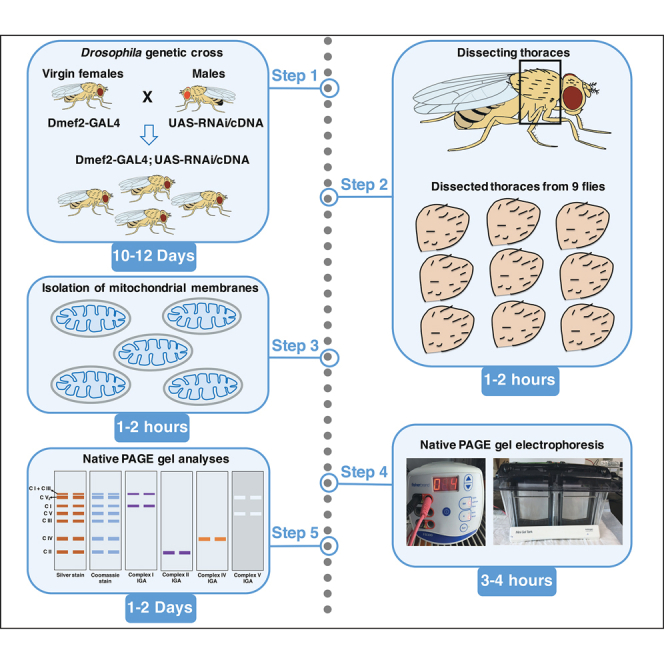
Highlights
-
•
Blue native polyacrylamide gel electrophoresis (BN PAGE) separates OXPHOS complexes
-
•
OXPHOS assembly in Drosophila flight muscles can be assessed with BN PAGE
-
•
OXPHOS activities can be assessed by in-gel activity assays
Drosophila flight muscles are highly enriched with mitochondria and have emerged as a powerful genetic system for studying how oxidative phosphorylation (OXPHOS) complexes are assembled. Here, we describe a series of protocols for analyzing the integrity of OXPHOS complexes in Drosophila via blue native polyacrylamide gel electrophoresis (BN PAGE). We have also included protocols for the additional steps that are typically performed after OXPHOS complexes are separated by BN PAGE, such as Coomassie staining, silver staining, and in-gel OXPHOS activities.
Before you begin
The flight muscles in Drosophila have frequently been exploited to study mitochondrial dynamics and how various mitochondrial proteins influence overall organismal physiology and aging (Deng et al., 2008; Garcia et al., 2017; Owusu-Ansah et al., 2013; Rana et al., 2017; Rera et al., 2011; Ulgherait et al., 2014). Here, we describe a series of approaches for studying the assembly of OXPHOS complexes in Drosophila flight muscles. As Drosophila thoraces are enriched with mitochondria, we use mitochondria from 10 fly thoraces to check the integrity of OXPHOS complexes by BN-PAGE, followed by silver staining and Coomassie staining. For in-gel activity assays of OXPHOS complexes, we use mitochondria isolated from 20 fly thoraces. While we focus on the flight muscles, the techniques we describe can be adapted for use in other tissues, such as the brain, gut, and larval muscles. This protocol assumes the reader has a working knowledge of Drosophila genetics and the OXPHOS system.
Fly husbandry
Timing: Approximately 2 weeks
-
1.
Collect virgins of the appropriate genotype.
-
2.
Keep the virgins in a fly incubator at 25°C for 3–5 days prior to setting up the crosses.
CRITICAL: Before setting up the crosses, check that the female flies to be used are actually virgins, by ensuring that no larvae are present in the vials housing the female flies.
-
3.
Set up each cross with approximately 9 virgins (3–5 days old) and 3 adult male flies of the appropriate genotype in a fly food vial. For best results, use male flies that are 2–3 days old.
-
4.
Set up the crosses at 25°C in a Forma environmental chamber or any suitable fly incubator with a 12 h day/12 h night cycle.
-
5.
Let the flies lay eggs for at least 48 h at 25°C.
-
6.
Transfer the flies into fresh fly food and return both vials (both the new and previous vial that was used to start the cross) to the 25°C incubator.
-
7.
Repeat steps 5 and 6 to obtain 3 sets of crosses.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Sucrose | Fisher Scientific | S5-500 |
| Magnesium Chloride (MgCl2) | Fisher Scientific | M33-500 |
| Tris Base | Fisher Scientific | BP152-25 |
| Bis-Tris | Fisher Scientific | BP301-100 |
| Tricine | Fisher Scientific | BP315-100 |
| Sodium Phosphate Monobasic Anhydrous (NaH2PO4) | Fisher Scientific | S397-500 |
| Sodium Phosphate Dibasic Anhydrous (Na2HPO4) | Fisher Scientific | S375-500 |
| Hydrochloric Acid (HCl) | Fisher Scientific | SA55 |
| Methanol | Fisher Scientific | A412-4 |
| Glacial Acetic Acid | Fisher Scientific | A38-500 |
| Digitonin (−20°C) | Sigma-Aldrich | D141-500MG |
| G-250 Sample Additive (Blue Dye) (4°C) | Invitrogen | BN2004 |
| Sample Buffer (4°C) | Invitrogen | BN2003 |
| 20× Cathode Buffer Additive | Invitrogen | BN2002 |
| Halt Protease Inhibitor (4°C) | Thermo Scientific | PI78430 |
| Bovine Serum Albumin (4°C) | Fisher Scientific | BP9703100 |
| Nitrotetrazolium Blue Chloride (NBT) (4°C) | Sigma-Aldrich | N6876-500MG |
| Nicotinamide Adenine Dinucleotide Hydrate (NADH) (−20°C) | Sigma-Aldrich | N7004-1G |
| Adenosine Triphosphate (ATP) (4°C) | Acros Organics | AC102800100 |
| Disodium Succinate | Sigma-Aldrich | W327700 |
| Phenazine Methosulfate (−20°C) | Sigma-Aldrich | P9625-1G |
| Diaminobenzidine (DAB) (4°C) | Sigma-Aldrich | D5637-5G |
| Cytochrome C (−20°C) | Sigma-Aldrich | C2506-100MG |
| Glycine | Fisher Scientific | G46-500 |
| Magnesium Sulfate Anhydrous (MgSO4) | Fisher Scientific | M65-500 |
| Lead Nitrate [Pb(NO3)2] | Sigma-Aldrich | 228621-100G |
| Bradford 1× Dye Reagent | Bio-Rad Laboratories | 5000205 |
| Experimental models: Organisms/strains | ||
| D. melanogaster strain expressing GAL4 in muscles under the control of the Mef2 promoter. yw[1118]; Dmef2-Gal4 | (Ranganayakulu et al., 1996) | Dmef2-gal4 |
| D. melanogaster strain expressing temperature-sensitive GAL80 under the control of the alphaTub84B promoter. w [∗]; P{w{+mC]=tubPGAL80[ts]}10; TM2/TM6, Tb[1] | Bloomington Drosophila Stock Center | BDSC 7108; Flybase: FBti0027799 |
| D. melanogaster strain expressing a transgenic RNAi construct to dErv1 (Alr). y1 sc∗ v1 sev21; P {TRiP.HMC03697}attP40 | Bloomington Drosophila Stock Center | BDSC 65088; Flybase: FBst0065088 |
| D. melanogaster strain expressing a transgenic RNAi construct to AIF. w1118; P{GD822}v2544 | Vienna Drosophila Resource Center | VDRC v2544; Flybase: FBst0455887 |
| D. melanogaster strain, wild type used as a control. w1118 | Bloomington Drosophila Stock Center | BDSC 3605; Flybase: FBst0003605 |
| Critical commercial assays | ||
| Colloidal Blue Staining Kit | Invitrogen | LC6025 |
| SilverXpress Silver Staining Kit | Invitrogen | LC6100 |
| Other | ||
| Light Contrast Microscope | Olympus | SZ61 |
| Forceps | Fine Science Tools | 11251-10 |
| Power Supply | Fisher Scientific | FB300 |
| Bullet Blender | Next Advance | BBX24B |
| Zirconium Oxide Beads | Next Advance | ZROB05 |
| NativePAGE 3%–12% Bis-Tris Gel | Invitrogen | BN1003BOX |
| Forma Environmental Chamber | Fisher Scientific | 13-067-066 |
| ChemiDoc Imaging System | Bio-Rad Laboratories | 17001401 |
| Mini Gel Tank | Invitrogen | A25977 |
| LaserJet 200 color MFP | HP | CF145A#BGJ |
Materials and equipment
Preparing mitochondria isolation medium
| Mitochondria isolation medium (MIM) | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 1 M Sucrose | 250 mM | 250 mL |
| 0.1 M MgCl2 | 0.15 mM | 1.5 mL |
| 1 M Tris-HCl | 10 mM | 10 mL |
| ddH2O | 738.5 mL | |
| Total | 1000 mL | |
Aliquot in 50 mL portions in a falcon tube. Store up to 1 year @ −80°C.
Preparing 20× BN PAGE running buffer
| Composition of the 20× BN PAGE running buffer | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Bis-Tris | 1 M | 209.2 g |
| Tricine | 1 M | 179.2 g |
| ddH2O | Adjust volume to 1000 mL | |
| Total | 1000 mL | |
Store up to 6 months @ room temperature (25°C).
Preparing 1× BN PAGE anode buffer
| Composition of the 1× BN PAGE anode buffer | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 20× Running Buffer | 50 mM | 50 mL |
| ddH2O | 950 mL | |
| Total | 1000 mL | |
Store up to 6 months @ 4°C.
Preparing 1× BN PAGE cathode buffer
| Composition of the 1× BN PAGE cathode buffer | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 20× Running Buffer | 50 mM | 50 mL |
| 20× Cathode Buffer Additive | 0.1× | 5 mL |
| ddH2O | 945 mL | |
| Total | 1000 mL | |
Store up to 6 months @ 4°C.
Preparing BN PAGE fixative solution
| Composition of the BN PAGE fixative solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Methanol | 40% | 40 mL |
| Glacial Acetic Acid | 10% | 10 mL |
| ddH2O | 50 mL | |
| Total | 100 mL | |
Store up to 6 months @ room temperature (25°C).
Preparing BN PAGE staining solution
| Composition of the BN PAGE staining solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Methanol | 20% | 20 mL |
| Stainer A (from Colloidal Blue Staining Kit) | 20% | 20 mL |
| Stainer B (from Colloidal Blue Staining Kit) | 5% | 5 mL |
| ddH2O | 55 mL | |
| Total | 100 mL | |
Store up to 1 month @ room temperature (25°C).
Preparing BN PAGE destaining solution
| Composition of the BN PAGE destaining solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Glacial Acetic Acid | 8% | 80 mL |
| ddH2O | 920 mL | |
| Total | 1000 mL | |
Store up to 6 months @ room temperature (25°C).
Preparing 1% digitonin buffer
| Composition of the 1% digitonin buffer (D) | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 4× Sample Buffer | 1× | 62.5 μL |
| 5% Digitonin | 1% | 50 μL |
| 100× Protease Inhibitor | 1× | 2.5 μL |
| Ultrapure Water | 135 μL | |
| Total | 250 μL | |
Storage conditions: Make just before you begin, and keep on ice.
Preparing G-250 blue dye
| Composition of the blue dye (B) | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 4× Sample Buffer | 2.67× | 100 μL |
| 5% G-250 Sample Additive | 0.42% | 12.5 μL |
| Ultrapure Water | 37.5 μL | |
| Total | 150 μL | |
Storage conditions: Make just before you begin, and keep on ice.
Preparing sensitizer solution
| Composition of the sensitizer solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Sensitizer (from SilverXpress Silver Staining Kit) | Unknown | 5 mL |
| Methanol | 47.6% | 100 mL |
| Ultrapure Water | 105 mL | |
| Total | 210 mL | |
Storage conditions: Make just before you begin, and keep @ room temperature (25°C).
Preparing silver staining solution
| Composition of the silver staining solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Stainer A (from SilverXpress Silver Staining Kit) | Unknown | 5 mL |
| Stainer B (from SilverXpress Silver Staining Kit) | Unknown | 5 mL |
| Ultrapure Water | 90 mL | |
| Total | 100 mL | |
Storage conditions: Make just before you begin, and keep @ room temperature (25°C).
Preparing developer solution
| Composition of the developer solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Developer (from SilverXpress Silver Staining Kit) | Unknown | 5 mL |
| Ultrapure Water | 95 mL | |
| Total | 100 mL | |
Storage conditions: Make just before you begin, and keep @ room temperature (25°C).
Preparing 0.1 M sodium phosphate dibasic solution
| Composition of the 0.1 M sodium phosphate dibasic solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Sodium Phosphate Dibasic | 0.1 M | 7.1 g |
| Ultrapure Water | Adust volume to 500 mL | |
| Total | 500 mL | |
Store up to 1 year @ room temperature (25°C).
Preparing 0.1 M sodium phosphate monobasic solution
| Composition of the 0.1 M sodium phosphate monobasic solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| Sodium Phosphate Monobasic | 0.1 M | 6.0 g |
| Ultrapure Water | Adust volume to 500 mL | |
| Total | 500 mL | |
Store up to 1 year @ room temperature (25°C).
Preparing 0.1 M sodium phosphate buffer pH 7.2
| Composition of the 0.1 M sodium phosphate buffer pH 7.2 | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 0.1 M Sodium Phosphate Dibasic Solution | 0.1 M | 100 mL |
| 0.1 M Sodium Phosphate Monobasic Solution | 0.1 M | Adjust pH to 7.2 by adding drops of the 0.1 M sodium phosphate monobasic solution to 100 mL of the 0.1 M sodium phosphate dibasic solution |
Store up to 1 year @ room temperature (25°C).
Preparing complex I in-gel activity solution
| Composition of the complex I in-gel activity solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 10 mg/mL Nitrotetrazolium Blue | 2.5 mg/mL | 1.25 mL |
| 40 mM NADH | 0.136 mM | 0.017 mL |
| 10 mM Tris-HCl, pH 7.4 | 5 mM | 2.5 mL |
| Ultrapure Water | 1.233 mL | |
| Total | 5 mL | |
Storage conditions: Make just before you begin, and keep on ice.
Preparing complex II in-gel activity solution
| Composition of the complex II in-gel activity solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 1 M Sodium Succinate | 20 mM | 0.1 mL |
| 20 mM Phenazine Methosulfate | 0.2 mM | 0.05 mL |
| 10 mg/mL Nitrotetrazolium Blue |
2.5 mg/mL | 1.25 mL |
| 10 mM Tris-HCl, pH 7.4 | 5 mM | 2.5 mL |
| Ultrapure Water | 1.1 mL | |
| Total | 5 mL | |
Storage conditions: Make just before you begin, and keep on ice.
Preparing complex IV in-gel activity solution
| Composition of the complex IV in-gel activity solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 1% Diaminobenzidine | 0.05% | 0.25 mL |
| 0.83 mM Cytochrome C | 0.05 mM | 0.3 mL |
| 100 mM Sodium Phosphate, pH 7.2 | 50 mM | 2.5 mL |
| Ultrapure Water | 1.95 mL | |
| Total | 5 mL | |
Storage conditions: Make just before you begin, and keep on ice.
Preparing complex V in-gel activity solution
| Composition of the complex V in-gel activity solution | ||
|---|---|---|
| Reagent | Final concentration | Amount |
| 1 M Magnesium Sulfate | 14 mM | 0.07 mL |
| 10% Lead Nitrate | 0.2% | 0.1 mL |
| 100 mM Adenosine Triphosphate | 8 mM | 0.4 mL |
| 35 mM Tris Base + 270 mM Glycine, pH 8.4 | 4.43 mL | |
| Total | 5 mL | |
Storage conditions: Make just before you begin, and keep on ice.
Step-by-step method details
Collection and aging of flies
Timing: Approximately 1–2 days (excluding the variable period during which the flies are aged)
-
1.
Following eclosure of the flies (after approximately 10 days at 25°C), collect male and female offspring.
-
2.
Maintain collected flies in vials at a density of less than 25 flies per vial, and age them at 25°C until they reach a suitable time point.
-
3.
While aging the flies, transfer them into new food 2–3 times per week. Discard the old vials.
Dissection of Drosophila thoraces
Timing: Approximately 1 h
-
4.
Once the flies reach the desired age, anesthetize the flies on a CO2 pad, and dissect 10 fly thoraces under a microscope using suitable dissection forceps.
Note: Anesthetized flies are dissected by severing both head and abdomen from the thorax. Save the thorax. Flies do not need to be in a buffer during dissection, and wings or legs do not have to be removed.
-
5.
Place the dissected fly thoraces in an Eppendorf tube (without any buffer). Store immediately at −80°C until you are ready to perform the experiment.
Pause point: The frozen thoraces can be stored for at least a year at −80°C.
Isolation of mitochondrial membranes from thoraces
Timing: Approximately 1 h
-
6.
Thaw an aliquot of the MIM (see materials and equipment) by incubating it in a 37°C water bath until it is almost completely defrosted. Remove and keep on ice until it is completely thawed .
CRITICAL: Check the MIM periodically to ensure it does not fully defrost while in the 37°C water bath. When there is little ice left take it out and place it in the ice bucket. Vortex briefly to mix thoroughly.
-
7.
Obtain dissected thoraces from −80°C and keep on ice to defrost (Figure 1A).
-
8.
Place the sample buffer, protease inhibitor, and blue dye in the ice bucket.
-
9.
Add one scoop of Zirconium Oxide beads (equivalent to a bead volume of about 0.2 mL) and 0.5 mL of MIM containing 1× of Halt Protease inhibitors to each sample (Figure 1B).
-
10.
Homogenize the samples in the bullet blender (BB) for 2 min, at a speed of 7 on the BB in the cold room (Figure 1C).
CRITICAL: Analyze the samples to see if chunks of the thorax remain in suspension. For optimal yield, the thorax must be homogenized to the point where the remains of the thorax are barely/slightly visible in the liquid.
-
11.
Centrifuge the homogenized samples at 500 g (500 rcf) for 5 min at 4°C.
-
12.
In the meantime, start defrosting the 5% digitonin suspension.
-
13.
Following centrifugation for 5 min, transfer the supernatant into new Eppendorf tubes.
-
14.
Centrifuge the supernatant again at 500 g for 5 min at 4°C.
-
15.
Transfer the supernatant into new Eppendorf tubes and centrifuge at 5000 g for 5 min at 4°C.
-
16.
Discard the supernatant and save the mitochondria-enriched pellet (Figure 1D).
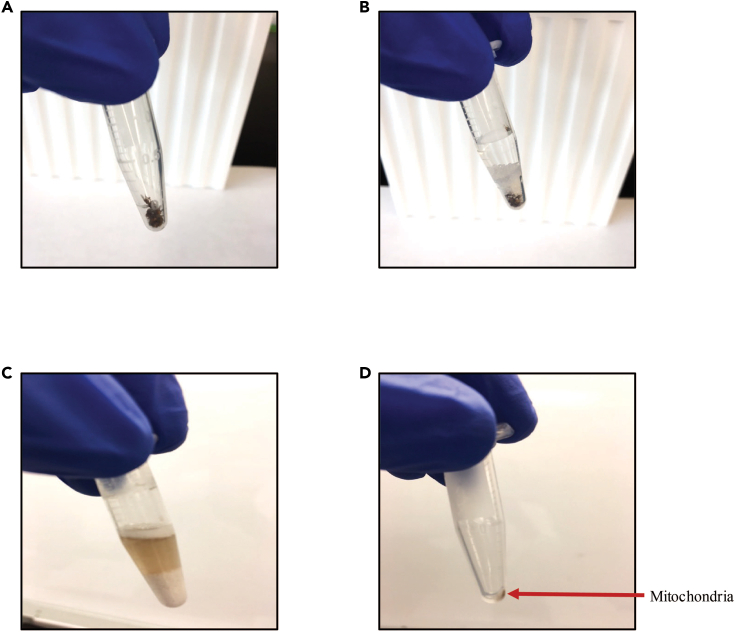
Figure 1.
Mitochondria isolation from Drosophila flight muscles
Images showing fly thoraces before homogenization (A), fly thoraces with one scoop of beads, protease inhibitor and MIM (B), homogenized fly thoraces (C), and isolated mitochondrial pellet (D).
BN PAGE gel tank preparation
Timing: Approximately 10–20 min
-
17.Prepare the gel tank as follows:
-
a.Obtain gel plates (3%–12% gradient native gels) and the gel tank (Figure 2). Unwrap the plates, remove the comb and wash with Milli-Q water, and then with cathode buffer. Peel off the white strip and place it in the gel chamber and lock it.
-
b.Add the anode buffer to the outer chamber, and cathode buffer to the inner chamber. Be sure to leave enough room for the wells to be visible. Using a 1000 μL pipette, add the cathode buffer to the wells.
-
c.After setting up the gel tank, place the gel tank in the cold room or any 4°C incubator.
-
a.
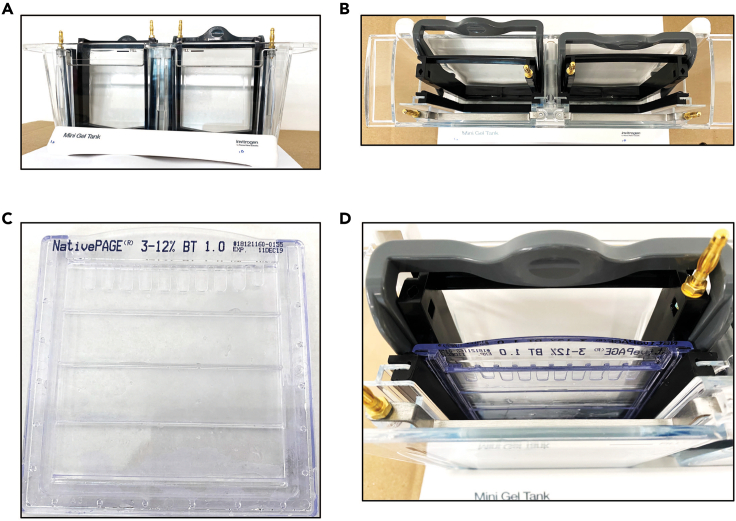
Figure 2.
Setting up the gel tank for gel electrophoresis
Images showing gel tank prior to assembling the gel plate (side view) (A), gel tank prior to assembling the gel plate (top view) (B), 3%–12% gradient Native PAGE gel plate (C), one half of the gel tank after assembling the gel plate (D).
Sample preparation for BN PAGE silver staining
Timing: Approximately 4 h
-
18.
Solubilize the mitochondrial pellet (from step 16) by adding 25 μL of 1% digitonin buffer. Solubilize by pipetting up-and-down (see materials and equipment).
-
19.
Incubate the solubilized mitochondrial pellet on ice for 15 min.
-
20.
Centrifuge the samples at 20,000 g for 20 min at 4°C.
-
21.
Save the supernatant, and transfer it into new Eppendorf tubes. Keep on ice.
Note: The supernatant will have the extracted OXPHOS complexes, while what remains of the membranes will be in the pellet.
-
22.
Measure the protein concentration by Bradford method and normalize the samples to ascertain how much (volume) of each sample needs to be loaded in the gel for equal loading.
-
23.
In a new Eppendorf tube, add 15 μL G-250 blue dye (see materials and equipment), 10 μL 1× sample buffer and 5 μL sample; mix gently.
-
24.
Load the samples in wells based on the amounts given by the Bradford method calculations.
-
25.
Run the gel using the FB300 Fisher Scientific power supply at 4°C.
-
26.
For two gels, run at 300 V and 4 mA (keep milliamps constant) for approximately 3 h.
Silver staining of native gel
This method allows the OXPHOS complexes to be visualized. It is more sensitive than Coomassie staining.
Timing: Approximately 1–2 h
Note: All steps are at room temperature.
-
27.
After gel electrophoresis is complete, remove the gel from the cassette, place in a container with the silver staining fixative solution (50% methanol and 10% acetic acid) and rotate on a shaker for 10 min.
-
28.
Remove the fixative solution and add 100 mL of the sensitizer solution (105 mL of ultrapure water, 100 mL of methanol and 5 mL of Sensitizer) and keep on the shaker for 10 min.
-
29.
Following sensitization, wash the gel twice with 200 mL of ultrapure water for 5 min each.
-
30.
Then incubate the gel in the silver staining solution (90 mL of ultrapure water, 5 mL of Stainer A and 5 mL of Stainer B) for 15 min on the shaker.
-
31.
Wash the gel twice with 200 mL of ultrapure water for 5 min each.
-
32.
Develop the gel by adding the developer (95 mL of ultrapure water and 5 mL of Developer). Stop the reaction by adding the stopper solution once bands are visible.
-
33.
After about 10 min, capture the image with the ChemiDoc Imaging System (Figures 3A–3C and 4A).
Figure 3.
Imaging of a silver-stained native gel by a Bio-Rad ChemiDoc Imaging system
Images of the Bio-Rad ChemiDoc instrument (A), program setting to capture a silver-stained native gel by the ChemiDoc Imager (B), and a silver-stained native gel captured by the ChemiDoc Imager (C).
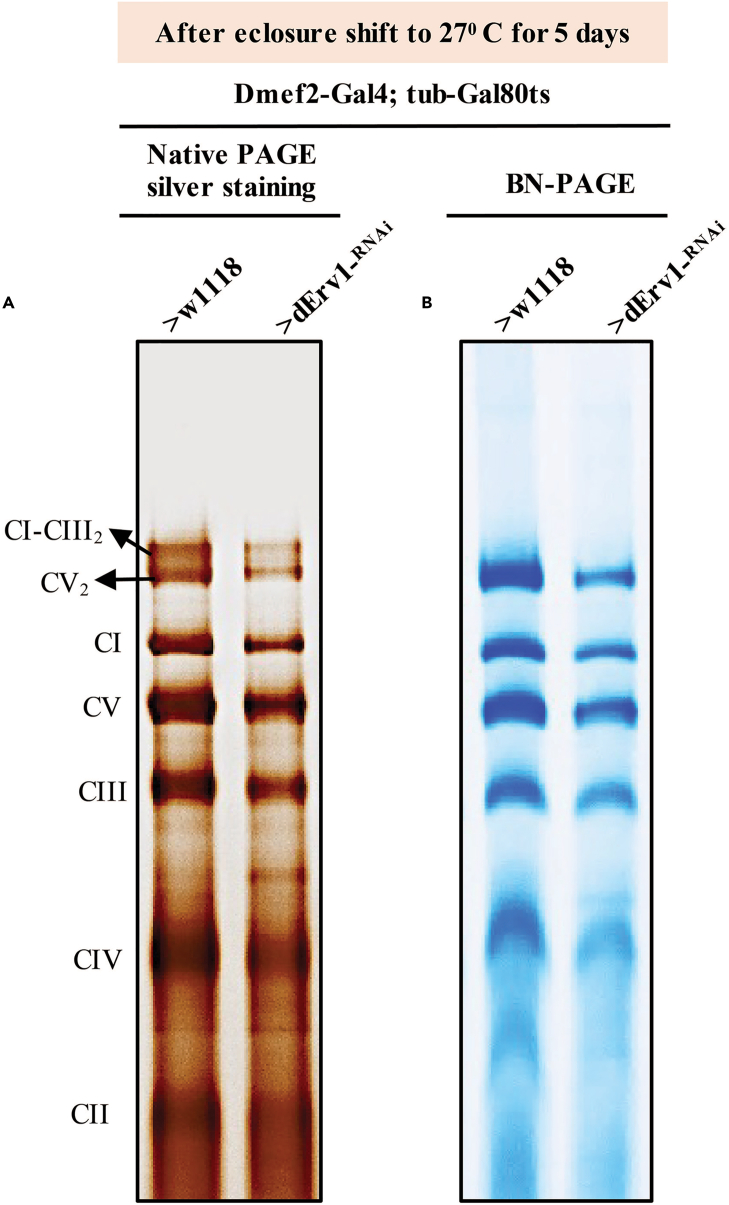
Figure 4.
Analysis of OXPHOS complexes integrity by BN PAGE and silver staining of native gels
BN PAGE followed by silver staining of native gels containing OXPHOS complexes isolated from mitochondria from flight muscles of flies with the genotype indicated (A). BN PAGE and Coomassie staining analysis of native gels containing OXPHOS complexes isolated from mitochondria from flight muscles of flies with the genotypes indicated (B).
Sample preparation for BN PAGE Coomassie staining
Timing: Approximately 4 h
-
34.
In a new Eppendorf tube add 7.5 μL of the blue dye and 15 μL of sample and mix gently.
-
35.
Load the samples in wells based on the amounts given by the Bradford method calculations.
-
36.
Run the gel using the FB300 Fisher Scientific power supply at 4°C. For four gels, run at 300 V and 8 mA (keep milliamps constant) for approximately 3 h.
Coomassie staining of native gel
(see materials and equipment).
This method allows the OXPHOS complexes to be visualized. It is less sensitive than silver staining.
Timing: Approximately 16 h
Note: All steps are at room temperature.
-
37.
Following gel electrophoresis, remove the gel from the cassette, place in a container with the BN PAGE fixative solution (see materials and equipment) and rotate on the shaker for 30 min.
-
38.
Remove the fixative solution, add 100 mL of the BN PAGE staining solution (see materials and equipment) and rotate on the shaker overnight (approximately 16 h).
-
39.
Remove the staining solution and de-stain the gel by adding 100 mL of the de-stainer (see materials and equipment).
-
40.
After the bands are visible, capture the image with the ChemiDoc Imaging System (Figures 5A–5C and 4B).
Figure 5.
Imaging of a Coomassie-stained native gel by a Bio-Rad ChemiDoc Imaging System
Images of the Bio-Rad ChemiDoc instrument (A), program setting to capture the Coomassie-stained native gel by the ChemiDoc Imager (B), and a Coomassie-stained native gel captured by the ChemiDoc Imager (C).
Because Coomassie does not denature proteins it is suitable for characterizing enzyme activities. In-gel enzyme assays make it feasible to identify enzymes in gels based solely on enzymatic activity. Typically, after a gel is run it is soaked in a buffer containing substrates for the enzyme to be detected. Following incubation for a suitable period and a series of washes, the enzyme being assayed can be detected as a distinct colored band in the gel. Here, we describe protocols for visualizing CI, CII, CIV and CV activities of Drosophila OXPHOS complexes within blue native gels.
Sample preparation for in-gel activities of native gels (complex I, complex II, complex IV and complex V)
Timing: 2 days
Here, we describe the sample preparation and the specific steps for analyses of Complex I, II, IV, and V activities in native gels.
-
41.
After steps 1–22, in a new Eppendorf tube add 20 μL of blue dye and 40 μL of sample and mix gently.
-
42.
Load the samples in wells in four sets for CI, CII, CIV and CV in-gel activities based on the amounts given by the Bradford method calculations.
-
43.
Run the gel using the FB300 Fisher Scientific power supply at 4°C. For two gels, run at 300 V and 4 mA (keep milliamps constant).
-
44.After the gel electrophoresis is complete, remove the gel from the cassette.
-
a.Complex I: Place the first gel in a container with Complex I in-gel activity solution (see materials and equipment), and place it on the shaker overnight (approximately 16 h) at 4°C. After the Complex I band is clearly visible, capture the image with a HP LaserJet scanner (Figure 6A).
-
b.Complex II: Place the second gel in a container with Complex II in-gel activity solution (see materials and equipment), and place it on the shaker overnight (approximately 16 h) at 4°C. After the Complex II band is clearly visible, capture the image with a HP LaserJet scanner (Figure 6B).
-
c.Complex IV: Place the third gel in a container with Complex IV in-gel activity solution (see materials and equipment), and place on the shaker overnight (approximately 16 h) at room temperature. After the Complex IV band is clearly visible, capture the image with a HP LaserJet scanner (Figure 6C).
-
d.Complex V: Place the fourth gel in a container with Complex V in-gel activity solution (see materials and equipment), and place on the shaker overnight (approximately 16 h) at room temperature. After the Complex V band is clearly visible, capture the image with a HP LaserJet scanner (Figure 6D).
-
a.
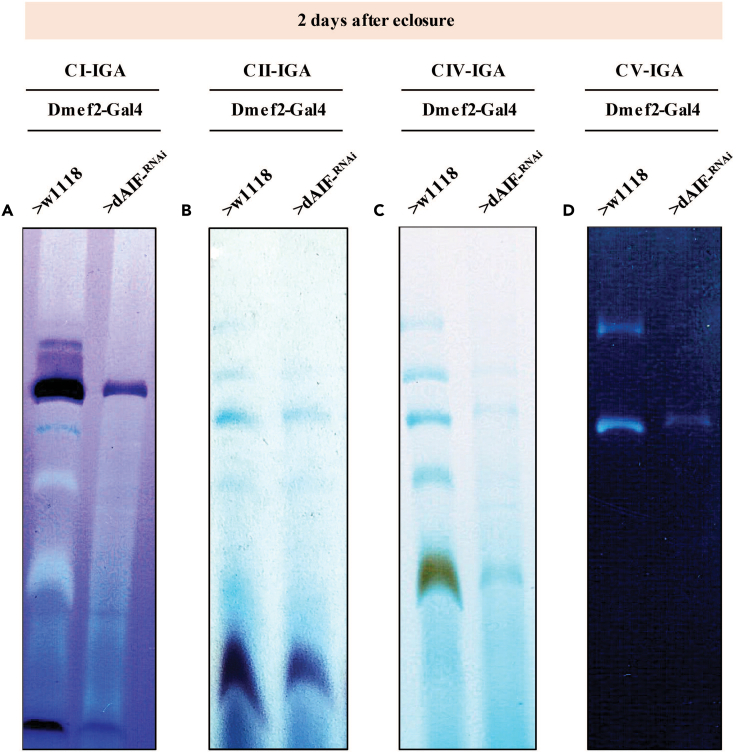
Figure 6.
In-gel activity analyses of OXPHOS complexes in native gels
Complex I (A), Complex II (B), Complex IV (C), and Complex V (D) in-gel activity assays of OXPHOS complexes isolated from flight muscles with the genotypes indicated.
Expected outcomes
This protocol provides the methodology for analyzing the integrity and activities of OXPHOS complexes in Drosophila tissues. Because BN PAGE preserves complexes in their native state it allows OXPHOS complexes and supercomplexes to be visualized in gels. As demonstrated here, BN PAGE can be followed by additional techniques such as Coomassie blue staining, silver staining and in-gel enzyme activity assays to expand its applications.
Limitations
The predominant factor that determines whether a BN PAGE experiment will be successful is the extent to which the OXPHOS complexes are extracted from the mitochondrial membranes. This in turn depends largely on the amount and type of detergent used. The optimal detergent should be able to extract the OXPHOS complexes sufficiently, but not denature the complexes. While the protocol we have described has been optimized for flight muscles (thoraces), modifications may be required for other tissues such as the brain (head extracts), gut, fat body and even larval muscles. This may require adjusting the detergent:protein ratio or even using other detergents such as n-Dodecyl-B-D-Maltoside (DDM). We also note that in-gel OXPHOS activity assays are semi-quantitative at best. Accordingly, for more quantitative results, additional mitochondrial assays such as using microplate readers to measure the specific activity of each of the OXPHOS enzymes spectrophotometrically should be used to confirm the in-gel activity results. Alternatively, mitochondrial respiration can be measured by high resolution respiratory units to validate any results obtained with the in-gel activity assays.
Troubleshooting
Problem 1
Gel electrophoresis takes too long to run (step 26).
Potential solution
Check the electrophoresis wire connections to ensure there are no loose connections to the power pack.
Problem 2
Protein streaks on the gel (step 33).
Potential solution
During step 14 collect only the supernatant and discard the pellet. If you see any particles in the sample (collected supernatant) re-centrifuge at high speed to collect only the supernatant.
Problem 3
Reduced or no in-gel activity (Complex I, II, IV and V) observed after electrophoresis (step 44 a- 44 d).
Potential solution
Ensure samples are processed at 4°C and perform the gel electrophoresis at 4°C. Processing the samples at room temperature and performing gel electrophoresis at room temperature causes protein denaturation and ultimately impairs activity of OXPHOS complexes.
Problem 4
Poorly resolved electrophoretic bands (step 33 & 40).
Potential solution
Add the appropriate amount of the blue dye to the protein sample during sample preparation.
Problem 5
Weak detection of higher molecular weight complexes in silver-stained gels (step 33).
Potential solution
Increase the duration of incorporation with the developer solution in step 32. However, while this makes the higher molecular weight complexes more visible, it comes at the expense of an increase in background staining in the vicinity of the lower molecular weight complexes.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Edward Owusu-Ansah (eo2364@cumc.columbia.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgements
We thank past and present members of the Owusu-Ansah lab for general discussions and critiques. We acknowledge the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for various fly strains. Work in the Owusu-Ansah lab is supported by an Irma Hirschl award and NIH grants AR077312 (R21) and GM124717 (R35).
Author contributions
A. Murari and E. Owusu-Ansah worked together to organize and compose this protocol.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Anjaneyulu Murari, Email: am4821@cumc.columbia.edu.
Edward Owusu-Ansah, Email: eo2364@cumc.columbia.edu.
Data and code availability
This study did not generate any new datasets or code.
References
- Deng H., Dodson M.W., Huang H., Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C.J., Khajeh J., Coulanges E., Chen E.I., Owusu-Ansah E. Regulation of mitochondrial complex I biogenesis in Drosophila flight muscles. Cell Rep. 2017;20:264–278. doi: 10.1016/j.celrep.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murari A., Rhooms S.K., Goparaju N.S., Villanueva M., Owusu-Ansah E. An antibody toolbox to track complex I assembly defines AIF's mitochondrial function. J. Cell Biol. 2020;219 doi: 10.1083/jcb.202001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W., Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A., Oliveira M.P., Khamoui A.V., Aparicio R., Rera M., Rossiter H.B., Walker D.W. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun. 2017;8:448. doi: 10.1038/s41467-017-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G., Schulz R.A., Olson E.N. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 1996;176:143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
- Rera M., Bahadorani S., Cho J., Koehler C.L., Ulgherait M., Hur J.H., Walker D.W. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulgherait M., Rana A., Rera M., Graniel J., Walker D.W. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 2014;8:1767–1780. doi: 10.1016/j.celrep.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any new datasets or code.